Abstract
The 5′ leader (Ω) of tobacco mosaic viral RNA functions as a translational enhancer. Sequence analysis of a 102-kD protein, identified previously as a specific Ω RNA-binding protein, revealed homology to the HSP101/HSP104/ClpB family of heat shock proteins and its expression in yeast complemented a thermotolerance defect caused by a deletion of the HSP104 gene. Up to a 50-fold increase in the translation of Ω-luc, but not luc mRNA was observed in yeast expressing the tobacco HSP101 whereas Ω failed to enhance translation in the absence of HSP101. Therefore, HSP101 and Ω comprise a two-component translational regulatory mechanism that can be recapitulated in yeast. Analysis of HSP101 function in yeast translation mutants suggested that the initiation factor (eIF) 3 and specifically one (TIF4632) of the two eIF4G proteins were required for the HSP101-mediated enhancement. The RNA-binding and translational regulatory activities of HSP101 were inactive in respiring cells or in cells subject to nutrient limitation, but its thermotolerance function remained unaffected. This is the first identification of a protein required for specific translational enhancement of capped mRNAs, the first report of a translational regulatory function for any heat-shock protein, and the first functional distinction between the two eIF4G proteins present in eukaryotes.
Keywords: HSP101, translation initiation, protein synthesis, RNA-binding protein
There is now substantial evidence showing the important regulatory role that an mRNA 5′ leader can play during translation in eukaryotes. In addition to structural features, such as its length (Kozak 1988; Gallie and Walbot 1992) and higher order structure (Pelletier and Sonenberg 1985), the presence of cis-acting regulatory elements in a leader have been shown to influence protein synthesis including those present within ferritin mRNA (for review, see Rouault et al. 1996), GCN4 mRNA (for review, see Hinnebusch 1996) and picornaviruses (for review, see Ehrenfeld 1996). In each of these examples, the 5′ leader serves as a regulator that enables translation to occur only under certain cellular conditions.
Tobacco mosaic virus (TMV) is a single-stranded, positive-sense RNA virus for which the genomic RNA serves as an efficiently translated mRNA. The 68-nucleotide 5′ leader (known as Ω) is responsible, in part, for the efficient translation of TMV mRNA and is distinguished by the absence of guanosine residues and a central poly(CAA) region required for the stimulation of translation (Gallie et al. 1987a,b; Gallie and Walbot 1992). A model frequently proposed to explain the translational enhancement afforded by Ω posits that the lack of secondary structure within the leader, in itself, facilitates 40S subunit scanning and thereby reduces the requirement for initiation factors (eIFs), such as eIF4E or eIF4A (Sleat et al. 1988; Altmann et al. 1990a,b, 1997; Kozak 1991, 1992, 1994). This model predicts that Ω functions passively by presenting no barrier to 40S ribosomal subunits as they scan the 5′ leader in search of the initiation codon. In contrast, an active mechanism requiring a trans-acting factor that would specifically bind Ω and actively mediate the regulation from this sequence was suggested by the identification of the 25 nucleotide poly(CAA) sequence within Ω as the essential subsequence for translational enhancement (Gallie and Walbot 1992). As Ω does not require any other viral sequence or protein to enhance translation, such a trans-acting factor would have to be cellular in origin. Previously, we reported that a 102-kD protein (p102) was an RNA-binding protein that binds to Ω within the poly(CAA) subsequence responsible for translational enhancement (Leathers et al. 1993; Tanguay and Gallie 1996). RNA-binding assays and Western analysis revealed that p102 is a conserved RNA-binding protein throughout the higher plant kingdom (Tanguay and Gallie 1996).
In this study, we show that the 102-kD Ω-binding protein is the heat shock protein, HSP101. Expression of wheat or tobacco cDNAs encoding HSP101 not only complemented the thermotolerance defect in a yeast hsp104 null mutant but also mediated up to a 50-fold increase in translation specifically from Ω-containing mRNAs. The translation initiation factor eIF3 and specifically one of the two yeast eIF4Gs (i.e., TIF4632) was required for the HSP101-mediated enhancement of translation. HSP101 activity was repressed following transient or severe amino acid starvation, in respiring cells, or during late-exponential growth/early stationary phase when the reduced availability of nutrients results in metabolic changes required for the successful entry into the stationary phase. As amino acid starvation or respiration failed to support HSP101 translational regulatory activity, these data suggest that the function of HSP101 during translation is itself subject to regulation by the nutrient status of the cell.
Results
Identification of p102 from wheat and tobacco as HSP101
p102 was purified from wheat and tobacco on the basis of its ability to bind Ω RNA as described previously (Tanguay and Gallie 1996). Several regions of amino acid sequence were obtained through peptide sequencing that were identical or highly similar to HSP101 from Arabidopsis (Schirmer et al. 1994), soybean (Lee et al. 1994), and to a lesser extent, to yeast Hsp104 (Parsell et al. 1991). Regions designated with asterisks in Figure 1 represent the five peptide sequences obtained. Full-length cDNAs encoding the wheat and tobacco 102-kD proteins were obtained by expression screening of wheat and tobacco heat shock cDNA libraries by use of anti-p102 antibodies. The predicted amino acid sequence from each cDNA was aligned with known homologs (Fig. 1). The tobacco HSP101 (101,141 kD; encoded by NtHSP101: Nicotiana tabacum HSP101) is 84.8%, 86.3%, and 46.5% identical to the Arabidopsis, soybean, and yeast homologs, whereas the wheat HSP101 (100,337 kD; encoded by TaHSP101: Triticum aestivum HSP101) is 68.4%, 67.1%, and 39.8% identical to these same proteins, respectively, and is 68.5% identical to the tobacco HSP101. The peptide sequences obtained from the purified p102 proteins were identical to the corresponding regions in the predicted sequence of the tobacco or wheat HSP101 proteins.
Figure 1.
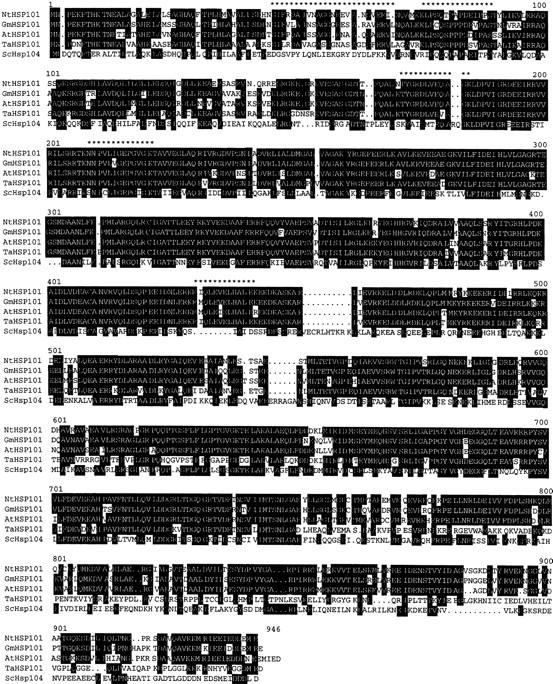
The predicted amino acid sequence of the tobacco and wheat cDNAs encoding HSP101 and their homology with Arabidopsis (Schirmer et al. 1994), soybean (Lee et al. 1994), and yeast (Parsell et al. 1991) homologs. Regions of homology between the proteins are shaded in black when conserved in at least three of the five proteins. The peptide sequences obtained from amino acid sequencing of the purified wheat and tobacco p102 proteins are indicated by asterisks. The tobacco and wheat cDNA sequences were deposited into GenBank as accession nos. AF083343 and AF083344, respectively.
To determine whether the isolated cDNAs encoded functional HSP101, each was placed under the control of the GAL1 promoter in the yeast expression vector, pYES2, the resulting constructs introduced into the Δhsp104 strain, SL304A, and early exponential cells grown in synthetic galactose medium (SGM) were tested for thermotolerance as described previously for yeast Hsp104 (Parsell et al. 1991). SL304A(pYS104), containing the wild-type yeast HSP104, was used as the positive control, whereas SL304A(pYES2) served as the negative control. NtHSP101 complemented the Δhsp104 mutant (Fig. 2A) to the same extent observed for Arabidopsis and soybean HSP101 (Lee et al. 1994; Schirmer et al. 1994). TaHSP101 also partially complemented the Δhsp104 mutant (Fig. 2A). NtHSP101 expression was required for thermotolerance as complementation was observed in SL304A(pGAL1–NtHSP101) grown in SGM but not in glucose [synthetic medium containing dextrose (SDM)] or raffinose-containing media, which repress GAL1 promoter activity (Fig. 2B). Expression of NtHSP101 also conferred a high degree of thermotolerance in late-exponential/early stationary cells (Fig. 2C). These data show that the tobacco and wheat cDNAs encode functional HSP101.
Figure 2.
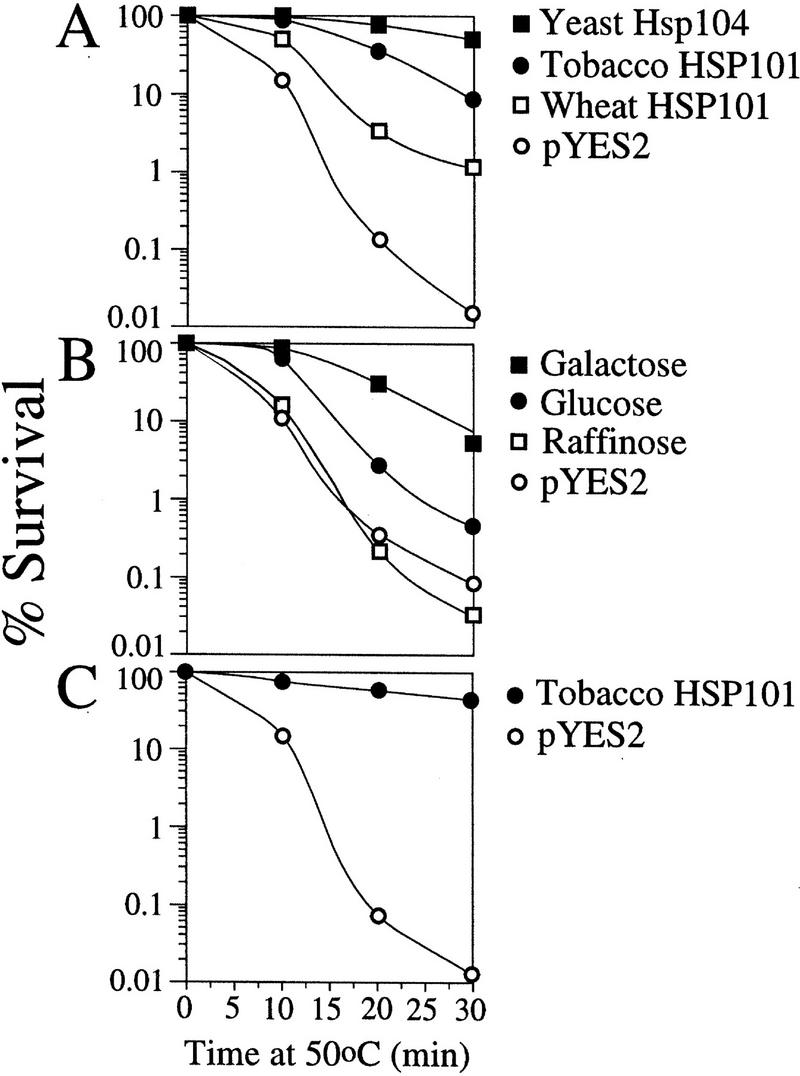
p102 from tobacco and wheat is a functional HSP101 that complements a thermotolerance defect in yeast. (A) SL304A, the hsp104 mutant containing pGAL1–NtHSP101 or pGAL1–TaHSP101, was grown to an early exponential stage in SGM (an OD of 0.06 or 1.8 × 106 cells/ml) prior to assaying for thermotolerance. The expression vector, pYES2, was used as a negative control and pYS104, containing HSP104, was used as a positive control. The percentage of survival at 50°C was plotted as a function of the length of treatment. (B) SL304A(pGAL1–NtHSP101) was grown in galactose, glucose, or raffinose prior to assaying for thermotolerance. SL304A(pYES2) grown in galactose was included as a negative control. (C) Thermotolerance conferred by tobacco HSP101 in late-exponential yeast (an OD of 1.2 or 3.6 × 107 cells/ml) was examined.
HSP101 specifically enhances translation from Ω-containing mRNA
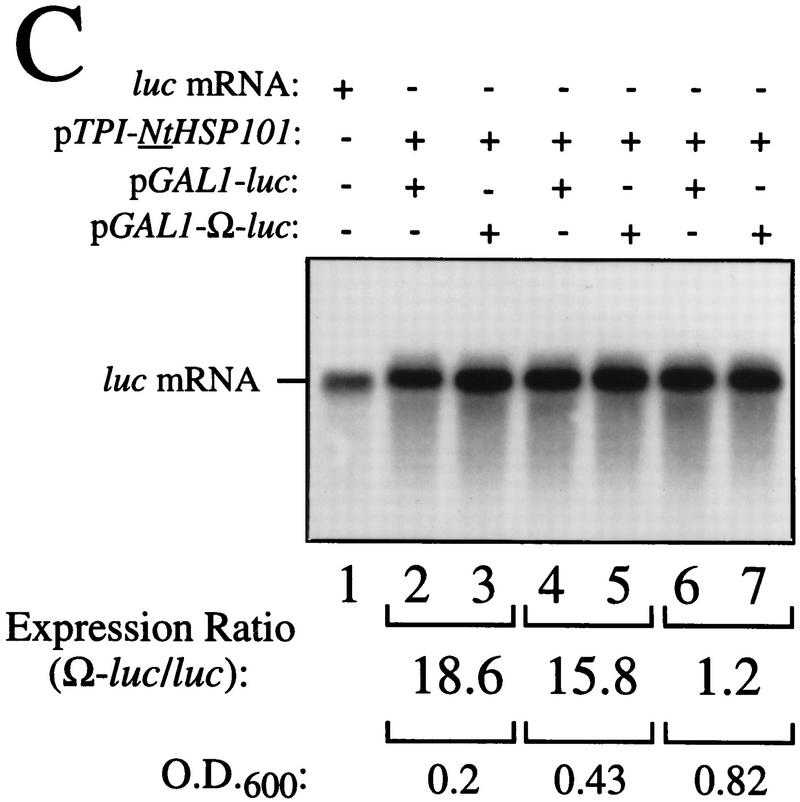
As Ω does not enhance translation in yeast (Everett and Gallie 1992; van den Heuval and Raue 1992), we examined whether NtHSP101 was sufficient to mediate the translational enhancement from Ω in this species. pGAL1–NtHSP101 (NtHSP101 under the control of the GAL1 promoter) and either pTPI–luc or pTPI–Ω-luc (luc and Ω-luc under the control of the constitutively active triose phosphate isomerase (TPI) promoter) were introduced into SL304A (Fig. 3A). SL304A(pYES2) with either pTPI–luc or pTPI–Ω-luc served as a negative control (Fig. 3A, bottom). Luciferase assays were performed to measure the level of expression from pTPI–luc and pTPI–Ω-luc in the presence or absence of NtHSP101. Expression during growth was measured as light units per OD unit of yeast to normalize the luciferase expression to the same number of cells regardless of the growth phase. To quantitate the degree of NtHSP101 translational regulatory function, the expression ratio of translation from the Ω-luc versus luc construct (i.e., Ω-luc/luc) was calculated and included below each graph (Fig. 3A). Therefore, the expression ratio is a measure of NtHSP101 activity independent of any differences in absolute expression caused by differences in strains, medium conditions, or growth phase. Although expression from the luc construct appears low when plotted together with expression from the Ω-luc construct (Fig. 3A), it is typically 10,000–20,000 light units per sample depending on the medium conditions, and there is no detectable background luciferase activity in yeast. In addition to the graphical representation of Ω-luc and luc expression, the actual expression values from each used to calculate the expression ratio are included below each graph. As stationary cells were used to initiate the experiment, a characteristic lag phase in cell growth (observed in the OD of the growth curves in Fig. 3A) during the first hour after transfer to fresh medium was observed. Expression from Ω-luc was substantially higher (up to 19-fold) than the luc control in exponentially growing cells grown in galactose medium (NtHSP101 expression induced) (Fig. 3A, top) but not in cells grown in glucose (NtHSP101 expression repressed) (Fig. 3A, middle). Expression from the control luc mRNA remained unaffected by NtHSP101 expression (compare expression from luc mRNA in Fig. 3A, top and bottom) and expression from Ω-luc and luc were equivalent in the absence of NtHSP101 (Fig. 3A, bottom). NtHSP101 expression (Fig. 3B, middle) and RNA-binding activity (Fig. 3B, top) were observed in yeast grown in SGM (GAL1 promoter active) but not SDM (GAL1 promoter inactive), which correlate with the presence or absence of translational enhancement observed from Ω-luc (Fig. 3A, top and middle, respectively). Only one shifted complex was observed in each case, whereas binding of purified TaHSP101 to Ω resulted in the appearance of two shifted bands, which is typically observed when a high level of this protein is used (Fig. 3B, bottom; Tanguay and Gallie 1996).
Figure 3.
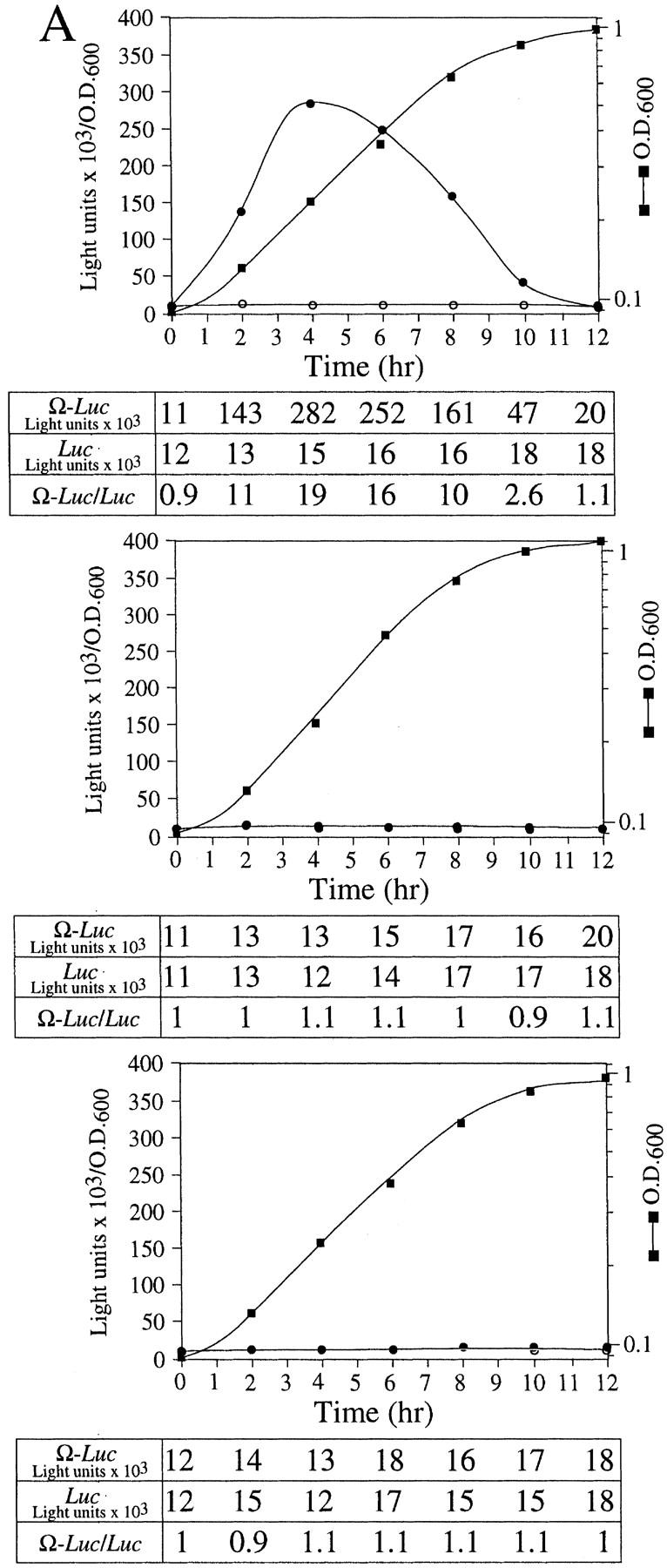
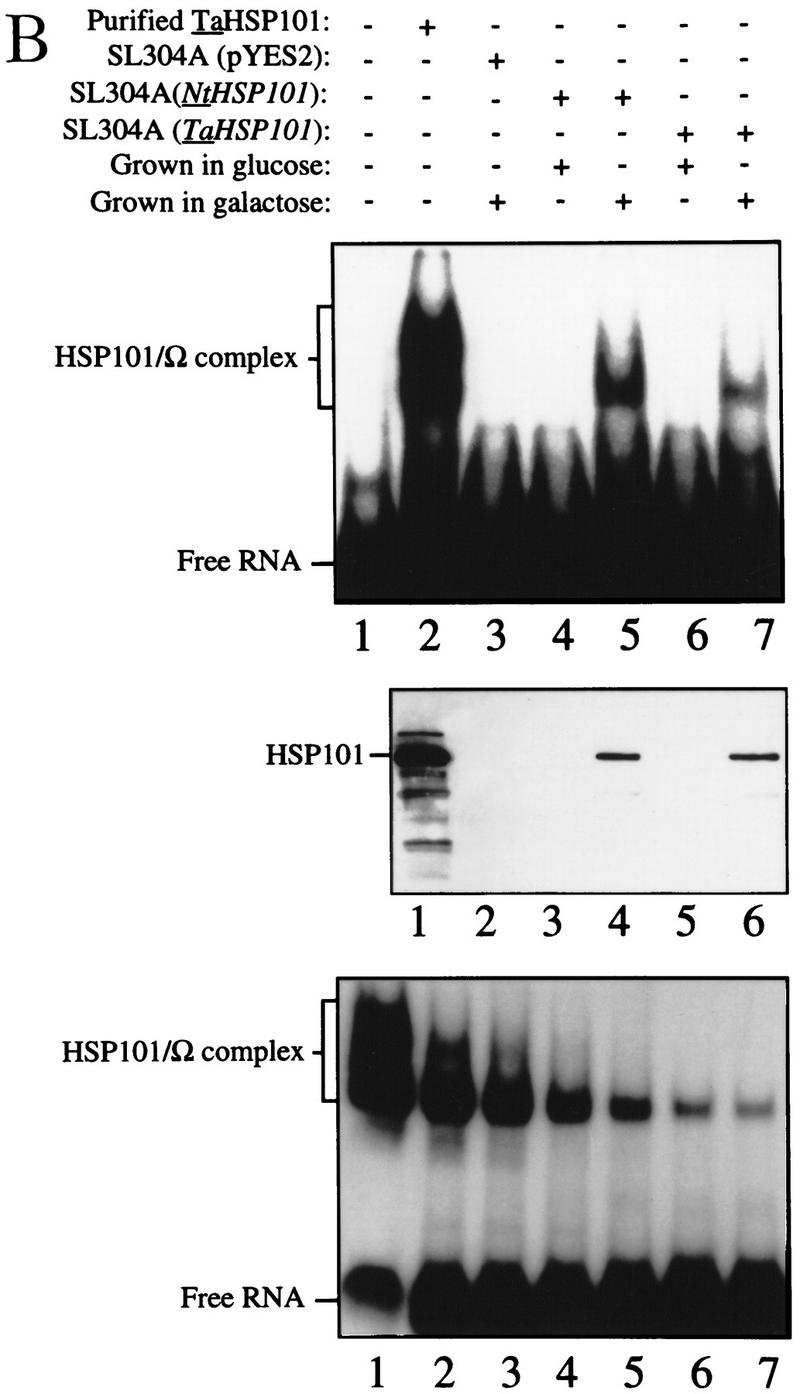
NtHSP101 specifically enhancesexpression from an mRNA when Ω is present as the 5′ leader. (A) Expression from pTPI–Ω-luc (•) or pTPI–luc (○) was followed in SL304A transformed with pGAL1–NtHSP101 (top and middle) or pYES2 (bottom) to examine the regulatory role of NtHSP101 during translation. Transformants were first grown to late-exponential stage (whereupon expression from pTPI–Ω-luc and pTPI–luc was equivalent) and then inoculated into synthetic galactose medium (top and bottom) or synthetic dextrose medium (middle) at the zero time point. Luciferase expression was measured at time points during the growth cycle, normalized to the OD (right) during growth. The expression from pGAL1–Ω-luc, pGAL1–luc, and their expression ratio (i.e., NtHSP101 activity as indicated by the ratio, Ω-luc/luc) are shown below each panel. (B) Tobacco and wheat HSP101 retain RNA-binding activity when expressed in yeast. (Top) SL304A containing pGAL1–NtHSP101 or pGAL1–TaHSP101 were grown in galactose or glucose and crude extracts used in RNA gel shift binding assays with radiolabeled Ω RNA. SL304A(pYES2) was used as a negative control and purified HSP101 was included as a positive control. (Middle) Western analysis of crude extracts from SL304A (containing either pGAL1–NtHSP101, pGAL1–TaHSP101, or pYES2) by use of anti-p102 (i.e., anti-HSP101) antibodies. The lanes correspond to the extracts as indicated at top. The lower molecular weight bands in lane 1 are degradation products of HSP101 that are observed when a high level of the purified protein is analyzed. (Bottom) Titration of HSP101 protein in the Ω RNA gel shift binding assay. The concentration of purified HSP101 used in each binding reaction was decreased twofold in each succeeding lane, whereas the concentration of Ω RNA was held constant.
The preferential translation of Ω-luc mRNA observed in Figure 3A initiated as the cells left stationary phase, persisted at an elevated level until the mid-exponential phase, whereupon its preferential translation progressively decreased as cell growth slowed during the late-exponential/early stationary phase of growth. This slower growth during the late-exponential phase is caused by depletion from the medium of nutrients that are required for growth. As synthetic medium contains only limited nutrients, growth under these conditions slows substantially once an OD of 0.7 is reached. To examine whether the preferential translation of Ω-luc mRNA mediated by NtHSP101 is regulated by nutrient availability, SL304A(pTPI–NtHSP101) or SL304A(pYX232) containing either pGAL1–Ω-luc or pGAL1–luc (which contains the 5′ leader of GAL1 instead of Ω) were grown in a synthetic (SGM) or rich [yeast extract/peptone/galactose (YPG)] medium (Fig. 4). Exponentially growing cells were used to inoculate each medium to eliminate the lag phase to show that the preferential translation from Ω-luc mRNA in cells expressing NtHSP101 can be observed from the earliest time point following transfer to fresh medium (Fig. 4, top). In SGM, the expression ratio (i.e., translation from Ω-luc versus luc mRNA) declined to 1 as the OD reached 0.9 as observed in Figure 3A, top, whereas in YPG this did not occur until the OD reached 7. Expression from pGAL1–luc in yeast expressing NtHSP101 was equivalent to expression from this same construct in the absence of NtHSP101 (Fig. 4, bottom). The richer nature of the YPG medium permitted cell growth to continue to a higher density, but as observed with growth in SGM, the loss in the preferential translation from Ω-luc mRNA in cells expressing NtHSP101 occurred prior to the onset of slower growth because of the eventual depletion of nutrients from the medium. In YPG, translation from Ω-luc dropped >60-fold between 9 and 17 hr of growth (Fig. 4, top right), whereas less than an ∼2-fold drop in expression was observed from luc during the same period. The decline in expression from pGAL1–Ω-luc during late-exponential growth in SGM was not observed for pGAL1–luc, from which expression actually increased (see Fig. 3A or Fig. 4, left). These data suggest that the decline in expression from Ω-luc mRNA in late-exponential/early stationary cells was largely specific to Ω and suggest that NtHSP101 may itself be regulated by nutrient availability.
Figure 4.
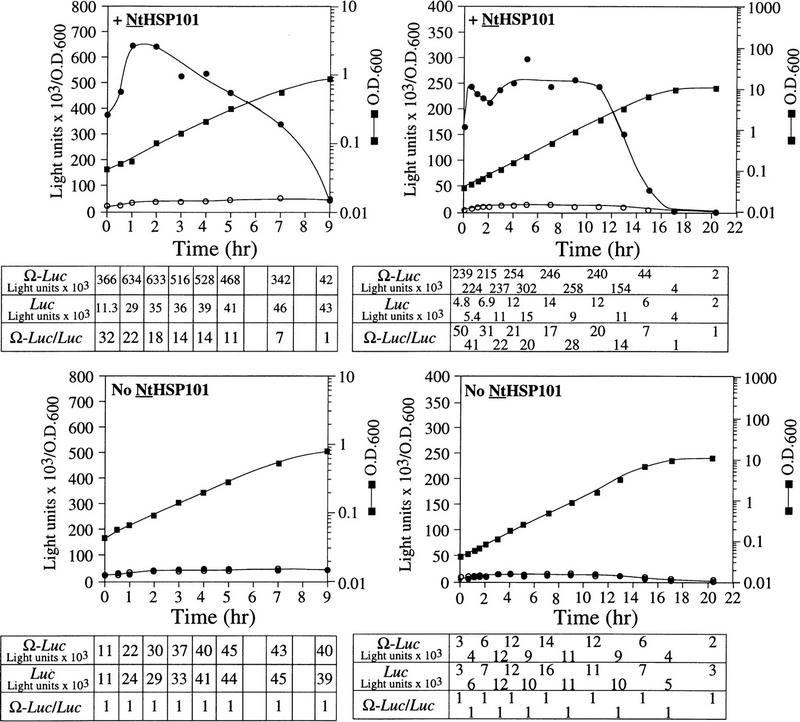
Growth in rich medium supports HSP101 activity to higher cell density. (Top) SL304A(pTPI–NtHSP101) and (bottom) SL304A(pYX232) containing either pGAL1–Ω-luc (•) or the GAL1 5′ leader construct, pGAL1–luc (○), were grown in SGM (left) or YPG medium (right). The OD of the cultures is indicated at right. Luciferase expression was normalized to the OD and plotted as a function of the growth phase. The expression from pGAL1–Ω-luc, pGAL1–luc, and their expression ratio (i.e., NtHSP101 activity as indicated by the ratio, Ω-luc/luc) are shown below each panel.
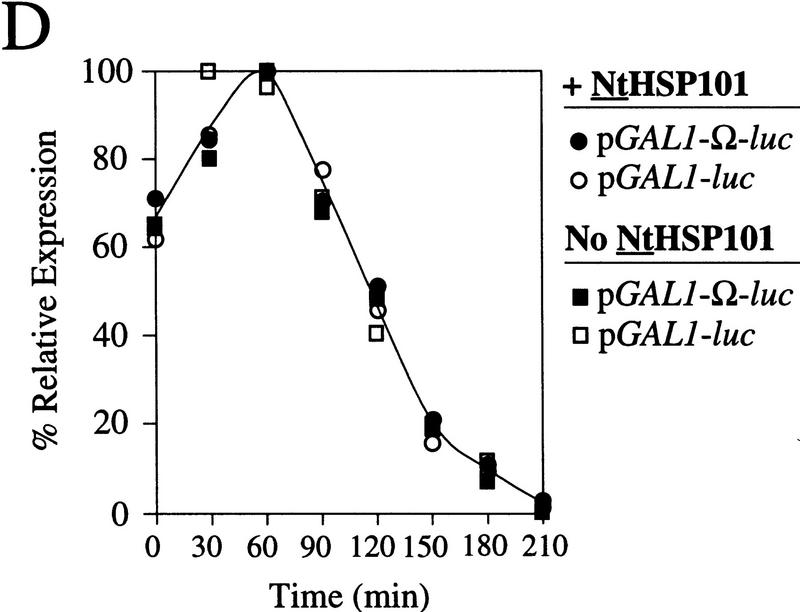
The loss in preferential translation from Ω-luc mRNA during late-exponential growth might result from a reduction in the level of NtHSP101, a reduction in its activity, or a selective reduction in the level of Ω-luc mRNA relative to luc mRNA. Western analysis of yeast expressing NtHSP101 at different points in the growth cycle ranging from an OD of 0.2 to 3.0 revealed no significant change in the level of NtHSP101 (Fig. 5A) showing that the loss in the NtHSP101-mediated enhancement during late-exponential growth cannot be explained by a change in the level of NtHSP101 expression. Moreover, Northern analysis of the level of Ω-luc and luc mRNAs in yeast expressing NtHSP101 during stages in the growth cycle revealed no differences in their relative amounts (Fig. 5C).
Figure 5.
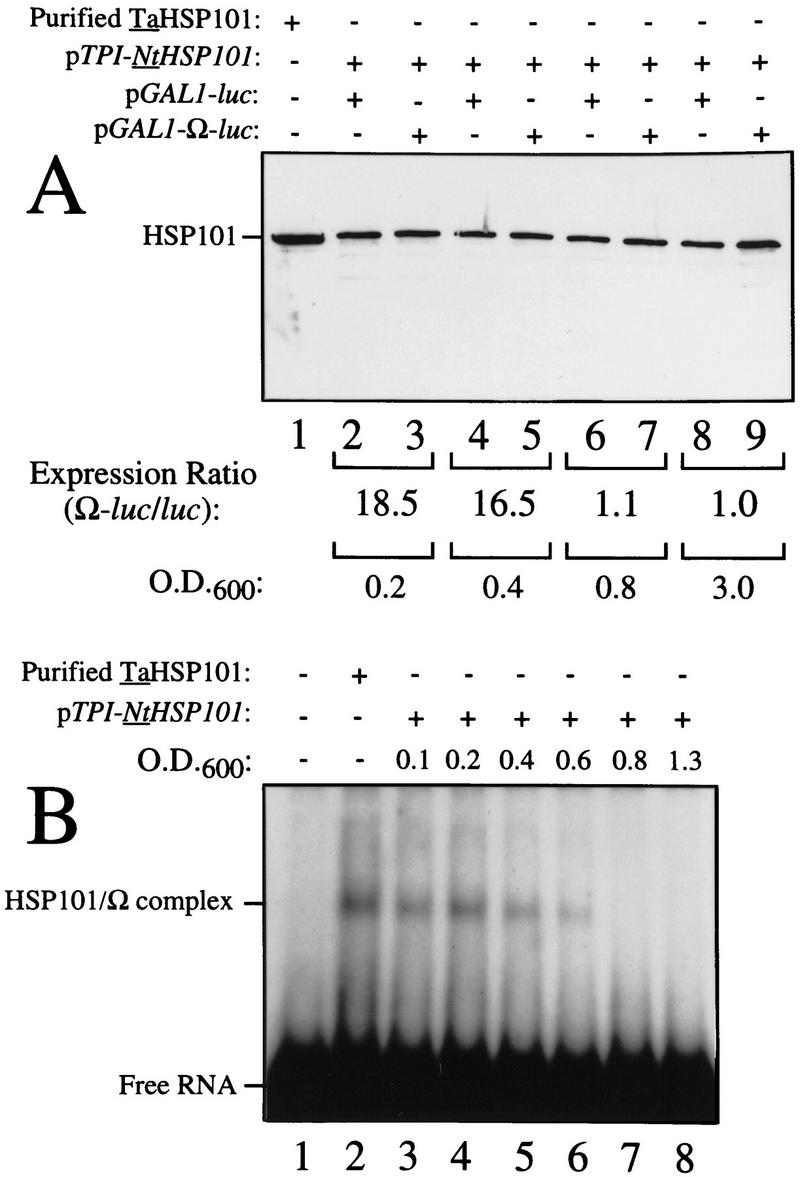
HSP101 RNA-binding activity is regulated during yeast growth without corresponding changes in luciferase mRNA or protein stability. (A) Western analysis of extracts from early, mid, or late-exponential or stationary cells of SL304A(pTPI–NtHSP101) containing either pGAL1–luc or pGAL1–Ω-luc grown in SGM. The expression ratio of Ω-luc/luc at each OD indicated is shown below each pair of lanes. Purified HSP101 from wheat germ (TaHSP101) was loaded in lane 1 as a positive control. (B) NtHSP101 RNA binding to radiolabeled Ω was assayed by RNA gel shift analysis by use of crude extracts of early, mid-, or late exponential SL304A(pTPI–NtHSP101) cells grown in SGM to the OD indicated. Purified TaHSP101, whose RNA-binding properties were characterized previously (Tanguay and Gallie 1996), was included in lane 2 as a positive control. (C) Northern analysis of luc mRNA levels from either pGAL1–Ω-luc or pGAL1–luc in early, mid-, or late-exponential SL304A(pTPI–NtHSP101) cells grown in SGM. Total RNA was used for the analysis and in vitro-synthesized luc mRNA (lane 1) was included to indicate the size of the mRNA. The OD of the cells and expression ratio (i.e., translation from Ω-luc relative to luc mRNA) is indicated below each pair of lanes representing each growth stage examined. (D) The effect of NtHSP101 on luciferase protein stability was determined by switching early exponential SL304A(pTPI–NtHSP101, ○,•) or SL304A(pYX232, □,█) containing either pGAL1–Ω-luc or pGAL1–luc from SGM to SDM and the level of luciferase measured at time points following the transcriptional repression of the GAL1 promoter. Equal volumes of cell culture were used for the measurements to account for the dilution of luciferase protein during cell growth and division. The amount of luciferase activity is shown relative to the level (defined as 100%) present in the cells immediately following introduction into SDM.
The enhancing effect of NtHSP101 on expression from Ω-luc mRNA cannot be explained by a selective, chaperone-mediated alteration of the stability of luciferase protein because the protein produced from Ω-luc and luc mRNA is identical. Moreover, expression from luc mRNA is unaffected by NtHSP101 (see Figs. 3A and 4) showing that NtHSP101 does not alter luciferase protein stability, activity, or affect overall cellular translational activity through any indirect chaperone-mediated function. That NtHSP101 does not affect luciferase turnover synthesized from either Ω-luc and luc mRNAs was confirmed by the analysis of the stability of luciferase when synthesized in the presence or absence of NtHSP101 (Fig. 5D). Expression from pGAL1–luc or pGAL1–Ω-luc was measured in either SL304A(pTPI–NtHSP101) or SL304A(pYX232) following their shift from galactose to glucose medium to repress the further synthesis of Ω-luc and luc mRNAs. No difference in the rate of loss of luciferase activity following transcriptional repression was detected between either mRNA regardless of whether NtHSP101 was expressed or not. However, these data do show that luciferase protein has a relatively short half-life (a maximum value of 35 min), showing the utility of luc as a reporter gene in examining rapid changes in gene expression in yeast.
Although no alteration in NtHSP101 protein level was observed during growth, the loss of translational enhancement mediated by NtHSP101 did correlate with a progressive loss in its RNA-binding activity (Fig. 5B). RNA-binding activity was observed in extracts prepared from early exponential SL304A(pTPI–NtHSP101) cells grown in SGM (Fig. 5B, lane 3). RNA-binding activity decreased in cells approaching the late-exponential stage: at an OD of 0.4, only 50% of RNA-binding activity remained relative to the activity present in cells at an OD of 0.1 and only 10% remained at an OD of 0.6 (Fig. 5B, lanes 3–6). No binding activity was observed following further growth (Fig. 5B, lanes 7,8). As the loss in NtHSP101 RNA-binding activity at an OD corresponding to the late-exponential phase correlated with the loss of preferential translation at this same stage from Ω-luc mRNA in cells expressing NtHSP101 (see Fig. 4, top, left), these data suggest that the loss in NtHSP101 regulatory activity during late-exponential growth may be a result of the loss of RNA-binding activity (also see below).
HSP101 functions through a poly(A) tail-independent mechanism
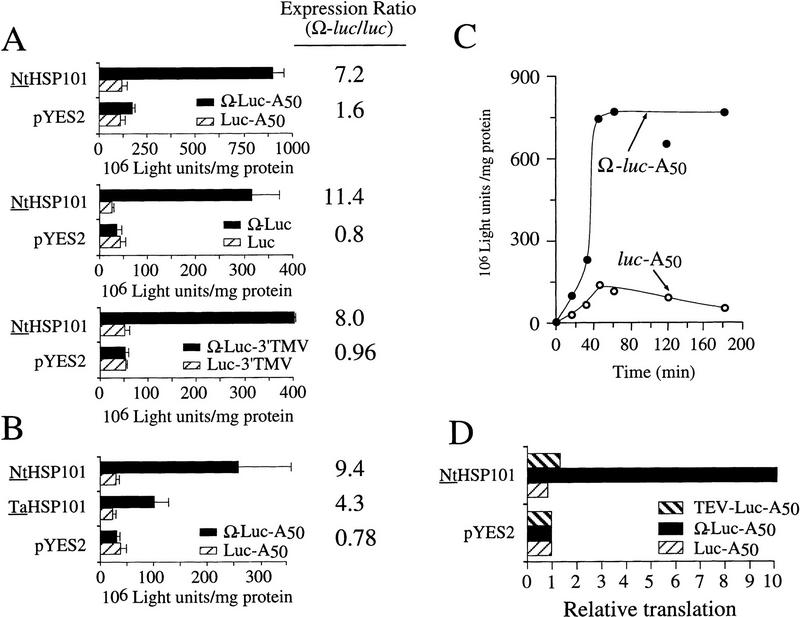
The role of the poly(A)-binding protein (PABP)/poly(A) tail during translational initiation has been shown in yeast and higher eukaryotes (Le et al. 1997; Tarun et al. 1997; Craig et al. 1998). To examine whether NtHSP101 regulatory function requires the poly(A) tail, translation from poly(A)+ versus poly(A)− luc and Ω-luc mRNAs was compared in SL304A(pGAL1–NtHSP101). As poly(A)− mRNAs cannot be produced in vivo, in vitro-synthesized, capped Ω-luc-A50, luc-A50, Ω-luc, and luc mRNAs were delivered by use of electroporation. The presence of a poly(A) tail increased expression as expected (Fig. 6A, cf. top and middle and note the difference in scale) and NtHSP101 increased expression from Ω-luc regardless of whether the mRNA was polyadenylated or not (Fig. 6A) or terminated in the 204 nucleotide 3′UTR of TMV (Fig. 6A, bottom), which itself serves as a translational enhancer (Gallie and Walbot 1990; Leathers et al. 1993). These data suggest that the mechanism by which NtHSP101 enhances translation is independent of 3′ regulatory elements.
Figure 6.
HSP101 enhancement is specific to Ω-containing mRNAs and is not dependent on a poly(A) tail. (A) In vitro-synthesized luc mRNA constructs terminating with or without a poly(A) tail or with the TMV 3′UTR (indicated in each panel) were electroporated into SL304A (containing either pGAL1–NtHSP101 or pYES2 as indicated at left) and expression measured following the completion of translation. The expression ratio is shown at right. (B) Ω-luc-A50 and luc-A50 mRNAs were introduced into SGM-grown, SL304A(pGAL1–NtHSP101), SL304A(pGAL1–TaHSP101), or SL304A(pYES2) by electroporation and expression measured following the completion of translation. The expression ratio is shown at right. (C) NtHSP101 increases the rate of translation from an Ω-containing mRNA. Ω-luc-A50 and luc-A50 mRNAs were introduced into SGM-grown, SL304A(pGAL1–NtHSP101) cells by electroporation, luciferase expression measured following RNA delivery, and the data plotted as a function of the time of translation. (D) NtHSP101 does not regulate translation from TEV 5′ leader-containing mRNA. TEV-luc-A50, Ω-luc-A50, or luc-A50 mRNAs were electroporated into SL304A containing either pGAL1–NtHSP101 or pYES2. Expression is shown relative to the level obtained from each construct in SL304A(pYES2).
To determine whether the translational regulatory function of NtHSP101 is unique to this protein, the wheat homolog (TaHSP101) was expressed from the GAL1 promoter in the pYES2 vector in yeast. Capped Ω-luc-A50 and luc-A50 mRNAs were then delivered by use of electroporation and the degree of enhancement conferred by TaHSP101 was compared with that mediated by NtHSP101 (Fig. 6B). TaHSP101 enhanced translation specifically from Ω-luc-A50 mRNA as observed with NtHSP101, although the degree of enhancement was reduced compared with NtHSP101. This reduced enhancement conferred by wheat HSP101 is in good agreement with our previous observation that Ω enhances translation less in monocot versus dicot plants (Gallie et al. 1989) and also correlated with its lower-binding affinity for Ω relative to NtHSP101 (see Fig. 3B, lane 7, top). The translation regulatory activity exhibited by TaHSP101 shows that this function is not unique to NtHSP101.
Delivery of Ω-luc-A50 and luc-A50 mRNAs to yeast also allows analysis of NtHSP101 translational regulatory function independent of nuclear steps during gene expression. Following delivery of the mRNAs into SGM-grown, SL304A(pGAL1–NtHSP101) cells, the rate of luciferase accumulation functions as a measure of their respective translation efficiencies and the period of time over which the mRNAs are translationally active is a measure of their relative stabilities (Fig. 6C). The rate of translation from Ω-luc-A50 mRNA was sevenfold higher than for luc-A50 mRNA, whereas both mRNAs were translationally active for the same period of time (∼45 min). The extent of enhancement of the rate of translation correlates well with the end-point expression measurements (Fig. 6A).
As shown in Figure 4, NtHSP101 failed to enhance translation from the control luc construct containing the GAL1 5′ leader. To examine whether NtHSP101 mediates translational regulation from a second, but unrelated, viral 5′ leader sequence known to regulate translation, a luc construct containing the leader from tobacco etch virus (TEV), which is naturally uncapped and confers cap-independent translation (Carrington and Freed 1990; Gallie et al. 1995), was delivered as uncapped mRNA to SL304A(pGAL1–NtHSP101) and SL304A(pYES2). Translation from the TEV–luc mRNA was virtually unaffected by NtHSP101 (Fig. 6D) as observed for the luc control construct, whereas translation increased 10-fold when Ω was present as the leader of uncapped luc mRNA in cells expressing NtHSP101. The failure of NtHSP101 to regulate translation from the TEV 5′ leader correlated with the low-binding affinity that NtHSP101 exhibited for this leader (data not shown). These data show that NtHSP101 does not enhance translation from a second but unrelated viral 5′ leader. Moreover, these data show that NtHSP101 regulatory function is not dependent on the presence of a 5′ cap as translational enhancement was observed from the Ω construct even when uncapped.
NtHSP101 activity is regulated by the nutrient status of the cell
The loss in NtHSP101 RNA-binding activity and the corresponding loss in preferential translation of Ω-luc mRNA during late-exponential/early stationary phase growth suggests that NtHSP101 activity may be regulated by either the depletion of an essential nutrient from the medium or a result of the change from fermentation to respiration that characterizes the transition to the stationary phase. To examine whether the onset of the loss in NtHSP101 activity can be triggered prematurely by the limitation of an essential nutrient, SL304A(pTPI–NtHSP101) and either pGAL1–luc or pGAL1–Ω-luc were grown to early exponential stage in SGM to induce NtHSP101 regulatory activity and then transferred to SGM in which histidine was present at either the normal concentration (95 μm), or at 19, 5, or 0 μm. SL304A is auxotrophic for histidine resulting in a strong starvation response following histidine depletion that cannot be relieved through induction of the histidine biosynthetic pathway. Early exponential cells, in which NtHSP101 was highly active, and consequently, the expression ratio was already high (Fig. 7A, see zero time point), were used to initiate this experiment so that the effect of severe amino acid starvation on the timing of the decline in NtHSP101 activity could be determined prior to its normal decline in activity observed above an OD of 0.6. Immediately following transfer to fresh SGM supplemented with the various levels of histidine, the expression ratio was ∼23–24 (Fig. 7A). Growth in the absence of histidine resulted in a complete loss of NtHSP101 activity within 2 hr as determined by the reduction in the expression ratio. Growth in 5 or 19 μm histidine also resulted in a loss of NtHSP101 activity at a lower cell density than that observed in 95 μm histidine (Fig. 7A). It should be noted that expression from pGAL1–luc, which contains the 5′ leader of GAL1 in place of Ω, was not similarly affected in the reduced or histidine-free medium (Fig. 7A). The level of NtHSP101 protein and the level of Ω-luc or luc mRNAs in histidine-starved cells were identical to those in cells grown in 95 μm histidine over the time course of this experiment as revealed by Western and Northern analysis, respectively (data not shown). Thus, the limitation of an essential amino acid was sufficient to repress NtHSP101 activity in cells at low density such as that observed during late-exponential/early stationary phase in unmodified SGM.
Figure 7.
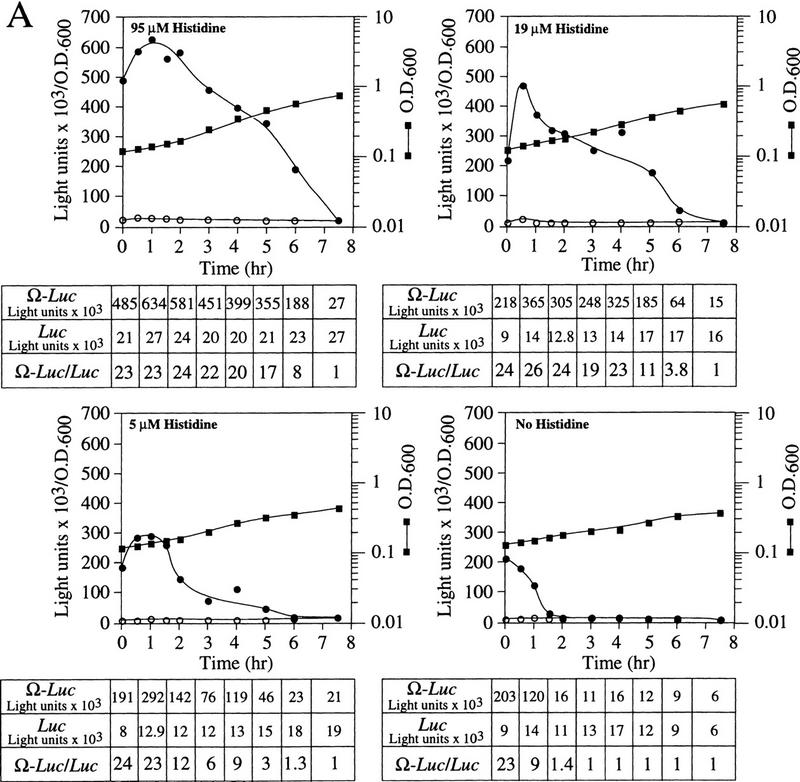
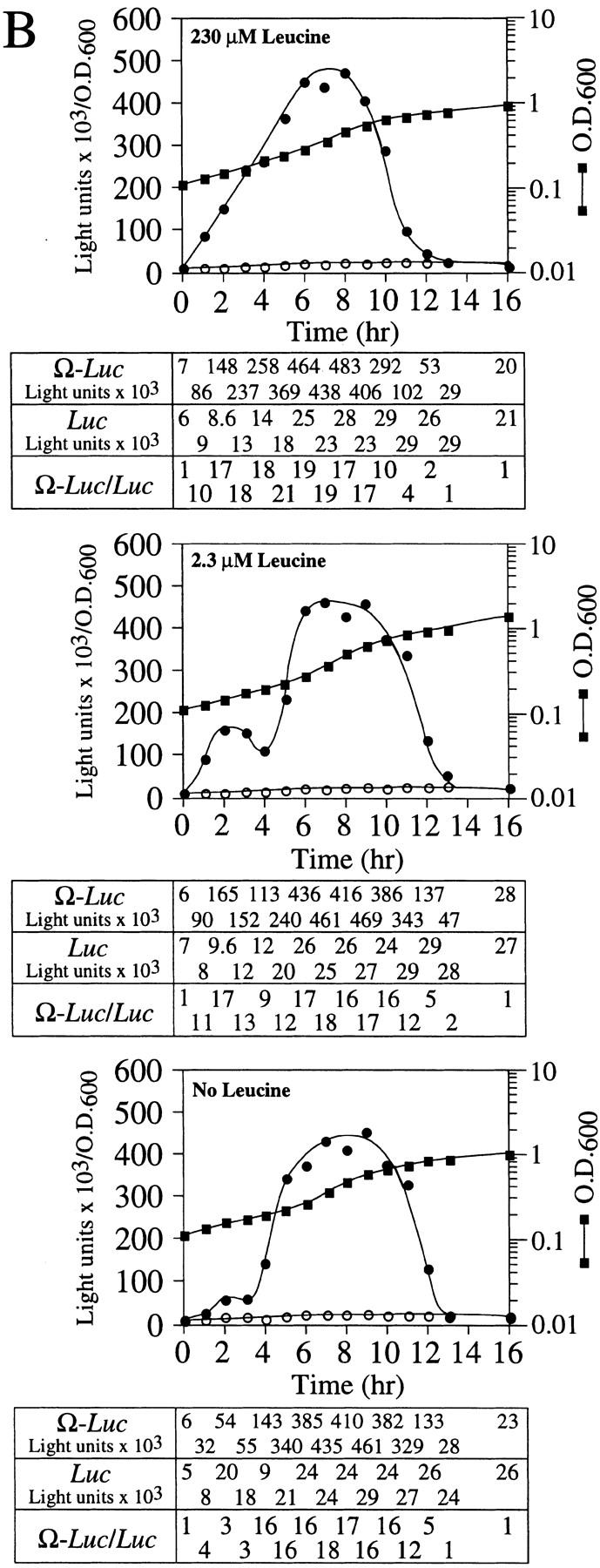
NtHSP101 activity is repressed by amino starvation. SL304A(pTPI–NtHSP101) containing either pGAL1–Ω-luc (•) or pGAL1–luc (○) was grown to an early exponential stage in SGM before introduction into SGM containing (A) 95 μm, 19 μm, 5 μm, or no histidine or (B) 230 μm, 2.3 μm, or no leucine. The expression from pGAL1–Ω-luc, pGAL1–luc, and their expression ratio (i.e., NtHSP101 activity as indicated by the ratio, Ω-luc/luc) are shown below each panel. See A for symbol designation.
The exhaustion of an exogenously supplied amino acid for which the yeast are prototrophic should provoke a transient starvation response prior to the induction of its biosynthetic pathway. To examine whether a transient starvation response affects NtHSP101 activity, the same yeast used in the prior experiment, which are prototrophic for leucine, were grown in SGM in which leucine was present at either the normal concentration (230 μm), or at 2.3, or 0 μm leucine. In this experiment, late-exponential cells in which NtHSP101 activity was fully repressed and the expression ratio of Ω-luc to the luc construct was equivalent (Fig. 7B, see zero time point) were used to examine the effect of transient starvation on the induction of NtHSP101 activity during early exponential growth. The expression ratio rapidly increased in cells grown in 230 μm leucine, remained high for several hours, and then decreased during late-exponential/early stationary phase to equivalence by 13 hr of growth (Fig. 7B, top). The period of time required for the activation and subsequent decline of NtHSP101 activity was longer in this experiment (13 hr) than observed previously in the same medium when early exponential cells were used to initiate growth (9 hr in SGM, see Fig. 4, top, left) as late-exponential cells require extra time to pass through lag phase before entering exponential growth. Consequently, growth for 13 hr was required in this experiment to reach a similar OD as that achieved in 9 hr in SGM in Figure 4. The expression ratio in cells grown in 2.3 μm leucine increased initially but then reproducibly suffered a transient drop from the second to fourth hours of growth (Fig. 7B, middle) before it increased to a level similar to that observed in cells in 230 μm leucine. In the absence of leucine, the expression ratio rose only to a low level during the first 3 hr of growth (Fig. 7B, bottom) before increasing again to the level observed for cells grown in 230 or 2.3 μm leucine. The effect of limiting leucine on expression was specific to the Ω-luc construct as no effect was observed for the GAL1 5′ leader luc construct. This is best illustrated by the expression ratio calculated for time points during the first 5 hr of each experiment in Figure 7B. The timing and rate of decline in NtHSP101 activity during the late-exponential/early stationary phase was similar in each case.
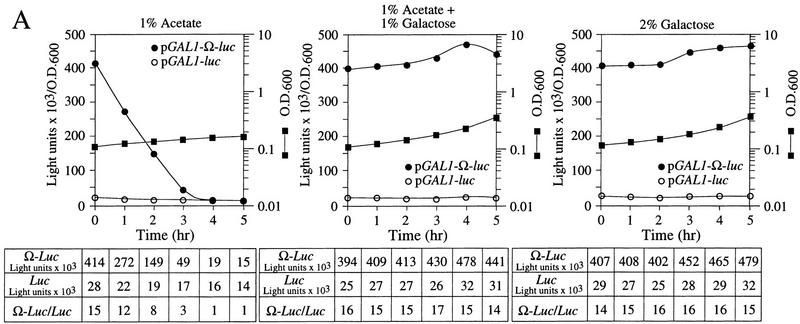
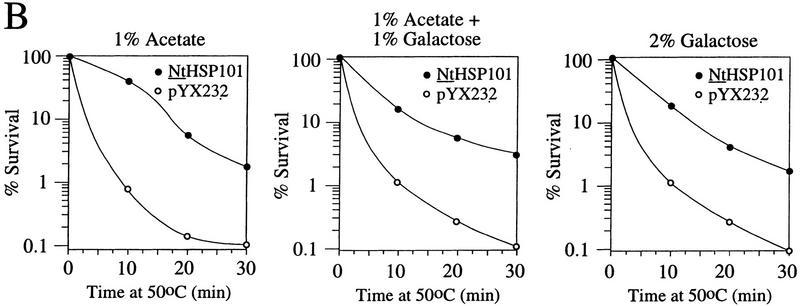
To examine whether NtHSP101 activity is repressed in respiring cells, the same yeast used above were grown in SGM for 3 hr to induce NtHSP101 activity before transferring the cells to synthetic medium containing 1% potassium acetate, a nonfermentable carbon source that forces the cells to switch from fermentation to respiration. NtHSP101 activity was progressively lost during growth in the acetate-only medium as determined by the reduction in the expression ratio (Fig. 8A, left). Although expression from pGAL1–luc (containing the GAL1 5′ leader) decreased to 50% of its initial value following 5 hr of growth in acetate-only medium, showing a decrease in GAL1 promoter activity, the decrease in expression from pGAL1–Ω-luc was disproportionately greater so that expression from pGAL1–Ω-luc and pGAL1–luc reached parity in <4 hr of growth in the acetate-only medium (Fig. 8A, left). It was not the presence of acetate per se that caused the repression in NtHSP101 activity as its activity remained as high in cells grown in synthetic medium supplemented with 1% acetate plus 1% galactose (which supports growth by fermentation) as in SGM (see the expression ratio, Fig. 8A, middle and right, respectively). The level of NtHSP101 protein was similar in cells grown in each medium (Fig. 8C, top) suggesting that the loss of NtHSP101 activity in respiring cells was not a consequence of a reduction in the level of NtHSP101 expression. In contrast, NtHSP101 RNA-binding activity was abolished in respiring cells: NtHSP101 binding activity to Ω was observed with extract from SL304A(pTPI–NtHSP101) cells grown in synthetic medium containing either galactose or acetate plus galactose (Fig. 8C, lanes 2 and 4, respectively, bottom), but not when grown in acetate-only medium (Fig. 8C, lane 6, bottom). These data support the correlation between NtHSP101 RNA-binding activity and translational regulatory activity and show that both are repressed in respiring cells.
Figure 8.
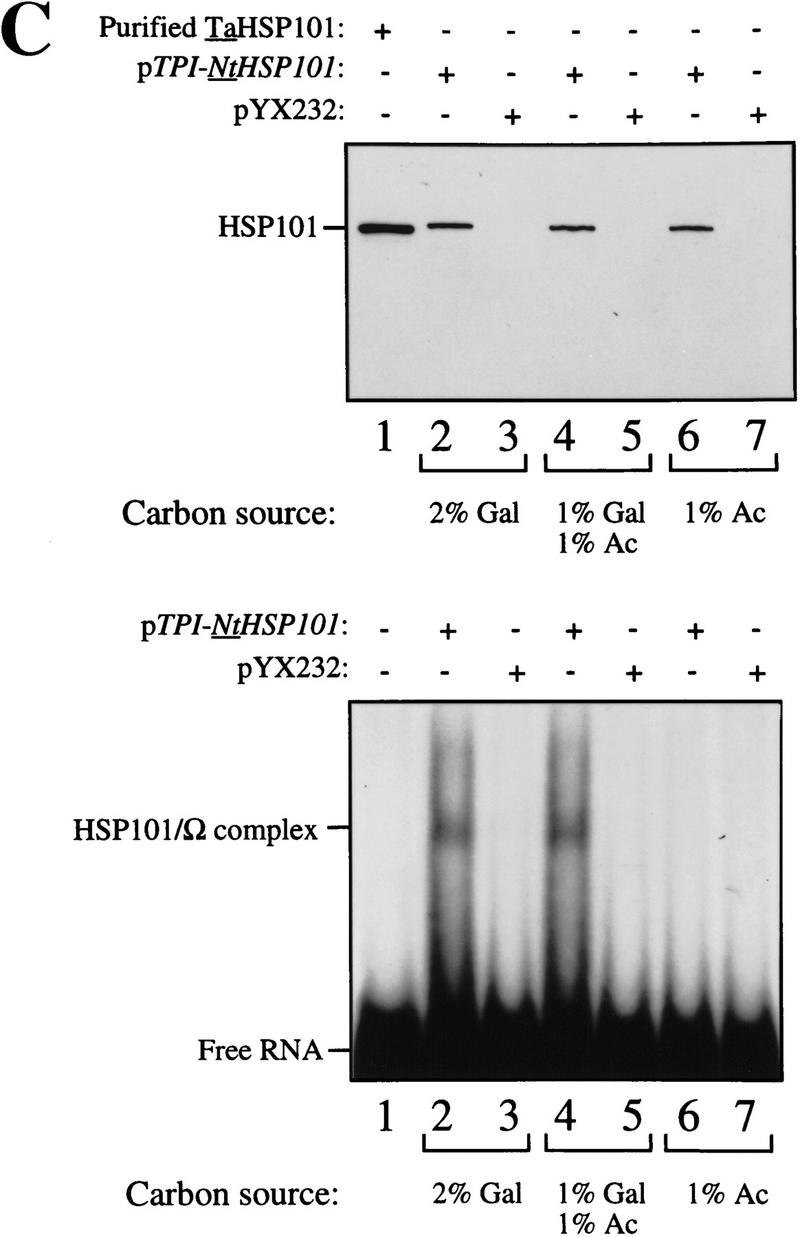
HSP101 translational regulatory and RNA-binding activity are repressed in respiring cells but retain thermotolerance activity. SL304A(pTPI–NtHSP101) containing either pGAL1–Ω-luc or pGAL1–luc was grown to an early exponential stage in SGM, transferred to synthetic medium with the indicated carbon source supplements and tested for (A) translational regulatory activity by measuring luciferase expression as described in Fig. 4; (B) thermotolerance as described in Fig. 2; or (C) RNA-binding activity as described in Fig. 3B. In C, Western analysis of NtHSP101 expression (top) and NtHSP101 RNA-binding activity (bottom) as revealed by RNA gel shift analysis.
To examine whether the loss of NtHSP101 activity as a translational regulator in respiring cells is accompanied by a similar reduction in its ability to confer thermotolerance, SL304A(pTPI–NtHSP101) was grown in synthetic medium supplemented with 1% acetate for 5 hr to ensure that the translational regulatory activity of NtHSP101 was fully repressed (Fig. 8A, left) and the cells tested in the thermotolerance assay. No loss in the ability of NtHSP101 to confer thermotolerance was observed in acetate-grown cells (Fig. 8B, left) compared with growth in either galactose or acetate plus galactose (Fig. 8B, right and middle, respectively). These data show that growth by respiration selectively abolishes the translational regulatory function and RNA-binding activity of NtHSP101 without affecting its ability to confer thermotolerance.
Loss of eIF4G or eIF3 function reduces HSP101-mediated translational enhancement
To determine whether NtHSP101 regulatory activity specifically requires the involvement of one or more translation initiation factors, the degree of NtHSP101-mediated enhancement of translation from Ω-luc constructs was examined following the introduction of pTPI–NtHSP101 and either pGAL1–luc or pGAL1–Ω-luc into yeast mutants with null or temperature-sensitive mutations in genes encoding specific initiation factors. eIF4F is composed of the cap-binding subunit, that is, eIF4E (encoded by a single gene), and eIF4G (encoded by two genes) (Merrick and Hershey 1996), whose interaction to form eIF4F is prevented by the binding of CAF20 to eIF4E (Altmann et al. 1997; de la Cruz et al. 1997). eIF4A, a factor that exhibits ATP-dependent RNA helicase activity and is essential for viability (Linder and Slonimski 1989; Schmid and Linder 1991), and the nonessential eIF4B that assists eIF4A (Coppolecchia et al. 1993) are two additional factors that participate in eIF4F function. In wild-type CW04, translational enhancement was observed from pGAL1–Ω-luc but not from pGAL1–luc (Fig. 9A) similar to that observed in SL304A in Figure 4. The time required to complete the cycle of the nutrient-induced increase and subsequent starvation-induced decrease in NtHSP101 translational regulatory function was longer for CW04 (18 hr) than for SL304A (12 hr) because of the slower growth rate of CW04. Nevertheless, the changes in the expression ratio occurred at similar stages in the growth cycle as determined by the OD of the culture. Null mutations affecting eIF4B (Fig. 9B) or CAF20 (Fig. 9C) had no substantial effect on NtHSP101 function as measured by the expression ratios during the growth cycle, suggesting that neither was required for the NtHSP101-mediated regulation. In contrast, tif4632, an eIF4G null mutant (Goyer et al. 1993), substantially failed to support NtHSP101 activity (Fig. 9E). Although overall translation was reduced in this mutant as evidenced by the lower level of luciferase expression from pGAL1–luc, expression from pGAL1–Ω-luc was reduced disproportionately, resulting in an expression ratio (i.e., Ω-luc/luc) of only 3 during early to mid-exponential growth (Fig. 9E) compared with 13 in CW04 (Fig. 9A). Interestingly, the loss of TIF4632 did not substantially reduce the growth rate of this mutant. A null mutation (tif4631) affecting the second gene encoding eIF4G, whose gene product (TIF4631) is expressed to the same level as TIF4632 (Tarun et al. 1997) but is only 53% identical to TIF4632 (Goyer et al. 1993), exhibited a near normal level of NtHSP101-mediated translational enhancement (Fig. 9D), data implicating TIF4632 to be involved in the NtHSP101-mediated regulation. Although 50 hr were required to complete the cycle of regulation of NtHSP101 activity in the tif4631 mutant because of its very slow growth phenotype, the changes in NtHSP101 activity occurred at similar stages in the growth cycle (determined by OD) as observed for wild-type strains.
Figure 9.
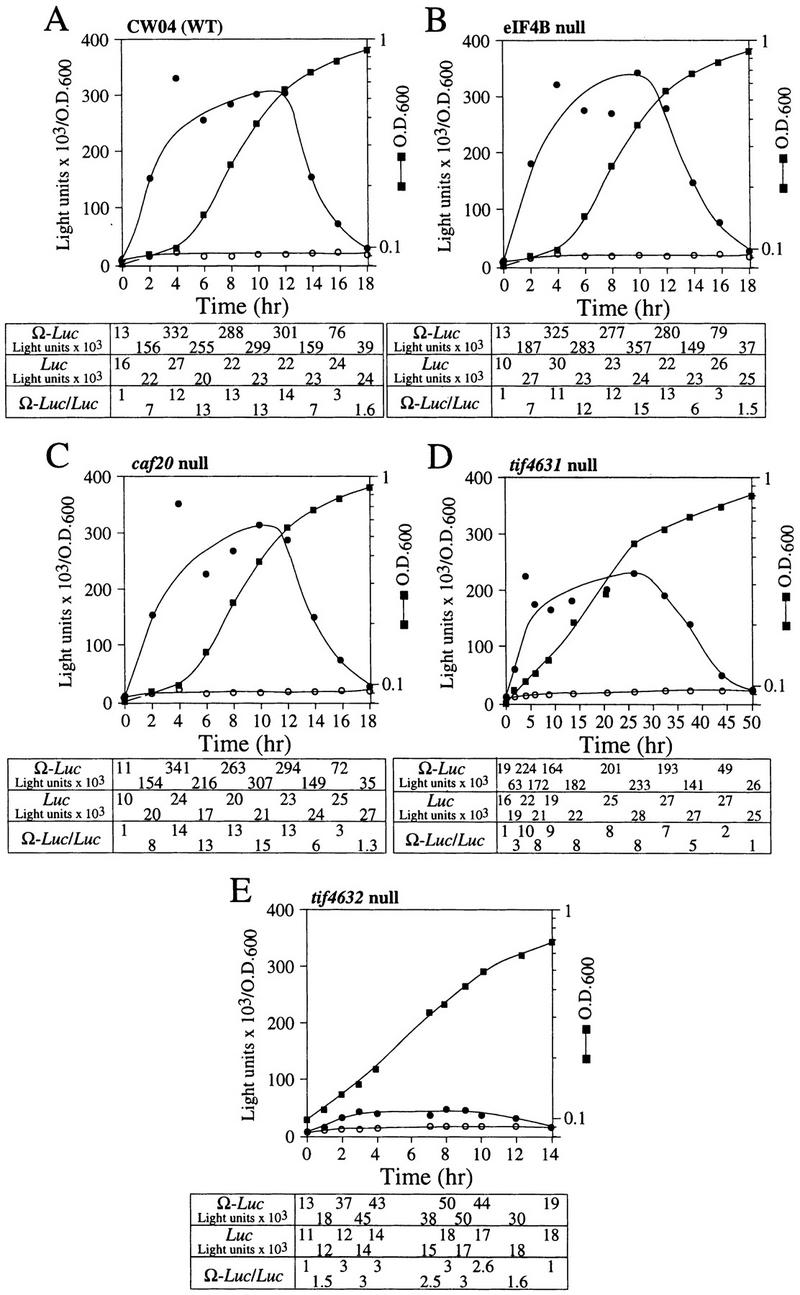
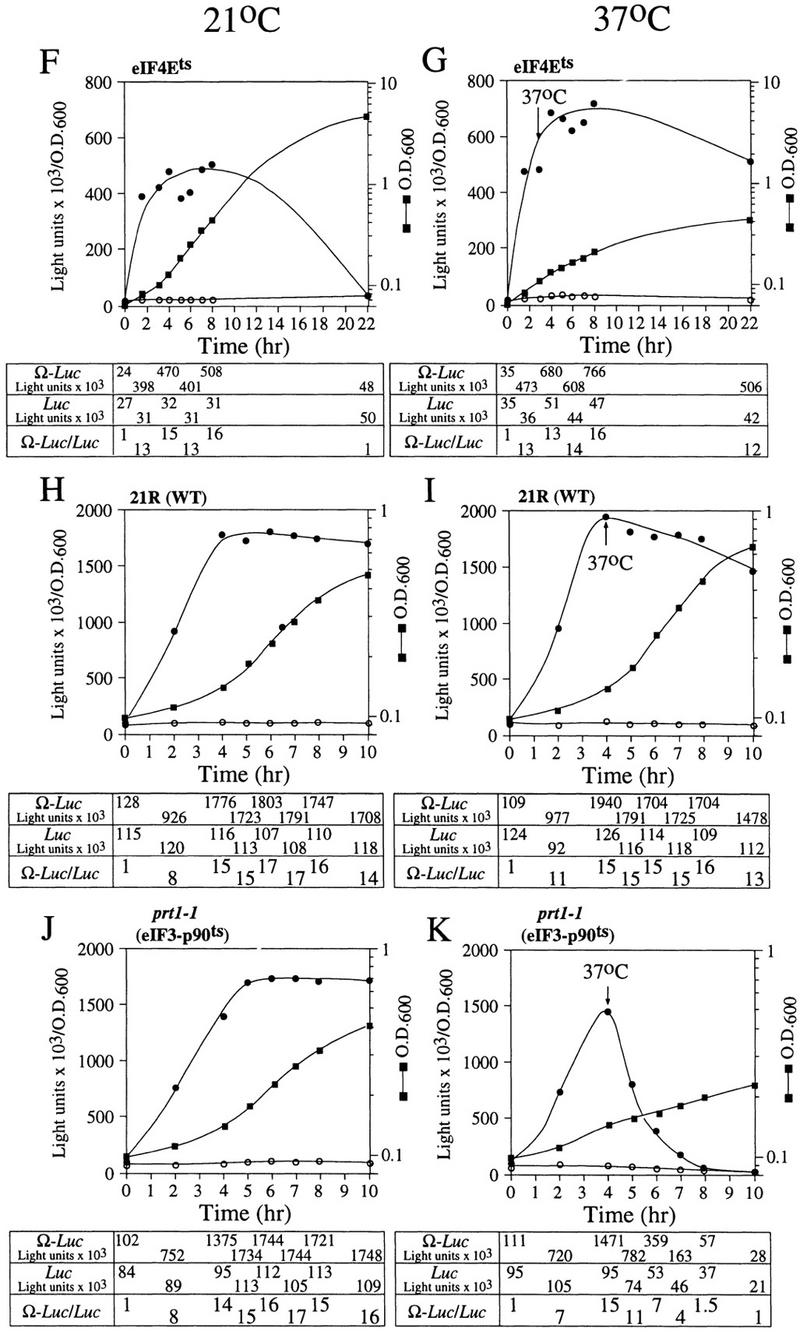
Analysis of NtHSP101 translational regulatory activity in yeast mutants affecting individual translation initiation factors. pTPI–NtHSP101 and either pGAL1–Ω-luc (•) or pGAL1–luc (○) were introduced into null and temperature-sensitive mutants affecting translation initiation factors. Transformants first grown to late-exponential stage (whereupon expression from pGAL1–Ω-luc and pGAL1–luc was equivalent) were inoculated into fresh SGM at the zero time point. The expression from pGAL1–Ω-luc, pGAL1–luc, and their expression ratio (i.e., NtHSP101 activity as indicated by the ratio, Ω-luc/luc) are shown below each panel. Analysis of the temperature-sensitive mutants was performed at the permissive (21°C) (F,H,J) and nonpermissive (37°C) (G,I,K) temperatures, where the arrow indicates the time point at which each culture was shifted to 37°C. (WT) parent strain. Strains used for the analysis: CW04 is wild-type for TIF3, CAF20, TIF4631, TIF4632 (A); RCB1-1A is the eIF4B null mutant (tif3) (B); CDK36-1A is the caf20 null mutant (C); YCG318 is the tif4632 null mutant (E); YCG165 is the tif4631 null mutant (D); 4-2 is the eIF4Ets mutant (F,G); 21R is the PRT1 parent strain (H,I); and TP11B-4-1(prt1-1) is an eIF3-p90ts mutant (J,K).
NtHSP101 activity was unaffected by inactivation of eIF4E following a shift of an eIF4Ets mutant (Altmann et al. 1989) to the nonpermissive temperature that substantially inhibited further growth (Fig. 9F,G). Note that although NtHSP101 activity at the permissive temperature (as measured by the expression ratio) was completely lost by 22 hr when further growth could not be supported because of nutrient limitation (OD of 4.5) (Fig. 9F), NtHSP101 activity remained high for the duration of the experiment following a shift to the nonpermissive temperature as cell growth was largely inhibited and the culture reached an OD of only 0.42 by 22 hr (Fig. 9G). This observation suggests that the mechanism by which NtHSP101 enhances translation does not specifically require eIF4E. Although it is possible that any residual eIF4E activity present after transfer to the nonpermissive temperature may be sufficient for NtHSP101 function, the complete absence of effect on NtHSP101 activity when growth was so dramatically reduced would argue against this. The observation that NtHSP101 enhanced translation from uncapped Ω-luc mRNA (see Fig. 6D) to at least the same extent as from capped mRNA (see Fig. 6A) supports the results obtained with the eIF4Ets mutant indicating that neither a cap nor eIF4E are specifically required for the NtHSP101-mediated regulation. These data also suggest that NtHSP101 can remain active in cells exhibiting a reduced growth rate as long as the reduction in growth is not a result of nutrient limitation. Inactivation of eIF4A following the shift of a eIF4A-conditional mutant to the nonpermissive temperature significantly reduced overall protein synthesis but had no effect on the NtHSP101 activity (Fig. 10A). Note that because of the lack of sufficient selectable markers in the mutant needed to support plasmid-based constructs, it was necessary to deliver the constructs as capped Ω-luc-A50 and luc-A50 mRNAs.
Figure 10.
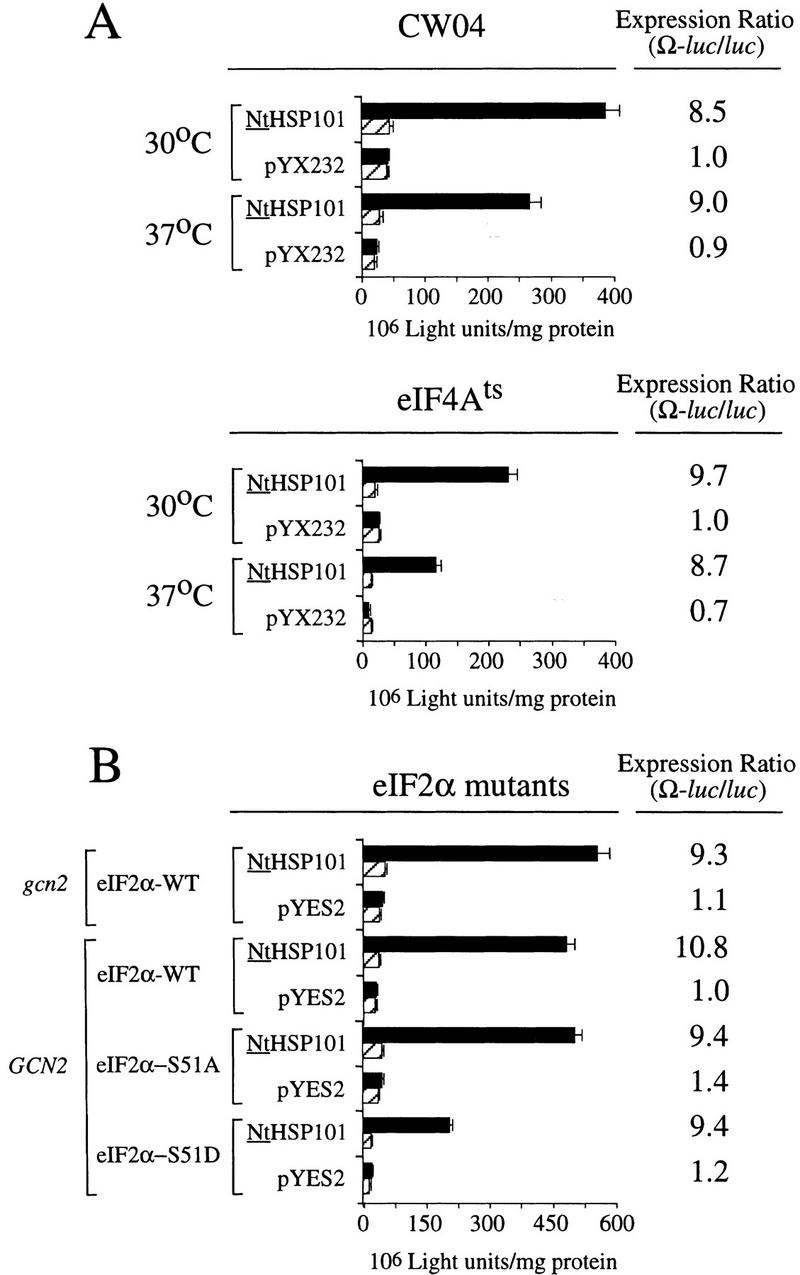
The NtHSP101-mediated regulation is not affected following the inactivation of eIF4A in the conditional mutant, SS8-3B(pSSC120), or in yeast expressing eIF2α mutants affecting factor activity. Translation from in vitro-synthesized capped Ω-luc-A50 and luc-A50 mRNAs was measured following RNA delivery into early exponential (A) wild-type CW04 and eIF4Ats mutant cells grown at the permissive (30°C) or nonpermissive (37°C) temperatures or (B) gcn2 containing wild-type eIF2α, i.e., gcn2(eIF2α-WT), GCN2 containing wild-type eIF2α, i.e., GCN2(eIF2α-WT), GCN2 containing the eIF2α-S51A mutant, or GCN2 containing the eIF2α-S51D mutant, each containing either pGAL1–NtHSP101 or pYES2. The expression ratio is shown to the right of each pair of histograms. (Solid bars) Ω-luc-A50; (hatched bars) luc-A50.
Phosphorylation of eIF2α at serine 51 by GCN2, for example, in response to amino acid starvation, reduces eIF2 activity (Hinnebusch 1996). Mutation of Ser-51 to alanine (eIF2α-S51A) prevents phosphorylation and results in constitutive activity, whereas mutation of the serine to aspartic acid (eIF2α-S51D) mimics phosphorylation and results in constitutively reduced activity (Dever et al. 1992). To examine whether changes in eIF2α function affect NtHSP101 activity, expression from Ω-luc-A50 and luc-A50 mRNAs was examined in mutants expressing eIF2α containing either the S51A or S51D mutations. Translation from Ω-luc-A50 mRNA was 10.8-fold higher than luc-A50 mRNA when NtHSP101 was expressed in a wild-type eIF2α/GCN2 background but virtually identical to luc-A50 mRNA in the same yeast not expressing NtHSP101 (Fig. 10B). Similar results were observed in a eIF2α-S51A/GCN2 or in wild-type eIF2α/gcn2 background in which eIF2α is constitutively active as a result of either the mutation of Ser-51 to alanine or the loss of GCN2 kinase activity, respectively. As expected, in a eIF2α-S51D/GCN2 background in which the mutation of Ser-51 to aspartic acid mimics a constitutive phosphorylated state resulting in reduced eIF2α activity, the level of expression from both luc mRNAs was reduced by more than twofold (Fig. 10B). However, the degree of enhancement from Ω-luc-A50 mRNA relative to luc-A50 remained at 9.4-fold in yeast expressing NtHSP101 (Fig. 10B). These data suggest that the mechanism by which NtHSP101 functions to enhance translation is unaffected by the difference in eIF2α activity caused by the mutations at Ser-51.
eIF3 contains a core of five subunits (Phan et al. 1998), binds eIF4G (Lamphear et al. 1995), and is required for 40S ribosomal subunit binding to an mRNA. prt1-1 is an eIF3 mutant containing a temperature-sensitive mutation in the eIF3–p90 subunit that results in the inactivation of eIF3 at the nonpermissive temperature (Naranda et al. 1994). NtHSP101 activity was observed in the parent strain (i.e., 21R) at the normal (Fig. 9H) and nonpermissive temperature (Fig. 9I) and in the prt1-1 mutant under normal conditions (Fig. 9J). As expected, following the shift of the prt1-1 mutant to the nonpermissive temperature, the loss of Prt1p function affected expression from both the luc and Ω-luc constructs (Fig. 9K). However, expression from the Ω-luc construct was lost preferentially: A 26-fold drop in expression from the Ω-luc construct was observed within 4 hr following inactivation of Prt1p when only a 2.6-fold decrease in expression from the luc construct containing the GAL1 5′ leader occurred in the same period. The preferential loss in translation from the Ω-luc construct is also revealed by the drop in the expression ratio from a value of 15 at the time of the shift to the nonpermissive temperature to a value of 1.5 within 4 hr following inactivation of Prt1p (Fig. 9K). The loss in NtHSP101 activity was not caused by nutrient limitation as it was observed at an OD of only 0.15.
Discussion
With this work, we show that Ω increases translational efficiency through an HSP101-mediated mechanism that can be recapitulated in yeast. The presence of Ω was a prerequisite for the HSP101-mediated enhancement from an mRNA, as no increase in expression was observed from constructs containing either the GAL1 or TEV 5′ leaders. Moreover, HSP101 enhanced translation from Ω-containing constructs under conditions of rapid fermentative growth, but not following amino acid starvation or during respiration, showing that the enhancement conferred by Ω is not an inherent property of this sequence. Instead, a tight correlation was observed between HSP101 RNA-binding activity and HSP101-mediated translational enhancement from an Ω-containing construct, suggesting that HSP101 binding is a prerequisite for translational regulation. Previous work that had identified the HSP101-binding site within Ω as the 25-nucleotide poly(CAA) subsequence (Tanguay and Gallie 1996) that is responsible for translational enhancement (Gallie and Walbot 1992) supports this conclusion.
Wheat HSP101 also enhanced translation from an Ω-containing construct showing that the translational regulatory role is not unique to NtHSP101. However, wheat HSP101 exhibited both a lower affinity for Ω (see Fig. 3B, lane 7, top) and a reduced capacity (Fig. 6B) to enhance translation from an Ω-containing mRNA than that observed for NtHSP101, supporting the correlation between RNA binding and translational regulation. The difference in RNA binding between the two plant HSP101s may indicate evolutionary divergence in RNA specificity between these proteins. It is not altogether surprising that tobacco HSP101 exhibits a higher affinity for Ω than wheat HSP101, as tobacco is the host species for TMV. The observation that yeast Hsp104 does not enhance translation from Ω-luc mRNA (D. Gallie, unpubl.) supports the conclusion that RNA-binding activity or specificity may differ between members of this heat shock protein family. Although it remains to be determined whether yeast Hsp104 can function as a translational regulator and what the client mRNAs may be, the conservation of a mechanism in yeast that regulates the RNA-binding and translational regulatory functions of plant HSP101 in response to nutrient availability supports the possibility of conserved function.
The effect of HSP101 on expression is specific to translation: Only the rate of translation of Ω-containing mRNA was affected without a corresponding change in reporter mRNA or protein stability. A screen of translation initiation factor mutants supported a translational role for HSP101. Although inactivation of Prt1p had a global effect on protein synthesis, its inactivation disproportionally affected translation from Ω-containing mRNA: The loss of eIF3 function following growth of the prt1p-conditional mutation at the nonpermissive temperature resulted in 26-fold drop in translation from Ω-luc mRNA, but only a 2.6-fold reduction from luc mRNA and, consequently, resulted in a virtually equivalent level of translation from both mRNAs within 4 hr of eIF3 inactivation. Taking into account the time required for the turnover of the elevated level of luciferase protein made from Ω-luc mRNA prior to the inactivation of Prt1p at the nonpermissive temperature, the actual loss in HSP101 translational regulatory function was considerably faster than that indicated by the data in Figure 9K.
The two genes encoding eIF4G (TIF4631 and TIF4632) are expressed to similar levels (Tarun et al. 1997) but are only 53% identical in yeast (Goyer et al. 1993). Although the presence of either gene is sufficient for viability, their redundancy, divergence, and difference in growth phenotype when absent (Fig. 9D,E; Goyer et al. 1993) suggests some degree of functional specialization. The loss of TIF4632 specifically affected the HSP101-mediated translational enhancement from Ω-luc mRNA as expression from luc mRNA (containing the GAL1 5′ leader) was not similarly affected. Moreover, the loss of HSP101-mediated enhancement in tif4632, but not the tif4631, mutant suggests that TIF4632 is specifically required for HSP101 regulatory function. Loss of TIF4631 substantially reduced the growth rate, suggesting that it may have adopted a more general translational role or at least be required for the translation of those mRNAs required for growth. In contrast, the absence of TIF4632 had little effect on the growth rate, suggesting that it may possess a more specific translational function. The fact that the loss of TIF4632 (but not TIF4631) specifically affected the translational regulatory function of HSP101 shows that the requirement for TIF4632 was not a consequence of the growth phenotype. The continued activity of HSP101 in the eIF4Ets mutant following its virtual cessation of growth at the nonpermissive temperature (Fig. 9G) supports the conclusion that rapid growth is not a prerequisite for the translational regulatory function of HSP101. As no effect on HSP101-mediated translational enhancement was observed in null, conditional, or point mutations affecting eIF4A, eIF4B, eIF4E, CAF20, or eIF2, these data suggest that HSP101 functionally requires only TIF4632 and eIF3. HSP101 is present in yeast in multiple isoelectric states and only the most acidic isoforms are found associated with polysomes even in the absence of Ω-containing mRNA (H. Le and D. Gallie, unpubl.). This finding suggests that HSP101 can bind some yeast mRNAs, or that the ability of HSP101 to associate with either eIF3 or eIF4G (i.e., TIF4632) is independent from its RNA-binding function.
The functional consequences of the tif4632 and prt1p mutations on HSP101 function suggests that HSP101 may facilitate the recruitment of TIF4632 and eIF3 to an mRNA when HSP101 is bound to the 5′ leader or promote the interaction between TIF4632 and eIF3 that is thought necessary for 40S ribosomal subunit binding. Such an interaction could be related to its chaperone activity, which involves protein–protein interactions with non-native substrates and other chaperones (Glover and Lindquist 1998). However, if the chaperone function of HSP101 has been adapted to promote recruitment or interaction between eIF4G and eIF3 during the translation of those mRNAs containing a high-affinity HSP101-binding site, its chaperone function during translation must be distinct from its chaperone function required for conferring thermotolerance as the deletion of either the amino- or carboxy-terminal domains of HSP101 abolishes its thermotolerance function without abolishing its translation regulatory function (D. Gallie, unpubl.).
The correlation between the RNA-binding activity and the translational regulatory function of HSP101 under different growth conditions suggests that its activity as a translational regulator may be controlled through the regulation of its RNA-binding activity. A loss of HSP101 activity early in the growth phase of yeast could be triggered by limiting the availability of amino acids in the medium. Even a transient state of starvation induced by the depletion of an amino acid for which the yeast was prototrophic was sufficient to cause a transient reduction in the activity of HSP101. The loss of HSP101 activity in late-exponential cells or following the depletion of a limiting nutrient could not be explained by a change in the cellular level of HSP101 or by a change in the relative stability of either control or Ω-containing mRNAs, but correlated with a loss in HSP101 RNA-binding activity to Ω. These conclusions were supported with observations that used respiring cells in which a loss in HSP101 translational regulatory activity and RNA-binding activity was rapidly lost. These data suggest that the RNA-binding activity and the translational regulatory function of HSP101 are manifested only in cells engaged in fermentation whose growth is not subjected to nutrient limitation. In contrast, its role in thermotolerance appears to be active regardless of the growth conditions. This may reflect the paramount role that its thermotolerance function plays in mounting a response to elevated temperatures.
The evolution of the TMV 5′ leader as a high-affinity HSP101-binding site suggests that variants of this sequence may be present in cellular mRNAs. The identification of cellular mRNAs subject to control by HSP101 will be an important step in understanding not only the basis for the evolution of the translational regulatory function of this heat shock protein, but also in determining whether the translational role of HSP101 is specific for a limited set of client mRNAs, or whether its regulatory function may be more global.
Materials and methods
Protein purification, protein sequencing, and cDNA library screening
The purification of HSP101 to near homogeneity has been described previously (Tanguay and Gallie 1996). For protein sequencing, purified wheat HSP101 protein was digested with Lys-C, the resulting peptides separated by HPLC, and selected individual peptides sequenced by William Lane (Harvard University, Boston, MA). Purified tobacco HSP101 was resolved on an 8% SDS–polyacrylamide gel, transferred to PVDF membrane, and following staining with Ponceau S, the HSP101-containing portion of the membrane subjected to digestion with Lys-C and the peptides separated and sequenced as described for the wheat HSP101.
Seven-day-old tobacco and five-day-old wheat seedlings were subjected to a 37°C heat shock for 90 min, and following a 60-min recovery at room temperature, total RNA from each was isolated. First-strand cDNA synthesis was initiated from poly(A)+ RNA by use of an oligo(dT)–NotI primer (Promega) and the second strand synthesis by use of a Boehringer cDNA Kit. EcoRI adapters were added to the cDNA, which was then digested with NotI and introduced into the EcoRI–NotI sites of λgt11 (Promega). The DNA was packaged with Gigapack II Gold packaging extract (Stratagene) and the resulting phage used to transfect Escherichia coli strain Y1090. Expression screening was performed with anti-p102 antibodies (Tanguay and Gallie 1996).
In vitro RNA synthesis, gel shift assay, and RNA electroporation
pT7-Ω-luc-A50, pT7-luc-A50, pT7-luc-3′TMV, and T7-based constructs containing Ω control sequences have been described previously (Leathers et al. 1993). In vitro transcription and gel shift assays were carried out as described previously (Tanguay and Gallie 1996).
For mRNA delivery by electroporation, SL304A (leu2-3, 112 trp1-1 ura3-1 ade2-1 his3-11,15 lys2Δ can1-100 hsp104::LEU2) (Schirmer et al. 1994), a Δhsp104 yeast mutant (generously provided by Dr. Susan Lindquist, University of Chicago, IL), containing either pGAL1–NtHSP101 or pGAL1–TaHSP101, was grown to mid-log phase in synthetic galactose (2%) medium, electroporated as described (Everett and Gallie 1992), and incubated for 4 hr before being harvested for analysis. Luciferase assays were performed as described (Tanguay and Gallie 1996). Ten replicate samples of each mRNA delivered to yeast were assayed in duplicate.
Western analysis
Yeast extracts were prepared by boiling 6 × 106 cells in 2× SDS-loading buffer, the protein resolved on a 10% SDS–polyacrylamide gel, transferred to nitrocellulose membrane, blocked for 30 min in TPBS with 5% reconstituted dry milk, and incubated with anti-HSP101 antibodies diluted in TPBS with 1% milk for 1.5 hr. The blots were then washed with TPBS, incubated with goat anti-rabbit horseradish peroxidase antibodies for 1 hr, and the HSP101 signal detected by enhanced chemiluminescence.
Plasmid construction, translational analysis, and thermotolerance assay
The tobacco or wheat HSP101 cDNAs were introduced into the BamHI–NotI sites or EcoRI–XbaI sites, respectively, of pYES2 (Invitrogen) in which HSP101 expression is under the control of the GAL1 promoter. Each cDNA was also introduced into the BamHI site of pYX232 (Novagen), in which HSP101 expression is under the control of the TPI promoter. luc or Ω-luc was introduced into the HindIII–BamHI sites of pYES2. SL304A was transformed with plasmid constructs by the polyethylene glycol/LiCl method as described (Hill et al. 1991; Gietz et al. 1992).
SL304A was transformed with either pTPI–NtHSP101 (tobacco HSP101 in pYX232) or pTPI–TaHSP101 (wheat HSP101 in pYX232) and either pGAL1–Ω-luc or pGAL1–luc and grown in synthetic medium containing the appropriate supplements. For translation studies, overnight yeast cultures grown in either SDM or SGM were diluted to the indicated OD into either synthetic medium containing the appropriate supplements or YPG medium and samples taken at time intervals during their subsequent growth for luciferase assays.
The yeast strains, CW04, RCB1-1A (tif3), and CDK36-1A (caf20) (Coppolecchia et al. 1993; de la Cruz et al. 1997); YCG318 (tif4632) and YCG165 (tif4631) (Goyer et al. 1993); T93C and 4-2 (eIF4Ets), (Altmann et al. 1989); and 21R and TP11B-4-1 (prt1-1) (Hanic-Joyce et al. 1987) were transformed with pTPI–NtHSP101 and either pGAL1–luc or pGAL1–Ω-luc. SGM was inoculated from an overnight culture and the appropriate transformed cells and luciferase expression from pGAL1–luc or pGAL1–Ω-luc was measured during growth. For 21R and TP11B-4-1 (prt1-1), the shift to the nonpermissive temperature (resulting in a loss of eIF3 function caused by the temperature-sensitive mutation of prt1-1) occurred after 4 hr of growth in SGM, whereas for T93C and 4-2 (eIF4Ets), the shift occurred after 3 hr of growth. Because of limited selectable markers, the eIF4Ats mutant, SS8-3B(pSSC120) (Schmid and Linder 1991) was transformed only with pTPI–NtHSP101. Transformants containing pYX232 served as the control. Early exponential cells of each transformant were divided and treated at the permissive (30°C) or nonpermissive (37°C) temperatures for 1.5 hr, whereupon Ω-luc-A50 and luc-A50 mRNAs were delivered by electroporation to both sets of cells. The cells were maintained at their respective temperatures for the duration of the experiment prior to assaying for luciferase. The eIF2α mutant strains, H1816, H1896, H1897, and H1898 were transformed only with pGAL1–NtHSP101. Each was transformed with pYES2 to serve as a control.
SL304A was transformed with pGAL1–NtHSP101 (tobacco HSP101 in pYES2) or pGAL1–TaHSP101 (wheat HSP101 in pYES2). Complementation of Hsp104 function in yeast was determined in a thermotolerance assay as described (Sanchez and Lindquist 1990; Parsell et al. 1991).
Acknowledgments
We thank Dr. Susan Lindquist for SL304A and pYS104; Dr. Patrick Linder for CW04, RCB1-1A, CDK36-1A, and SS8-3B; Dr. Nahum Sonenberg for YCG318 and YCG165; Dr. Michael Altmann for T93C and 4-2; Dr. Gerald Johnston for 21R and TP11B-4-1; and Dr. Alan Hinnebusch for the eIF2a mutant strains, H1816, H1896, H1897 and H1898. This work was supported by a grant (NRICGP 96-35301-3144) from the U.S. Department of Agriculture.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL drgallie@citrus.ucr.edu; FAX (909) 787-3590.
References
- Altmann M, Sonenberg N, Trachsel H. Translation in Saccharomyces cerevisiae: Initiation factor 4E-dependent cell-free system. Mol Cell Biol. 1989;9:4467–4472. doi: 10.1128/mcb.9.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M, Blum S, Pelletier J, Sonenberg N, Wilson TMA, Trachsel H. Translation initiation factor-dependent extracts from Saccharomyces cerevisiae. Biochim Biophys Acta. 1990a;1050:155–159. doi: 10.1016/0167-4781(90)90158-x. [DOI] [PubMed] [Google Scholar]
- Altmann M, Blum S, Wilson TMA, Trachsel H. The 5′-leader sequence of tobacco mosaic virus RNA mediates initiation-factor-4E-independent, but still initiation-factor-4A-independent translation in yeast extracts. Gene. 1990b;91:127–129. doi: 10.1016/0378-1119(90)90173-o. [DOI] [PubMed] [Google Scholar]
- Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Freed DD. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppolecchia R, Buser P, Stotz A, Linder P. A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J. 1993;12:4005–4011. doi: 10.1002/j.1460-2075.1993.tb06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AWB, Haghighat A, Yu ATK, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 549–574. [Google Scholar]
- Everett JG, Gallie DR. RNA Delivery in Saccharomyces cerevisiae using electroporation. Yeast. 1992;8:1007–1014. doi: 10.1002/yea.320081203. [DOI] [PubMed] [Google Scholar]
- Gallie DR, Walbot V. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes & Dev. 1990;4:1149–1157. doi: 10.1101/gad.4.7.1149. [DOI] [PubMed] [Google Scholar]
- ————— Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 1992;20:4631–4638. doi: 10.1093/nar/20.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Sleat DE, Watts JW, Turner PC, Wilson TMA. The 5′-leader sequence of tobacco mosaic virus RNA enhances the expression of foreign gene transcripts in vitro and in vivo. Nucleic Acids Res. 1987a;15:3257–3273. doi: 10.1093/nar/15.8.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A comparison of eukaryotic viral 5′-leader sequences as enhancers of mRNA expression in vivo. Nucleic Acids Res. 1987b;15:8693–8711. doi: 10.1093/nar/15.21.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Lucas WJ, Walbot V. Visualizing mRNA expression in plant protoplasts: Factors influencing efficient mRNA uptake and translation. Plant Cell. 1989;1:301–311. doi: 10.1105/tpc.1.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Tanguay RL, Leathers V. The tobacco etch viral 5′ leader and poly(A) tail are synergistic regulators of translation. Gene. 1995;165:233–238. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Goyer C, Altmann M, Lee HS, Blanc A, Deshmukh M, Woolford JL, Trachsel H, Sonenberg N. TIF4631 and TIF4632: Two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanic-Joyce PJ, Johnston GC, Singer RA. Regulated arrest of cell proliferation mediated by yeast prt1 mutations. Exp Cell Res. 1987;172:134–145. doi: 10.1016/0014-4827(87)90100-5. [DOI] [PubMed] [Google Scholar]
- Hill J, Donald KA, Griffiths DE. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational control of GCN4: Gene-specific regulation by phosphorylation of eIF2. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 199–244. [Google Scholar]
- Kozak M. Leader length and secondary structure modulate mRNA function under conditions of stress. Mol Cell Biol. 1988;8:2737–2744. doi: 10.1128/mcb.8.7.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— An analysis of vertebrate mRNA sequences: Intimations of translational control. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A consideration of alternative models for the initiation of translation in eukaryotes. Crit Rec Biochem Mol Biol. 1992;27:385–402. doi: 10.3109/10409239209082567. [DOI] [PubMed] [Google Scholar]
- ————— Determinants of translational fidelity and efficiency in vertebrate mRNAs. Biochimie. 1994;76:815–821. doi: 10.1016/0300-9084(94)90182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Le H, Tanguay RL, Balasta ML, Wei C-C, Browning KS, Metz AM, Goss DJ, Gallie DR. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- Leathers V, Tanguay R, Kobayashi M, Gallie DR. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol Cell Biol. 1993;13:5331–5347. doi: 10.1128/mcb.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-RJ, Nagao RT, Key JL. A soybean 101-kD heat shock protein complements a yeast HSP104 deletion mutant in acquiring thermotolerance. Plant Cell. 1994;6:1889–1897. doi: 10.1105/tpc.6.12.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Slonimski PP. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missence mutation. Proc Natl Acad Sci. 1989;86:2286–2290. doi: 10.1073/pnas.86.7.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Hershey JWB. In: Translational control. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. p. 31. [Google Scholar]
- Naranda T, MacMillan SE, Hershey JWB. Purified yeast translational initiation factor eIF-3 is an RNA-binding protein complex that contains the PRT1 protein. J Biol Chem. 1994;269:32286–32292. [PubMed] [Google Scholar]
- Parsell DA, Sanchez Y, Stitzel JD, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher H-P, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA, Klausner RD, Harford JB. Translational control of ferritin. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 335–362. [Google Scholar]
- Sanchez Y, Lindquist S. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Lindquist S, Vierling E. An arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SR, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: Analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleat DE, Hull R, Turner PC, Wilson TMA. Studies on the mechanism of translational enhancement by the 5′-leader sequence of tobacco mosaic virus RNA. Eur J Biochem. 1988;175:75–86. doi: 10.1111/j.1432-1033.1988.tb14168.x. [DOI] [PubMed] [Google Scholar]
- Tanguay RL, Gallie DR. Isolation and characterization of the 102-kilodalton RNA-binding protein that binds to the 5′ and 3′ translational enhancers of tobacco mosaic virus RNA. J Biol Chem. 1996;271:14316–14322. doi: 10.1074/jbc.271.24.14316. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Heuval JJ, Raue HA. Translational efficiency of mRNA in yeast cells is not increased by plant viral leader sequences. Appl Microbiol Biotechnol. 1992;37:84–87. [Google Scholar]