Abstract
In about half of all human cancers, the tumor suppressor p53 protein is either lost or mutated, frequently resulting in the expression of a transcriptionally inactive mutant p53 protein. Loss of p53 function is well known to influence cell cycle checkpoint controls and apoptosis. But it is now clear that p53 regulates other key stages of metastatic progression, such as cell migration and invasion. Moreover, recent data suggests that expression of mutant p53 is not the equivalent of p53 loss, and that mutant p53s can acquire new functions to drive cell migration, invasion, and metastasis, in part by interfering with p63 function.
Introduction
The p53 protein is probably the best known of all tumor suppressors. In about half of all human cancers, p53 is either lost or mutated in a way that compromises its function. It is well established that loss or inhibition of p53 prevents cellular senescence and apoptosis. And more recently, it has become clear that in situations where p53 function is compromised, other key processes that impact tumor progression, such as cell migration and invasion, are altered. In various cancers, mutations have been found throughout Tp53, the gene that encodes p53. Many of these mutations give rise to mutant p53 proteins that are highly expressed. Recent data indicate that some of the most common mutant p53 proteins have, in addition to losing transcriptional function, acquired a gain of function: these mutants drive tumor cell migration and metastasis that is in part a result of their ability to interfere with another p53 family member, p63.
Loss of p53 is associated with an EMT-like phenotype
Normally, p53 levels are suppressed by the activity of the E3 ubiquitin ligase MDM2 (mouse double minute 2; Haupt et al., 1997; Kubbutat et al., 1997). However, cellular stresses, such as DNA damage or oncogene activation, can alleviate this suppression, leading to p53 stabilization. Thus, in tumors where p53 expression is lost, the consequent inability of cells to engage apoptosis or senescence after stressful insults contributes to tumor formation. However, in order to metastasize, tumor cells must disseminate from the primary tumor, invade the stroma to reach blood or lymph vessels, and then extravasate to colonize other tissues. In addition to affecting apoptosis and cellular senescence, p53 loss has recently been shown to influence cell motility in a way that can contribute to a tumor’s invasive and metastatic potential.
The Slug/Snail and Twist families of transcription factors are master regulators of key events in embryonic development, in particular the epithelial–mesenchymal transition (EMT; Shiota et al., 2008). These transcriptional programs are characterized by the loosening of cell–cell junctions and the acquisition of a more motile mesenchymal phenotype. More recently, Slug/Snail and Twist proteins have been shown to down-regulate molecules involved in stabilizing cell–cell junctions (such as E-cadherin), and to up-regulate components of the migratory machinery in order to become invasive (Bolós et al., 2003; Shih et al., 2005). Clearly, there is an important interaction between these master regulators of EMT and p53 (Fig. 1; Wang et al., 2009b). It is thought that in order to drive EMT, Twist opposes p53 function, indicating that p53 maintains a transcriptional program to prevent EMT and that loss of this suppression may contribute to the induction of an EMT-like phenotype in p53-null tumors (Shiota et al., 2008; Smit and Peeper, 2008). The reciprocal relationship between p53 and the execution of an EMT-like program is further exemplified by recent observations that expression of p53 can promote MDM2-mediated degradation of Slug to enhance E-cadherin expression (Wang et al., 2009b). Consistent with the paradigm that carcinogenesis carries many characteristics of EMT, loss of E-cadherin is frequently observed in cancer, and this has been shown to be capable of driving metastasis in animal models (Derksen et al., 2006). In p53+/− mice, most of the chemically induced papillomas with undetectable p53 expression display E-cadherin loss from the cell membrane, whereas papillomas that retain wild-type p53 expression display normal E-cadherin expression (Cano et al., 1996). Increased Slug expression can also be identified in many p53-null tumors, and this correlates with loss of E-cadherin (Shih et al., 2005; Castro Alves et al., 2007), which indicates that p53 loss can activate a more mesenchymal phenotype in human cancer. However, as not all tumors with p53 loss can be regarded as having undergone EMT, relatively straightforward relationships between p53 expression and EMT regulators that have been identified in ex vivo and animal cancer models may not be considered as axioms with respect to the human disease.
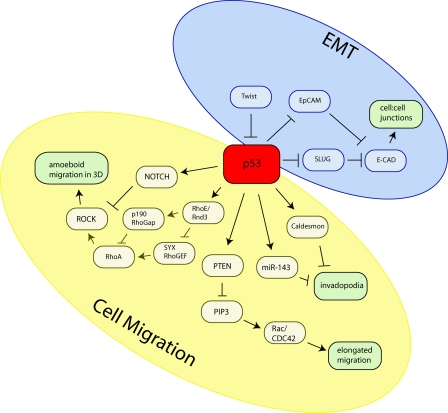
Figure 1.
p53 opposes EMT and cell migration to prevent metastasis. p53 plays a role in opposing EMT and cell migration. A hallmark of EMT is loss of E-cadherin, and p53 can prevent this by inhibiting Slug or the adhesion molecule EpCam expression. Furthermore, loss of p53 or decreased p53 activity after Twist expression can therefore drive EMT. p53 can also inhibit invasive migration. This can be mediated via increased expression of Caldesmon or miRNA-143 to oppose invadopodia formation. By transactivating PTEN, p53 can reduce PIP3 (and thereby Rac) levels, resulting in inhibition of mesenchymal/elongated motility. p53 can also inhibit amoeboid cell motility by preventing activation of ROCK, either by inducing Notch or by promoting RhoE (Rnd3)-mediated inhibition of RhoA. E-CAD, E-cadherin.
p53 and cell interactions with the extracellular matrix
After their dissemination from tumors, metastatic cells acquire the capacity to actively migrate and invade through the stroma. Indeed, several studies have shown that deletion of p53 can change the morphology and polarity of fibroblasts, and that this allows them to migrate faster in scratch-wound assays and through 3D matrices, such as Matrigel (Alexandrova et al., 2000; Gadéa et al., 2002; Guo et al., 2003; Guo and Zheng, 2004; Gadea et al., 2007. Moreover, studies linking loss of p53 to increased cell motility are not restricted to fibroblasts; similar observations have been made in other cell types including keratinocytes (Lefort et al., 2007), epithelial cancer lines (HCT116 and H1299; Sablina et al., 2003; Xia and Land, 2007), and neurons where increased growth cone motility has been associated with compromised p53 function (Qin et al., 2009). It has been known for some time that p53-regulated genes include components of the adhesive machinery that are known to contribute to cell motility through the stroma. Expression of fibronectin and collagens, ECM proteins that are intrinsically associated with the acquisition and implementation of an elongated mesenchymal migratory phenotype, are strongly increased after p53 loss (Guo and Zheng, 2004), potentially enhancing the interaction between cell and ECM to allow for cell motility.
It is clear that both the synthesis/deposition of ECM proteins and their degradation by extracellular proteases are important during the invasive process. Two recent studies have revealed a role for p53 in suppressing invadopodia that can trigger degradation of components of the ECM and basal membrane to allow cells to access the stroma. In one of these studies, the actin-binding protein Caldesmon was identified as a transcriptional target of p53. Caldesmon has been shown to inhibit podosome formation after oncogenic transformation by activated forms of Src (Mukhopadhyay et al., 2009). Additionally, p53 has been shown to regulate the microRNA (miRNA) 143 that might target various components of the invadopodia formation machinery and therefore inhibit podosome formation (Fig. 1; Quintavalle et al., 2010).
Loss of p53 activates Rho GTPases and their associated signaling pathways
Although p53’s ability to control expression of cytoskeletal and ECM proteins is undoubtedly important, it is also evident that signaling pathways modulating cell migration and the chemotactic responses are influenced by p53. Most signals leading to altered cell migration and invasion converge on the Rho family of small GTPases. Members of this family, including Rac, cdc42, and Rho, control actin dynamics and are integral to cytoskeletal changes accompanying tumor cell migration and invasion (Bishop and Hall, 2000; Heasman and Ridley, 2008). The upstream control of RhoGTPase activity is primarily mediated by molecules that influence their GTP loading, including RhoGAPs (Rho GTPase-activating proteins), which turn them off by increasing GTP hydrolysis, and RhoGEFs (Rho guanine nucleotide exchange factors) that activate them (Sanz-Moreno and Marshall, 2009).
The balance between the activities of Rac and Rho has been well-established to influence the way in which tumor cells migrate (Sahai and Marshall, 2003; Sanz-Moreno and Marshall, 2009). When Rac predominates, migration proceeds with the elongated morphology characteristic of tumor cells that have assumed a mesenchymal phenotype. Thus, given p53’s ability to suppress Slug and its attendant EMT-like program, it might be expected that p53-deficient cells would have increased levels of active Rac. Possible links between p53 and Rac can be garnered by considering findings that the PTEN gene, a lipid phosphatase that reduces levels of PIP3, leading to inhibition of PIP3-dependent GEFs for Rac and cdc42, can be transactivated by p53 (Stambolic et al., 2001). However, although Rac and cdc42 activation was initially reported in p53-null cells, most subsequent studies have focused on increased activity of RhoA after loss of p53 (Table I and Fig. 1; Guo et al., 2003; Sablina et al., 2003; Guo and Zheng, 2004; Roger et al., 2006; Gadea et al., 2007; Lefort et al., 2007; Xia and Land, 2007; Qin et al., 2009).
Table I.
Loss of p53 function in cell migration
| Primary transcriptional targets | Posttranscriptional effects | Secondary transcriptional targets | Downstream consequences | References | |
| Cellular level | Tissue level | ||||
| Unknown | Slug ↑ (due to reduced mdm2-mediated degradation) | E-Cadherin ↓ | Cell–cell junctions ↓ | EMT-like phenotype | Cano et al., 1996; Shiota et al., 2008; Wang et al., 2009b |
| EpCAM ↑ | Unknown | NA | Cell–cell junctions ↓ | EMT-like phenotype | Sankpal et al., 2009 |
| RhoE ↓ | ROCK ↑ | NA | Rho signaling ↑ | Cell migration (2D and 3D) ↑ | Ongusaha et al., 2006; Riento and Ridley, 2006 |
| RhoE ↓ | p190RhoGAP ↓ | NA | Rho signaling ↑ | Cell migration (2D and 3D) ↑ | Wennerberg et al., 2003 |
| RhoE ↓ | Syx RhoGEF ↑ | NA | Rho signaling ↑ | Cell migration (2D and 3D) ↑ | Goh and Manser, 2010 |
| ROCK ↑ | NA | NA | Rho signaling ↑ | Cell migration (2D and 3D) ↑ | Guo and Zheng, 2004; Gadea et al., 2007; Lefort et al., 2007; Qin et al., 2009 |
| Notch ↓ | NA | MRCKα↑, ROCK↑ | Rho signaling ↑ | Cell migration (2D and 3D) ↑ | Lefort et al., 2007 |
| Fibronectin ↑ | NA | NA | Rho signaling ↑ | Cell migration (2D and 3D) ↑ | Guo and Zheng, 2004 |
| Collagens ↑ | NA | NA | Rho signaling ↑ | Cell migration (2D and 3D) ↑ | Zhao et al., 2000 |
| LASP1 ↑ | NA | NA | Rho signaling ↑ | Cell migration (2D and 3D) ↑ | Wang et al., 2009a |
| PTEN ↓ | Various | Rac/ Cdc42↑ | Rac and PKB/Akt signalling ↑ | Cell migration (2D and 3D) ↑ | Stambolic et al., 2001; Guo et al., 2003; Guo and Zheng, 2004; Gadea et al., 2007 |
| MMP2 ↑ | NA | NA | NA | ECM degradation ↑ | Delassus et al., 2010 |
| miRNA-143 ↑ | Various | NA | Invadopodia ↑ | ECM degradation ↑ | Quintavalle et al., 2010 |
| Caldesmon ↑ | NA | NA | Invadopodia ↑ | ECM degradation ↑ | Mukhopadhyay et al., 2009 |
Selected list of genes with an involvement in EMT, cell migration, and ECM degradation that are regulated by p53. p53 status, suppression or loss of wild-type p53.
RhoA acts through its main effector Rho-associated coiled-coil containing protein kinase (ROCK) to promote contractility, favoring rounded amoeboid migration as opposed to the elongated mode favored by Rac (Sahai and Marshall, 2003). It is not clear how amoeboid migration fits into an EMT paradigm for tumor progression, but it certainly drives very rapid translocation of tumor cells through Matrigel and collagen gels, and intravital imaging indicates that this is how many tumor cells migrate in vivo (Pinner and Sahai, 2008). There is good consensus in the literature that loss of p53 leads to increased levels of GTP-bound, active RhoA and activated ROCK, although the molecular details of how this occurs and conclusions as to whether this represents the sole route from p53 loss to altered migration varies somewhat between studies (Guo and Zheng, 2004; Gadea et al., 2007; Lefort et al., 2007; Qin et al., 2009). Indeed, Gadéa and coworkers have reported that the morphological changes occurring in p53−/− fibroblasts can be caused by activation of cdc42 (Gadéa et al., 2002) and/or RhoA/ROCK signaling (Gadea et al., 2007). However, given that the enhanced amoeboid migration of p53-null fibroblasts can be opposed by pharmacological inhibition of ROCK (Gadea et al., 2007), it is likely that RhoA is an important effector of p53 with regard to this type of tumor cell migration. It is not clear how p53 loss leads to ROCK activation, but recent studies have suggested two plausible mechanisms by which this might occur. In keratinocytes, the expression of ROCK is repressed by p53 via down-regulation of Notch1 (Lefort et al., 2007). Additionally, the atypical Rho protein RhoE (or Rnd3) is a p53 target gene that opposes activation of RhoA (Ongusaha et al., 2006). Indeed, this can be mediated via RhoE’s ability both to inhibit Syx (a GEF that activates RhoA; Goh and Manser, 2010) and to activate p190-RhoGAP (a key RhoA inactivator; Wennerberg et al., 2003). Furthermore, loss of p53 has been shown to reduce phosphorylation of p190-RhoGAP on Tyr1105 in Ras-transformed cells (Xia and Land, 2007) to oppose RhoGAP activity (Fig. 1). These observations collectively provide evidence for signaling mechanisms through which p53 loss can drive increased activity of the RhoA/ROCK axis to promote cell motility and invasion during tumor progression.
Mutant p53 proteins drive an aggressive cancer phenotype by gain of function
Although p53 knockout mice are highly tumor prone, these lesions do not metastasize frequently, nor do they generally display invasive pathology (Attardi and Jacks, 1999). This suggests that p53 loss is not, in itself, sufficient to drive invasive migration in vivo. However, in many human tumors, p53 expression is not lost, but the gene acquires point mutations that disrupt the ability of the p53 protein to bind to DNA. Two p53 mutants that are commonly found in human cancer and that have been extensively used to study p53’s role in cell migration are R273H (R270H in mice), which directly compromises DNA binding, and R175H (R172H in mice), which causes a global conformational distortion of p53 (Cho et al., 1994). These mutations inhibit p53’s ability to act as a transcription factor, accounting for their reduced ability to function as tumor suppressors. Moreover, these mutant p53 proteins are often expressed at very high levels in cancer cells, and a growing body of evidence now supports additional gain-of-function roles for mutant p53s in the context of tumorigenesis, cancer invasion, and metastasis (Oren and Rotter, 2010) Indeed, introduction of mutant p53s (R172H or R270H) leads to a marked increase in the incidence of highly metastatic carcinomas in various mouse models (Liu et al., 2000; Lang et al., 2004; Olive et al., 2004; Heinlein et al., 2008; Doyle et al., 2010; Morton et al., 2010).
Mutant p53 proteins and cell migration
Wild-type p53 can be inhibited in trans by p53 mutants under conditions of high mutant p53 expression, so it is only possible to assign bona fide gain of function to mutant p53s when they are expressed in a p53-null background. Using both cell-based approaches and knock-in mice, mutant p53s have been shown to acquire new functions in processes such as proliferation, resistance to apoptosis, genomic instability, somatic cell reprogramming, and cell migration and invasion (Li et al., 1998; Gurtner et al., 2010; Liu et al., 2010; Oren and Rotter, 2010; Sarig et al., 2010; Stambolsky et al., 2010). Although many of these processes will likely contribute to tumor aggressiveness, here we will focus on the gain of function of mutant p53 in driving tumor cell migration.
Gain of function of mutant p53 in driving cell migration has been shown across different contexts, including cell movement on 2D surfaces, invasive-type migration through 3D matrices (such as Matrigel), and metastasis of mutant p53 cells implanted as xenografts (Dhar et al., 2008; Junk et al., 2008; Adorno et al., 2009; Muller et al., 2009). To gain insight into how this gain of function is achieved by mutant p53, several groups have addressed the possibility that mutant p53 expression changes the transcriptional profile of cells in a p53-null background. Interestingly, many mutant p53-regulated genes are ones that would be expected to influence cell migration either directly or indirectly (Table II). Indeed, mutant p53s can act via Slug or Twist to induce partial EMT-like transitions that are reflected in their ability to suppress E-cadherin synthesis (Wang et al., 2009b; Kogan-Sakin et al., 2010), or this can be mediated via suppression of the anti-invasive gene CCN-5/WISP2 (Dhar et al., 2008). Many cancer cells invade the stroma using an amoeboid migratory mode that is promoted by increased RhoA/ROCK signaling, and several studies have found components of this pathway to be transcriptionally regulated by mutant p53s. Indeed, recent studies indicate that both the guanine nucleotide dissociation inhibitor (RhoGDI; Bossi et al., 2008) and an exchange factor for RhoA (Rho GEF-H1; Mizuarai et al., 2006) are up-regulated by mutant p53s, and more work will be needed to determine how the combination of these changes impacts on GTP-RhoA levels and the acquisition of amoeboid-type migration in cancer.
Table II.
Mutant p53-regulated events that impact cell migration
| “Direct” target genes | Via p63 inhibition | Secondary machinery | Downstream consequences | References | |
| Cellular level | Tissue level | ||||
| RhoGEF-H1 ↑ | Unknown | NA | Integrin and Rho GTPase signalling ↑ | Cell growth and migration ↑ | Mizuarai et al., 2006 |
| RhoGDI ↑ | Unknown | NA | Integrin and Rho GTPase signalling ↑ | Cell growth and migration ↑ | Bossi et al., 2008 |
| IGF1R ↑ | Unknown | NA | Integrin and Rho GTPase signalling ↑ | Cell growth and migration ↑ | Bossi et al., 2008 |
| Paxillin-β ↑ | Unknown | NA | Integrin and Rho GTPase signalling ↑ | Cell growth and migration ↑ | Bossi et al., 2008 |
| WISP2/CCN5 ↓ | Unknown | NA | Integrin and Rho GTPase signalling ↑ | Cell growth and migration ↑ | Dhar et al., 2008 |
| Sharp-1 ↓ | Yes | NA | Unknown | Tumor cell migration and invasion ↑ | Adorno et al., 2009 |
| Cyclin G2 ↓ | Yes | NA | Unknown | Tumor cell migration and invasion ↑ | Adorno et al., 2009 |
| Unknown | Yes | RCP-dependent trafficking | α5β1 integrin/EGFR recycling | Tumor cell migration and invasion ↑ | Muller et al., 2009 |
| Twist ↑ | Unknown | NA | Unknown | EMT | Kogan-Sakin et al., 2010 |
Selected list of genes/proteins that affect EMT, cell migration, invasion, and metastasis that are regulated by mutant p53 in a gain-of-function manner. p53 status, expression of mutant p53s in p53−/− background.
Mechanisms of mutant p53’s gain of function
Overall, two main mechanisms that could underlie mutant p53’s gain of function have been intensively studied by many groups. First, mutant p53 has been proposed to be a transcription factor in its own right. Even though the majority of mutations within p53 occur in the DNA binding domain, mutant p53s might still influence promoter activity via the N-terminal transactivation domain, and, as the expression of mutant p53 is often very high in many cancers, this may be sufficient to drive the expression of its own set of target genes (Weisz et al., 2004; Zalcenstein et al., 2006). Indeed, the ability of mutant p53 to oppose apoptosis is ablated by removal of its N-terminal transactivation domain (Lin et al., 1995; Matas et al., 2001). However, mutant p53 is also known to operate by interfering with and/or modifying the function of other proteins. For instance, mutant p53 can interact with Mre11 to prevent ataxia telangiectasia mutated (ATM)-dependent DNA repair (Song et al., 2007; Liu et al., 2010). Importantly, p53 is also known to impose upon the function of other transcription factors, including the well-studied p53 family members, p63 and p73, that share a homologous transactivation domain, a DNA binding domain, and an oligomerization domain. Indeed, both cell-based assays (Di Como et al., 1999; Strano et al., 2000, 2002) and knock-in mice expressing mutant forms of p53 support a mechanism by which mutant p53s interact with p63 and p73, and negatively regulate their function (Lang et al., 2004; Li and Prives, 2007). For example, knockdown of either p63 or p73 is as effective as expression of a mutant p53 in increasing the colony-forming potential of fibroblasts from p53−/− mice. Furthermore, knockdown of p63 or p73 does not further increase colony formation in cells expressing a mutant p53, which indicates the likelihood that these three p53 family members may be acting on similar pathways to tumorigenesis (Lang et al., 2004). Moreover, removal of the transactivation domain from mutant p53s does not interfere with its invasive capabilities, suggesting that gain of function in driving invasion is predominantly mediated by inhibition of p63 and/or p73 (Adorno et al., 2009; Muller et al., 2009).
p63 is an effector of mutant p53
p63 expression is driven by two alternative promoters, resulting in a full-length TA isoform or an N-terminally truncated ΔN form that lacks the transactivation domain. Furthermore, ΔNp63 and TAp63 transcripts can be spliced to yield α, β, or γ variants with alternative C termini. Not only do these isoforms and variants have different expression patterns according to cell type and differentiation status, they have been reported to possess different and sometimes opposing functions, which complicates the understanding of p63 function. cDNA microarray data indicates that overexpression of ΔNp63 or TAp63 results in largely nonoverlapping gene expression profiles, which suggests that these two p63 isoforms could have different target genes (Wu et al., 2003; Carroll et al., 2006). However, observations that ΔNp63 opposes TAp63 as an inducer of apoptosis suggest that it might exert inhibition over the TA isoform, nominate TAp63 as a tumor suppressor, and nominate ΔNp63 to be a tumor promoter (Candi et al., 2007).
To be a suppressor of tumor growth and metastasis, one might expect p63 to be mutated or lost in human cancers. Although mutations in p63 have rarely been found, loss of p63 is seen in a range of tumor types correlating closely with the invasiveness of these cancers (Park et al., 2000; Urist et al., 2002; Wang et al., 2002; Koga et al., 2003; Chen et al., 2004). With regard to invasiveness, p63 loss may be restricted to the TAp63 isoform, reinforcing the view that TAp63 functions as a suppressor of metastasis (Park et al., 2000; Chen et al., 2004). Furthermore, p63+/− mice can spontaneously develop solid malignant lesions, which primarily include squamous cell and histiocytic sarcomas that are highly metastatic (Flores et al., 2005). Interestingly, most tumor formation in p63+/− mice is accompanied by loss of the remaining p63 allele, and in tumors where this does not occur, it is primarily the ΔNp63 isoform that remains (Flores et al., 2005). More recent data obtained in TAp63-specific knockout mice indicate that TAp63 is required to drive Ras-dependent senescence and that its loss results in rapid p53-independent tumorigenesis, which further supports a role for TAp63 in tumor suppression (Guo et al., 2009). At the cellular level, it is clear that siRNA knockdown of p63 increases invasive migration of H1299 nonsmall lung cell carcinoma cells (Adorno et al., 2009; Muller et al., 2009). H1299 cells predominantly express TAp63 (Muller et al., 2009), and expression of mutant p53 recapitulates the effects of TAp63 loss, which indicates that mutant p53’s gain of function may operate by specifically inhibiting the TA isoform of p63 (Adorno et al., 2009; Muller et al., 2009). These data also further demonstrate that the TAp63 isoform functions as an inhibitor of invasion.
So how do mutant p53s inhibit p63? Physical interactions between mutant p53s and p63 that directly impair its transcriptional functions have been described previously (Gaiddon et al., 2001; Strano et al., 2002; Li and Prives, 2007). However, the relationship between mutant p53’s ability to bind p63 and its capacity to functionally inhibit it to promote invasion is not straightforward. Although both the R175H and R273H mutants of p53 are equally capable of inhibiting TAp63 and promoting invasion, p53R175H binds more tightly to TAp63α in coimmunoprecipitation experiments (Gaiddon et al., 2001; Strano et al., 2002; Li and Prives, 2007). Furthermore, a C-terminally truncated mutant p53 does not drive invasion, but still interacts with TAp63α, which suggests that mutant p53’s inhibition of p63 is not dictated exclusively by the capacity of the two proteins to associate with one another. Moreover, there are likely other members of the mutant p53–p63 complex that influence the way in which the two proteins associate. Adorno et al. (2009) have shown a role for TGF-β signaling in promoting the ability of mutant p53 to bind to and inhibit p63. These investigators demonstrated that SMAD2 phosphorylated downstream of TGF-β can serve as a platform for assembly of the mutant p53–p63 complex. This pathway illustrates an important mechanism by which TGF-β can control invasion. However, the relationships between TGF-β signaling and mutant p53 are not straightforward, and it would seem that TGF-β signaling is not always required for mutant p53 to function (Kalo et al., 2007; Muller et al., 2009). It is well known that TGF-β signaling has many complex and opposing roles in cancer progression (Jakowlew, 2006), and the involvement of phospho-SMAD2 in the regulation of p63 and cell migration by mutant p53 sheds some interesting light on at least some of the functions of TGF-β in cancer progression.
Downstream of p63 to drive invasion and metastasis
Transcriptome studies indicate that knockdown of p63 increases expression of known mediators of motility, invasion, and metastasis (Barbieri et al., 2006; Carroll et al., 2006; Gu et al., 2008). To identify factors downstream of p63 that were responsible for invasion, Adorno et al. (2009) investigated genes that were coregulated by mutant p53 and TGF-β, and identified Sharp-1 and Cyclin G2 as important targets. Suppression of either Sharp-1 or Cyclin G2 mimicked mutant p53’s capacity to drive cell migration, and low expression of Sharp-1 and Cyclin G2 was associated with poor prognosis and recurrence in breast cancer. Collectively, these data indicate that Sharp-1 and Cyclin G2 are targets of p63 in regard to its ability to suppress tumor cell migration and metastasis (Fig. 2 A; Adorno et al., 2009).
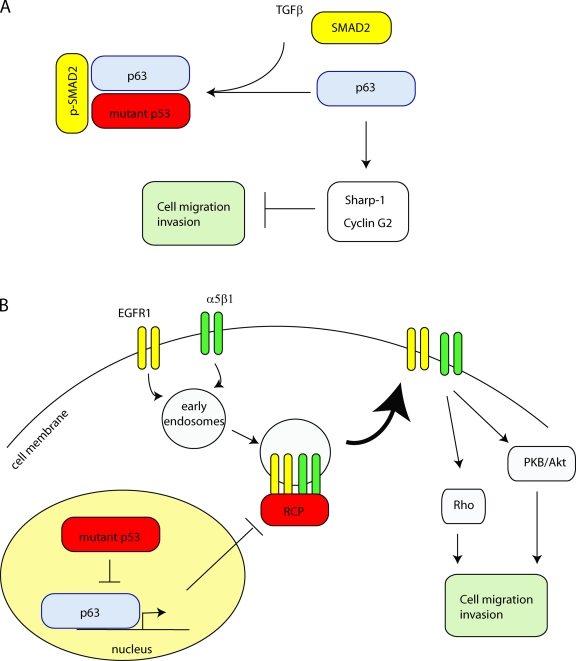
Figure 2.
Mutant p53 regulates cell migration and invasion by inhibiting p63. (A) Upon TGF-β induction, SMAD2 is phosphorylated and promotes binding of mutant p53 to p63, alleviating p63-mediated suppression of Sharp-1 and Cyclin G2 to allow for cell migration and invasion. (B) p63 inhibits activation of RCP (through transcriptional targets that are currently unknown) to prevent α5β1 integrin and EGFR recycling to the plasma membrane. Upon expression of mutant p53, p63 activity is suppressed, resulting in enhanced RCP-driven recycling of α5β1 integrin and EGFR. This activates Rho and PKB/Akt to promote cell migration and invasion.
But what are the components of the cell’s intracellular signaling network and/or migratory machinery that respond to p63 inhibition to engage tumor cell migration and invasion? It is now accepted that the cell’s main receptor for fibronectin, α5β1 integrin, is a key contributor to metastatic migration of cancer cells. Indeed, Muller et al. (2009) showed that the mutant p53-driven (and p63-inhibitable) component of tumor cell migration can depend on α5β1 integrin function and the presence of its ligand, fibronectin (Muller et al., 2009). The way in which integrins are internalized into endosomal compartments and returned (or recycled) to the plasma membrane influences their function, particularly with regard to their communication with Rho-GTPases and their ability to influence the recycling of other receptors such as EGFR1 or VEGFR2 (Caswell et al., 2009). The Rab11 effector protein Rab-coupling protein (RCP) associates with α5β1 integrin and plays an important role in guiding integrin transport from recycling endosomes to the plasma membrane to promote movement of tumor cells in fibronectin-rich 3D microenvironments (Caswell et al., 2008). Moreover, the importance of RCP in cancer progression is further illustrated by the finding that it is frequently overexpressed in human breast cancers (Zhang et al., 2009). It is now clear that the association of RCP with α5β1 integrin is inhibited by transcriptionally active TAp63. This inhibition is relieved after expression of mutant p53s, allowing RCP to bind to internalized α5β1 integrin and rapidly transport it to the plasma membrane, thus driving tumor cell migration and invasion (Fig. 2 B; Muller et al., 2009).
Although RCP-dependent integrin recycling is a key effector pathway downstream of p63, the transcriptional targets of p63 that regulate RCP’s recruitment to α5β1 integrin are unknown. It is interesting to speculate how this may be achieved, and there are signs that miRNAs and the machinery that processes them may be involved. In a computational genomic analysis to identify genes that may regulate components of the miRNA processing complex, an RNase III endonuclease, Dicer, was identified as a possible target of TAp63 (Boominathan, 2010). Conformation that Dicer is indeed a target gene of TAp63 was recently provided by an elegant study showing that TAp63 regulates Dicer to prevent metastasis (Su et al., 2010). p63-null mice die soon after birth because of severe skin defects. To parallel this, the epidermis-specific knockout of Dicer leads to impaired development of hair follicles (Andl et al., 2006). Down-regulation of miRNA processing is known to occur in a range of cancers (Lu et al., 2005; Kumar et al., 2009), and reduced levels of enzymes such as Dicer correlate with a poor prognosis (Grelier et al., 2009). Moreover, it has recently been shown that reduced levels of Dicer can promote tumorigenesis and invasion (Kumar et al., 2009; Han et al., 2010; Lambertz et al., 2010; Martello et al., 2010; Su et al., 2010). Furthermore, decreased Dicer levels correlate with increased activation of PKB/Akt, which suggests that it might act on the same pathway to induce invasion as mutant p53 and p63 (Han et al., 2010). Hopefully, future work will resolve the molecular details of the connection between p63 suppression, miRNA processing, and invasion. In particular, it will be interesting to determine whether components of the trafficking machinery that transport α5β1 integrin from recycling endosomes to the cell surface represent key targets of p63-regulated miRNAs.
Mutant p53, p63, and growth factor receptor trafficking
In addition to controlling recycling of α5β1 integrin, RCP can physically link the integrin to the EGF receptor (EGFR), thus mediating co-trafficking of these two important regulators of cell migration (Caswell et al., 2008). Moreover, because this process leads to increased return of internalized EGFR1 to the plasma membrane (thus diverting it from the route to lysosomal degradation), it also enhances signaling downstream of EGFR1 (Caswell et al., 2008). Consistent with this, mutant p53-expressing cells have enhanced levels of active PKB/Akt (Fig. 2 B; Dong et al., 2009; Muller et al., 2009), and the mutant p53 status of human and mouse cancers correlates with the degree of PKB/Akt activation (Muller et al., 2009; Blanco-Aparicio et al., 2010). PKB/Akt is an important prosurvival kinase that more recently has been established to play a key role in promoting cell migration during the invasive program. This indicates the likelihood that mutant p53-driven recycling conveys its signals to the cell’s invasive machinery, at least in part, via PKB/Akt. Another member of the EGFR family, ErbB2, can heterodimerize with EGFR1, and is known to cooperate with mutant p53 to increase tumorigenesis in mice (Li et al., 1997). Furthermore, in breast cancers, mutant p53 status in combination with high ErbB2 expression is associated with a very poor prognosis (Rahko et al., 2003). Alternative trafficking routes for EGFR1/ErbB2 heterodimers are thought to influence EGF signaling in cancer (Lenferink et al., 1998), and the possibility that the α5β1 integrin–RCP complex recruits and controls recycling of EGFR1/ErbB2 heterodimers and other prometastatic receptors with known connections to mutant p53, such as IGF1R and c-Met, should be considered.
Conclusions
By comparison with p53 loss, expression of mutant p53s is associated with more invasive and metastatic cancers, which suggests that these mutants not only lose the ability to function as tumor suppressors, but that they also gain prometastatic functions, some of which are related to increased cell migration and invasion. In general, it seems that loss of p53 is important in the loosening of cell–cell junctions and the loss of epithelial integrity, which would be expected to contribute to dissemination of cells from solid tumors. However, most in vivo work indicates that this event in itself is insufficient to generate invasive or metastatic tumors. Mutant p53s, however, are very potent inducers of the metastatic phenotype. This owes, at least in part, to their ability to act via p63 to drive invasive-type migration. The contribution of TGF-β signaling and the trafficking of integrins and growth factor receptors to mutant p53-driven invasion suggests some interesting new candidates for antimetastatic drug development. However, many questions remain as to how the elements of this pro-invasive program are connected to one another. In particular, we anticipate that the link between p63 and the cell’s migratory and trafficking machinery will provide fertile ground for those interested in targeting the metastatic process.
Acknowledgments
The work in J.C. Norman’s and K.H. Vousden’s laboratories was funded by Cancer Research UK, and we gratefully acknowledge this generous support. P.A.J. Muller is funded by a postdoctoral Rubicon fellowship from Netherlands Organisation for Scientific Research.
Footnotes
Abbreviations used in this paper:
- EGFR
- EGF receptor
- EMT
- epithelial–mesenchymal transition
- GAP
- GTPase-activating protein
- GEF
- guanine nucleotide exchange factor
- RCP
- Rab-coupling protein
- ROCK
- Rho-associated coiled-coil containing protein kinase
References
- Adorno M., Cordenonsi M., Montagner M., Dupont S., Wong C., Hann B., Solari A., Bobisse S., Rondina M.B., Guzzardo V., et al. 2009. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 137:87–98 10.1016/j.cell.2009.01.039 [DOI] [PubMed] [Google Scholar]
- Alexandrova A., Ivanov A., Chumakov P., Kopnin B., Vasiliev J. 2000. Changes in p53 expression in mouse fibroblasts can modify motility and extracellular matrix organization. Oncogene. 19:5826–5830 10.1038/sj.onc.1203944 [DOI] [PubMed] [Google Scholar]
- Andl T., Murchison E.P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J.W., Andl C.D., Seykora J.T., Hannon G.J., Millar S.E. 2006. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 16:1041–1049 10.1016/j.cub.2006.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardi L.D., Jacks T. 1999. The role of p53 in tumour suppression: lessons from mouse models. Cell. Mol. Life Sci. 55:48–63 10.1007/s000180050269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri C.E., Tang L.J., Brown K.A., Pietenpol J.A. 2006. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 66:7589–7597 10.1158/0008-5472.CAN-06-2020 [DOI] [PubMed] [Google Scholar]
- Bishop A.L., Hall A. 2000. Rho GTPases and their effector proteins. Biochem. J. 348:241–255 10.1042/0264-6021:3480241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Aparicio C., Cañamero M., Cecilia Y., Pequeño B., Renner O., Ferrer I., Carnero A. 2010. Exploring the gain of function contribution of AKT to mammary tumorigenesis in mouse models. PLoS One. 5:e9305 10.1371/journal.pone.0009305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolós V., Peinado H., Pérez-Moreno M.A., Fraga M.F., Esteller M., Cano A. 2003. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116:499–511 10.1242/jcs.00224 [DOI] [PubMed] [Google Scholar]
- Boominathan L. 2010. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 5:e10615 10.1371/journal.pone.0010615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi G., Marampon F., Maor-Aloni R., Zani B., Rotter V., Oren M., Strano S., Blandino G., Sacchi A. 2008. Conditional RNA interference in vivo to study mutant p53 oncogenic gain of function on tumor malignancy. Cell Cycle. 7:1870–1879 10.4161/cc.7.12.6161 [DOI] [PubMed] [Google Scholar]
- Candi E., Dinsdale D., Rufini A., Salomoni P., Knight R.A., Mueller M., Krammer P.H., Melino G. 2007. TAp63 and DeltaNp63 in cancer and epidermal development. Cell Cycle. 6:274–285 [DOI] [PubMed] [Google Scholar]
- Cano A., Gamallo C., Kemp C.J., Benito N., Palacios J., Quintanilla M., Balmain A. 1996. Expression pattern of the cell adhesion molecules. E-cadherin, P-cadherin and alpha 6 beta 4 intergrin is altered in pre-malignant skin tumors of p53-deficient mice. Int. J. Cancer. 65:254–262 [DOI] [PubMed] [Google Scholar]
- Carroll D.K., Carroll J.S., Leong C.O., Cheng F., Brown M., Mills A.A., Brugge J.S., Ellisen L.W. 2006. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8:551–561 10.1038/ncb1420 [DOI] [PubMed] [Google Scholar]
- Castro Alves C., Rosivatz E., Schott C., Hollweck R., Becker I., Sarbia M., Carneiro F., Becker K.F. 2007. Slug is overexpressed in gastric carcinomas and may act synergistically with SIP1 and Snail in the down-regulation of E-cadherin. J. Pathol. 211:507–515 10.1002/path.2138 [DOI] [PubMed] [Google Scholar]
- Caswell P.T., Chan M., Lindsay A.J., McCaffrey M.W., Boettiger D., Norman J.C. 2008. Rab-coupling protein coordinates recycling of α5β1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J. Cell Biol. 183:143–155 10.1083/jcb.200804140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell P.T., Vadrevu S., Norman J.C. 2009. Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10:843–853 10.1038/nrm2799 [DOI] [PubMed] [Google Scholar]
- Chen Y.K., Hsue S.S., Lin L.M. 2004. Expression of p63 (TA and deltaN isoforms) in human primary well differentiated buccal carcinomas. Int. J. Oral Maxillofac. Surg. 33:493–497 10.1016/j.ijom.2003.10.023 [DOI] [PubMed] [Google Scholar]
- Cho Y., Gorina S., Jeffrey P.D., Pavletich N.P. 1994. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 265:346–355 10.1126/science.8023157 [DOI] [PubMed] [Google Scholar]
- Delassus G.S., Cho H., Hoang S., Eliceiri G.L. 2010. Many new down- and up-regulatory signaling pathways, from known cancer progression suppressors to matrix metalloproteinases, differ widely in cells of various cancers. J. Cell. Physiol. 224:549–558 10.1002/jcp.22157 [DOI] [PubMed] [Google Scholar]
- Derksen P.W., Liu X., Saridin F., van der Gulden H., Zevenhoven J., Evers B., van Beijnum J.R., Griffioen A.W., Vink J., Krimpenfort P., et al. 2006. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 10:437–449 10.1016/j.ccr.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Dhar G., Banerjee S., Dhar K., Tawfik O., Mayo M.S., Vanveldhuizen P.J., Banerjee S.K. 2008. Gain of oncogenic function of p53 mutants induces invasive phenotypes in human breast cancer cells by silencing CCN5/WISP-2. Cancer Res. 68:4580–4587 10.1158/0008-5472.CAN-08-0316 [DOI] [PubMed] [Google Scholar]
- Di Como C.J., Gaiddon C., Prives C. 1999. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 19:1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P., Xu Z., Jia N., Li D., Feng Y. 2009. Elevated expression of p53 gain-of-function mutation R175H in endometrial cancer cells can increase the invasive phenotypes by activation of the EGFR/PI3K/AKT pathway. Mol. Cancer. 8:103 10.1186/1476-4598-8-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B., Morton J.P., Delaney D.W., Ridgway R.A., Wilkins J.A., Sansom O.J. 2010. p53 mutation and loss have different effects on tumourigenesis in a novel mouse model of pleomorphic rhabdomyosarcoma. J. Pathol. 222:129–137 [DOI] [PubMed] [Google Scholar]
- Flores E.R., Sengupta S., Miller J.B., Newman J.J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. 2005. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 7:363–373 10.1016/j.ccr.2005.02.019 [DOI] [PubMed] [Google Scholar]
- Gadéa G., Lapasset L., Gauthier-Rouvière C., Roux P. 2002. Regulation of Cdc42-mediated morphological effects: a novel function for p53. EMBO J. 21:2373–2382 10.1093/emboj/21.10.2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea G., de Toledo M., Anguille C., Roux P. 2007. Loss of p53 promotes RhoA-ROCK-dependent cell migration and invasion in 3D matrices. J. Cell Biol. 178:23–30 10.1083/jcb.200701120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiddon C., Lokshin M., Ahn J., Zhang T., Prives C. 2001. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21:1874–1887 10.1128/MCB.21.5.1874-1887.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh L.L., Manser E. 2010. The RhoA GEF Syx is a target of Rnd3 and regulated via a Raf1-like ubiquitin-related domain. PLoS One. 5:e12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelier G., Voirin N., Ay A.S., Cox D.G., Chabaud S., Treilleux I., Léon-Goddard S., Rimokh R., Mikaelian I., Venoux C., et al. 2009. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br. J. Cancer. 101:673–683 10.1038/sj.bjc.6605193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Coates P.J., Boldrup L., Nylander K. 2008. p63 contributes to cell invasion and migration in squamous cell carcinoma of the head and neck. Cancer Lett. 263:26–34 10.1016/j.canlet.2007.12.011 [DOI] [PubMed] [Google Scholar]
- Guo F., Zheng Y. 2004. Rho family GTPases cooperate with p53 deletion to promote primary mouse embryonic fibroblast cell invasion. Oncogene. 23:5577–5585 10.1038/sj.onc.1207752 [DOI] [PubMed] [Google Scholar]
- Guo F., Gao Y., Wang L., Zheng Y. 2003. p19Arf-p53 tumor suppressor pathway regulates cell motility by suppression of phosphoinositide 3-kinase and Rac1 GTPase activities. J. Biol. Chem. 278:14414–14419 10.1074/jbc.M300341200 [DOI] [PubMed] [Google Scholar]
- Guo X., Keyes W.M., Papazoglu C., Zuber J., Li W., Lowe S.W., Vogel H., Mills A.A. 2009. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat. Cell Biol. 11:1451–1457 10.1038/ncb1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner A., Starace G., Norelli G., Piaggio G., Sacchi A., Bossi G. 2010. Mutant p53-induced up-regulation of mitogen-activated protein kinase kinase 3 contributes to gain of function. J. Biol. Chem. 285:14160–14169 10.1074/jbc.M109.094813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Zhang A., Zhou X., Xu P., Wang G.X., Pu P.Y., Kang C.S. 2010. Downregulation of Dicer enhances tumor cell proliferation and invasion. Int. J. Oncol. 37:299–305 [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature. 387:296–299 10.1038/387296a0 [DOI] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9:690–701 10.1038/nrm2476 [DOI] [PubMed] [Google Scholar]
- Heinlein C., Krepulat F., Löhler J., Speidel D., Deppert W., Tolstonog G.V. 2008. Mutant p53(R270H) gain of function phenotype in a mouse model for oncogene-induced mammary carcinogenesis. Int. J. Cancer. 122:1701–1709 10.1002/ijc.23317 [DOI] [PubMed] [Google Scholar]
- Jakowlew S.B. 2006. Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 25:435–457 10.1007/s10555-006-9006-2 [DOI] [PubMed] [Google Scholar]
- Junk D.J., Vrba L., Watts G.S., Oshiro M.M., Martinez J.D., Futscher B.W. 2008. Different mutant/wild-type p53 combinations cause a spectrum of increased invasive potential in nonmalignant immortalized human mammary epithelial cells. Neoplasia. 10:450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo E., Buganim Y., Shapira K.E., Besserglick H., Goldfinger N., Weisz L., Stambolsky P., Henis Y.I., Rotter V. 2007. Mutant p53 attenuates the SMAD-dependent transforming growth factor beta1 (TGF-beta1) signaling pathway by repressing the expression of TGF-beta receptor type II. Mol. Cell. Biol. 27:8228–8242 10.1128/MCB.00374-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga F., Kawakami S., Fujii Y., Saito K., Ohtsuka Y., Iwai A., Ando N., Takizawa T., Kageyama Y., Kihara K. 2003. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin. Cancer Res. 9:5501–5507 [PubMed] [Google Scholar]
- Kogan-Sakin I., Tabach Y., Buganim Y., Molchadsky A., Solomon H., Madar S., Kamer I., Stambolsky P., Shelly A., Goldfinger N., et al. 2010. Mutant p53(R175H) upregulates Twist1 expression and promotes epithelial-mesenchymal transition in immortalized prostate cells. Cell Death Differ. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat M.H., Jones S.N., Vousden K.H. 1997. Regulation of p53 stability by Mdm2. Nature. 387:299–303 10.1038/387299a0 [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Pester R.E., Chen C.Y., Lane K., Chin C., Lu J., Kirsch D.G., Golub T.R., Jacks T. 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23:2700–2704 10.1101/gad.1848209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertz I., Nittner D., Mestdagh P., Denecker G., Vandesompele J., Dyer M.A., Marine J.C. 2010. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 17:633–641 10.1038/cdd.2009.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C., et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 119:861–872 10.1016/j.cell.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Lefort K., Mandinova A., Ostano P., Kolev V., Calpini V., Kolfschoten I., Devgan V., Lieb J., Raffoul W., Hohl D., et al. 2007. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 21:562–577 10.1101/gad.1484707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenferink A.E., Pinkas-Kramarski R., van de Poll M.L., van Vugt M.J., Klapper L.N., Tzahar E., Waterman H., Sela M., van Zoelen E.J., Yarden Y. 1998. Differential endocytic routing of homo- and hetero-dimeric ErbB tyrosine kinases confers signaling superiority to receptor heterodimers. EMBO J. 17:3385–3397 10.1093/emboj/17.12.3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Prives C. 2007. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 26:2220–2225 10.1038/sj.onc.1210311 [DOI] [PubMed] [Google Scholar]
- Li B., Rosen J.M., McMenamin-Balano J., Muller W.J., Perkins A.S. 1997. neu/ERBB2 cooperates with p53-172H during mammary tumorigenesis in transgenic mice. Mol. Cell. Biol. 17:3155–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Sutphin P.D., Schwartz D., Matas D., Almog N., Wolkowicz R., Goldfinger N., Pei H., Prokocimer M., Rotter V. 1998. Mutant p53 protein expression interferes with p53-independent apoptotic pathways. Oncogene. 16:3269–3277 10.1038/sj.onc.1201867 [DOI] [PubMed] [Google Scholar]
- Lin J., Teresky A.K., Levine A.J. 1995. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene. 10:2387–2390 [PubMed] [Google Scholar]
- Liu G., McDonnell T.J., Montes de Oca Luna R., Kapoor M., Mims B., El-Naggar A.K., Lozano G. 2000. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc. Natl. Acad. Sci. USA. 97:4174–4179 10.1073/pnas.97.8.4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.P., Song H., Xu Y. 2010. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 29:949–956 10.1038/onc.2009.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. 2005. MicroRNA expression profiles classify human cancers. Nature. 435:834–838 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- Martello G., Rosato A., Ferrari F., Manfrin A., Cordenonsi M., Dupont S., Enzo E., Guzzardo V., Rondina M., Spruce T., et al. 2010. A MicroRNA targeting dicer for metastasis control. Cell. 141:1195–1207 10.1016/j.cell.2010.05.017 [DOI] [PubMed] [Google Scholar]
- Matas D., Sigal A., Stambolsky P., Milyavsky M., Weisz L., Schwartz D., Goldfinger N., Rotter V. 2001. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 20:4163–4172 10.1093/emboj/20.15.4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuarai S., Yamanaka K., Kotani H. 2006. Mutant p53 induces the GEF-H1 oncogene, a guanine nucleotide exchange factor-H1 for RhoA, resulting in accelerated cell proliferation in tumor cells. Cancer Res. 66:6319–6326 10.1158/0008-5472.CAN-05-4629 [DOI] [PubMed] [Google Scholar]
- Morton J.P., Timpson P., Karim S.A., Ridgway R.A., Athineos D., Doyle B., Jamieson N.B., Oien K.A., Lowy A.M., Brunton V.G., et al. 2010. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 107:246–251 10.1073/pnas.0908428107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay U.K., Eves R., Jia L., Mooney P., Mak A.S. 2009. p53 suppresses Src-induced podosome and rosette formation and cellular invasiveness through the upregulation of caldesmon. Mol. Cell. Biol. 29:3088–3098 10.1128/MCB.01816-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., Caswell P.T., Doyle B., Iwanicki M.P., Tan E.H., Karim S., Lukashchuk N., Gillespie D.A., Ludwig R.L., Gosselin P., et al. 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell. 139:1327–1341 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 119:847–860 10.1016/j.cell.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Ongusaha P.P., Kim H.G., Boswell S.A., Ridley A.J., Der C.J., Dotto G.P., Kim Y.B., Aaronson S.A., Lee S.W. 2006. RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr. Biol. 16:2466–2472 10.1016/j.cub.2006.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Oren M., Rotter V. 2010. Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2:a001107 10.1101/cshperspect.a001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.J., Lee S.J., Kim J.I., Lee S.J., Lee C.H., Chang S.G., Park J.H., Chi S.G. 2000. Frequent alteration of p63 expression in human primary bladder carcinomas. Cancer Res. 60:3370–3374 [PubMed] [Google Scholar]
- Pinner S., Sahai E. 2008. Imaging amoeboid cancer cell motility in vivo. J. Microsc. 231:441–445 10.1111/j.1365-2818.2008.02056.x [DOI] [PubMed] [Google Scholar]
- Qin Q., Baudry M., Liao G., Noniyev A., Galeano J., Bi X. 2009. A novel function for p53: regulation of growth cone motility through interaction with Rho kinase. J. Neurosci. 29:5183–5192 10.1523/JNEUROSCI.0420-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintavalle M., Elia L., Condorelli G., Courtneidge S.A. 2010. MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J. Cell Biol. 189:13–22 10.1083/jcb.200912096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahko E., Blanco G., Soini Y., Bloigu R., Jukkola A. 2003. A mutant TP53 gene status is associated with a poor prognosis and anthracycline-resistance in breast cancer patients. Eur. J. Cancer. 39:447–453 10.1016/S0959-8049(02)00499-9 [DOI] [PubMed] [Google Scholar]
- Riento K., Ridley A.J. 2006. Inhibition of ROCK by RhoE. Methods Enzymol. 406:533–541 10.1016/S0076-6879(06)06041-1 [DOI] [PubMed] [Google Scholar]
- Roger L., Gadea G., Roux P. 2006. Control of cell migration: a tumour suppressor function for p53? Biol. Cell. 98:141–152 10.1042/BC20050058 [DOI] [PubMed] [Google Scholar]
- Sablina A.A., Chumakov P.M., Kopnin B.P. 2003. Tumor suppressor p53 and its homologue p73alpha affect cell migration. J. Biol. Chem. 278:27362–27371 10.1074/jbc.M300547200 [DOI] [PubMed] [Google Scholar]
- Sahai E., Marshall C.J. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711–719 10.1038/ncb1019 [DOI] [PubMed] [Google Scholar]
- Sankpal N.V., Willman M.W., Fleming T.P., Mayfield J.D., Gillanders W.E. 2009. Transcriptional repression of epithelial cell adhesion molecule contributes to p53 control of breast cancer invasion. Cancer Res. 69:753–757 10.1158/0008-5472.CAN-08-2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V., Marshall C.J. 2009. Rho-GTPase signaling drives melanoma cell plasticity. Cell Cycle. 8:1484–1487 10.4161/cc.8.10.8490 [DOI] [PubMed] [Google Scholar]
- Sarig R., Rivlin N., Brosh R., Bornstein C., Kamer I., Ezra O., Molchadsky A., Goldfinger N., Brenner O., Rotter V. 2010. Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J. Exp. Med. 207:2127–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J.Y., Tsai M.F., Chang T.H., Chang Y.L., Yuan A., Yu C.J., Lin S.B., Liou G.Y., Lee M.L., Chen J.J., et al. 2005. Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin. Cancer Res. 11:8070–8078 10.1158/1078-0432.CCR-05-0687 [DOI] [PubMed] [Google Scholar]
- Shiota M., Izumi H., Onitsuka T., Miyamoto N., Kashiwagi E., Kidani A., Hirano G., Takahashi M., Naito S., Kohno K. 2008. Twist and p53 reciprocally regulate target genes via direct interaction. Oncogene. 27:5543–5553 10.1038/onc.2008.176 [DOI] [PubMed] [Google Scholar]
- Smit M.A., Peeper D.S. 2008. Deregulating EMT and senescence: double impact by a single twist. Cancer Cell. 14:5–7 10.1016/j.ccr.2008.06.012 [DOI] [PubMed] [Google Scholar]
- Song H., Hollstein M., Xu Y. 2007. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat. Cell Biol. 9:573–580 10.1038/ncb1571 [DOI] [PubMed] [Google Scholar]
- Stambolic V., MacPherson D., Sas D., Lin Y., Snow B., Jang Y., Benchimol S., Mak T.W. 2001. Regulation of PTEN transcription by p53. Mol. Cell. 8:317–325 10.1016/S1097-2765(01)00323-9 [DOI] [PubMed] [Google Scholar]
- Stambolsky P., Tabach Y., Fontemaggi G., Weisz L., Maor-Aloni R., Siegfried Z., Shiff I., Kogan I., Shay M., Kalo E., et al. 2010. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell. 17:273–285 10.1016/j.ccr.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S., Munarriz E., Rossi M., Cristofanelli B., Shaul Y., Castagnoli L., Levine A.J., Sacchi A., Cesareni G., Oren M., Blandino G. 2000. Physical and functional interaction between p53 mutants and different isoforms of p73. J. Biol. Chem. 275:29503–29512 10.1074/jbc.M003360200 [DOI] [PubMed] [Google Scholar]
- Strano S., Fontemaggi G., Costanzo A., Rizzo M.G., Monti O., Baccarini A., Del Sal G., Levrero M., Sacchi A., Oren M., Blandino G. 2002. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J. Biol. Chem. 277:18817–18826 10.1074/jbc.M201405200 [DOI] [PubMed] [Google Scholar]
- Su X., Chakravarti D., Cho M.S., Liu L., Gi Y.J., Lin Y., Leung M.L., El-Naggar A.K., Creighton C.J., Suraokar M.B., et al. 2010. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 467:986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist M.J., Di Como C.J., Lu M.L., Charytonowicz E., Verbel D., Crum C.P., Ince T.A., McKeon F.D., Cordon-Cardo C. 2002. Loss of p63 expression is associated with tumor progression in bladder cancer. Am. J. Pathol. 161:1199–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Mori I., Tang W., Nakamura M., Nakamura Y., Sato M., Sakurai T., Kakudo K. 2002. p63 expression in normal, hyperplastic and malignant breast tissues. Breast Cancer. 9:216–219 10.1007/BF02967592 [DOI] [PubMed] [Google Scholar]
- Wang B., Feng P., Xiao Z., Ren E.C. 2009a. LIM and SH3 protein 1 (Lasp1) is a novel p53 transcriptional target involved in hepatocellular carcinoma. J. Hepatol. 50:528–537 10.1016/j.jhep.2008.10.025 [DOI] [PubMed] [Google Scholar]
- Wang S.P., Wang W.L., Chang Y.L., Wu C.T., Chao Y.C., Kao S.H., Yuan A., Lin C.W., Yang S.C., Chan W.K., et al. 2009b. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat. Cell Biol. 11:694–704 10.1038/ncb1875 [DOI] [PubMed] [Google Scholar]
- Weisz L., Zalcenstein A., Stambolsky P., Cohen Y., Goldfinger N., Oren M., Rotter V. 2004. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res. 64:8318–8327 10.1158/0008-5472.CAN-04-1145 [DOI] [PubMed] [Google Scholar]
- Wennerberg K., Forget M.A., Ellerbroek S.M., Arthur W.T., Burridge K., Settleman J., Der C.J., Hansen S.H. 2003. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol. 13:1106–1115 10.1016/S0960-9822(03)00418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Nomoto S., Hoque M.O., Dracheva T., Osada M., Lee C.C., Dong S.M., Guo Z., Benoit N., Cohen Y., et al. 2003. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 63:2351–2357 [PubMed] [Google Scholar]
- Xia M., Land H. 2007. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat. Struct. Mol. Biol. 14:215–223 10.1038/nsmb1208 [DOI] [PubMed] [Google Scholar]
- Zalcenstein A., Weisz L., Stambolsky P., Bar J., Rotter V., Oren M. 2006. Repression of the MSP/MST-1 gene contributes to the antiapoptotic gain of function of mutant p53. Oncogene. 25:359–369 [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu X., Datta A., Govindarajan K., Tam W.L., Han J., George J., Wong C., Ramnarayanan K., Phua T.Y., et al. 2009. RCP is a human breast cancer-promoting gene with Ras-activating function. J. Clin. Invest. 119:2171–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Gish K., Murphy M., Yin Y., Notterman D., Hoffman W.H., Tom E., Mack D.H., Levine A.J. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981–993 10.1101/gad.827700 [DOI] [PMC free article] [PubMed] [Google Scholar]