Abstract
An innate immune cell can sense a pathogen, either from a distance by recognizing chemoattractant stimuli or by direct physical contact. The pathogen is subsequently neutralized, which usually occurs through its phagocytic internalization. By investigating chemotaxis and phagocytosis from an immunophysical single-cell perspective, it now appears that the demarcation between these two processes is less distinct than originally thought. Several lines of evidence support this notion. First, chemotactic stimulation does not cease at the moment of initial contact between the cell and the pathogenic target. Second, even when classical chemotaxis of neutrophils is suppressed, the early cell response to contact with typical chemoattractant targets, such as zymosan, fungal spores or chemokine-coated particles, can still involve morphological attributes of chemotaxis. Recognizing that the changing morphology of motile cells is inextricably linked to physical cell behavior, this Commentary focuses on the mechanical aspects of the early response of innate immune cells to chemotactic and phagocytic stimuli. On the basis of this perspective, we propose that the combined study of chemotaxis and phagocytosis will, potentially, not only advance our grasp of the mechanisms underlying immune-cell motility but also open new lines of research that will promote a deeper understanding of the innate recognition of pathogens.
Key words: Chemotaxis, Phagocytosis, Cell motility, Immunophysics, Micropipette manipulation, Innate immune recognition
Introduction
The ability of certain types of white blood cell, including neutrophils, monocytes and macrophages, to actively change their shape and ‘crawl’ on substrates is a cornerstone of the host innate immune defense (Box 1 and Fig. 1) (Alberts et al., 2007; Murphy et al., 2007). It is also a key reason why the interest in these cell types spans diverse areas of cell science. For example, whereas mainstream immunobiological research focuses primarily on the molecular basis of pathogen recognition and subsequent intracellular signaling reactions, innate immune cell behavior also provides a highly instructive cross-disciplinary window into eukaryotic cell motility.
Box 1. The innate immune-cell response: a cross-disciplinary feat
The innate immune response is a pre-programmed, non-specific and immediate reaction of the host to invading pathogens. It is distinct from the later adaptive immune response, which involves the positive identification and the development of a ‘memory’ of specific threats and provides long-lasting protection against them (Murphy et al., 2007). By contrast, the innate immune system detects pathogens early by recognizing generic ‘non-self’ features on the surface of microbial invaders. This recognition is accompanied by a process called opsonization, that is, tagging of microorganisms with components of the host serum, such as complement proteins or immunoglobulins (also referred to as antibodies). Although some serum factors might attack microorganisms directly, most serve to mediate and substantially enhance the cellular host response. For example, serum enzymes, after binding to a pathogen, cleave small quickly diffusing peptides called anaphylatoxins from complement proteins. The exquisite sensitivity of innate immune cells to minuscule concentrations of anaphylatoxins facilitates the timely activation and recruitment of the motile cells to sites of infection. The resulting cell migration is an example of chemotaxis, that is, the directed movement of cells along a chemical concentration gradient. Chemotaxis can also be induced by cytokines that are secreted from previously activated immune cells or by biochemical cues that emanate directly from a pathogen. Once an innate immune cell makes contact with an opsonized target, it usually engulfs the target by phagocytosis. Some types of phagocytic cells then present antigens, which identify encountered pathogens, to the adaptive immune system. Other phagocytes neutralize the engulfed pathogen by poisoning it.
Although this brief overview cannot do justice to the enormous complexity of the immune defense of the host, it highlights the multitude of cross-disciplinary tasks that individual innate immune cells can undertake. Not only are these cells a source and ultrasensitive detector of chemical signals, they also coordinate an intricate spectrum of mechanical ‘pushing’ and ‘pulling’ forces and autonomous deformations, that is, morphological changes that are generated by the cell itself (in contrast to externally imposed shape changes) as it migrates towards a chemoattractant source or engulfs a pathogenic target (Fig. 1A,B; Fig. 2). However, although the study of biochemical reaction networks has long been adopted as an integral part of mainstream cell science, conspicuous gaps remain in our understanding of physical cell behavior.
Fig. 1.
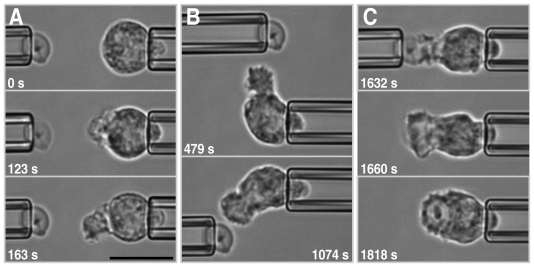
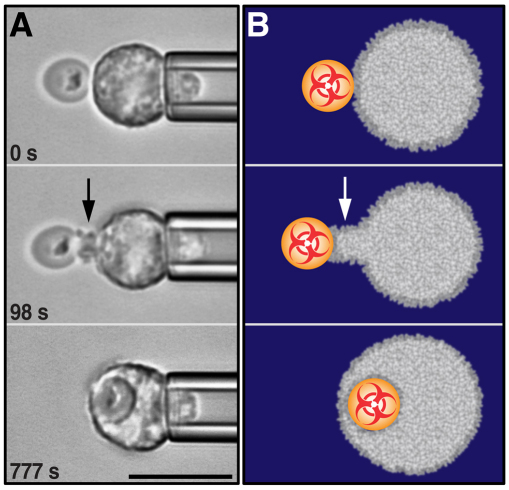
Interactions between individual innate immune cells and their targets. (A) Pseudocolor scanning electron microscopy images of fixed J774 macrophages engulfing antibody-coated beads: five 10 μm beads (upper image) and a 30 μm bead (lower image). Analysis of such images reveals that macrophages can expand their surface area by five-to-six times during the phagocytosis of large targets (Lam et al., 2009). (B) Live-cell studies using micropipette-held, initially quiescent human neutrophils that are brought into well-controlled contact with antibody-coated beads using a second pipette (not shown). This configuration eliminates interference from cell–substrate adhesion and, by imposing an essentially axisymmetric geometry, is much more amenable to quantitative analysis. Such analysis provides a wealth of information about the immunophysical behavior of innate immune cells, for example, the timelines of the cortical tension, cell–surface area and target position. The relative recording time of each image is included. (C) Computer simulations of ‘virtual cells’ are an integral part of an immunophysical analysis, enabling us to corroborate or discard hypotheses about the mechanoregulation of the responses of innate immune cells to pathogenic targets. The examples shown are from a simulation that reproduced not only the overall morphology of phagocytosis at the proper length scale (the scale bar represents 10 μm) but also the dynamics of this response (the simulation times are included), such as the time-dependent cortical tension, surface area and bead position. Moreover, the model predicted the density distribution of the cytoskeleton (encoded by color, with blue representing the lowest density and magenta the highest density). Scale bars: 10 μm.
Cell motility generally encompasses the recognition of stimuli, the processing of these signals through biochemical reaction networks, and the resulting physical cell responses, which coordinate adhesion, morphology changes and mechanical forces (Bray, 2000; Howard, 2001; Pollard, 2003; Herant and Dembo, 2010; Insall, 2010; Wong, 2011). In the case of motile leukocytes, the cells can alter their shape and move fast enough to facilitate real-time live-cell investigations of their autonomous deformation. A prominent example of this is the movie of a neutrophil ‘chasing’ Staphylococcus aureus (recorded by David Rogers at Vanderbilt University, Nashville, TN in the 1950s; for details, see the virtual library of biochemistry, molecular biology and cell biology website at www.biochemweb.org/neutrophil.shtml). Furthermore, even though these white cells lack undulipodia – intrinsically motile intracellular structures, such as flagella or cilia – as well as any other specialized propelling devices, they can still produce a fascinating spectrum of motions, as illustrated in supplementary material Movie 1 and reported elsewhere (Herant et al., 2006; Lee et al., 2011).
The coordination of these cellular motions is inextricably linked to the physiological functions of the cells, including chemotaxis, phagocytosis, wound healing and the inflammatory response. As the first line of the immune defense, host cells can undertake a remarkably diverse range of tasks: detecting and identifying invaders, migrating towards the site of infection or trauma, engulfing objects that are recognized as non-self and post-processing of such targets (e.g. through chemical neutralization or through the presentation of antigens to the adaptive immune system; Box 1). This cross-disciplinary behavior of motile immune cells provides exciting opportunities for collaborative research, but it also highlights the challenges of trying to establish a comprehensive and rigorous understanding of innate immunity. Nonetheless, in recent years, the number of studies that examine innate immune cell function appears to have noticeably increased and one key factor seems to be an emerging alliance between biological intuition and physical rigor (Herant et al., 2006; Discher et al., 2009; Wolgemuth, 2011). Indeed, part of the success of many recent works on innate immune cells can be traced to a growing integration of immunophysical concepts and tools. In this Commentary, we use an immunophysical perspective to address similarities and differences in the physical behavior of innate immune cells during phagocytosis and chemotaxis.
Chemotaxis and phagocytosis by innate immune cells
At a first glance, the distinction between phagocytosis and chemotaxis appears straightforward. On the one hand, classical chemotaxis is the directed movement of cells along a concentration gradient of soluble chemicals emanating from a distant source (Box 2). Phagocytosis, on the other hand, is the enveloping motion by which cells engulf and internalize particles (Fig. 1A,B; Box 2). However, when studying the mechanisms that govern these immune functions, it is the ‘perspective’ of the cell itself that matters, that is, there is no ‘prior knowledge’ about the type of stimulus that is encountered by the cell. This view raises several questions. Given that cell stimulation starts with specific ligand–receptor interactions at the cell surface, how does a cell distinguish between chemotactic and phagocytic ligands? Is there a clear-cut division between purely chemotactic and phagocytic cell-surface receptors, in which case a stimulus could be identified by the ligand–receptor biochemistry alone? Or, are the mechanosensing abilities of the cell subtle enough to discriminate between soluble and surface-bound ligands? If so, how does this mechanorecognition work? Consider, for example, a thought experiment in which a cell expressing a highly specialized chemotactic receptor (that does not distinguish between soluble and immobilized ligand molecules) encounters and binds a freely suspended particle that is coated with the ligand for the receptor. In this case, the only stimulus ‘sensed’ by the cell is chemotactic. The logical cell response would be to crawl in the direction that is defined by the attached particle, pushing the particle along rather than engulfing it. Under certain conditions, neutrophils indeed seem to exhibit such behavior, as discussed below. First, we give a brief overview of ligand recognition and subsequent cellular processes in phagocytosis and chemotaxis.
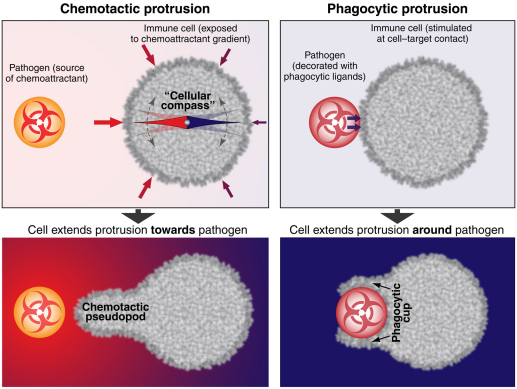
Box 2. How to steer protrusion?
The initial protrusive deformation of an innate immune cell follows a common principle in both chemotactic and phagocytic interactions with a pathogenic target: stimulation of cell-surface receptors activates intracellular signaling, which leads to cytoskeletal remodeling. A higher actin density underneath the cell-surface region that faces the target generates a pushing force that drives local protrusion. The protrusion extends towards the target in chemotaxis and around the target in phagocytosis (see figure). This process involves two important cellular tasks. First, the cell must ‘decide’ on the location of protrusion. In phagocytosis this is simply the site of cell–target contact. In chemotaxis, the ‘compass’ of the cell needs to detect – by means that are not completely understood – where on the cell surface the density of engaged receptors is highest. Second, the cell must implement the proper protrusive response – either a chemotactic pseudopod or a phagocytic cup. The key to this distinction appears to be the suppression of protrusion directly underneath the cell–target contact region in phagocytosis; as a result, the cell can only protrude parallel to the target surface. Together with strong cell–target adhesion, this results in the typical phagocytic cup. Recent findings suggest that this suppression of protrusion is biomechanical in nature, that is, it is realized by structural linkages between the adherent membrane and the cytoskeleton (Herant et al., 2011; Lee et al., 2011).
On the basis of the above distinction, we propose that the formation of a chemotactic-like pseudopod in cases where the cell is in physical contact with a target particle (and thus pushes the particle outward at the tip of the pseudopod before eventually engulfing it) signifies a hybrid chemotactic and phagocytic response.
Receptors
There appears to be little overlap between known chemotactic and phagocytic receptors. Chemotactic stimulation of eukaryotic cells primarily occurs through the large class of G-protein-coupled receptors (GPCRs) (Parent and Devreotes, 1999; Bjarnadóttir et al., 2006; Dorsam and Gutkind, 2007; Monk et al., 2007). The ligands of these receptors are soluble peptides (anaphylatoxins and formyl peptides) or small proteins classified as chemokines (Box 1). The small size of these chemoattractant ligands facilitates their fast diffusion. After binding a ligand, a single GPCR can activate many G-proteins (Roberts and Waelbroeck, 2004). Along with positive feedback in the subsequent signaling pathways (Charest and Firtel, 2006; Swaney et al., 2010), this serves to amplify chemotactic stimuli and provides a molecular basis for the exquisite sensitivity of innate immune cells to chemoattractants. Remarkably, despite this high sensitivity, GPCRs that specifically interact with anaphylatoxins (C5a, C3a and C4a) appear to remain impervious to encounters with the precursor complement proteins (C5, C3 and C4) from which the anaphylatoxins are cleaved (see also Box 1).
Phagocytosis, however, is mediated by several classes of receptor that recognize the opsonins that tag a pathogen (Box 1) and/or native constituents of the pathogen surface itself (Underhill and Ozinsky, 2002a; Stuart and Ezekowitz, 2005; Goodridge et al., 2009; Jaumouille and Grinstein, 2010; Romani, 2011). Most notable among the opsonin-dependent receptors are receptors for the constant (Fc) region of immunoglobulins (Swanson and Hoppe, 2004; Boross et al., 2008; Nimmerjahn and Ravetch, 2008) and for complement proteins (Fearon, 1984; Nielsen et al., 1997; van Bruggen et al., 2009). Interestingly, Fc receptors can distinguish between soluble antibodies and antibodies that are attached to a pathogen. Phagocytic receptors that recognize pathogens directly include Toll-like receptors (Underhill and Ozinsky, 2002b; Netea et al., 2007), glycan receptors [e.g. the mannose receptor, and the β-glucan receptors dectin-I and dectin-II (Brown et al., 2003; Goodridge and Underhill, 2008; Hollmig et al., 2009)] and scavenger receptors (Vera et al., 2009). For many cell–pathogen interactions, a detailed understanding of the relative physiological significance of phagocytic receptors, in addition to their complex sequence of action and possible cooperativity, has yet to be established. Such an understanding would have to account for factors such as the host species, the type of immune cell, the state of cell activation and the timecourse of the interaction.
Post-receptor processing
Once the above receptor–ligand interactions have positively identified surface stimuli as either chemotactic or phagocytic, how does an innate immune cell propagate this information in a manner that leads to the appropriate physical response? Three early tasks in this ‘processing’ can be classified as immunophysical: (1) discernment of the distribution of engaged receptors over the cell surface, (2) regulation of cell–substrate and cell–target adhesion, and (3) determination of the initial direction of cellular protrusion. The first task involves the stimulus-dependent establishment of cell polarity (Box 2). In phagocytosis, the location of the immediate response of the cell is defined by the region of cell–target contact, whereas the mechanisms by which a cell determines the direction of a chemoattractant gradient remain under active investigation (Parent and Devreotes, 1999; Devreotes and Janetopoulos, 2003; Iglesias and Devreotes, 2008; Kay et al., 2008; Thelen and Stein, 2008; Petrie et al., 2009; Insall, 2010; Swaney et al., 2010). The need to interpret a continuously varying spatial distribution of surface stimuli is specific to chemotaxis, therefore, it seems reasonable to assume that it is closely linked to GPCR-mediated signaling. Indeed, phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3] appears to be a key player in polarization, although neutrophils with inhibited phosphoinositide 3-kinases (PI3Ks) are still capable of chemotaxis (Van Haastert and Veltman, 2007; Afonso and Parent, 2011). Clearly, single-cell experiments (using non-adherent, initially quiescent cells) in which a well-defined chemotactic stimulus can be applied and relocated at will (e.g. alternated between opposite sides of a given cell as demonstrated in Fig. 2 and supplementary material Movie 1) provide a promising way to study the dynamics of this intriguing cellular ‘decision making’.
Fig. 2.
A chemotactic (A,B) and a predominantly phagocytic (C) response of a human neutrophil to a zymosan particle. This image series (see also supplementary material Movie 1) of single-live-cell–single-target interactions in the presence of serum was enabled by using highly controllable dual-micropipette manipulation. (A) For the pipette-held, non-adherent and initially quiescent neutrophil, chemotaxis takes the form of a pseudopodial protrusion towards the target particle (held in a second pipette on the left). (B) The cell reacts rapidly to relocation of the target by retracting the former pseudopod and extending a new one towards the particle. (C) After the zymosan particle has been ‘handed over’ to the cell, phagocytosis completes rapidly. Relative times are included in each image. Scale bar: 10 μm.
A second task that depends on the type of cell–surface stimulation is the coordination of adhesive interactions. In chemotactic migration, adhesion provides dynamic bracing support on a substrate, whereas in phagocytosis it enables the cell to anchor itself to a substrate (Fig. 1A), hold on to a particulate pathogen and surround the particle (Fig. 1A,B). It is unclear whether these diverse processes engage the same adhesion molecules. An important question, in phagocytosis in particular, is: to what extent are the receptors that recognize specific stimuli capable of supporting adhesive strength in their ligand bonds? This type of questioning is immunophysical at heart and is addressed by the young field of dynamic force spectroscopy (DFS) of single-molecule interactions (Bell, 1978; Evans and Ritchie, 1997; Evans and Calderwood, 2007). A key insight of DFS is that knowledge of the affinity of a receptor–ligand bond is insufficient to predict the behavior of the bond under mechanical stress. With a few exceptions, we have currently barely touched the surface of a rigorous quantitative understanding of such adhesive molecular interactions. Fortunately, by devising suitable single-cell experiments (see below) it is possible to separate essential phagocytic and chemotactic processes from cell–substrate adhesion.
Recent investigations of the third cellular task – control of the direction of initial protrusion – have yielded intriguing clues as to how a cell might implement the distinction between movement towards and around a stimulus that is sensed at its surface (Box 2). We discuss this distinction in more detail in a separate section below.
Finally, we note that cell-signaling biochemistry is but one of several cornerstones in an interdisciplinary view of phagocytosis and chemotaxis. A primary goal of this Commentary is to call attention to other, often under-represented, immunophysical processes that might advance our understanding in areas in which progress has been slow. We note that despite the differences in receptors and initial signaling reactions (Chimini and Chavrier, 2000; Swanson and Hoppe, 2004; Stuart and Ezekowitz, 2005; Hall et al., 2006; Van Haastert and Veltman, 2007; Swaney et al., 2010; Romani, 2011), the biochemistry of chemotaxis and phagocytosis appears to converge during signaling to the cytoskeleton during the formation of protrusions. For example, the role of Rho GTPases (Burridge and Wennerberg, 2004) appears to be very similar at the stage in which actin rearrangements produce either chemotactic pseudopods or phagocytic cups (Chimini and Chavrier, 2000; Hall et al., 2006; Insall and Machesky, 2009). Indeed, local activation of Rac has been shown to be sufficient to drive protrusion in a migratory cell (Levskaya et al., 2009) and during phagocytosis (Castellano et al., 2000). The same applies to actin-nucleation factors, including the actin-related protein 2/3 (Arp2/3) complex or formin (Chhabra and Higgs, 2007; Insall and Machesky, 2009), and multiple actin-adapter proteins, such as coronin (Humphries et al., 2002; Yan et al., 2005; Cai et al., 2007) and profilin (Pearson et al., 2003; Bae et al., 2010). For example, it has been reported that in the same Dictyostelium cell, both a phagocytic cup and the leading edge of a pseudopod compete for coronin (Maniak et al., 1995). Overall, however, “little is known about the mechanisms by which signaling events regulate the actin cytoskeleton” (Swaney et al., 2010) and much work remains to be performed in order to establish the information that a quantitative immunophysical analysis of the complex signaling networks requires. Such information would have to include, for example, complete chemical reaction schemes, spatiotemporal distributions of reactants, values of kinetic rate constants and the values of Michaelis–Menten constants of the involved enzymes.
To address many of the open questions mentioned above, a reductionist immunophysical strategy is very useful. Rather than settling for a qualitative description of an enormously complicated physiological process, this strategy compartmentalizes the full problem into parts that can be studied separately in greater depth and then reintegrates the results in a meaningful manner, gradually building up a rigorous bottom-up understanding of cellular behavior.
Single-cell immunophysics
Immunophysics is not limited to the study of cells but encompasses a wide range of subjects and scales: from structure–function relationships of individual molecules (Somers et al., 2000; Kim et al., 2006) and the dynamic strengths of receptor–ligand interactions (Williams et al., 2000; Marshall et al., 2003; Evans and Calderwood, 2007) to population dynamics of pathogens (McQueen, 2010; Vynnycky and White, 2010) and mathematical models of pandemics (Lemey et al., 2009; Chan et al., 2010). At the cellular scale, processes such as phagocytosis and chemotaxis exemplify both the necessity and benefits of an interdisciplinary analysis (Box 1). This analysis cannot be based on standard bulk assays alone, such as flow cytometry (Bassoe, 2002) or two-chamber chemotaxis methods (Haddox and Pfister, 1993; Wilkinson, 1998; Jin and Hereld, 2009; Muinonen-Martin et al., 2010). Instead, careful inspection of individual live cells (Figs 1, 2) is required to reveal subtle nuances that otherwise would be obscured by cell-to-cell baseline variability and to quantify the causal sequences and detailed timelines of immunophysical events that control these functions. Modern experimental techniques designed to characterize single cells with continually improving resolution include confocal microscopy, atomic force microscopy, optical tweezers, micro- and nano-fabrication and automated micropipette manipulation (Dewitt and Hallett, 2002; Zhelev et al., 2004; Suzuki et al., 2006; Jaumouille and Grinstein, 2010; Lomakina and Waugh, 2010; Lee et al., 2011). In addition, growing computational power is advancing all areas of physical immunology (Herant et al., 2006; Zhang and Morikis, 2006; Onsum and Rao, 2009; Herant et al., 2011; Liu et al., 2011). Perhaps most importantly, integrating essential physical insight with immunobiology allows us to define tighter constraints on possible explanations of cell and molecular behavior. Physical criteria for the validity of biological hypotheses include, for example, the requirement that the timing of dynamic processes is compatible with known kinetic parameters and, when signaling and structural molecules (including chemoattractants) traverse distances, with the known mobilities of the molecules in the respective environments. Likewise, a rigorous immunophysical analysis ensures that any proposed mechanistic scheme properly balances mechanical forces and satisfies the applicable conservation laws (of energy, mass, momentum, etc.).
Finally, a successful immunophysical research strategy also feeds back into the large body of mainstream immunology. For example, the already-daunting catalog of chemotactic and phagocytic receptors (as described in the above section) is continually expanding. However, it often remains unclear which receptor dominates a particular stage of the immune recognition of a specific target by a certain type of immune cell in a given host species and how an innate immune cell coordinates receptor-based target recognition with the physical processes involved in neutralizing the specific threat that the target might pose. A fresh mechanistic view that focuses on the time-dependent cell morphology and mechanics during single-cell–single-target interactions can perhaps guide future classifications of receptors, as well as subsequent signaling reactions, on the basis of their participation in temporally or spatially distinct immunophysical processes.
Pure chemotactic compared with pure phagocytic response by non-adherent cells
An instructive approach to study the single-cell behavior of innate immune cells is based on micropipette manipulation [for a tutorial see Heinrich and Rawicz (Heinrich and Rawicz, 2005)]. This technique provides unparalleled control over the contacts between cells and their targets. For example, pipette manipulation of the target particle overcomes the difficulty “of presenting the stimulus to the cell at a defined time and at a defined location” (Dewitt and Hallett, 2002). The addition of a second micropipette, which can be used to partially aspirate the cell itself and lift it above the chamber bottom, enables the study of chemotaxis and phagocytosis without interference from intracellular processes that would otherwise coordinate adhesion to a substrate [alternatively, a laser-optical trap can be used at a low laser power to suspend phagocytes above the chamber bottom (Suzuki et al., 2006)]. Following stimulation, initially quiescent, pipette-held cells exhibit pronounced phagocytic (Fig. 1B) or chemotactic (Fig. 2A,B) activity, as demonstrated by Zhelev and co-workers in pioneering studies that used two pipettes: one to manipulate the cell and the other to apply a stimulus (Evans et al., 1993; Zhelev et al., 1996; Chodniewicz and Zhelev, 2003; Zhelev et al., 2004). To mimic chemotactic stimulation, these authors use a microneedle to release the bacterial formyl peptide fMLP (formyl-methionyl-leucyl-phenylalanine) (Zhelev et al., 1996) and other GPCR ligands (Zhelev et al., 2004) towards a pipette-held neutrophil. The cell reacts by forming a pseudopod – a localized cellular protrusion – that is directed towards the chemoattractant source, which is similar to the response shown in Fig. 2A (see also Box 2). Clearly, this cell morphology is the characteristic shape of a ‘pure’ (i.e. adhesion-free) chemotactic response. A more physiological situation is mimicked in the experiment shown in Fig. 2A,B and supplementary material Movie 1 in which the left pipette holds a fungal particle, such as zymosan (an insoluble fraction from yeast cell walls often used to model fungal infection) or a fungal spore. In this case, chemoattractant anaphylatoxins (mainly C5a) are produced at the particle surface by the complement system (Box 1). The swift reaction of the cell to displacements of the particle provides evidence that the latter is indeed the chemoattractant source (Fig. 2B; supplementary material Movie 1).
These experiments corroborate, and allow us to study, the predominant role of actin-based protrusive deformation (May and Machesky, 2001; Chhabra and Higgs, 2007; Insall and Machesky, 2009; Pollard and Cooper, 2009; Clarke et al., 2010) in leukocyte chemotaxis. A minimalistic immunophysical framework of the ‘mechanistic response program’ of the cell at the onset of pure chemotaxis thus includes the following steps (Fig. 3A). First, soluble chemoattractant ligands bind to their cell-surface receptors. The surface distribution of the engaged receptors mirrors the local concentration of the ligand. Thus, the density of occupied receptors is highest at the side of the cell that faces the source of the chemoattractant gradient. Second, receptor ligation induces intracellular signaling. Although the biochemical reactions of the signaling network can be diverse and complex, a common feature is that early reaction steps trigger local actin polymerization or cross-linking. Third, the denser actin near the ligand–receptor-binding site generates a net pushing force that effectively displaces the cell membrane outwards. As this local protrusion is a consequence of receptor ligation, it is most pronounced towards the chemoattractant source (irrespective of the specific details that are governing this type of cell polarization).
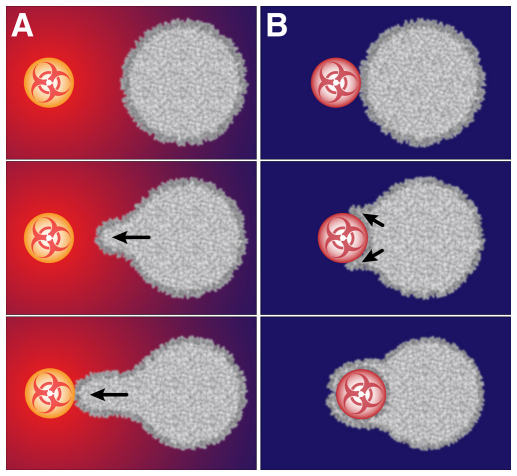
Fig. 3.
Distinctive attributes of pure chemotaxis and pure phagocytosis. An early morphological sign of cell activation is the formation of a local protrusion. Depending on the type of stimulus sensed at the cell surface, this protrusion proceeds in different directions (indicated by arrows), either towards a chemoattractant source or around a phagocytic particle (see Box 2). (A) The radial red-to-blue color gradient depicts the concentration gradient of a chemoattractant molecule that is produced at, and diffuses away from, the surface of the pathogenic target. Sensing the direction of this gradient, the non-adherent cell extends a chemotactic pseudopod toward the target. (B) A ‘purely phagocytic’ target (i.e. one that does not produce chemoattractants) is brought into contact with an initially quiescent, non-adherent cell. In this case, protrusion directly underneath the cell–target contact region is suppressed. Instead, a pseudopodial lamella proceeds parallel to the target surface, forming a phagocytic cup until the particle is fully engulfed.
The ultimate duty of a neutrophil during encounters with pathogens is not just to reach the pathogenic target by chemotaxis but also to neutralize it by phagocytosis. However, the definition of the cell response becomes ambiguous as soon as the protrusive pseudopod makes physical contact with a target that produces chemoattractants. Before examining this situation, it is instructive to establish a mechanistic baseline of pure phagocytosis (Fig. 1B; Fig. 3B) (Swanson, 2008; Clarke et al., 2010). A suitable single-cell approach is to confront pipette-held neutrophils with antibody-coated targets (Fc targets), which do not release chemoattractants. In this case, the observed steps of pure phagocytosis (i.e. without a chemotactic component) comprise the following (Fig. 3B). First, ligands immobilized on the surface of a pathogenic particle bind to phagocytic cell-surface receptors. At the same time, strong adhesion between the cell and the target is established. Adhesion could be supported by the phagocytic receptors themselves or by additional adhesion molecules that are not specific to phagocytic ligands (as described above). Second, receptor ligation triggers intracellular signaling pathways that stimulate local actin polymerization and actin cross-linking. Third, the increased actin density generates a local protrusive force. Fourth, protrusion directly underneath the cell–target contact region is suppressed. As a result, the outward deformation of the cell is most pronounced at the rim of the contact area, which leads to a lamellar pseudopod that grows parallel to the target surface. Fifth, gradual growth of the contact region advances the primary signaling source of actin polymerization. The resulting local protrusion, in concert with cell–target adhesion, guides the lamellar pseudopod around the target.
Schemes, such as these, are valuable guides for systematic cross-disciplinary enquiries into the mechanistic nature of pure chemotaxis and phagocytosis. A comprehensive immunophysical analysis strives to translate such schemes into mathematical models and to test hypotheses by simulating the behavior of these models on a computer. Although such modeling cannot account for all details of complex processes, such as chemotaxis or phagocytosis, it helps to establish the minimal conceptual frameworks that capture essential immune cell behavior (see Fig. 1C). On the basis of the pioneering modeling work of Herant and Dembo, the schemes outlined above have undergone and passed such theoretical validation. A continuum–mechanical model (Herant et al., 2003) successfully reproduced the time-dependent morphology of chemotactic pseudopods (Zhelev et al., 1996). The same computational framework has been adapted to model phagocytosis and has accurately reproduced the morphology and timecourse of the phagocytosis of Fc targets of different sizes (Herant et al., 2006). Recently, this quantitative model also reproduced mechanical variations in the phagocytosis of different target types (Herant et al., 2011). Such agreement between experimental observations and model predictions reinforces the conclusion that the above mechanistic framework is not only biologically plausible but also physically realistic.
Directional difference in initial protrusion
Before comparing the onset of cellular protrusion in chemotaxis and phagocytosis in more detail, we will briefly touch on two prominent hypotheses that have been proposed to explain how polymerizing cytoskeletal filaments could generate protrusive force, that is, ‘Brownian ratchets’ and ‘end-tracking motors’. Similar to mathematical modeling, such conceptualizations are an integral part of an immunophysical quest for a quantitative understanding of cell and molecular behavior. Among others, these hypotheses can assist in the interpretation of experimental observations, provide useful clues for the discovery of new functional roles of proteins or even guide searches for hitherto unidentified biomolecular components. For example, the thermodynamic and kinetic analysis of actin-filament elongation recently led Breitsprecher and co-workers to anticipate that all Ena/VASP proteins are potent filament elongators in vivo (Breitsprecher et al., 2011).
Common to Brownian ratchet models is the assumption that the membrane and the cytoskeleton undergo thermal fluctuations relative to one another (Hill, 1981; Peskin et al., 1993; Howard, 2001; Mogilner and Oster, 2003). Such fluctuations create transient spaces that allow for the incorporation of free globular (G)-actin monomers into growing actin filaments. As filaments that interact with a fluctuating membrane grow in length, they prevent the membrane from returning to its former position. This rectifies the fluctuations and generates a net repulsive force between the membrane and the cytoskeleton. By contrast, end-tracking proteins are assumed to maintain a physical link between the membrane and the ends of growing actin filaments at all times (Dickinson et al., 2004; Dickinson, 2009). These membrane-linked motor proteins facilitate the addition of G-actin monomers to the actin filament while continually ‘tracking’ (i.e. binding to) the most recently added monomer. Importantly, the continuum–mechanical computer model by Herant, as described in the previous section, encompasses both of these molecular-scale notions of force generation.
Regardless of the detailed molecular mechanisms, cells generate outwards motion during both chemotaxis and phagocytosis by a common method whereby local stimulation at the cell surface leads to localized actin reorganization and subsequent cell protrusion (Box 2). Thus, it is remarkable that the primary direction of protrusion is different in the two cases, that is, along the chemoattractant gradient in chemotaxis, but parallel to the target surface in phagocytosis (see arrows in Fig. 3 and sketch in Box 2). This difference does not appear to be related to the loci of receptor ligation, because the strongest cell-surface stimulation occurs in the same region – opposite the target – during both chemotaxis and the early stages of phagocytosis. Instead, the discrimination between motion towards a chemoattractant target and around a phagocytic particle (rather than simply pushing the latter along) is likely to be programmed into the mechanical properties of molecular or subcellular structures.
A key component of the explanation of this directional difference is the suppression of protrusion underneath the cell–target contact region in pure phagocytosis. Because fresh contact between the cell and the target stimulates local protrusion, some ‘swelling’ of the cell underneath the adherent target might generally be expected during the initial stage of phagocytosis. By contrast, antibody-coated targets rarely cause outwards protrusions before their internalization by normal neutrophils (Herant et al., 2006; Lee et al., 2011). Surprisingly though, if the structure of the cytoskeleton is perturbed by mild actin inhibition with latrunculin A or cytochalasin D, neutrophils tend to form small, but distinct, pedestal-like protrusions that at first push adherent Fc targets outwards (Lee et al., 2011). These results demonstrate that the suppression of protrusion beneath an adherent Fc target requires an intact actin cytoskeleton and might be dependent on the formation of structural linkages between the cytoskeleton and the membrane that is adherent to the target. Strong support for this hypothesis was provided by the results of recent computer simulations (Herant et al., 2011). Presumably these ‘cytoskeletal membrane anchors’ are molecular linkages between actin and the cytoplasmic domains of Fc receptors or transmembrane proteins that are associated with the receptors.
A blurred transition from chemotactic chase to phagocytic consumption
Encounters between an immune cell and particulate pathogens that are completely devoid of chemotactic activity are probably rare. Targets immersed in normal serum are more likely to be sources of at least some chemoattractants. Once an immune cell makes physical contact with such a target and starts forming a pseudopodial cup, the interaction is commonly classified as phagocytosis. Yet, as long as the pathogen surface remains partially exposed (Fig. 2C, top two panels; Fig. 4), it continues to release chemoattractants that stimulate the unobstructed chemotactic receptors on the cell surface. Therefore, the moment of first contact between an immune cell and a chemoattractant particle does not mark a sharp transition between chemotactic cell motion and phagocytic uptake of the particle. Instead, phagocytosis and chemotaxis continue to proceed in parallel throughout the entire period from first contact to closure of the pseudopodial cup. During this period, the primary source from which chemoattractant molecules can freely diffuse is the part of the target surface that is not in contact with the phagocyte. Therefore, one might still define a spatial separation between the stimuli of chemotaxis and phagocytosis. Other than that, the mixed signals processed by the intracellular machinery at this stage obscure the distinction between chemotactic and phagocytic responses.
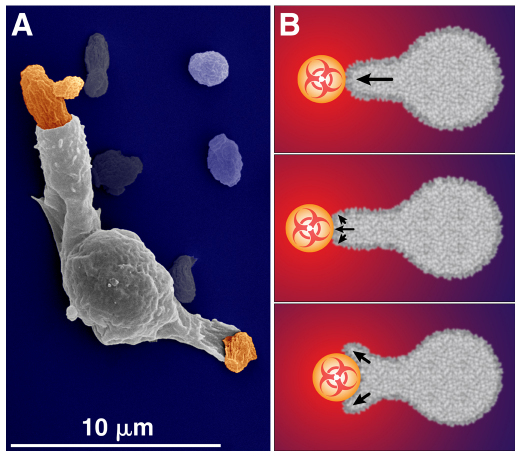
Fig. 4.
Blurred transition from a chemotactic to a phagocytic response. (A) Pseudocolor scanning electron microscopy image showing a human neutrophil in the process of engulfing two zymosan particles. (B) Once a cell makes contact with a chemoattractant particle, it starts to engulf the particle by phagocytosis. However, this does not mean that chemotactic stimulation ceases. Instead, chemoattractant molecules continue to emanate from the part of the target surface that is not yet in contact with the cell. Therefore, the pseudopod is likely to continue to grow and push an otherwise free particle along for some time, while, at the same time, starting to surround it. Eventually, as the cell begins ‘pulling in’ the target, the direction of target movement reverses. The immunophysical analysis of single-cell experiments has revealed that inwards movement of the target is mainly driven by the cortical tension of the cell (in cooperation with cell–target adhesion).
Even though the line between phagocytosis and chemotaxis can become blurred, some elements of the morphological or mechanical cell response can still be categorized as chemotactic or phagocytic. For example, the adhesive contact between the cell membrane and target is an attribute of phagocytosis. By contrast, the front of the chemotactic pseudopod is unlikely to reverse direction immediately after touching the target. Instead, its continued outward motion will initially push the chemoattractant particle away from the main cell body, as indicated in Fig. 4B (upper two panels). Transient displacement of the target outwards thus appears to be a sign of chemotactic behavior of normal cells.
Hybrid chemotactic and phagocytic response to fungal and other targets
At a first glance, it seems unlikely that chemotactic cell behavior can occur in the absence of soluble chemoattractants. Surprisingly, however, such behavior is in fact observed when neutrophils are brought into contact with fungal targets (zymosan or fungal spores) under conditions that obviate classical chemotaxis (Lee et al., 2011). Experimentally, this can be achieved by using a buffer that contains a supplement of heat-treated serum (heat treatment incapacitates the production of anaphylatoxins by the complement system). In this case, neutrophils indeed fail to recognize fungal particles over a distance.
However, once such a particle makes contact with an initially quiescent neutrophil, in these conditions, it usually adheres to and activates the cell, eliciting an intriguing response that appears to be a hybrid between chemotaxis and phagocytosis (Fig. 5). Despite the lack of priming by anaphylatoxins, the neutrophil will ingest not just one but multiple fungal particles. In contrast to pure phagocytosis of Fc targets, however, the cell initially forms a pedestal-like protrusion that pushes the fungal particle outwards by some distance (Fig. 5). As discussed above, this protrusion resembles a pseudopod growing towards a chemoattractant stimulus – except that the fungal particle is already attached to the pseudopod. Remarkably, under certain conditions, neutrophils have been observed to exhibit this characteristic protrusive response also following contact with interleukin-8 (IL8)-coated beads (Elena B. Lomakina and Richard E. Waugh, personal communication), which indicates that this hybrid chemotactic and phagocytic neutrophil behavior is not exclusive to interactions with fungal particles.
Fig. 5.
Hybrid chemotactic and phagocytic response of human neutrophils to fungal targets. (A) The cell is brought into contact with a normally chemoattractant particle in conditions that obviate classical chemotaxis. The top image depicts a pipette-held neutrophil immediately after attachment of a fungal particle (using a second pipette, not shown). The particle is zymosan here, but a similar cell behavior is observed with fungal spores. The first change to the cell morphology is the formation of a pedestal-like protrusion that resembles a chemotactic pseudopod and pushes the particle outwards (labeled by an arrow in the middle image). Later, the neutrophil switches from this chemotactic-like protrusion to phagocytic target internalization and completes phagocytosis within a few minutes (relative times are included in each image). (B) Illustration of the characteristic neutrophil behavior during the mixed chemotactic and phagocytic response to a fungal particle. Even though the cell is stimulated by local physical contact with a particulate target, its initial reaction is to protrude a chemotactic pseudopod (see also Box 2). The pseudopod eventually ‘overflows’ and thereby internalizes the target. The cell completes this phagocytic phase by returning to a spherical shape that now contains the target. Scale bar: 10 μm.
These observations suggest that the initial contact of a quiescent neutrophil with either a fungal or IL8-coated particle, or soluble chemoattractants, will first trigger a chemotactic response. We speculate that, while this response is underway, the neutrophil prepares the mechanistic program for phagocytosis, which might involve the shuttling of phagocytic and adhesive receptors to the cell surface, as well as the lateral redistribution and activation of these receptors. However, this preparatory phase requires time. If the cell makes ‘premature’ physical contact with the target, the chemotactic response will continue until the phagocytic response is engaged. Then the motion of the particle into the cell is similar to what one observes during the antibody-mediated ‘pure’ phagocytosis.
Compared with the swift phagocytosis of similarly sized Fc targets, the substantially slower uptake of fungal targets – mainly owing to the initial protrusive ‘detour’ in cell deformation – marks the latter as the less efficient form of target internalization in itself. We thus speculate that a multipurpose functionality that tightly interweaves motion towards a fungal particle, with its subsequent phagocytic neutralization, has been a dominant criterion in the evolutionary optimization of the neutrophil response to fungi. By contrast, the straightforward target engulfment and lack of chemotactic activity in neutrophil interactions with antibody-tagged pathogens indicate that, in this case, a high speed of target uptake presented a crucial evolutionary advantage.
The immune detection of fungi is generally viewed as a prototype for the recognition of a pattern of surface-immobilized ligands that are native to a pathogen (van de Veerdonk et al., 2008; Mogensen, 2009; Romani, 2011). Puzzlingly, we find it problematic to fit the results discussed above on early processing of cell-surface stimuli into the framework of this paradigm. Judging by the morphology of the initial cell response (as resolved by single-cell experiments), neutrophils appear unable, at first, to distinguish between particulate fungal targets and soluble chemoattractants. It seems peculiar that a supposedly highly specialized pattern-dependent recognition of such particles should be followed by a much more generic and slow uptake – especially as neutrophils respond more efficiently to other targets, such as antibody-coated pathogens. Although our reasoning is largely speculative at this point, these findings indicate that there is a need to carefully revisit the exact mechanistic nature of innate fungal recognition, including the time-dependent engagement of various types of receptor.
Concluding remarks and perspectives
Chemotaxis and phagocytosis have traditionally been studied separately. The present discussion demonstrates that, from an immunophysical viewpoint, these vital processes appear to be more interrelated than previously thought. Integration of thorough single-cell experiments and computer modeling has laid the groundwork for the combined study of chemotactic and phagocytic cell behaviors and calls for further inspection of their similarities and differences. A promising future direction will be to examine the response of individual live cells to encounters with particles coated with (controlled densities of) chemoattractants. Innate immune cells indeed have been shown to react to immobilized chemoattractants by increasing cell adhesiveness (Shamri et al., 2005; Hyduk et al., 2007; Woolf et al., 2007; Lomakina and Waugh, 2010). Future studies of single-cell interactions with chemoattractant-coated particles will not only establish whether a chemotactic response requires soluble ligands (the results discussed above with fungal targets suggest that it does not) but also allow us to quantify the dependence of the cell response on the number and spacing of the encountered chemoattractant molecules. Moreover, single-cell approaches can reveal intriguing new insights into the dynamics of the immune cell processing of chemoattractant stimuli, including the timelines of cell signaling reactions (Lomakina and Waugh, 2010). Although, in most cases, such stimulation will eventually lead to the phagocytic uptake of the target particle, an initial cellular protrusion that pushes the particle outwards serves as a telltale sign of chemotactic behavior, as discussed above. Overall, it is our belief that the combined study of chemotaxis and phagocytosis will deepen our fundamental understanding of innate immunity while, additionally, elucidating universal mechanisms of eukaryotic cell motility.
Acknowledgements
We thank Patricia Kysar for instruction, and Jonathan Lam for his assistance in acquiring the scanning electron microscopy images in Fig. 1A and Fig. 4A. We are grateful to Elena B. Lomakina and Richard E. Waugh for their permission to discuss their unpublished observation. Scanning electron microscopy was carried out in the Electron Microscopy Laboratory of the Department of Medical Pathology and Laboratory Medicine (School of Medicine, University of California at Davis).
Footnotes
Funding
This work was supported by National Institutes of Health [grant R01 A1072391]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/18/3041/DC1
References
- Afonso P. V., Parent C. A. 2011. PI3K and chemotaxis: a priming issue? Sci. Signal. 4, pe22 [DOI] [PubMed] [Google Scholar]
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. 2007. Molecular Biology of the Cell. New York: Garland Science; [Google Scholar]
- Bae Y. H., Ding Z., Das T., Wells A., Gertler F., Roy P. 2010. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc. Natl. Acad. Sci. USA 107, 21547-21552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassoe C. F. 2002. Assessment of phagocyte functions by flow cytometry. Curr. Protoc. Cytom. Chapter 9, Unit 9 19 [DOI] [PubMed] [Google Scholar]
- Bell G. I. 1978. Models for the specific adhesion of cells to cells. Science 200, 618-627 [DOI] [PubMed] [Google Scholar]
- Bjarnadóttir T. K., Gloriam D. E., Hellstrand S. H., Kristiansson H., Fredriksson R., Schiöth H. B. 2006. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics 88, 263-273 [DOI] [PubMed] [Google Scholar]
- Boross P., van de Poel K., Van de Winkel J. G., Leusen J. H. 2008. Fc Receptors. In Encyclopedia of Life Sciences (ELS). Chichester: John Wiley & Sons; doi: 10.1002/9780470015902.a000916.pub2 [Google Scholar]
- Bray D. 2000. Cell Movements: From Molecules to Motility. New York, NY: Garland Publishing; [Google Scholar]
- Breitsprecher D., Kiesewetter A. K., Linkner J., Vinzenz M., Stradal T. E., Small J. V., Curth U., Dickinson R. B., Faix J. 2011. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 30, 456-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. D., Herre J., Williams D. L., Willment J. A., Marshall A. S., Gordon S. 2003. Dectin-1 mediates the biological effects of beta-glucans. J. Exp. Med. 197, 1119-1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. 2004. Rho and Rac take center stage. Cell 116, 167-179 [DOI] [PubMed] [Google Scholar]
- Cai L., Marshall T. W., Uetrecht A. C., Schafer D. A., Bear J. E. 2007. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 128, 915-929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano F., Montcourrier P., Chavrier P. 2000. Membrane recruitment of Rac1 triggers phagocytosis. J. Cell Sci. 113, 2955-2961 [DOI] [PubMed] [Google Scholar]
- Chan J., Holmes A., Rabadan R. 2010. Network analysis of global influenza spread. PLoS Comput. Biol. 6, e1001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest P. G., Firtel R. A. 2006. Feedback signaling controls leading-edge formation during chemotaxis. Curr. Opin. Genet. Dev. 16, 339-347 [DOI] [PubMed] [Google Scholar]
- Chhabra E. S., Higgs H. N. 2007. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9, 1110-1121 [DOI] [PubMed] [Google Scholar]
- Chimini G., Chavrier P. 2000. Function of Rho family proteins in actin dynamics during phagocytosis and engulfment. Nat. Cell Biol. 2, E191-E196 [DOI] [PubMed] [Google Scholar]
- Chodniewicz D., Zhelev D. V. 2003. Novel pathways of F-actin polymerization in the human neutrophil. Blood 102, 2251-2258 [DOI] [PubMed] [Google Scholar]
- Clarke M., Engel U., Giorgione J., Muller-Taubenberger A., Prassler J., Veltman D., Gerisch G. 2010. Curvature recognition and force generation in phagocytosis. BMC Biol. 8, 154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P., Janetopoulos C. 2003. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem. 278, 20445-20448 [DOI] [PubMed] [Google Scholar]
- Dewitt S., Hallett M. B. 2002. Cytosolic free Ca(2+) changes and calpain activation are required for beta integrin-accelerated phagocytosis by human neutrophils. J. Cell Biol. 159, 181-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson R. B. 2009. Models for actin polymerization motors. J. Math. Biol. 58, 81-103 [DOI] [PubMed] [Google Scholar]
- Dickinson R. B., Caro L., Purich D. L. 2004. Force generation by cytoskeletal filament end-tracking proteins. Biophys. J. 87, 2838-2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D., Dong C., Fredberg J. J., Guilak F., Ingber D., Janmey P., Kamm R. D., Schmid-Schonbein G. W., Weinbaum S. 2009. Biomechanics: cell research and applications for the next decade. Ann. Biomed. Eng. 37, 847-859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsam R. T., Gutkind J. S. 2007. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 7, 79-94 [DOI] [PubMed] [Google Scholar]
- Evans E., Ritchie K. 1997. Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541-1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. A., Calderwood D. A. 2007. Forces and bond dynamics in cell adhesion. Science 316, 1148-1153 [DOI] [PubMed] [Google Scholar]
- Evans E., Leung A., Zhelev D. 1993. Synchrony of cell spreading and contraction force as phagocytes engulf large pathogens. J. Cell Biol. 122, 1295-1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon D. T. 1984. Structure and function of the human C3b receptor. Fed. Proc. 43, 2553-2557 [PubMed] [Google Scholar]
- Goodridge H. S., Underhill D. M. 2008. Fungal recognition by TLR2 and Dectin-1. Handb. Exp. Pharmacol. 183, 87-109 [DOI] [PubMed] [Google Scholar]
- Goodridge H. S., Wolf A. J., Underhill D. M. 2009. Beta-glucan recognition by the innate immune system. Immunol. Rev. 230, 38-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddox J. L., Pfister R. R. 1993. Evaluation of the methodology of polymorphonuclear leukocyte chemotaxis. J. Immunol. Methods 163, 273-275 [DOI] [PubMed] [Google Scholar]
- Hall A. B., Gakidis M. A., Glogauer M., Wilsbacher J. L., Gao S., Swat W., Brugge J. S. 2006. Requirements for Vav guanine nucleotide exchange factors and Rho GTPases in FcgammaR- and complement-mediated phagocytosis. Immunity 24, 305-316 [DOI] [PubMed] [Google Scholar]
- Heinrich V., Rawicz W. 2005. Automated, high-resolution micropipet aspiration reveals new insight into the physical properties of fluid membranes. Langmuir 21, 1962-1971 [DOI] [PubMed] [Google Scholar]
- Herant M., Dembo M. 2010. Form and function in cell motility: from fibroblasts to keratocytes. Biophys. J. 98, 1408-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herant M., Marganski W. A., Dembo M. 2003. The mechanics of neutrophils: synthetic modeling of three experiments. Biophys. J. 84, 3389-3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herant M., Heinrich V., Dembo M. 2006. Mechanics of neutrophil phagocytosis: experiments and quantitative models. J. Cell Sci. 119, 1903-1913 [DOI] [PubMed] [Google Scholar]
- Herant M., Lee C.-Y., Dembo M., Heinrich V. 2011. Protrusive push versus enveloping embrace: computational model of phagocytosis predicts key regulatory role of cytoskeletal membrane anchors. PLoS Comput. Biol. 7, e1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T. L. 1981. Microfilament or microtubule assembly or disassembly against a force. Proc. Natl. Acad. Sci. USA 78, 5613-5617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmig S. T., Ariizumi K., Cruz P. D., Jr 2009. Recognition of non-self-polysaccharides by C-type lectin receptors dectin-1 and dectin-2. Glycobiology 19, 568-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- Humphries C. L., Balcer H. I., D'Agostino J. L., Winsor B., Drubin D. G., Barnes G., Andrews B. J., Goode B. L. 2002. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol. 159, 993-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyduk S. J., Chan J. R., Duffy S. T., Chen M., Peterson M. D., Waddell T. K., Digby G. C., Szaszi K., Kapus A., Cybulsky M. I. 2007. Phospholipase C, calcium, and calmodulin are critical for alpha4beta1 integrin affinity up-regulation and monocyte arrest triggered by chemoattractants. Blood 109, 176-184 [DOI] [PubMed] [Google Scholar]
- Iglesias P. A., Devreotes P. N. 2008. Navigating through models of chemotaxis. Curr. Opin. Cell Biol. 20, 35-40 [DOI] [PubMed] [Google Scholar]
- Insall R. H. 2010. Understanding eukaryotic chemotaxis: a pseudopod-centred view. Nat. Rev. Mol. Cell Biol. 11, 453-458 [DOI] [PubMed] [Google Scholar]
- Insall R., Machesky L. 2009. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev. Cell 17, 310-322 [DOI] [PubMed] [Google Scholar]
- Jaumouille V., Grinstein S. 2010. Receptor mobility, the cytoskeleton, and particle binding during phagocytosis. Curr. Opin. Cell Biol. 23, 22-29 [DOI] [PubMed] [Google Scholar]
- Jin T., Hereld D. 2009. Chemotaxis: Methods and Protocols. Vol. 571 (eds Jin T., Hereld D.). New York, NY: Humana Press; [Google Scholar]
- Kay R. R., Langridge P., Traynor D., Hoeller O. 2008. Changing directions in the study of chemotaxis. Nat. Rev. Mol. Cell Biol. 9, 455-463 [DOI] [PubMed] [Google Scholar]
- Kim S. V., Mehal W. Z., Dong X., Heinrich V., Pypaert M., Mellman I., Dembo M., Mooseker M. S., Wu D., Flavell R. A. 2006. Modulation of cell adhesion and motility in the immune system by Myo1f. Science 314, 136-139 [DOI] [PubMed] [Google Scholar]
- Lam J., Herant M., Dembo M., Heinrich V. 2009. Baseline mechanical characterization of J774 macrophages. Biophys. J. 96, 248-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-Y., Herant M., Heinrich V. 2011. Target-specific mechanics of phagocytosis: protrusive neutrophil response to zymosan differs from the uptake of antibody-tagged pathogens. J. Cell Sci. 124, 1106-1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Suchard M., Rambaut A. 2009. Reconstructing the initial global spread of a human influenza pandemic: a Bayesian spatial-temporal model for the global spread of H1N1pdm. PLoS Curr. Influenza 1, PRN1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A., Weiner O. D., Lim W. A., Voigt C. A. 2009. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zhang J., Tan P. Y., Hsu D., Blom A. M., Leong B., Sethi S., Ho B., Ding J. L., Thiagarajan P. S. 2011. A computational and experimental study of the regulatory mechanisms of the complement system. PLoS Comput. Biol. 7, e1001059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakina E. B., Waugh R. E. 2010. Signaling and dynamics of activation of LFA-1 and Mac-1 by immobilized IL-8. Cell. Mol. Bioeng. 3, 106-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniak M., Rauchenberger R., Albrecht R., Murphy J., Gerisch G. 1995. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell 83, 915-924 [DOI] [PubMed] [Google Scholar]
- Marshall B. T., Long M., Piper J. W., Yago T., McEver R. P., Zhu C. 2003. Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190-193 [DOI] [PubMed] [Google Scholar]
- May R. C., Machesky L. M. 2001. Phagocytosis and the actin cytoskeleton. J. Cell Sci. 114, 1061-1077 [DOI] [PubMed] [Google Scholar]
- McQueen P. 2010. Population dynamics of a pathogen: the conundrum of vivax malaria. Biophys. Rev. 2, 111-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen T. H. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22, 240-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A., Oster G. 2003. Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys. J. 84, 1591-1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk P. N., Scola A. M., Madala P., Fairlie D. P. 2007. Function, structure and therapeutic potential of complement C5a receptors. Br. J. Pharmacol. 152, 429-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muinonen-Martin A. J., Veltman D. M., Kalna G., Insall R. H. 2010. An improved chamber for direct visualisation of chemotaxis. PLoS ONE 5, e15309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. P., Travers P., Walport M. 2007. Janeway's Immunobiology. New York: Garland Science; [Google Scholar]
- Netea M., Meer J., Kullberg B. 2007. Recognition of fungal pathogens by Toll-like receptors. In Immunology of Fungal Infections (ed. Brown G. D., Netea M. G.), pp. 259-272 Dordrecht, Netherlands: Springer; [Google Scholar]
- Nielsen C. H., Antonsen S., Matthiesen S. H., Leslie R. G. 1997. The roles of complement receptors type 1 (CR1, CD35) and type 3 (CR3, CD11b/CD18) in the regulation of the immune complex-elicited respiratory burst of polymorphonuclear leukocytes in whole blood. Eur. J. Immunol. 27, 2914-2919 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F., Ravetch J. V. 2008. Fc[gamma] receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34-47 [DOI] [PubMed] [Google Scholar]
- Onsum M. D., Rao C. V. 2009. Calling heads from tails: the role of mathematical modeling in understanding cell polarization. Curr. Opin. Cell Biol. 21, 74-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent C. A., Devreotes P. N. 1999. A cell's sense of direction. Science 284, 765-770 [DOI] [PubMed] [Google Scholar]
- Pearson A. M., Baksa K., Rämet M., Protas M., McKee M., Brown D., Ezekowitz R. A. B. 2003. Identification of cytoskeletal regulatory proteins required for efficient phagocytosis in Drosophila. Microbes Infect. 5, 815-824 [DOI] [PubMed] [Google Scholar]
- Peskin C. S., Odell G. M., Oster G. F. 1993. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 65, 316-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R. J., Doyle A. D., Yamada K. M. 2009. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 10, 538-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. 2003. The cytoskeleton, cellular motility and the reductionist agenda. Nature 422, 741-745 [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Cooper J. A. 2009. Actin, a central player in cell shape and movement. Science 326, 1208-1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. J., Waelbroeck M. 2004. G protein activation by G protein coupled receptors: ternary complex formation or catalyzed reaction? Biochem. Pharmacol. 68, 799-806 [DOI] [PubMed] [Google Scholar]
- Romani L. 2011. Immunity to fungal infections. Nat. Rev. Immunol. 11, 275-288 [DOI] [PubMed] [Google Scholar]
- Shamri R., Grabovsky V., Gauguet J. M., Feigelson S., Manevich E., Kolanus W., Robinson M. K., Staunton D. E., von Andrian U. H., Alon R. 2005. Lymphocyte arrest requires instantaneous induction of an extended LFA-1 conformation mediated by endothelium-bound chemokines. Nat. Immunol. 6, 497-506 [DOI] [PubMed] [Google Scholar]
- Somers W. S., Tang J., Shaw G. D., Camphausen R. T. 2000. Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell 103, 467-479 [DOI] [PubMed] [Google Scholar]
- Stuart L. M., Ezekowitz R. A. 2005. Phagocytosis: elegant complexity. Immunity 22, 539-550 [DOI] [PubMed] [Google Scholar]
- Suzuki T., Yanai M., Kubo H., Kanda A., Sasaki H., Butler J. P. 2006. Interaction of non-adherent suspended neutrophils to complement opsonized pathogens: a new assay using optical traps. Cell Res. 16, 887-894 [DOI] [PubMed] [Google Scholar]
- Swaney K. F., Huang C. H., Devreotes P. N. 2010. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu. Rev. Biophys. 39, 265-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. A. 2008. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 9, 639-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. A., Hoppe A. D. 2004. The coordination of signaling during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 76, 1093-1103 [DOI] [PubMed] [Google Scholar]
- Thelen M., Stein J. V. 2008. How chemokines invite leukocytes to dance. Nat. Immunol. 9, 953-959 [DOI] [PubMed] [Google Scholar]
- Underhill D. M., Ozinsky A. (2002a). Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20, 825-852 [DOI] [PubMed] [Google Scholar]
- Underhill D. M., Ozinsky A. (2002b). Toll-like receptors: key mediators of microbe detection. Curr. Opin. Immunol. 14, 103-110 [DOI] [PubMed] [Google Scholar]
- van Bruggen R., Drewniak A., Jansen M., van Houdt M., Roos D., Chapel H., Verhoeven A. J., Kuijpers T. W. 2009. Complement receptor 3, not Dectin-1, is the major receptor on human neutrophils for beta-glucan-bearing particles. Mol. Immunol. 47, 575-581 [DOI] [PubMed] [Google Scholar]
- van de Veerdonk F. L., Kullberg B. J., van der Meer J. W., Gow N. A., Netea M. G. 2008. Host-microbe interactions: innate pattern recognition of fungal pathogens. Curr. Opin. Microbiol. 11, 305-312 [DOI] [PubMed] [Google Scholar]
- Van Haastert P. J. M., Veltman D. M. 2007. Chemotaxis: navigating by multiple signaling pathways. Sci. STKE 2007, pe40 [DOI] [PubMed] [Google Scholar]
- Vera J., Fenutria R., Canadas O., Figueras M., Mota R., Sarrias M. R., Williams D. L., Casals C., Yelamos J., Lozano F. 2009. The CD5 ectodomain interacts with conserved fungal cell wall components and protects from zymosan-induced septic shock-like syndrome. Proc. Natl. Acad. Sci. USA 106, 1506-1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vynnycky E., White R. G. 2010. An Introduction to Infectious Disease Modelling. Oxford: Oxford University Press; [Google Scholar]
- Wilkinson P. C. 1998. Assays of leukocyte locomotion and chemotaxis. J. Immunol. Methods 216, 139-153 [DOI] [PubMed] [Google Scholar]
- Williams T. E., Nagarajan S., Selvaraj P., Zhu C. 2000. Concurrent and independent binding of Fcgamma receptors IIa and IIIb to surface-bound IgG. Biophys. J. 79, 1867-1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolgemuth C. W. 2011. Does cell biology need physicists? Physics 4, 4 [Google Scholar]
- Wong W. 2011. Focus issue: moving in the right direction. Sci. Signal. 4, eg4 [DOI] [PubMed] [Google Scholar]
- Woolf E., Grigorova I., Sagiv A., Grabovsky V., Feigelson S. W., Shulman Z., Hartmann T., Sixt M., Cyster J. G., Alon R. 2007. Lymph node chemokines promote sustained T lymphocyte motility without triggering stable integrin adhesiveness in the absence of shear forces. Nat. Immunol. 8, 1076-1085 [DOI] [PubMed] [Google Scholar]
- Yan M., Collins R., Grinstein S., Trimble W. 2005. Coronin-1 function is required for phagosome formation. Mol. Biol. Cell 16, 3077-3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Morikis D. 2006. Immunophysical properties and prediction of activities for vaccinia virus complement control protein and smallpox inhibitor of complement enzymes using molecular dynamics and electrostatics. Biophys. J. 90, 3106-3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelev D. V., Alteraifi A. M., Hochmuth R. M. 1996. F-actin network formation in tethers and in pseudopods stimulated by chemoattractant. Cell Motil. Cytoskeleton 35, 331-344 [DOI] [PubMed] [Google Scholar]
- Zhelev D. V., Alteraifi A. M., Chodniewicz D. 2004. Controlled pseudopod extension of human neutrophils stimulated with different chemoattractants. Biophys. J. 87, 688-695 [DOI] [PMC free article] [PubMed] [Google Scholar]