While ESCRT-0 is ubiquitinated by the Rsp5 E3 ligase, loss of Rsp5 does not disrupt monoubiquitin-dependent sorting into multivesicular bodies.
Abstract
ESCRTs (endosomal sorting complexes required for transport) bind and sequester ubiquitinated membrane proteins and usher them into multivesicular bodies (MVBs). As Ubiquitin (Ub)-binding proteins, ESCRTs themselves become ubiquitinated. However, it is unclear whether this regulates a critical aspect of their function or is a nonspecific consequence of their association with the Ub system. We investigated whether ubiquitination of the ESCRTs was required for their ability to sort cargo into the MVB lumen. Although we found that Rsp5 was the main Ub ligase responsible for ubiquitination of ESCRT-0, elimination of Rsp5 or elimination of the ubiquitinatable lysines within ESCRT-0 did not affect MVB sorting. Moreover, by fusing the catalytic domain of deubiquitinating peptidases onto ESCRTs, we could block ESCRT ubiquitination and the sorting of proteins that undergo Rsp5-dependent ubiquitination. Yet, proteins fused to a single Ub moiety were efficiently delivered to the MVB lumen, which strongly indicates that a single Ub is sufficient in sorting MVBs in the absence of ESCRT ubiquitination.
Introduction
Many ESCRT (endosomal sorting complex required for transport) components have Ubiquitin (Ub)-binding domains (UBDs) that contribute to their ability to sort ubiquitinated membrane proteins into multivesicular bodies (MVBs). Like other proteins that contain UBDs, ESCRTs can be ubiquitinated by a variety of mechanisms mediated by the UBD itself in a process termed “coupled ubiquitination” (Haglund and Stenmark, 2006). Moreover, ESCRT-0 and ESCRT-I associate with Ub-ligases and deubiquitinating enzymes that might influence ESCRT activity by modulating ESCRT ubiquitination (Kato et al., 2000; Amit et al., 2004; McCullough et al., 2004; Kim et al., 2007; Ren et al., 2007). Coupled ubiquitination has been proposed to drive critical aspects of ESCRT function by altering intermolecular interactions between ESCRT proteins that house UBDs, or triggering cargo release by causing intramolecular association between the UBD and attached Ub (Hicke et al., 2005; Komada and Kitamura, 2005; Piper and Katzmann, 2007; Williams and Urbé, 2007; Roxrud et al., 2010). However, definitive evidence that such UBD-dependent ubiquitination drives a physiologically relevant aspect of ESCRT function has been difficult to obtain. In addition, it remains unclear what type of Ub-based sorting motif actually serves as the signal for incorporation into MVBs. Structural studies showing tandem arrays of UBDs within ESCRTs as well as functional studies in cells deficient in polyubiquitin assembly suggest that short K63-linked chains of Ub constitute the operative sorting signal recognized by ESCRT UBDs (Erpapazoglou et al., 2008; Kulathu et al., 2009; Lauwers et al., 2009; Paiva et al., 2009; Ren and Hurley, 2010). However, limitations from these correlative experiments complicate such conclusions about potential regulatory mechanisms that control how cargo is identified, recognized, and processed by the endocytic system. Using a variety of approaches, we found that ubiquitination of the ESCRT apparatus is not required for its ability to sort monoubiquitinated cargo into the MVB.
Results and discussion
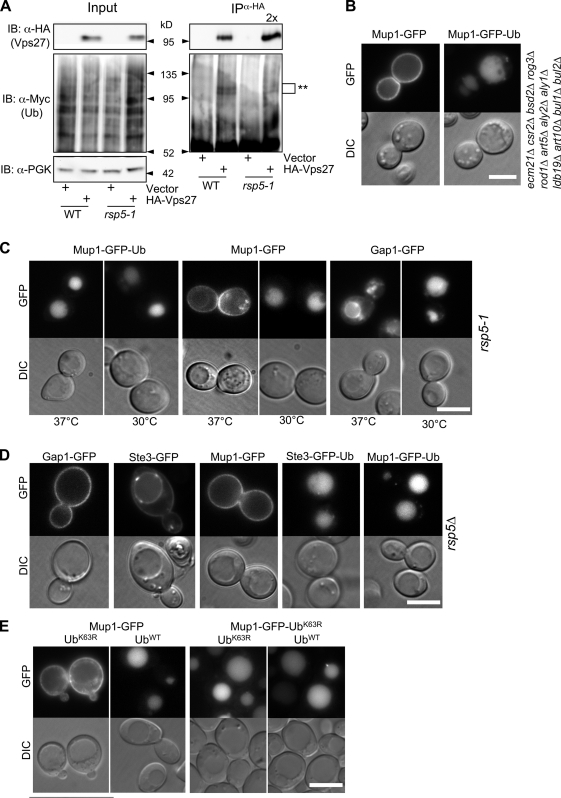
We found that the yeast Hrs homologue Vps27 is ubiquitinated by immunoprecipitating HA-Vps27 from denatured lysates and immunoblotting for Ub. The major proportion of HA-Vps27 migrated at ∼96 kD, whereas a less prominent higher molecular weight band (∼105 kD) corresponded to ubiquitinated HA-Vps27 (Fig. 1 A).
Figure 1.
Vps27 lacking lysine residues is not ubiquitinated but still functions in MVB cargo sorting. (A) Ubiquitination of Vps27 was assessed in cells expressing HA-Vps27 and myc-Ub. Vps27 immunoprecipitates (IP) from denatured cell lysates were immunoblotted (IB) for HA-Vps27 (anti-HA) or Ub (anti-myc). Input represents a 5% equivalent. Black lines indicate that dividing lanes have been spliced out. (B) Domain organization of yeast ESCRT-0 protein Vps27. Blue lines show positions of the 47 lysines. (C) Ubiquitination of wild-type and lysine-less Vps27 (HA-Vps27WT and HA-Vps27K>R) was assessed in ubp2Δ cells expressing myc-Ub. Anti-HA (Vps27) immunoprecipitates (IP) were immunoblotted (IB) for HA-Vps27 (anti-HA) or Ub (anti-myc). Input represents a 5% equivalent. *, expected molecular weight; **, ubiquitinated forms. (D) Sorting of Ste3-GFP in vps27Δ, vps27Δ ubp2Δ, or vps27Δ hse1Δ (ESCRT-0Δ) cells transformed with plasmids expressing HA-Vps27WT or HA-Vps27K>R. Bar, 5 µm.
To assess whether ESCRT-0 ubiquitination is required for MVB sorting, we generated a Vps27K>R mutant lacking all 47 lysine residues in the full-length protein (Fig. 1 B). All of the Lys residues were altered because coupled ubiquitination can promiscuously use a variety of lysines (Polo et al., 2002; Miller et al., 2004; Woelk et al., 2006; Uchiki et al., 2009). Ubiquitination of Vps27K>R was undetectable, even in cells lacking the deubiquitinating enzyme Ubp2, which antagonizes the HECT-type Ub ligase Rsp5 responsible for a wide variety of ubiquitination events within the endocytic pathway (Kee et al., 2005, 2006; Rotin and Kumar, 2009). In contrast, a sizable proportion of wild-type Vps27 was ubiquitinated in ubp2Δ cells because ∼20% was found in the higher ∼105-kD form (Fig. 1 C). We also found that both wild type and the Vps27K>R protein were equally stable (Fig. S1 A).
To determine if loss of Vps27 ubiquitination impairs MVB sorting, we examined the localization of Ste3-GFP, a GFP-tagged G protein–coupled receptor normally targeted to MVBs. Cells lacking Vps27 accumulated Ste3-GFP on the vacuole limiting membrane and in endosomes. However, cells with either wild-type Vps27 or Vps27K>R sorted Ste3-GFP into the vacuole interior (Fig. 1 D). Both Vps27 proteins functioned equally in cells also lacking Ubp2, which implies that although wild-type Vps27 is ubiquitinated to a greater extent in these cells, the MVB sorting process is normal. Under conditions where MVB sorting is less efficient, the Vps27K>R mutant performed like wild type, which indicates that loss of Vps27 ubiquitination does not increase sorting efficiency (Fig. S1 B).
Yeast ESCRT-0 contains both Vps27 and the STAM homologue Hse1 (Bilodeau et al., 2002). STAM also undergoes ubiquitination (Row et al., 2006), raising the possibility that ubiquitination of Hse1 may regulate ESCRT-0 activity. To test this, we used the SEY6210 parental yeast strain, which requires Vps27 but not Hse1 for MVB sorting (unpublished data and Emr, S.D., personal communication). We found that Ste3-GFP sorting was normal in hse1Δ vps27K>R cells where all the lysines of ESCRT-0 are eliminated (Fig. 1 D).
Hrs associates with HECT-type Ub ligases such as Nedd4 and AIP4, which promote ubiquitination of Hrs both in vivo and in vitro (Katz et al., 2002; Marchese et al., 2003; Woelk et al., 2006). The yeast homologue of Nedd4 is Rsp5, which interacts with ESCRT-0 (Ren et al., 2007), and is a prime candidate for the ligase that ubiquitinates Vps27. Vps27 ubiquitination was severely attenuated in rsp5-1 temperature-sensitive cells at 37°C to ∼10%, the level observed in wild-type cells (loss of Ub-Vps27 was 91% ± 2.7% SD, n = 3; Fig. 2 A). These data indicated that Rsp5 is the major Ub ligase responsible for ESCRT-0 ubiquitination, which suggests that Rsp5 inhibition might be a means of assessing the role of ESCRT ubiquitination.
Figure 2.
Cells lacking Rsp5 activity still sort ubiquitinated cargo proteins into vacuoles. (A) Wild-type and temperature-sensitive rsp5-1 cells expressing HA-Vps27 and Myc-Ub were shifted to 37°C for 1 h. Input lysates and Vps27 immunoprecipitates (IP) were immunoblotted (IB) for HA (Vps27) and Myc (Ub). Twice the amount of the anti-HA IP from rsp5-1 cells was loaded. *, expected molecular weight; **, ubiquitinated forms. (B) Localization of Mup1-GFP and Mup1-GFP-Ub in mutant EN67 cells lacking multiple Rsp5 adaptors. Cells were grown in YPD and copper for 4 h to produce Mup1 fusion proteins from the CUP1 promoter. (C) Sorting of Gap1-GFP, Mup1-GFP, and Mup1-GFP-Ub after Rsp5 inactivation. Cells (rsp5-1) were shifted to 37°C for 1 h and grown in YPD in the presence of 100 µM CuCl2 to induce production of GFP-tagged proteins under control of the CUP1 promoter. (D) Localization of Gap1-GFP, Ste3-GFP, Mup1-GFP, Ste3-GFP-Ub, or Mup1-GFP-Ub in rsp5Δ-null cells grown in YPD + 25 µM CuCl2. (E) Localization of Mup1-GFP or Mup1-GFP-UbK63R in cells expressing wild-type (WT) Ub (SUB492) or K63R Ub (SUB493) as their sole source of Ub. Bars, 5 µm.
We next examined MVB sorting of Mup1-GFP in cells with compromised Rsp5 activity. We first examined sorting in cells lacking many of the adaptors Rsp5 requires to modify cargo (Nikko and Pelham, 2009), and in which we found ubiquitination of Mup1-GFP to be severely compromised (Fig. 2 B and S1 C). We also assessed sorting in rsp5-1 cells at 37°C (Fig. 2 C). Either method of inhibiting Rsp5 dramatically blocked sorting of Gap1-GFP and Mup1-GFP, which is consistent with previous studies demonstrating a requirement for Rsp5-mediated ubiquitination (Rotin and Kumar, 2009). However, simply adding Ub in frame restored sorting of Mup1-GFP, indicating that ESCRT-dependent MVB sorting operates normally as long as the requirement for cargo ubiquitination is satisfied.
Previous studies indicate that rsp5-1 cells might still harbor residual Rsp5 activity at 37°C (Kee et al., 2006), perhaps enough to mediate a low level of ESCRT ubiquitination. RSP5 is essential; however; its essential function can be bypassed by expressing a truncated version of the Mga2 transcription factor that undergoes Rsp5- and proteasome-dependent cleavage (Hoppe et al., 2000; Chellappa et al., 2001). We generated rsp5Δ cells, which grew slowly at 30°C and were inviable at 37°C (Fig. S1 D). Sorting of proteins such as Mup1-GFP, Ste3-GFP, and Gap1-GFP along the MVB pathway was completely blocked in rsp5Δ cells. In contrast, fusion of Ub onto the C terminus of either Ste3-GFP or Mup1-GFP restored sorting (Fig. 2 D), which indicates that ESCRT ubiquitination by Rsp5 is not required for MVB sorting.
Rsp5 favors formation of K63-linked polyubiquitin chains (Kee et al., 2005, 2006), and several studies suggest that K63-linked Ub chains, rather than a single mono-Ub, serve as a necessary signal for entry into the MVB sorting pathway (Lauwers et al., 2010). Indeed, cargo fused to a single Ub, which induces MVB sorting, might still undergo additional K63 ubiquitination (Reggiori and Pelham, 2001; Urbanowski and Piper, 2001). However, our data demonstrate that fusion of a single Ub to a cargo protein is sufficient for MVB sorting in cells where Rsp5-dependent K63 polyubiquitination is absent. We further tested the role of K63 Ub chains in cells that were entirely incapable of generating K63 polyubiquitin chains (Fig. 2 E). The sole source of Ub in these cells was UbK63R, which was a condition previously shown to cause severe MVB sorting defects for membrane proteins such as Gap1, Jen1, and Sit1 (Erpapazoglou et al., 2008; Lauwers et al., 2009; Paiva et al., 2009). Similarly, we found that Mup1-GFP was localized exclusively to the cell surface and intracellular puncta in UbK63R cells. However, Mup1-GFP-UbK63R, containing a single in-frame Ub also lacking K63, sorted normally to the vacuole interior. Collectively, these results indicate that a single Ub is sufficient for cargo to sort into the MVB lumen. They also imply that the sorting defects resulting from depriving cells of K63-linked Ub chains may arise from global changes that disrupt trafficking pathways upstream of the MVB or perturb the dynamic cycle of cargo ubiquitination and deubiquitination, from which Mup1-GFP-Ub is immune. In addition, fusion of Ub may route cargo more efficiently to endosomes, circumventing many of the obstacles that other cargos might encounter in Ub-K63–deficient cells during their endocytic itinerary (see supplemental material). Our data clearly agree with previous studies establishing some type of role for K63-linked chains in regulating MVB sorting (Erpapazoglou et al., 2008; Lauwers et al., 2009; Paiva et al., 2009). However, our finding that fusing UbK63R is sufficient for MVB sorting suggests that K63-linked chains need not be on cargo themselves. It still remains possible that the sorting of some cargo proteins benefits from the added avidity such chains have for the ESCRT Ub receptors. K63-linked chains may allow cargo to resist the effects of deubiquitinating enzymes as they travel through the endocytic pathway.
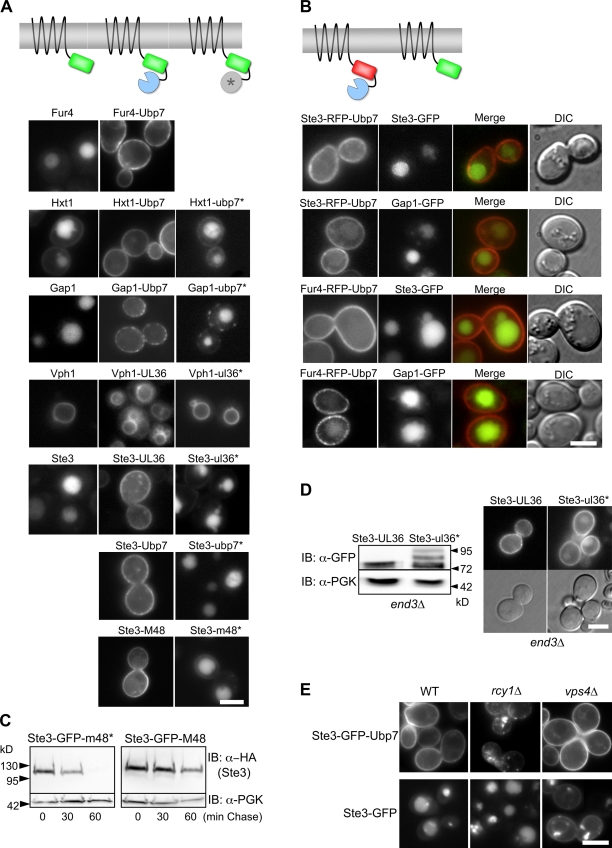
The preceding data indicate that neither removing ubiquitinatable lysine residues from ESCRT-0 nor eliminating the major Ub ligase responsible for the majority of ESCRT-0 ubiquitination critically interferes with ESCRT function. However, these approaches have caveats. First, although eliminating Rsp5 dramatically reduces ESCRT-0 ubiquitination, other Ub-ligases might still modify ESCRTs at undetectable levels. This is a distinct possibility because coupled ubiquitination in vitro can be mediated by many different ligases or by E2-conjugating enzymes alone (Hoeller et al., 2007; Uchiki et al., 2009). Second, although we eliminated all of the lysine residues within ESCRT-0, it is possible that another protein associated with the canonical ESCRT-0 may be a relevant Ub acceptor. Finally, Rsp5 is capable of ubiquitinating the N termini of proteins in vitro (Kim and Huibregtse, 2009), whereas other ligases can ubiquitinate cysteine, serine, and threonine residues (Cadwell and Coscoy, 2005; Wang et al., 2007). Thus, the prospect of removing all ubiquitinatable residues within ESCRT-0 and its associated proteins is daunting and leaves no way to control for structural perturbations caused by the mutations themselves. Therefore, we devised a different method to render proteins resistant to ubiquitination. Because in vivo removal of Ub is catalyzed by deubiquitinating peptidases (DUbs), we reasoned that it was possible to generate nonubiquitinatable proteins by fusing them to the catalytic domain of a DUb (Fig. 3 A). We chose three catalytic domains from cysteine-based DUbs that can be inactivated by mutating their catalytic cysteine (Reyes-Turcu et al., 2009): the C-terminal catalytic domain from yeast Ubp7, the UL36 Ub-specific protease domain from type I Herpes virus (Gredmark et al., 2007; Kattenhorn et al., 2005), and the M48 Ub-specific protease domain of murine cytomegalovirus (Schlieker et al., 2007).
Figure 3.
Fusing cargo to the catalytic domain from deubiquitinating enzymes blocks ubiquitination and trafficking to the vacuole. (A) Schematic of DUb catalytic domain fusion proteins (top). Localization of the indicated GFP-tagged proteins fused to either enzymatically active or inactive (*) catalytic domains of Ubp7, HSV-1 UL36, and mCMV M48 (bottom). (B) The effect of DUb fusion proteins (Ste3-RFP-Ubp7 or Fur4-RFP-Ubp7) on coexpressed Gap1-GFP or Ste3-GFP. (C) Cycloheximide chase with cells expressing HA-tagged Ste3-GFP-m48* and Ste3-GFP-M48, which carried active and inactive DUb domains, respectively. Aliquots were removed at the indicated times after cycloheximide addition (100 µg/ml) and immunoblotted for HA and PGK. (D) Ste3-GFP-UL36 or Ste3-GFP-ul36* (with an inactive catalytic domain) were expressed in end3Δ cells and immunoblotted with anti-GFP antibodies (left) or localized (right). (E) Localization of Ste3-GFP or Ste3-GFP-Ubp7 in wild type, rcy1Δ cells, and vps4Δ cells . Bars, 5 µm.
When these catalytic domains were fused to GFP-tagged membrane proteins that normally undergo MVB sorting, their localization to the vacuole interior was completely blocked, with localization primarily at the cell surface (Fig. 3 A). The effect on Fur4 and Gap1 was consistent with the strong plasma membrane localization of Fur4 and Gap1 lacking their ubiquitination sites, as described in previous studies (Blondel et al., 2004; Lauwers et al., 2009). Fusion of a DUb domain did not adversely affect the normal localization of Vph1 to the vacuole limiting membrane. In addition, coexpressing DUb-tagged membrane proteins did not affect the sorting of other proteins that normally follow a Ub-dependent pathway to the vacuole interior (Fig. 3 B). As expected, the stability of Ste3-GFP fused to an active DUb was substantially higher than when fused to a catalytically inactive DUb (Figs. 3 C and S1 E). Similarly, fusion of Ste3 to active UL36 catalytic domain abolished detectable ubiquitination (Fig. 3 D). Despite its steady-state localization to the cell surface, we found that Ste3-GFP-DUb was still internalized (Fig. 3 E) because it accumulated in endosomal compartments of rcy1Δ mutants where recycling is blocked (Wiederkehr et al., 2000; Galan et al., 2001). However, Ste3-GFP-DUb did not accumulate in the late endosome–like class E compartments that accumulate in vps4Δ cells (Babst et al., 1997). These data imply that Ste3 internalizes in a Ub-independent manner and that without Ub, it quickly recycles to the cell surface via early endosomal compartments without reaching late endosomal compartments.
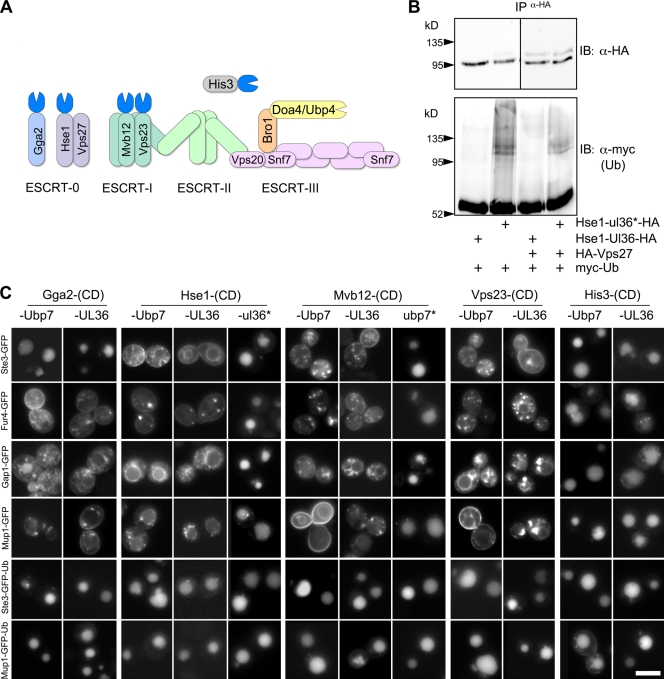
Equipped with a new method to immunize proteins against ubiquitination, we reexamined the role of ESCRT ubiquitination (Fig. 4). Fusing DUb catalytic domains to ESCRTs should not only prevent ubiquitination of the ESCRTs themselves, but if suitably robust, the DUb should also strip Ub from cargo proteins and block their entry into MVBs (Fig. 4 A). Moreover, if ESCRT ubiquitination is not required for the sorting process, cargo fused to Ub so that it cannot be removed by DUb activity should be able to sort normally. Fusion of UL36 catalytic domain to ESCRT-0 subunit Hse1 prevented accumulation of ubiquitinated Hse1 and Vps27 (Fig. 4 B). Expression of Hse1-DUb proteins in wild-type cells caused a dramatic block in the sorting of Ste3, Fur4, Gap1, and Mup1, all of which rely on Rsp5-dependent ubiquitination for sorting into the vacuole (Fig. 4 C). Importantly, both Ste3-GFP-Ub and Mup1-GFP-Ub, which contained a single “non-deubiquitinatable” Ub, were properly sorted to the vacuole interior. We also fused DUb catalytic domains to Vps23 and Mvb12, two ESCRT-I subunits thought to act as a downstream Ub cargo receptor and whose mammalian counterparts also undergo ubiquitination (Kim et al., 2007; Tsunematsu et al., 2010). The Vps23-DUb and Mvb12-DUb fusions also interrupted the Ub- and Rsp5-dependent sorting of membrane cargo to the MVB lumen yet did not block sorting of cargo with an in-frame fusion of Ub. All of the MVB sorting defects were abolished when the catalytic activity of the DUb domains was inactivated (Fig. 4 C), and no nonspecific defects in MVB sorting were observed when we expressed catalytic domains fused to His3, a cytosolic protein within the histidine biosynthesis pathway (Struhl and Davis, 1977). All of the DUb fusion proteins were expressed to comparable levels (Fig. S1, F and G), and none caused a vacuolar protein secretion (VPS) phenotype (Fig. S2 A), which is consistent with ESCRT function being maintained. We also examined the effect of fusing a DUb catalytic domain to Gga2, a Ub-binding clathrin adaptor protein thought to sort ubiquitinated proteins from the TGN to endosomes (Scott et al., 2004). GGA proteins may also contribute to Ub-dependent transport of proteins from the cell surface to endosomes in yeast; however, evidence for a clear role in this pathway is obscured because ESCRT-0 also operates in this pathway in parallel (Erpapazoglou et al., 2008; Deng et al., 2009; Lauwers et al., 2009). The dominant-acting Gga2-DUbs dramatically blocked sorting of Gap1, Mup1, and Fur4 but did not affect that of Ste3 to the vacuole, which indicates that yeast Gga proteins may also mediate Ub-specific sorting of a subset of cargos as they travel between the cell surface (Puertollano and Bonifacino, 2004).
Figure 4.
Fusion of DUbs to ESCRT proteins inhibits their ubiquitination but allows vacuolar sorting of Ub fusion cargo proteins. (A) Schematic of the ESCRT sorting apparatus showing subunits fused to different DUb catalytic domains. (B) Cells expressing myc-Ub and either Hse1-UL36-HA or Hse1-ul36*-HA (inactive) were transformed with vector alone or with plasmids expressing HA-Vps27. Lysates were immunoprecipitated (IP) with anti-HA antibodies. Immunoprecipitates were immunoblotted (IB) for HA (top) to confirm isolation of HA-tagged proteins; immunoprecipitates were immunoblotted with anti-myc to assess ubiquitination. Fusion of active UL36 abolished ubiquitination of either HA-tagged Hse1 or Vps27 proteins. Black lines indicate that dividing lanes have been spliced out. (C) ESCRT subunits, GGA, or His3 fused to the indicated active (Ubp7 or UL36) or inactive (ubp7* or ul36*) catalytic domains (CD) were expressed in wild-type cells. Cells coexpressed the indicated GFP-tagged MVB cargo. Bar, 6 µm.
The ESCRT-DUb fusion proteins could also functionally substitute for their wild-type counterparts in the MVB sorting process (Fig. 5, A–C). Neither cells with Hse1-UL36 as their sole source of Hse1 nor cells with Vps23-UL36 as their sole source of Vps23 sorted Ste3-GFP or Gap1-GFP into the vacuole lumen. However, both cell types were able to sort Ste3-GFP-Ub and Fur4-GFP-Ub fusion proteins. Mvb12-Ubp7 and Mvb12-UL36 were also able to functionally substitute for wild-type Mvb12 to promote the sorting of Ste3-GFP-Ub, but they effectively blocked sorting of Fur4-GFP (Fig. 5 A). Because loss of Mvb12 results in only mild MVB sorting defects (Curtiss et al., 2007), we assessed Mvb12-DUb function in cells that also had mutations in the UBDs of Vps23 and Vps36, which yield a severe synthetic class E VPS phenotype (Shields et al., 2009). These data indicate that the ESCRT-fused DUb domains are robust enough to deubiquitinate cargo and prevent ubiquitination of ESCRTs, yet are still able to sort Ub-tagged cargo.
Figure 5.
The effect of localizing DUb domains on ESCRTs and the Ub ligase Rsp5. (A) Complementation of mvb12Δ by Mvb12-UL36. The localization of Fur4-GFP and Ste3-GFP-Ub in mvb12Δ vps23ΔUb vps36ΔUb cells carrying vector plasmid alone or plasmids expressing Mvb12 fused to active (UL36, Ubp7) or inactive (ubp7*) DUb domains is shown. (B) Complementation of hse1Δ by Hse1-UL36 and Hse1-ul36*(inactive). The localization of Ste3-GFP and Ste3-GFP-Ub in hse1Δ cells carrying the indicated plasmids is shown. (C) Complementation of vps23Δ by Vps23-FP-UL36. Localization of Gap1-GFP and Fur4-RFP-Ub in vps23Δ cells carrying indicated plasmids is shown. (D) Wild-type cells expressing Rsp5 fused to active (Ubp7) or inactive (ubp7*) catalytic DUb domain and the indicated GFP-tagged MVB cargo proteins. Expression of Rsp5 fusions were from the CUP1 inducible promoter, induced 5 h before microscopy (for Rsp5-Ubp7) or overnight (for Rsp5-ubp7*). Bars, 5 µm.
We then evaluated whether fusion of Rsp5 to a DUb catalytic domain would create a dominant “reversal” enzyme that could deubiquitinate Rsp5 substrates in vivo. Expressing Rsp5 fused to the catalytic domain of Ubp7 effectively blocked sorting of many Rsp5 substrates (Fig. 5 D). No effect was observed with Rsp5 fused to an inactive catalytic domain (Fig. 5 D) expressed at comparable levels (Fig. S2 B). Expressing Rsp5-DUb also inhibited growth, which is consistent with the essential role of Rsp5 for viability (Fig. S2 C). Although Rsp5-DUb removed Ub from endogenous Rsp5 substrates and blocked their sorting, cargo such as Ste3-GFP-Ub and Mup1-GFP-Ub sorted into the vacuole normally, which supports the idea that their single in-frame Ub is sufficient for their sorting. Furthermore, these data indicate that any deubiquitination of ESCRTs by Rsp5-DUb does not hamper ESCRT sorting activity.
Potentially, the organization of the ESCRT apparatus could be regulated by ubiquitination by altering inter- and intramolecular interactions among its UBD-containing components. Cumulative loss of multiple UBDs within the ESCRTs results in a general loss of function that is not confined to cargo recognition alone (Shields et al., 2009). These data imply that UBDs may contribute to the productive organization of the ESCRT apparatus in a manner similar to what has been proposed for the internalization apparatus (Dores et al., 2010). Alternatively, ESCRT ubiquitination might mediate an intramolecular interaction that prevents interaction with Ub cargo, providing a mechanism for cargo release. However, ESCRTs as well as many other Ub-binding proteins undergo ubiquitination by virtue of simply having UBDs, and this may not reflect a genuine regulatory mechanism. We found that preventing ubiquitination of ESCRTs did not diminish or enhance their ability to sort Ub cargo or affect their ability to release cargo because cargo did not accumulate on endosomes, nor did nonubiquitinatable ESCRT-0 accumulate in the vacuole lumen (Fig. S2 D). It remains possible that future experiments will demonstrate that other ESCRT components also become ubiquitinated. However, the best candidate to mediate such ubiquitination is Rsp5, and we have taken several approaches to demonstrate that Rsp5-dependent ubiquitination is not required for the MVB sorting process once a single Ub modifies cargo. Determining whether such coupled ubiquitination is correlative or indeed causes a specific and physiologically relevant regulatory mechanism in vivo requires that the process of coupled ubiquitination be uncoupled. The multiple approaches used here, including the approach of tethering DUb activity to particular protein complexes, provides a comprehensive complementary strategy for assessing a physiological role for such ubiquitination events.
Materials and methods
Materials, yeast strains, and plasmids
Chemicals, growth methods, and other general procedures were used as described previously (Shields et al., 2009). Growth in the presence and absence of copper was done with plates containing 100 µM CuCl2 or bathocuproine disulfonate (BCS), respectively. Anti-CPY, GFP, HA, and Pho8 were used as described previously (Ren et al., 2007). Purchased antibodies were: anti-myc monoclonal (A00704; GenScript), anti-myc polyclonal (18826-01; QED Biosciences), anti-Ub (P4D1, sc-8017; Santa Cruz Biotechnology, Inc.), and anti-HA monoclonal (MMS-101R; Covance). Yeast strains are described in Table I and plasmids are described in Table II. The vps27Δ ubp2Δ strain was created by inserting the HIS5 gene (Gueldener et al., 2002) into VPS27 within the ubp2Δ mutant from the BY4742 MATα yeast deletion collection (Giaever et al., 2002). The hse1Δ vps27Δ strain was made by inserting the TRP1 gene into the HSE1 locus (Ren et al., 2007). The rsp5Δ-null strains were made from a parental W303 strain, which contained a disruption of the genomic RSP5 gene and contained a URA3-based low copy plasmid housing the wild-type RSP5 gene (pRS416-RSP5). This strain was transformed with a low copy plasmid expressing a truncated Mga2 protein (Chellappa et al., 2001), and an ADE2 selectable plasmid expressing a GFP-tagged reporter protein. Stains that lost the plasmid-borne RSP5 gene were selected on plates containing 5-fluoroorotic acid and oleic acid that were incubated at 25°C (Wang et al., 1999), and loss of RSP5 was confirmed by lack of growth at 37°C (Hoppe et al., 2000; Chellappa et al., 2001). DUb catalytic domains were residues 561–1,071 of yeast Ubp7, the N-terminal residues 15–260 (UL36) of the type I Herpes virus VP1/2 tegument protein (Kattenhorn et al., 2005; Gredmark et al., 2007), and residues 4–234 of M48 Ub-specific protease (Schlieker et al., 2007). M48 is active toward both K48- and K63-linked Ub chains, whereas UL36 was originally reported to show specificity to K48-linked Ub chains (Kattenhorn et al., 2005). However, recent studies demonstrate that UL36 cleaves both K48- and K63-linked chains (Kim et al., 2009), which is consistent with our own observations (Fig. S2 E).
Table I.
Plasmids used
| Plasmids | Description | Use | Source |
| pRS315pRS316 | Centromere containing low copy yeast shuttle plasmids (LEU2 and URA3) | Figs. 1 (A, C, and, D), 2 A, 5 (A–C), and S2 A | Sikorski and Hieter, 1989 |
| pAC376 | Yep351 (2µ LEU2) expressing myc-tagged Ub from CUP1 promoter | Figs. 1 (A and C), 2 A, and 4 B | A. Cooper (Garvan Institute, Sydney Australia) |
| pRSmga2NΔtm | HIS3 low copy plasmid containing a truncated MGA2 gene lacking C-terminal 329 codons that include the membrane-spanning domain. | Figs. 2 D and S1 D | Chellappa et al., 2001 |
| pYM-N36 | nourseothricin-resistance marker natMX | Janke et al., 2004 | |
| Genomic DNA | HSV-1 (strain F) | Ejercito et al., 1968 | |
| pPL4149 | pRS316 expressing HA-Vps27 from MET25 promoter | Figs. 1 (A, C, and D), 2 A, 4 B, and S1 (A and B) | This study |
| pPL4150 | pRS316 expressing HA-Vps27K>R (where all K residues are altered to R) from MET25 promoter. | Figs. 1 (C and D) and S1 (A and B) | This study |
| pPL967 | pRS315 expressing Ste3-GFP from STE3 promoter | Figs. 1 D, 2 D, 3 (A and E), 4 C, 5 (B and D), and S2 F | Urbanowski and Piper, 2001 |
| pPL991 | pRS313 expressing Ste3-GFP from STE3 promoter | Fig. 1 D | Urbanowski and Piper, 2001 |
| pPL2334 | pRS315 expressing Gap1-GFP from CUP1 promoter | Figs. 2 (C and D), 3 (A and B), 4 C, and 5 (C andD) | This study |
| pPL3484 | pRS315 expressing Ste3-GFP-Ub from STE3 promoter | Figs. 4 C and 5, A, B, and D | This study |
| pPL3797 | pRS315 expressing Fur4-GFP from CUP1 promoter | Figs. 3 A, 4 C, and 5, A and D | This study |
| pPL3931 | ADE2 conversion of pPL3484 (ADE2 replacement of LEU2) expressing Ste3-GFP-Ub from STE3 promoter | Fig. 2 D | This study |
| pPL3962 | pRS315 expressing Hxt1-GFP from HXT1 promoter | Fig. 3 A | This study |
| pPL4144 | pRS315 expressing Fur4-mCherry-Ub from CUP1 promoter | Fig. 5 C | This study |
| pPL4146 | pRS315 expressing Mup1-GFP from CUP1 promoter | Figs. 2 (B, C, and E), 4 C, and 5D | This study |
| pPL4147 | pRS315 expressing Mup1-GFP-Ub from CUP1 promoter | Figs. 2 (B–D), 4 C, 5 D, and S2 F | This study |
| pPL4214 | ADE2 conversion of pPL967 (ADE2 replacement of LEU2) expressing Ste3-GFP from STE3 promoter | Fig. 2 D | This study |
| pPL4215 | ADE2 conversion of pPL2334 (ADE2 replacement of LEU2) expressing Gap1-GFP from CUP1 promoter | Fig. 2 D | This study |
| pPL4216 | ADE2 conversion of pPL4146 (ADE2 replacement of LEU2) expressing Mup1-GFP from CUP1 promoter | Fig. 2 D | This study |
| pPL4217 | ADE2 conversion of pPL4147 (ADE2 replacement of LEU2) expressing Mup1-GFP-Ub from CUP1 promoter | Fig. 2 D | This study |
| pPL4252 | natMX conversion of pPL4146 (natMX replacement of LEU2) expressing Mup1-GFP from CUP1 promoter | Figs. 2 E and S2 F | This study |
| pPL4320 | pRS315 expressing Mup1-GFP-UbK63R from CUP1 promoter | This study | |
| pPL4342 | natMX conversion of pPL4320 (natMX replacement of LEU2) expressing Mup1-GFP-UbK63R from CUP1 promoter | Fig. 2 E | This study |
| pPL3584 | pRS316 expressing Mvb12-Ubp7CD-3xHA from CUP1 promoter; contains residues 561-1071 of Ubp7 | Figs. 4 C, 5 A, and S1 F | This study |
| pPL3609 | pRS316 expressing Ste3-mCherry-Ubp7CD from STE3 promoter | Fig. 3 B | This study |
| pPL3669 | pRS316 expressing Fur4-GFP-Ubp7CD from CUP1 promoter | Fig. 3 B | This study |
| pPL3810 | pRS316 expressing Fur4-mCherry-Ubp7CD from CUP1 promoter | Fig. 3 B | This study |
| pPL3876 | pRS316 expressing Ste3-GFP-Ubp7CD from STE3 promoter | Figs. 3 A and S1 E | This study |
| pPL3900 | pRS316 expressing Gap1-GFP-Ubp7CD from CUP1 promoter | Fig. 3 A | This study |
| pPL3964 | pRS316 expressing Hxt1-GFP-Ubp7CD from HXT1 promoter | Fig. 3 A | This study |
| pPL3986 | pRS316 expressing Ste3-GFP-Ubp7CD(G>S) -3xHA from STE3 promoter; contains residues 561–1,071 of Ubp7 with mutations G932S | Figs. S1 E and S2B | This study |
| pPL3890 | pRS316 expressing Ste3-GFP-3xHA from STE3 promoter | Fig. S1 E | This study |
| pPL3722 | pRS316 expressing His3-Ubp7CD-3xHA from CUP1 promoter. Contains residues 561–1071 of Ubp7 followed by 3×HA epitope | Figs. 4C, S1 F, and S2, A and C | This study |
| pPL3723 | pRS316 expressing Hse1-Ubp7CD-3xHA from CUP1 promoter | Figs. 4 C, S1 F, and S2 A | This study |
| pPL3724 | pRS316 expressing Vps23-Ubp7CD-3xHA from CUP1 promoter | Figs. 4 C, S1 F, and S2 A | This study |
| pPL3742 | pRS316 expressing Rsp5-Ubp7CD-3xHA from CUP1 promoter | Figs. 5 D, and S2 B | This study |
| pPL3746 | pRS316 expressing Gga2-Ubp7CD-3xHA from CUP1 promoter | Figs. 4 C, S1 F, and S2 A | This study |
| pPL3643 | pRS316 expressing Ste3-GFP-ubp7CD* from STE3 promoter; contains residues 561–1071 of Ubp7 with C618S mutation | Fig. 3 A | This study |
| pPL3776 | pRS316 expressing Mvb12- ubp7CD*-3xHA from CUP1 promoter | Figs. 4 C, 5 A, S1 F, and S2 A | This study |
| pPL3901 | pRS316 expressing Gap1-GFP- ubp7CD* from CUP1 promoter | Fig. 3 A | This study |
| pPL4371 | pRS316 expressing Hxt1-GFP-ubp7CD* from HXT1 promoter | Fig. 3 A | This study |
| pPL3994 | pRS316 expressing Ste3-GFP-UL36CD from STE3 promoter; contains 561–591 of Ubp7 (Ubp7 linker)-and residues 15–260 of HSV1 UL36 | Fig. 3, A and D | This study |
| pPL4012 | pRS316 expressing Hse1-UL36CD-3xHA from CUP1 promoter | Figs. 4 (B and C), 5 B, S1 G, and S2 A | This study |
| pPL4013 | pRS316 expressing Mvb12-UL36CD-3xHA from CUP1 promoter | Figs. 4 C, 5 A, S1 G, and S2 A | This study |
| pPL4014 | pRS316 expressing His3-UL36CD-3xHA from CUP1 promoter | Figs. 4 C, S1 G, and S2 A | This study |
| pPL4015 | pRS316 expressing Vps23-GFP-UL36CD from VPS23 promoter | Fig. 5 C | This study |
| pPL4017 | pRS316 expressing Gga2-UL36CD-3xHA from CUP1 promoter | Figs. 4 C, S1 G, and S2 A | This study |
| pPL4140 | HIS3 conversion of pPL4012 expressing Hse1-UL36CD-3xHA from CUP1 promoter | Fig. 4 B | This study |
| pPL4247 | pRS316 expressing Vps23-UL36CD-3xHA from CUP1 promoter | Figs. 4 and S2, B and C | This study |
| pPL4058 | pRS316 expressing Ste3-GFP-ul36CD* from STE3 promoter; contains 561–591 of Ubp7 (Ubp7 linker) and residues 15–260 of HSV1 UL36 with mutation of C40S in UL36 | Fig. 3, B and D | This study |
| pPL4131 | pRS316 expressing Hse1-ul36CD*-3xHA from CUP1 promoter | Figs. 4 C, 5 B, and S2, B and C | This study |
| pPL4141 | HIS3 conversion of pPL4131 expressing Hse1-ul36CD*-3xHA from CUP1 promoter | Fig. 4 C | This study |
| pJLU40 | pRS316 expressing Vph1-GFP from VPH1 promoter | Fig. 3 A | This study |
| pPL4056 | pRS316 expressing Vph1-GFP- UL36CD from VPH1 promoter | Fig. 3 A | This study |
| pPL4202 | pRS316 expressing Vph1-GFP- ul36CD* from VPH1 promoter | Fig. 3 A | This study |
| pPL4218 | pRS316 expressing Ste3-GFP-M48CD from STE3 promoter; contains 561–591 of Ubp7 (Ubp7 linker) AGDRDIV-mCMV M48 (residues 4–234)-VAITHPLR. | Fig. 3 A | This study |
| pPL4219 | pRS316 expressing Ste3-GFP-m48CD* from STE3 promoter; contains linker residues 561–591 of Ubp7, residues 15–21 of UL36, residues 4–234 of mCMV M48, and the mutations C23A, L110A, Y113A, and Y192A followed by residues (253–260 of UL36) | Fig. 3 A | This study |
| pPL3974 | pET151D expressing UL36CD (residues 15–260 of HSV1 UL36) | Fig. S2 E | This study |
| pPL2554 | HIS3 conversion of pPL2356 (HIS3 replacement of URA3) expressing GFP-Cps1 from Cps1 promoter | Fig. S1 B | This study |
| pPL4500 | pRS316 expressing Ste3-GFP-M48-3xHA from STE3 promoter | Fig. 3 C | This study |
| pPL4502 | pRS316 expressing Rsp5-ubp7*-3xHA from CUP1 promoter | Figs. 5 D and S2 B | This study |
| pPL4513 | pRS316 expressing Ste3-GFP-m48*-3xHA from STE3 promoter | Fig. 3 C | This study |
| pPL4515 | pRS16 expressing Ste3-GFP-ubp7*-3xHA from STE3 promoter | Fig. S1 E | This study |
| pPL4253 | natMX conversion of pPL4147 (natMX replacement of LEU2) expressing Mup1-GFP-Ub from CUP1 promoter | Fig. S2 F | This study |
| pPL4248 | natMX conversion of pPL967 (natMX replacement of LEU2) expressing Ste3-GFP from STE3 promoter | Fig. S2 F | This study |
| pPL4249 | natMX conversion of pPL3484 (natMX replacement of LEU2) expressing Ste3-GFP-Ub from STE3 promoter | Fig. S2 F | This study |
Table II.
Yeast strains used
| Yeast strain | Description | Use | Source |
| BY4742 | MATα his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0 | Throughout | Brachmann et al., 1998 |
| SF838-9D | MATα leu2-3,112 ura3-52 his4-519 ade6 pep4-3 | Fig. S2 D | Raymond et al., 1992 |
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel | Throughout | Banta et al., 1988 |
| PLY3909 | MATα hse1Δ::TRP1 vps27Δ::LEU2 ura3-52 his3-Δ200 lys2-801 suc2-Δ9 pep4 | Fig. 1 D | This study |
| PLY1776 | MATα vps27Δ::LEU2 leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel | Fig. S1 B | This study |
| PLY2858 | MATα vps27Δ::LEU2 ura3-52 his3-Δ200 lys2-801 suc2-Δ9 pep4 | Fig. S1 B | This study |
| GW004 | MATα rsp5Δ::LEU2, pRS416-RSP5 leu2-3112,ade2-,his3-11,trp1-1,can1-100; derived from W303 | Fig. 2 D | Wang et al., 1999 |
| PLY441 vpl23-5 (vps27Δ) | MATα leu2-3,112, ura3-52, his4, ade6 vpl23-5 (vps27Δ) pep4-3 | Fig. S2 D | Raymond et al., 1992 |
| BY4742 YOR124C | MATα ubp2Δ:: Kanr his3Δ0, leu2ΔO, lys2ΔO, uraΔO | Fig. 1 C | Winzeler et al., 1999 |
| PLY3827 | MATα ubp2Δ:: Kanr vps27Δ::HIS3 leu2ΔO, lys2ΔO, uraΔO | Fig. 1 D | This study |
| RH1800 | MATa leu2 his4 ura3 bar1 | Fig. 2 A | Munn et al., 1999 |
| LHY23 | MATa rsp5-1 leu2 ura3 his3 trp1 lys2 bar1 GAL | Fig. 2, A and C | Dunn and Hicke, 2001 |
| KLY201 | MATα end3Δ::Kanr his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0 | Figs. 3 D and S2 F | Liu et al., 2007 |
| BY4742 YJL204C | MATα rcy1Δ::Kanr his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0 | Fig. 3 E | Winzeler et al., 1999 |
| vpl4-21 | MATα vps4Δ leu2-3,112, ura3-52, his4, ade6 (SF838-9D) | Fig. 3 E | Raymond et al., 1992 |
| PLY3623 | MATα mvb12Δ vps36-ΔNZF::TRP1::vps36Δ::HIS vps23-ΔUb (F52Q117W125) leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel | Fig. 5 A | Shields et al., 2009 |
| BY4742 YCL008C | MATα vps23Δ:: KanR his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0 | Fig. 5 C | Winzeler et al., 1999 |
| BY4742 YHL002W | MATα hse1Δ::KanR his3Δ0 leu2Δ0 lys2Δ0 ura3Δ0 | Fig. 5 B | Winzeler et al., 1999 |
| EN67 | MATa ecm21::G418 csr2::G418 bsd2 rog3::natMX rod1 ygr068c aly2 aly1 ldb19 ylr392c bul1 bul2::HIS his3 ura3 leu2 | Figs. 2 B and S1C | Nikko and Pelham, 2009 |
| SUB492 | MATa lys2-801 leu2-3, 112 ura3-52 his3-200 trp1-1[am] ubi1-1::TRP1 ubi2-2::ura3 ubi3-Δ ubi4-Δ::LEU2 [pUB39][pUB100]; pUB39 is 2µ plasmid with CUP1-Ub (wild type); pUB100 the is plasmid that expresses RPL40A tail | Fig. 2 E | Spence et al., 1995 |
| SUB493 | Isogenic to SUB492 except pUB197 replaces pUB39; pUB197 is 2µ plasmid with CUP1-Ub (K63R) | Fig. 2 E | Spence et al., 1995 |
| PLY3378 | MATα end3::KANR gga1Δ::TRP1 gga2Δ::HIS3 | Fig. S2 F | Bilodeau et al., 2004 |
For experiments requiring detection of intact Ubp7 fusions, we used an altered form of the Ubp7 catalytic domain. The catalytic domain of Ubp7 undergoes autocleavage, resulting in a clipped form that apparently remains associated and active. The catalytically dead Ubp7 C618S mutant does not undergo this cleavage (Fig. S2 B). This characteristic has been observed for other catalytic domains such as Usp1, but is not required for its enzymatic activity (Huang et al., 2006). Ubp7 has a GG motif that is localized in a predicted loop that would lie between β11 and β12 of the USP fold “fingers” subdomain of the USP catalytic domain. Mutation of G to S (corresponding to G932 in full-length Ubp7) abolished autocleavage while preserving enzyme activity (Fig. S2 B).
Fluorescence microscopy
Cells were grown to mid-log phase, briefly centrifuged, and resuspended in 0.2% NaN3, 0.2% NaFl, and 100 mM Tris, pH 8.0. GFP and mCherry in cells at 22°C were imaged with an epifluorescence microscope (BX60; Olympus) with a 100× objective lens, NA 1.4. Images were captured with a cooled charge-coupled device camera (Orca R2; Hamamatsu Photonics) using iVision-Mac software (Biovision Technology) and scaled to the brightest and darkest pixel intensity with Photoshop software (Adobe). For cells containing copper-inducible expression plasmids (containing the CUP1 promoter), cells were grown overnight, diluted in fresh media, and then induced with 100 µM CuCl2. Induction of CUP1-dependent expression in rsp5-1 cells was similar except cells were grown at permissive (30°C) and nonpermissive temperatures (37°) during copper-triggered induction lasting 2 h. Cells were grown in media replete with methionine (yeast peptone dextrose [YPD] or synthetic defined [SD] media with 20 µg/ml methionine [Met]) for all Mup1 localization experiments except where noted. Like other transporters such as Gap1, Mup1 can follow a Ub-dependent route from the cell surface into the MVB lumen that is triggered by the addition of its substrate methionine. Alternatively, Mup1 can also bypass the plasma membrane on its route from the Golgi to MVB/endosomes via a GGA-dependent sorting step if synthesized in the presence of methionine (Fig. S2 F). Unlike Gap1, however, fusion of Ub to the C terminus of Mup1-GFP still allowed for proper folding and exit out of the ER, thus providing a tool to bypass the need for ligase-mediated ubiquitination.
Immunoblotting and immunoprecipitation
Whole cell denatured lysates were generated by resuspending pelleted cells in 0.2 M NaOH. After 2 min, cells were pelleted and resuspended in 8 M urea, 5% SDS, 10% glycerol, and 50 mM Tris, pH 6.8, and heated at 70°C for 5 min as described previously (Ren et al., 2008). Samples for immunoblotting Mup1-GFP proteins were prepared similarly, although they were not heated. For immunoprecipitations, whole cell denatured lysates were diluted with 10 volumes of 100 mM Tris, pH 7.5, and 0.4% Triton X-100. Samples were incubated with polyclonal anti-HA for 90 min at 25°C. Immune complexes were captured on fixed Staphylococcus aureus cells (IgGSorb; Enzyme Center). Beads were washed thrice in 0.1 M Tris, pH 7.5, 0.4% Triton X-100, and 0.1% SDS, and analyzed by SDS-PAGE and immunoblotting. Cells carrying the CUP1-myc-Ub plasmid were grown overnight in 50 µM CuCl2 before lysis. Immunoprecipitation from rsp5-1 cells was performed as in Fig. 1 except that cell cultures were divided and grown at 37°C for 1 h before lysis. To assess ubiquitination of Ste3-GFP fused to active and inactive UL36, experiments were performed in end3Δ cells defective for internalization, which facilitated our comparison because they accumulate proteins in a ubiquitinated state at the cell surface (Kölling and Hollenberg, 1994).
Protease treatment
Recombinant UL36 catalytic domain was generated by expressing UL36 from pET151 in BL21 (DE3) star cells. Enzyme was purified from bacterial lysates (French press; Slm-Aminco) using Cobalt Talon resin (Takara Bio, Inc.). In vitro deubiquitinating assays were performed at 37°C for 30 min in PBS containing 0.2 mM DTT. Ub chains (1 µG of K48 or K63; Boston Biochem) were mixed with recombinant UL36CD in the presence and absence of 1 µM Ub-Aldehyde (Boston Biochem). Reactions were terminated by adjusting samples to 2% SDS and heating at 100°C for 10 min.
For protease protection assays, cells (∼15 OD) were grown to mid-log phase. Cells were spheroplasted in 1.4 M sorbitol, 50 mM Tris, pH 8, 10 mM NaN3, and 1 mM DTT containing Zymolyase (AMS Biotechnology) for 1 h. Cells were purified over a cushion of 1.4 M sucrose by centrifugation and lysed at 4°C with 12 passes through a 26-g needle in sorbitol and 50 mM Tris, pH 8.0. Debris was removed from lysates by centrifugation at 1,000 g for 5 min. The resulting supernatant was then incubated in the presence and absence of trypsin (Sigma-Aldrich). Samples were adjusted to 2.5% SDS and 4 M urea, and heated to 100°C for 5 min to halt protease activity.
Online supplemental material
Fig. S1 describes the additional effects of Vps27K>R, the effect of Rsp5 adaptors on ubiquitination of Mup1, and the expression levels of the ESCRT-DUb fusion proteins. Fig. S2 shows the effect of ESCRT-Dubs on CPY sorting, the autocleavage of the Ubp7 catalytic domain, the activity of UL36 toward both K48- and K63-linked Ub chains, and the intracellular itinerary of key cargo proteins used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008121/DC1.
Acknowledgments
This work was supported by an American Hearth Association predoctoral fellowship to D.K. Stringer and National Institutes of Health grant R01GM58202 to R.C. Piper.
Footnotes
Abbreviations used in this paper:
- DUb
- deubiquitinating peptidase
- ESCRT
- endosomal sorting complex required for transport
- MVB
- multivesicular body
- Ub
- Ubiquitin
- UBD
- Ub-binding domain
References
- Amit I., Yakir L., Katz M., Zwang Y., Marmor M.D., Citri A., Shtiegman K., Alroy I., Tuvia S., Reiss Y., et al. 2004. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 18:1737–1752 10.1101/gad.294904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Sato T.K., Banta L.M., Emr S.D. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831 10.1093/emboj/16.8.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banta L.M., Robinson J.S., Klionsky D.J., Emr S.D. 1988. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107:1369–1383 10.1083/jcb.107.4.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau P.S., Urbanowski J.L., Winistorfer S.C., Piper R.C. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534–539 [DOI] [PubMed] [Google Scholar]
- Bilodeau P.S., Winistorfer S.C., Allaman M.M., Surendhran K., Kearney W.R., Robertson A.D., Piper R.C. 2004. The GAT domains of clathrin-associated GGA proteins have two ubiquitin binding motifs. J. Biol. Chem. 279:54808–54816 10.1074/jbc.M406654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M.O., Morvan J., Dupré S., Urban-Grimal D., Haguenauer-Tsapis R., Volland C. 2004. Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell. 15:883–895 10.1091/mbc.E03-04-0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132 [DOI] [PubMed] [Google Scholar]
- Cadwell K., Coscoy L. 2005. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 309:127–130 10.1126/science.1110340 [DOI] [PubMed] [Google Scholar]
- Chellappa R., Kandasamy P., Oh C.S., Jiang Y., Vemula M., Martin C.E. 2001. The membrane proteins, Spt23p and Mga2p, play distinct roles in the activation of Saccharomyces cerevisiae OLE1 gene expression. Fatty acid-mediated regulation of Mga2p activity is independent of its proteolytic processing into a soluble transcription activator. J. Biol. Chem. 276:43548–43556 10.1074/jbc.M107845200 [DOI] [PubMed] [Google Scholar]
- Curtiss M., Jones C., Babst M. 2007. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol. Biol. Cell. 18:636–645 10.1091/mbc.E06-07-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Guo Y., Watson H., Au W.C., Shakoury-Elizeh M., Basrai M.A., Bonifacino J.S., Philpott C.C. 2009. Gga2 mediates sequential ubiquitin-independent and ubiquitin-dependent steps in the trafficking of ARN1 from the trans-Golgi network to the vacuole. J. Biol. Chem. 284:23830–23841 10.1074/jbc.M109.030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dores M.R., Schnell J.D., Maldonado-Baez L., Wendland B., Hicke L. 2010. The function of yeast Epsin and Ede1 ubiquitin-binding domains during receptor internalization. Traffic. 11:151–160 10.1111/j.1600-0854.2009.01003.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R., Hicke L. 2001. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol. Biol. Cell. 12:421–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejercito P.M., Kieff E.D., Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357–364 10.1099/0022-1317-2-3-357 [DOI] [PubMed] [Google Scholar]
- Erpapazoglou Z., Froissard M., Nondier I., Lesuisse E., Haguenauer-Tsapis R., Belgareh-Touzé N. 2008. Substrate- and ubiquitin-dependent trafficking of the yeast siderophore transporter Sit1. Traffic. 9:1372–1391 10.1111/j.1600-0854.2008.00766.x [DOI] [PubMed] [Google Scholar]
- Galan J.M., Wiederkehr A., Seol J.H., Haguenauer-Tsapis R., Deshaies R.J., Riezman H., Peter M. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21:3105–3117 10.1128/MCB.21.9.3105-3117.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Gredmark S., Schlieker C., Quesada V., Spooner E., Ploegh H.L. 2007. A functional ubiquitin-specific protease embedded in the large tegument protein (ORF64) of murine gammaherpesvirus 68 is active during the course of infection. J. Virol. 81:10300–10309 10.1128/JVI.01149-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23 10.1093/nar/30.6.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Stenmark H. 2006. Working out coupled monoubiquitination. Nat. Cell Biol. 8:1218–1219 10.1038/ncb1106-1218 [DOI] [PubMed] [Google Scholar]
- Hicke L., Schubert H.L., Hill C.P. 2005. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6:610–621 10.1038/nrm1701 [DOI] [PubMed] [Google Scholar]
- Hoeller D., Hecker C.M., Wagner S., Rogov V., Dötsch V., Dikic I. 2007. E3-independent monoubiquitination of ubiquitin-binding proteins. Mol. Cell. 26:891–898 10.1016/j.molcel.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H.D., Jentsch S. 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 102:577–586 10.1016/S0092-8674(00)00080-5 [DOI] [PubMed] [Google Scholar]
- Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D’Andrea A.D. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8:339–347 [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kato M., Miyazawa K., Kitamura N. 2000. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem. 275:37481–37487 10.1074/jbc.M007251200 [DOI] [PubMed] [Google Scholar]
- Kattenhorn L.M., Korbel G.A., Kessler B.M., Spooner E., Ploegh H.L. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell. 19:547–557 10.1016/j.molcel.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Katz M., Shtiegman K., Tal-Or P., Yakir L., Mosesson Y., Harari D., Machluf Y., Asao H., Jovin T., Sugamura K., Yarden Y. 2002. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic. 3:740–751 10.1034/j.1600-0854.2002.31006.x [DOI] [PubMed] [Google Scholar]
- Kee Y., Lyon N., Huibregtse J.M. 2005. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 24:2414–2424 10.1038/sj.emboj.7600710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y., Muñoz W., Lyon N., Huibregtse J.M. 2006. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J. Biol. Chem. 281:36724–36731 10.1074/jbc.M608756200 [DOI] [PubMed] [Google Scholar]
- Kim H.C., Huibregtse J.M. 2009. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29:3307–3318 10.1128/MCB.00240-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.Y., Olzmann J.A., Barsh G.S., Chin L.S., Li L. 2007. Spongiform neurodegeneration-associated E3 ligase Mahogunin ubiquitylates TSG101 and regulates endosomal trafficking. Mol. Biol. Cell. 18:1129–1142 10.1091/mbc.E06-09-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.T., Oh S.E., Lee Y.O., Gibson W., Ahn J.H. 2009. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J. Virol. 83:12046–12056 10.1128/JVI.00411-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölling R., Hollenberg C.P. 1994. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 13:3261–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M., Kitamura N. 2005. The Hrs/STAM complex in the downregulation of receptor tyrosine kinases. J. Biochem. 137:1–8 10.1093/jb/mvi001 [DOI] [PubMed] [Google Scholar]
- Kulathu Y., Akutsu M., Bremm A., Hofmann K., Komander D. 2009. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 16:1328–1330 10.1038/nsmb.1731 [DOI] [PubMed] [Google Scholar]
- Lauwers E., Jacob C., André B. 2009. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185:493–502 10.1083/jcb.200810114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. 2010. The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20:196–204 10.1016/j.tcb.2010.01.004 [DOI] [PubMed] [Google Scholar]
- Liu K., Hua Z., Nepute J.A., Graham T.R. 2007. Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol. Biol. Cell. 18:487–500 10.1091/mbc.E06-07-0592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A., Raiborg C., Santini F., Keen J.H., Stenmark H., Benovic J.L. 2003. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell. 5:709–722 10.1016/S1534-5807(03)00321-6 [DOI] [PubMed] [Google Scholar]
- McCullough J., Clague M.J., Urbé S. 2004. AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166:487–492 10.1083/jcb.200401141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.L., Malotky E., O’Bryan J.P. 2004. Analysis of the role of ubiquitin-interacting motifs in ubiquitin binding and ubiquitylation. J. Biol. Chem. 279:33528–33537 10.1074/jbc.M313097200 [DOI] [PubMed] [Google Scholar]
- Munn A.L., Heese-Peck A., Stevenson B.J., Pichler H., Riezman H. 1999. Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell. 10:3943–3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E., Pelham H.R. 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 10:1856–1867 10.1111/j.1600-0854.2009.00990.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva S., Vieira N., Nondier I., Haguenauer-Tsapis R., Casal M., Urban-Grimal D. 2009. Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter: role of lysine 63-linked ubiquitin chains. J. Biol. Chem. 284:19228–19236 10.1074/jbc.M109.008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper R.C., Katzmann D.J. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23:519–547 10.1146/annurev.cellbio.23.090506.123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S., Sigismund S., Faretta M., Guidi M., Capua M.R., Bossi G., Chen H., De Camilli P., Di Fiore P.P. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 416:451–455 10.1038/416451a [DOI] [PubMed] [Google Scholar]
- Puertollano R., Bonifacino J.S. 2004. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 6:244–251 10.1038/ncb1106 [DOI] [PubMed] [Google Scholar]
- Raymond C.K., Howald-Stevenson I., Vater C.A., Stevens T.H. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 3:1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R. 2001. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20:5176–5186 10.1093/emboj/20.18.5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Hurley J.H. 2010. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 29:1045–1054 10.1038/emboj.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Kee Y., Huibregtse J.M., Piper R.C. 2007. Hse1, a component of the yeast Hrs-STAM ubiquitin-sorting complex, associates with ubiquitin peptidases and a ligase to control sorting efficiency into multivesicular bodies. Mol. Biol. Cell. 18:324–335 10.1091/mbc.E06-06-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Pashkova N., Winistorfer S., Piper R.C. 2008. DOA1/UFD3 plays a role in sorting ubiquitinated membrane proteins into multivesicular bodies. J. Biol. Chem. 283:21599–21611 10.1074/jbc.M802982200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78:363–397 10.1146/annurev.biochem.78.082307.091526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D., Kumar S. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10:398–409 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- Row P.E., Prior I.A., McCullough J., Clague M.J., Urbé S. 2006. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 281:12618–12624 10.1074/jbc.M512615200 [DOI] [PubMed] [Google Scholar]
- Roxrud I., Stenmark H., Malerød L. 2010. ESCRT & Co. Biol. Cell. 102:293–318 10.1042/BC20090161 [DOI] [PubMed] [Google Scholar]
- Schlieker C., Weihofen W.A., Frijns E., Kattenhorn L.M., Gaudet R., Ploegh H.L. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell. 25:677–687 10.1016/j.molcel.2007.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott P.M., Bilodeau P.S., Zhdankina O., Winistorfer S.C., Hauglund M.J., Allaman M.M., Kearney W.R., Robertson A.D., Boman A.L., Piper R.C. 2004. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 6:252–259 10.1038/ncb1107 [DOI] [PubMed] [Google Scholar]
- Shields S.B., Oestreich A.J., Winistorfer S., Nguyen D., Payne J.A., Katzmann D.J., Piper R. 2009. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 185:213–224 10.1083/jcb.200811130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J., Sadis S., Haas A.L., Finley D. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15:1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Davis R.W. 1977. Production of a functional eukaryotic enzyme in Escherichia coli: cloning and expression of the yeast structural gene for imidazole-glycerolphosphate dehydratase (his3). Proc. Natl. Acad. Sci. USA. 74:5255–5259 10.1073/pnas.74.12.5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu T., Yamauchi E., Shibata H., Maki M., Ohta T., Konishi H. 2010. Distinct functions of human MVB12A and MVB12B in the ESCRT-I dependent on their posttranslational modifications. Biochem. Biophys. Res. Commun. 399:232–237 10.1016/j.bbrc.2010.07.060 [DOI] [PubMed] [Google Scholar]
- Uchiki T., Kim H.T., Zhai B., Gygi S.P., Johnston J.A., O’Bryan J.P., Goldberg A.L. 2009. The ubiquitin-interacting motif protein, S5a, is ubiquitinated by all types of ubiquitin ligases by a mechanism different from typical substrate recognition. J. Biol. Chem. 284:12622–12632 10.1074/jbc.M900556200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski J.L., Piper R.C. 2001. Ubiquitin sorts proteins into the intralumenal degradative compartment of the late-endosome/vacuole. Traffic. 2:622–630 10.1034/j.1600-0854.2001.20905.x [DOI] [PubMed] [Google Scholar]
- Wang G., Yang J., Huibregtse J.M. 1999. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol. Cell. Biol. 19:342–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Herr R.A., Chua W.J., Lybarger L., Wiertz E.J., Hansen T.H. 2007. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177:613–624 10.1083/jcb.200611063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., Avaro S., Prescianotto-Baschong C., Haguenauer-Tsapis R., Riezman H. 2000. The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149:397–410 10.1083/jcb.149.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.L., Urbé S. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8:355–368 10.1038/nrm2162 [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Woelk T., Oldrini B., Maspero E., Confalonieri S., Cavallaro E., Di Fiore P.P., Polo S. 2006. Molecular mechanisms of coupled monoubiquitination. Nat. Cell Biol. 8:1246–1254 10.1038/ncb1484 [DOI] [PubMed] [Google Scholar]