Abstract
In vertebrates, many cytokines and growth factors have been identified as activators of the JAK/STAT signaling pathway. In Drosophila, JAK and STAT molecules have been isolated, but no ligands or receptors capable of activating the pathway have been described. We have characterized the unpaired (upd) gene, which displays the same distinctive embryonic mutant defects as mutations in the Drosophila JAK (hopscotch) and STAT (stat92E) genes. Upd is a secreted protein, associated with the extracellular matrix, that activates the JAK pathway. We propose that Upd is a ligand that relies on JAK signaling to stimulate transcription of pair-rule genes in a segmentally restricted manner in the early Drosophila embryo.
Keywords: JAK, hopscotch, segmentation, ligand
The Janus kinase cascade is a ubiquitous intracellular signaling pathway required for response to many extracellular ligands. The pathway includes receptor-associated tyrosine kinases (JAKs) and their substrate transcription factors, signal transducers and activators of transcription (STATs). The JAK/STAT cascade has emerged as an essential reutilized facet of vertebrate signaling through a large number of cytokines and growth factors. These signals induce proliferation or differentiation and are crucial to the proper growth and development of mammalian tissues, particularly blood cell lineages. Both decreases and increases in activity of this signaling pathway have severe consequences. Loss of specific JAK functions can render cells unable to respond to interferons, interleukins, growth hormone, or other signaling molecules. In humans, defects in JAK3 have been linked to autosomal severe combined immune deficiency (SCID) (Macchi et al. 1995), causing a dramatic reduction in lymphoid cell development. Similarly, mice that are immunocompromised or have other hematopoietic defects have been generated by JAK or STAT knock-outs (Nosaka et al. 1995; Durbin et al. 1996; Meraz et al. 1996; Neubauer et al. 1998; Parganas et al. 1998; Rodig et al. 1998). On the other hand, constitutive activation of JAKs and/or STATs is correlated with several oncogenic pathways. Most directly, a fusion of JAK2 with the oligomerization domain of the TEL transcription factor causes acute lymphoblastic leukemia (Lacronique et al. 1997; Peeters et al. 1997). These data point to a significant involvement of the JAK/STAT cascade in both normal development and in oncogenesis.
JAKs are a novel class of nonreceptor tyrosine kinases, of which four mammalian members have been characterized (for review, see Ihle et al. 1997). The unique feature of these molecules is the presence of two tandem kinase-homologous domains. The carboxy-terminal domain of these proteins has been shown to catalyze tyrosine phosphorylation, whereas the more amino-terminal domain is apparently catalytically inactive. The major substrates for JAK tyrosine phosphorylation are the STATs. This family of molecules, with seven identified members in mammals, is able to bind to specific DNA sequences and activate transcription (Ihle 1996; Darnell 1997). Together, the JAKs and STATs comprise a simple intracellular signal relay that can be activated by a number of extracellular factors. In vertebrates, the cytokines are the largest class of molecules to utilize the cascade. Although the cytokines have significantly different amino acid sequences, they are divided into two categories on the basis of their structures. The larger class, type I cytokines, have a characteristic up–up–down–down helical structure, referring to the arrangement of alpha helices in the protein (Sprang and Bazan 1993). The receptors for these molecules are heteromultimeric subunits, with many cytokines using a common subunit in the receptor complex. Whereas the type II cytokines have different structure than the type I class, they also bind to heteromultimeric receptor complexes.
Mechanistically, the JAK/STAT pathway provides a direct means to respond to extracellular signals (for review, see Darnell 1997; Ihle et al. 1997). The first step in activation is binding of the extracellular ligand to the appropriate transmembrane receptor. Ligand binding typically induces receptor dimerization or multimerization. Current models hold that receptor dimerization brings their associated JAK molecules into close proximity, presumably facilitating trans-phosphorylation of the JAKs. Signals are then transmitted directly to the nucleus through the activation of appropriate STATs. Inactive STATs are found in the cytoplasm and, upon JAK activation, are recruited to the inner surface of the cell membrane. Here, they are tyrosine phosphorylated by the JAKs, then translocate to the nucleus and bind specific DNA sequences to regulate transcription. In this manner, direct response to signals is achieved via transcriptional activation of downstream target genes by activated STATs.
Our current understanding of JAK/STAT signaling pathways is deduced primarily from studies in mammals. Recently, a Drosophila paradigm has emerged in which a single JAK (hopscotch, hop) and STAT (stat92E; also called marelle) have been identified (for review, see Hou and Perrimon 1997). Drosophila hop was originally identified as a gene involved in embryonic pattern formation (Perrimon and Mahowald 1986). Embryos derived from females that do not express the hop gene product in their germ lines show novel embryonic defects characterized by the loss and/or fusion of a specific subset of body segments. This phenotype is unusual because it does not resemble any of the stereotypical classes of segmentation genes (gap, pair-rule, or segment polarity genes, see Nüsslein-Volhard and Weischaus 1980). This same embryonic phenotype is also seen in animals derived from females that lack stat92E gene activity.
Here we describe the characterization of a third gene, unpaired (upd), with the distinctive embryonic mutant phenotype of hop and stat92E. We provide genetic and biochemical evidence that upd encodes a secreted molecule that can activate the JAK cascade, suggesting that it encodes a ligand for the JAK/STAT pathway during Drosophila segmentation.
Results
The upd phenotype is similar to that of hop and stat92E
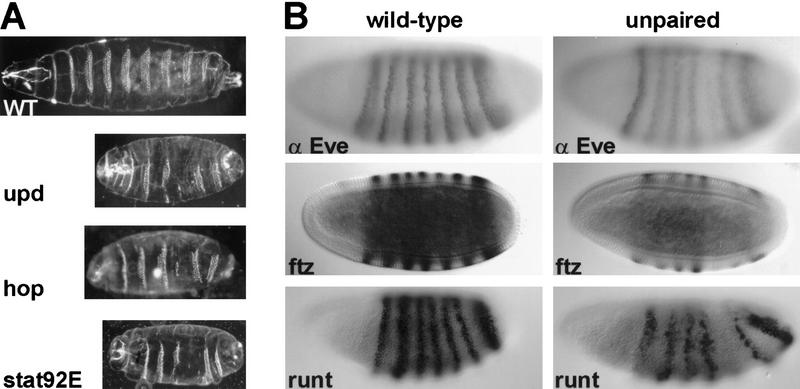
Loss of zygotic upd activity causes segmentation defects in the Drosophila embryo that resemble the phenotype of hop and stat92E mutant embryos (Wieschaus et al. 1984) (Fig. 1). These defects always include loss of the fifth abdominal denticle band and the posterior mid-ventral portion of the fourth band. Defects in other segments are variable, but often include reduction of the second thoracic and eighth abdominal denticle bands and fusion of the sixth and seventh bands. In contrast to hop or stat92E (Perrimon and Mahowald 1986; Hou et al. 1996), zygotic upd activity is essential but maternal activity is not, as evidenced by the lack of a maternal effect phenotype for upd mutants (Eberl et al. 1992). The similarity between embryos that lack zygotic upd and those that lack maternal hop or stat92E suggests that upd is a component of the JAK signaling pathway. This hypothesis is further supported by genetic interactions between these genes. It has been observed previously that certain allelic combinations of hop are viable, but have adult defects (Perrimon and Mahowald 1986). The partial loss of hop activity in such animals causes reduced viability, held-down wings, reduced production of mature eggs, and/or defects in eggs produced. Each of the heteroallelic combinations results in a consistent and predictable degree of severity with respect to these phenotypes. To test whether the hop and upd genes interact genetically, one copy of upd was removed from animals carrying allelic combinations of hop. Altering the dose of upd activity exacerbated the defects observed for these hop mutant combinations (see Table 1). Such enhancement is likely to occur if the two gene products are active in the same pathway.
Figure 1.
Embryonic effects of JAK pathway mutations. (A) Representative embryonic cuticle phenotypes are shown from wild-type, hopc111 germ-line mutant clone-derived (GLC) embryos, stat92EP1681 GLC embryos, and Df(1)os1A (upd mutant) embryos. The defects seen are similar for all three mutations and are described in the text. (B) The effects of loss of upd on the expression of three pair–rule genes is shown. Wild-type embryos (left) and Df(1)os1A/ Y embryos (right) are compared at cellular blastoderm for presence of Even-skipped (Eve) protein, fushi tarazu (ftz) RNA, and runt RNA. Each shows reduced expression of the fifth stripe in the upd mutant embryos (middle). Variable reductions in other stripes of expression are described in the text (not shown). In all pictures, anterior is to the left. Cuticles are shown in ventral views, whereas stainings are shown in lateral views.
Table 1.
Enhancement of hop mutant phenotypes by upd mutations
|
|
hopM4
|
hopVA275
|
hopGA32
|
hopVA85
|
hopM75
|
hopM38
|
hopmsvl
|
hopM637
|
hopM13
|
|---|---|---|---|---|---|---|---|---|---|
| Viability | |||||||||
| hopmsvl | 100% | 100% | 100% | 100% | 70% | 50% | 25% | 5% | 2% |
| hopmsvl updYM55 | 95% | 90% | 55% | 20% | 43% | 16% | 12% | 0% | 0% |
| Held-down wings | |||||||||
| hopmsvl | no | no | no | no | no | yes | yes | yes | yes |
| hopmsvl updYM55 | no | no | yes | yes | yes | yes | yes | N.A. | N.A. |
| Egg production | |||||||||
| hopmsvl | +++ | +++ | + | + | + | − | − | − | − |
| hopmsvl updYM55 | + | − | − | − | − | − | − | N.A. | N.A. |
Heteroallelic combinations of hopmsvland some other hop alleles result in viable adult females with various phenotypes. The severity of these heteroallelic phenotypes can be enhanced by introduction of the updYM55allele on the hopmsvlchromosome. Viability was scored relative to the hop/FM7 females from the same cross. The held-down wing phenotype was scored as present or absent from females. Egg production is indicated as follows: (−) for complete lack of any eggs deposited; (+) for production of small numbers of eggs, and (+++) for production of nearly normal numbers of eggs. Phenotypes that are not applicable are denoted N.A.
upd is required for stripe-specific expression of pair–rule genes
The similarity of the upd mutant phenotype with that of hop and stat92E, along with the genetic interactions described above, suggest that all three genes are involved in the same developmental process. A prediction of this hypothesis is that mutations in upd would affect the expression of segmentation genes in the same manner as hop and stat92E. Analysis of the expression of pair–rule genes in upd mutant embryos confirms this prediction. The removal of upd results in the stripe-specific loss of expression of several pair–rule genes (Fig. 1). Specifically, in upd mutants, the fifth stripes of expression of the genes even-skipped (eve), fushi tarazu (ftz), and runt are reduced or absent. Additionally, the third stripes of eve and ftz, and the second stripe of runt are variably reduced. These stripe-specific effects are identical to those described for maternal loss of hop and stat92E activities (Binari and Perrimon 1994; Hou et al. 1996).
The enhancer elements responsible for control of the third stripe of eve expression have been mapped to a 500-bp element upstream of the eve transcriptional start site (Small et al. 1996). A reporter construct carrying 5.2 kb of eve upstream sequence, including this enhancer region, fused to the lacZ gene drives expression of lacZ in the second, third, and seventh stripes of eve (Goto et al. 1989). Previous work has shown that this fragment contains sequences that bind Stat92E protein in vitro (Yan et al. 1996b). Removal of maternal activity of either hop or stat92E results in the loss of the third stripe from the reporter construct (Hou et al. 1996; Yan et al. 1996b). Similarly, zygotic mutation of upd also causes the specific loss of the third stripe, without affecting the second or seventh stripes (not shown).
The upd gene encodes a 2.2-kb transcript
Because of its likely involvement in JAK signaling, molecular identification of the upd gene was undertaken. Previous genetic mapping placed upd at ∼59, in polytene band 17A (Eberl et al. 1992). This is proximal to the Shaker complex but distal to and near CREB and porcupine (porc) (Kadowaki et al. 1996). Strong alleles of upd are embryonic lethal, but weaker alleles show an outstretched (os) phenotype, resulting in adult flies with wings held out away from the body. Allelism of upd and os is based on the failure of zygotic lethal upd alleles to complement the wing phenotype of os alleles (this study and Eberl et al. 1992). For example, combination of the embryonic lethal allele updYC43 with the viable allele oso results in viable adult flies with outstretched wings. To provide additional breakpoints for molecular mapping of the locus, new os alleles were generated by use of X-rays (see Materials and Methods).
A chromosomal walk from the Shaker region was generously made available to us by O. Pongs (Krah-Jentgens 1989). Southern blot analysis of various upd/os mutations and rearrangements in the region was used to delimit the extent of the upd gene (Fig. 2). The positions of these mutations indicated that upd must lie within the region designated as 370–415 kb of the chromosomal walk. Candidate transcription units were identified by Northern blot analysis by use of RNA isolated from staged embryo collections of wild-type flies probed with DNA from this 45-kb region. Three nonoverlapping RNAs were identified (Fig. 2). Only the 2.2-kb transcript is temporally expressed in a profile consistent with upd function. This RNA is expressed during early zygotic development, the stage at which early segmentation genes are required. Sequencing of the updYM55 and updYC43 mutant alleles has uncovered nonsense and frameshift mutations, respectively, in the ORF of the 2.2-kb transcript (Fig. 4, below; first noted by L. Sefton and T. Cline, pers. comm.). These data, as well as experiments described below, provide substantial evidence to conclude that the 2.2-kb transcript represents upd.
Figure 2.
Molecular map of the upd region. The map indicates the position of phages from the chromosomal walk, in kilobases. The positions of alleles that delimit the extent of the upd locus are shown beneath the chromosome, with bold lines indicating deleted DNA. Broken lines indicate uncertainties in the exact position of breakpoints. The position of transcription units is indicated at bottom. The 1-kb transcript has not been precisely mapped and falls within the indicated area. The 2.5-kb RNA is expressed only during late embryogenesis (after 13 hr), and the 1-kb RNA is present maternally, and again late in embryogenesis. The upd candidate transcript, 2.2 kb, is expressed during early zygotic stages (also see Fig. 3). CREB is shown for reference (porc is off the map at right). Proximal is to the right. Identification of single site lesions in two upd alleles, updYM55 and updYC43, confirms the assignment of these mutations to the same complementation group. Because these mutations fail to complement os alleles and only affect the protein encoded by the 2.2-kb transcript (Fig. 4), we conclude that os and upd are separable functions of the same locus. The breakpoints of os4-51-1, osc18, and os109 (all os−, upd+) map at least 13 kb from the 2.2-kb transcript, suggesting that the os phenotype may derive from the disruption of an enhancer that lies far from the 3′ end of the upd transcript.
Figure 4.
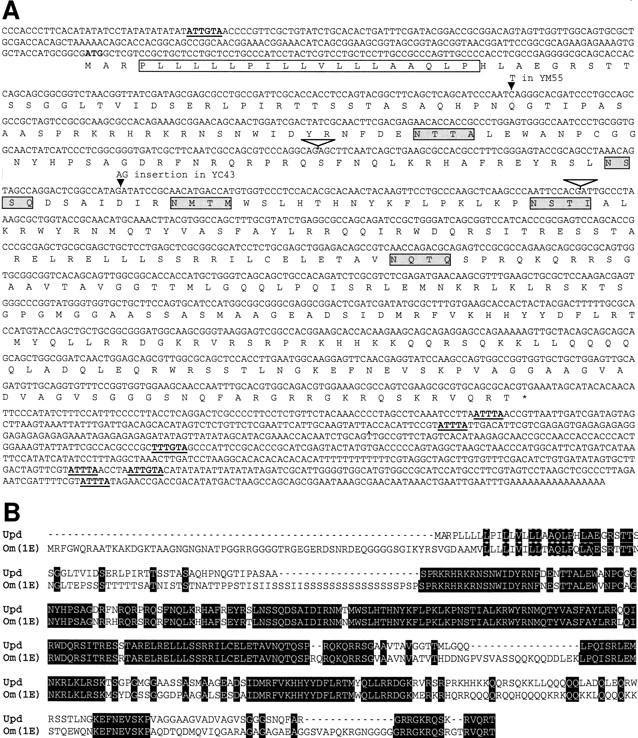
Sequence of upd. (A) The sequence of the upd cDNA (λKZ-GR) is shown. The conceptual ORF is shown below the DNA sequence. The presumed signal sequence is denoted by the open box, putative amino-linked glycosylation signals are indicated by the shaded boxes, and AU-rich sequences implicated in mRNA stability are underlined and bold. Intron positions are indicated by inverted open triangles. (B) The predicted protein sequence of D. melanogaster (Dm) Upd is aligned with the predicted protein sequence of the D. ananassae (Da) Om(1E). Amino acid identities are shown in black boxes.
upd embryonic expression is spatially restricted
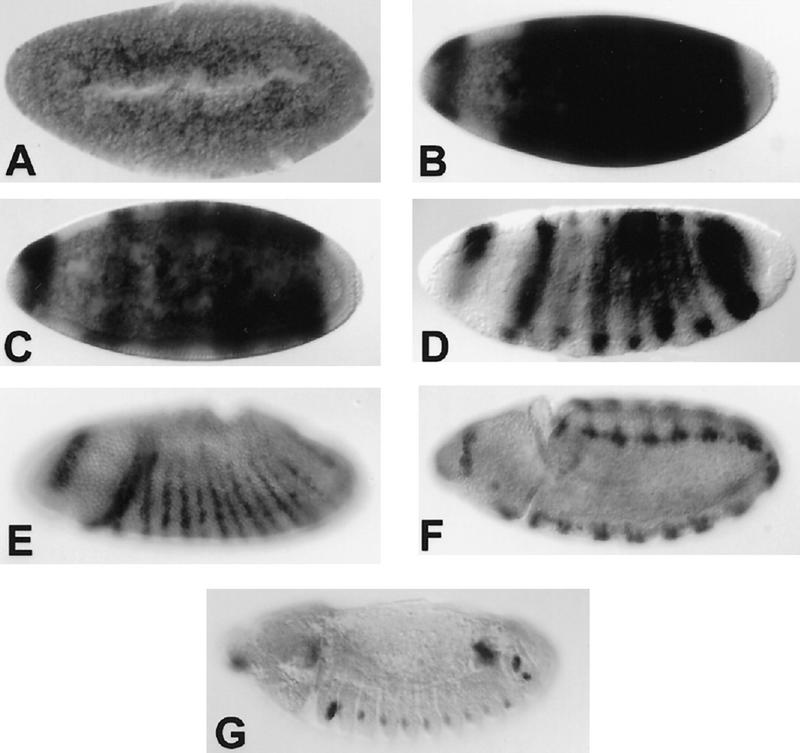
The spatial distribution of the 2.2-kb transcript is also consistent with a role in embryonic segmentation. In situ hybridization to RNA in whole mount embryos has revealed a dynamic and segmentally repetitive pattern of expression (Fig. 3). During the syncytial blastoderm stage, the 2.2-kb transcript is not expressed at levels that are detectable above background. Shortly before cellularization, the RNA becomes abundant, but is absent from the termini, and is restricted to only the trunk of the embryo and a single incomplete head stripe. At cellularization, the trunk expression resolves into ∼7 stripes, then into 14 stripes, a phenomenon seen in some pair–rule segmentation genes (Kilchherr et al. 1986; Macdonald et al. 1986).
Figure 3.
Embryonic expression of the upd transcript. Embryonic expression of the upd RNA is highly dynamic. No maternal product can be detected above background (A), but before cellularization, the RNA is expressed broadly throughout the trunk of the embryo and in a dorsal crescent in the head (B). As cellularization proceeds, expression resolves transiently into seven stripes (C,D). During early gastrulation, 14 stripes of expression appear (E). Later expression is largely restricted to the tracheal pits (F,G). Anterior is to the left and views are primarily lateral.
The functional relationship of Upd to Hop and Stat92E is not obvious from the pattern of upd expression. Hop and Stat92E are both maternally required and expressed throughout the blastoderm embryo. In contrast, upd is required zygotically, consistent with its segmental expression and lack of maternal effect phenotype. One possible model is that the hop and stat92E maternal gene products regulate the expression of zygotic upd. This was examined by analyzing the expression pattern of upd in embryos derived from either hop or stat92E germ-line clones. The blastoderm expression pattern of upd was unaltered in these embryos (data not shown) suggesting that Upd acts either upstream or in parallel to the JAK/STAT pathway.
The upd gene encodes a predicted extracellular protein
Sequencing of the largest recovered upd cDNA has identified an ORF that could encode a peptide of 46.8 kD (Fig. 4). The conceptual translation product is similar to only one other identified protein, the Om(1E) protein of Drosophila ananassae, a very closely related species (Juni et al. 1996). The proteins from the two species are ∼90% identical over two-thirds of their length (Fig. 4B) but diverge at the termini. Little is known about the developmental function of the Om(1E) gene, except that overexpression of the gene causes defects in the adult fly. In the developing eye imaginal disc, expression of Om(1E) causes overproliferation of cells. Also, overexpression in the developing wing imaginal disc causes ectopic bristles, outstretched wings, and deformations of the notum. Interestingly, these phenotypes are identical to defects caused by overexpression of the hop gene in Drosophila melanogaster (Harrison et al. 1995), thus lending further support to the hypothesis that upd is a component of JAK signaling in flies. The only striking structural features of the Om(1E) protein are a potential signal sequence at the amino terminus and several sites for possible amino-linked glycosylation, both of which suggest that the protein is extracellular. These features are also present in D. melanogaster Upd (Fig. 4). A simple model for the role of such a protein in JAK signaling is that it codes for a ligand that activates the JAK pathway.
In addition to the ORF, the cDNA contains 226-bp 5′ and 734-bp 3′ [excluding poly(A)] UTRs. Interestingly, these UTRs contain multiple copies of two sequence elements that have been shown to decrease mRNA stability. In the 3′ UTR there are four dispersed ATTTA motifs, which are found in a number of cytokines, lymphokines, and proto-oncogenes (Shaw and Kamen 1986). In addition, a A/TTTGTA motif that was identified in the pair-rule gene ftz (Riedl and Jacobs-Lorena 1996) is present in two copies in the 3′ UTR and one copy in the 5′ UTR of upd. These elements could contribute to the rapidly changing pattern of upd mRNA accumulation found in the early embryo.
Upd is a glycoprotein
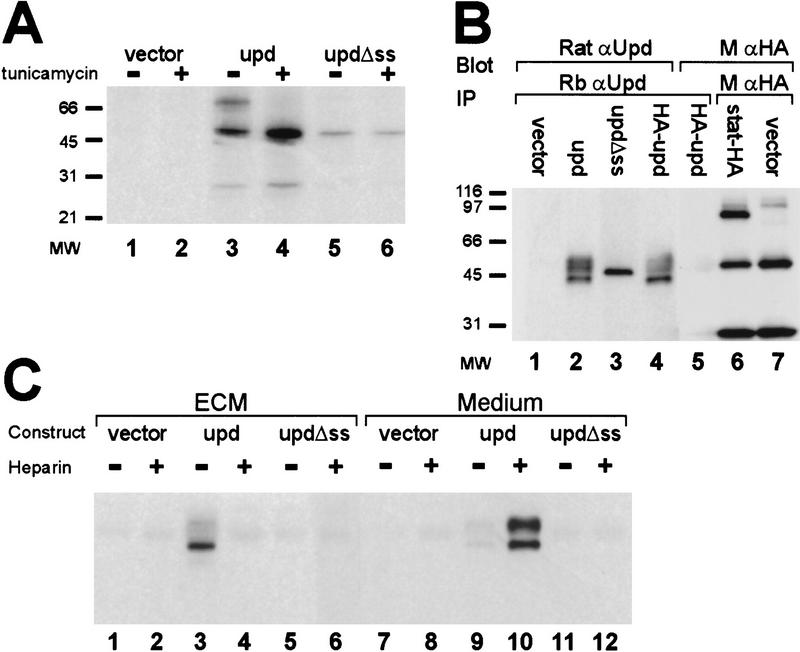
Examination of the upd transcript reveals the presence of three putative translation initiation codons. The first (at nucleotide 219) is unlikely to be used because it could only encode a peptide of 38 amino acids. The other two, at nucleotides 227 and 668, would result in proteins sharing the same reading frame but differing in their amino termini. To determine which translation initiation site is preferred in vivo, Drosophila Schneider 2 (S2) cells were transfected with a plasmid representing the entire upd transcript followed by immunoprecipitation and detection of Upd protein by use of antibodies directed against the common portion of the two putative Upd protein products. The only bands detected were 45 kD and larger (as seen in Fig. 5B, lane 2), suggesting that the translation initiation codon at nucleotide 227 is used, resulting in a protein with a putative signal sequence.
Figure 5.
Modification and localization of Upd. (A) The size of Upd protein was determined in the presence and absence of tunicamycin. Transfected constructs are empty vector (lanes 1,2), upd cDNA (lanes 3,4), and an amino-terminal truncation of upd that removes the predicted signal sequence (lanes 5,6). (B) The possible cleavage of Upd at the signal sequence was investigated by use of an amino-terminal HA epitope tag. Cells were transfected with empty vector (lane 1), upd cDNA (lane 2), amino-terminal truncation of upd that removes the signal sequence (lane 3), and upd cDNA tagged with a HA epitope at the amino terminus (lanes 4,5). Protein was immunoprecipitated with rabbit α-upd, electrophoresed, blotted, and detected with rat α-upd (lanes 1–4) or with α-HA (lane 5). A control shows HA antibody detection of HA-tagged Stat92E protein (lane 6) HA expression vector background (lane 7). (C) Cells were transfected with vector alone (lanes 1,2,7,8), upd cDNA (lanes 3,4,9,10), or upd missing the signal sequence (Δss) (lanes 5,6,11, 12). Upd protein was recovered from either the ECM (lanes 1–6) or the medium (lanes 7–12), in the presence or absence of heparin, by immunoprecipitation with rabbit α-upd and detection with rat α-upd.
The presence of a putative signal sequence and five amino-linked glycosylation sites in upd suggests that it is post-translationally modified in the secretory pathway. To examine the possibility of amino-linked glycosylation, a mammalian expression plasmid containing upd was transfected into human 293T cells. The cells were metabolically labeled with [35S]methionine in the presence or absence of tunicamycin, a potent inhibitor of amino-linked glycosylation. Upd protein was recovered by immunoprecipitation, and the sizes of resultant Upd proteins were compared. Several bands of 45–65 kD were seen in the untreated cells, whereas only the smallest 45-kD band was observed when the cells were treated with tunicamycin (Fig. 5A). The absence of the larger products in the treated cells suggests that the larger proteins are glycosylated forms. The presence of multiple forms of Upd in the untreated cells most likely reflects partially glycosylated intermediates. These results suggest that Upd contains a functional signal sequence that targets Upd to the endoplasmic reticulum for glycosylation and secretion. Therefore, removal of the putative signal sequence should eliminate targeting of the Upd protein to the endoplasmic reticulum and prevent glycosylation. As predicted, when the signal sequence was removed from Upd, only the 45-kD band was observed (Fig. 5A, lane 5).
Upd is localized to the extracellular matrix
Comigration of the smallest Upd protein species with upd lacking a signal sequence suggests that the signal sequence is normally cleaved in the mature protein. To examine cleavage of the signal sequence, a plasmid was constructed in which a hemagglutinin (HA) epitope tag was fused to the amino terminus of Upd. Previous work has shown that amino-terminal extensions on signal sequences do not abrogate in vivo function (Rottier et al. 1987). The plasmid encoding the tagged Upd was transfected into S2 cells and the resulting protein was precipitated with anti-Upd antisera. The precipitated protein was then detected by Western blotting by use of either anti-Upd or anti-HA antisera. As seen in Figure 5B, the protein was detectable with the anti-Upd serum (lane 4), but not with the anti-HA antisera (lane 5). Activity of the anti-HA antibody is shown in lane 6, where it is capable of detecting epitope-tagged Stat92E protein. These results suggest that the amino terminus of Upd, including the HA tag, is proteolytically removed.
The presence of modifications to Upd that take place in the secretory pathway are consistent with the hypothesis that Upd is normally secreted. However, in initial experiments, Upd could not be detected in the medium of transfected cells despite its high level of expression. Therefore, to determine the localization of Upd, culture medium and extracellular matrix (ECM) were harvested separately from upd-transfected S2 cells and tested for the presence of Upd. To avoid contamination of the ECM fraction with cells and cell fragments, culture dishes were washed extensively prior to harvesting. The majority of Upd protein was found to be associated specifically with the ECM, with only a small quantity free in the medium (Fig. 5C).
Binding of proteins to the ECM is often mediated by glycosaminoglycans, such as heparan sulfate. Therefore, free heparin was added to the medium from upd-transfected cells to determine whether it could prevent association of Upd with the ECM. Lanes 4 and 10 of Figure 5C show that addition of heparin releases nearly all of the Upd protein into the medium. Ability of heparin to compete with ECM suggests that Upd normally binds to the ECM through association with glycosaminoglycans. As expected, Upd lacking a signal sequence is not secreted and cannot be detected in either the medium or ECM (Fig. 5C, lanes 5,6,11,12).
Upd can activate hopscotch in Drosophila cells
The genetic and molecular data regarding upd and its relationship to Hop and Stat92E are all consistent with the predicted role of Upd as a ligand that activates the JAK signaling cascade. To directly investigate this hypothesis, upd was expressed in Drosophila cells, which were then assayed for tyrosine phosphorylation of Hop. The cell line chosen for this experiment is the Clone 8 (Cl.8) line, derived from developing wing imaginal disc (Peel and Milner 1992). For cells to respond to Upd by phosphorylating Hop, we hypothesize that some transmembrane receptor would be required to bind Upd by an extracellular domain, and be associated with Hop on the intracellular domain. As no such receptor has yet been identified in flies, we chose cells derived from a tissue known to be responsive to such a signal. It has been shown that Om(1E) overexpressed in the wing disc causes defects (Juni et al. 1996), suggesting a receptor for that Upd homolog must be present in the wing discs of D. ananassae. Thus, the D. melanogaster wing disc-derived Cl.8 cell line seemed a likely candidate to express a receptor for Upd.
To show Upd-dependent tyrosine phosphorylation of Hop, anti-Hop immunoprecipitates from upd-transfected cells were prepared and tested for reactivity with the anti-phosphotyrosine antibody 4G10. As shown in Figure 6, whereas Hop protein is detectable in all samples (middle), Hop is tyrosine phosphorylated only in immunoprecipitates prepared from upd-transfected cells (top, lane 2). Transfection of cells with upd lacking a signal sequence does not result in Hop phosphorylation (lane 3), consistent with the notion that Upd is required extracellularly for signaling to occur. Expression of transfected upd, as well as lack of endogenous upd expression, is shown at the bottom.
Figure 6.
Activation of Hop by Upd. The level of tyrosine phosphorylation on Hop is shown for Cl.8 cells transfected with upd or cocultured (CC) with S2 cells transfected with upd. Lane 1 shows the endogenous levels of Hop protein (middle), tyrosine-phosphorylated Hop (top), and Upd protein (bottom) in Cl.8 cells that were transfected with vector alone. When transfected with full-length Upd, Hop phosphorylation is stimulated (lane 2), whereas deletion of the Upd signal sequence eliminates Hop activation (lane 3). Similarly, S2 cells transfected with vector alone (lane 4) fail to activate Hop, whereas Cl.8 cells cocultured with S2 cells transfected with full-length Upd (lane 5) display activation of Hop.
To further prove that extracellularly provided Upd is necessary and sufficient to observe Hop phosphorylation, Cl.8 cells were cocultured with S2 cells transiently transfected with upd. After thorough removal of the nonadherent S2 cells, Hop immunoprecipitates were prepared from Cl.8 cell lysates and analyzed. Hop phosphorylation was only seen when Cl.8 cells were cultured in the presence of upd-transfected S2 cells (Fig. 6, lane 5). Upd protein produced by transfected S2 cells is shown in lane 4, bottom. Identical results were obtained when Cl.8 cells were grown in the presence of conditioned medium taken from upd-transfected 293T cells (not shown). These data are consistent with our hypothesis that Upd is an extracellular ligand that binds a membrane-bound receptor to activate the JAK signaling pathway.
Discussion
In this report, we have characterized Drosophila Upd as a ligand for the JAK/STAT signaling pathway during embryogenesis. We show that upd mutations are associated with phenotypes identical to mutations in hop and stat92E, Drosophila JAK and STAT, respectively. upd encodes a 47-kD core protein that is glycosylated and secreted. Secreted Upd is tightly associated with the ECM and can be released by heparin. The secreted Upd protein is able to stimulate the JAK pathway, as measured by the activation of Hop tyrosine phosphorylation in cultured cells. This leads us to propose that Upd is the embryonic ligand for the JAK pathway (Fig. 7).
Figure 7.

Model for JAK pathway activity in embryogenesis. We propose that Upd is the ligand for stimulation of the JAK pathway in early embryonic development. Upd protein is produced in a restricted set of embryonic cells, in which it is glycosylated and secreted, and diffusion is restricted by association with the ECM. Through binding of Upd to a yet unidentified receptor, the Hop JAK is stimulated, resulting in phosphorylation of Stat92E. Ultimately, transcription of specific pair–rule genes, such as eve, is activated (see text for details).
Upd is a secreted protein
The secreted nature of Upd, and its ability to stimulate JAK phosphorylation strongly suggest a model in which Upd encodes a ligand for the JAK/STAT pathway. Further, Upd is associated with the ECM, thus limiting diffusion and providing spatial restriction to pathway activity. Whereas a role for Upd as a ligand for the JAK pathway is the simplest explanation for these data, other models cannot be excluded. For example, it is possible that Upd is required to activate the true ligand for the pathway. However, Upd has no homology to proteases or other molecules that may be involved in such activation. Alternately, Upd could activate another signaling pathway that ultimately results in activation of Hop. The latter model seems unlikely because of the short time window between the onset of upd expression and the onset of eve stripes three and five expression in the embryo. Only minutes separate the zygotic expression of upd and eve, thus providing little time for the activation of an intermediate pathway. For these reasons, we favor the simplest model, in which Upd is the ligand for the JAK pathway, as diagrammed in Figure 7. Upd presumably binds to a yet unknown receptor that would associate with Hop. Receptor aggregation would stimulate Hop trans-phosphorylation, thereby activating Hop. The Stat92E latent transcription factor would then be recruited to the activated receptor complex, phosphorylated by activated Hop, dimerize, and translocate to the nucleus. The active dimers of Stat92E presumably then directly stimulate transcription of a set of downstream genes, including eve, in a spatially restricted manner. Previously, a regulatory fragment of the eve gene that controls expression of the third stripe was identified as a target of the JAK/STAT pathway. This fragment carries sequences similar to binding sites for mammalian STATs, which bind the Stat92E protein in vitro (Yan et al. 1996b). As proposed previously (Hou et al. 1996; Yan et al. 1996b), we assume that the effects of the pathway on eve expression arise from direct binding of Stat92E to eve enhancers, resulting in activation of transcription of the gene. Regulation of other pair-rule genes may be equally direct, though it is known that Eve protein also regulates transcription of other pair-rule genes. Thus, it is possible that effects of JAK pathway mutations on some of the pair-rule genes are secondary to a reduction in Eve activity.
The molecular nature of Upd is consistent with the previously reported local noncell autonomy associated with upd mutations (Gergen and Wieschaus 1986). Gergen and Wieschaus generated embryonic mosaics of upd, whereby patches of upd mutant cells were adjacent to wild-type territory. From their analyses they concluded that upd is largely cell autonomous, but they noticed that upd mutant cells immediately adjacent to the wild-type cells could generate a normal pattern. The requirement for close proximity of the rescuing cells indicates that the diffusible activity of the Upd product has a limited range. This is consistent with the tight association of the Upd protein with the ECM, tethering the ligand near its source. Anchoring of Upd to the ECM would provide a simple mechanism to spatially limit the activity of the protein and consequent activation of the JAK/STAT pathway. This situation is in contrast to other secreted ligands presumably found in the perivitelline space at about the same developmental stage. For example, the ligands of the Toll and Torso receptors have the ability to freely diffuse over long distances (Stein and Nüsslein-Volhard 1992; Casanova and Struhl 1993).
The role of Upd in embryonic segmentation
Extensive studies of the mechanisms underlying segmentation of the Drosophila embryo have led to a comprehensive model of the sequential steps involved in the establishment of the body plan (Ingham 1988; St Johnston and Nüsslein-Volhard 1992). Maternal activities have been shown to provide cues that direct the expression of specific factors along the anteroposterior (A/P) and dorsoventral axes. Along the A/P axis, these transcription factors, encoded by gap genes, regulate the spatial and temporal expression of other transcription factors encoded by pair-rule genes, which are subsequently involved in the regulation of segment polarity genes that define cell states within every segment. Upd in the JAK/STAT pathway is the earliest known secreted zygotic factor to act along the A/P axis. With the exception of a tenascin-like molecule that acts as a secondary pair–rule gene (Baumgartner et al. 1994), the next known secreted factors involved in A/P patterning are encoded by the segment polarity genes wingless (wg) and hedgehog (hh). Both Wg and Hh encode signaling molecules involved in the establishment and maintenance of the parasegmental boundaries of the developing embryo (for review, see Ingham 1988). Their expression is initiated after cellularization and they are required for maintenance of boundaries throughout embryogenesis.
Our findings that a secreted factor, Upd, activates the JAK/STAT signaling pathway to regulate the expression of pair rule genes such as eve, ftz, and runt, must be integrated into this model. To understand the function of the JAK/STAT pathway, we have used the regulation of eve stripe 3 expression as a paradigm (for review, see Hou and Perrimon 1997). The anterior and posterior borders of eve stripe 3 are set through repression by the Hb and Kni proteins, respectively. Because the Hb and Kni-binding sites are involved in defining the sharp on/off borders of gene expression, and the STAT-binding sites are involved in activating transcription, we have proposed that the JAK/STAT signaling pathway does not need to be spatially activated within this domain (Hou et al. 1996; for review, see Hou and Perrimon 1997). Thus, the JAK/STAT pathway may not be instructive per se but simply play a role in the up-regulation of specific genes, such as eve, that contain STAT binding sites in their regulatory regions. According to this model, the unusual mutant phenotype associated with loss of JAK/STAT activity would represent the cumulative defects in the expression of various segmentation genes.
Our analysis in the early embryo has shown that upd is initially expressed in a broad central domain immediately prior to cellularization, the stage when eve is regulated by the JAK/STAT pathway. This is consistent with the previously proposed model that the pathway is not instructive, but provides a ubiquitous system of activation. Later restriction of upd expression is somewhat enigmatic. The striped expression pattern of upd, together with the observations that Upd activity is mainly cell autonomous and that Upd binds to ECM suggest that the JAK/STAT pathway is locally activated in the blastoderm embryo. This suggests that perhaps, in contrast to the previous model, localized activation of this pathway is instrumental to proper segmentation. Analysis of Upd protein distribution and misexpression studies of Upd in the early embryo should provide critical tests to distinguish between these alternative hypotheses.
Specificity of the JAK pathway
In vertebrates, the JAK/STAT pathway can be activated by multiple ligand/receptor combinations. In addition to cytokine receptors, receptor tyrosine kinases such as the PDGFR, EGFR, and Eyk have been shown to regulate the activity of both JAK and STAT molecules (Vignais et al. 1996; Zong et al. 1996; Yamauchi et al. 1997). If the JAK/STAT pathway was activated by additional signaling pathways, one would expect that the phenotype of either hop or stat92E embryos would be more severe than the phenotype of upd mutant embryos. Because hop, stat92E, and upd mutants have similar phenotypes, we conclude that Upd is likely to be the predominant or sole activator of the JAK/STAT pathway during embryogenesis.
The JAK pathway in Drosophila has additional roles during zygotic development. The pathway is involved in blood cell development, differentiation of various adult structures, posture of the adult wing, spermatogenesis, and oogenesis (this work, Perirmon and Mahowald 1986; Harrison et al. 1995; Luo et al. 1995; Yan et al. 1996a). The genetic interaction studies described here suggest a role for Upd in adult wing posture and in oogenesis, but roles in other developmental processes have not yet been investigated. Further analyses will be required to determine whether Upd is the only ligand in all these tissues, or whether other activators can also stimulate the JAK/STAT pathway.
Upd and cytokine signaling
The list of secreted molecules that activate the JAK pathway in mammals has continued to grow over the past several years. Especially for cytokines, both the ligands and their receptors have been determined. This is not so for Drosophila; to date, no ligands or receptors involved in activation of JAK signaling have been identified. Thus, our characterization of Upd provides the first description of the molecules that utilize the JAK pathway in invertebrates.
The lack of sequence similarity between Upd and any known vertebrate activators is intriguing. But like cytokines, the predicted secondary structure for Upd includes several stretches of α-helical regions. Whereas Upd has no sequence homology with cytokines, perhaps the α-helices fold into a structure reminiscent of cytokines. There is precedent among mammalian ligands for structural similarities in molecules that do not have sequence homology with type I cytokines (e.g., Zhang et al. 1997). These molecules also utilize JAKs for signal transduction. Although there may not have been strong evolutionary conservation of specific sequences, there may be conservation of general structure in ligands because of functional requirement for binding to the appropriate receptor. On the other hand, some aspects of Upd structure are less consistent with a cytokine-type molecule. First, the Upd protein is extremely basic, with a predicted pI of nearly 12. Second, in contrast with many cytokines, Upd is associated with the ECM, which may limit the range of activity of the ligand. These characteristics suggest that Upd may be representative of a new family of ligands that stimulates the JAK pathway. Determination of the relationship between Upd and other JAK activating ligands must wait for the identification of Upd homologs in other species. Finally, characterization of the Upd receptor will reveal whether Upd signals through a conventional receptor molecule or whether it defines a novel mechanism by which the JAK/STAT pathway becomes activated.
Materials and methods
Fly stocks
The upd alleles used to examine embryonic lethality are Df(1)os1A, updYM55, and updYC43 (Wieschaus et al. 1984; Eberl et al. 1992). os1, oso, and oss are described previously (Lindsley and Zimm 1992). Additional os alleles (os4-51-1, osc18, osN1, and os109) were generated in an F1 screen by use of X-rays (N. Perrimon, unpubl.). We use the name upd in this work rather than os because it more accurately describes the null phenotype of the gene we have characterized.
Analysis of embryos
Embryos were collected from y w and Df(1)os1A/ FM7, and from females with germ-line mutant clones for hopc111 or stat92EP1681 generated as described previously (Binari and Perrimon 1994; Hou et al. 1996). Embryonic cuticles were prepared by the Hoyer’s mountant method (Ashburner 1989) and photographed by dark-field microscopy.
Localization of RNA by in situ hybridization and protein by immunohistochemical staining in y w and mutant strains updYM55, updYC43, and Df(1)os1A was performed as described previously (Binari and Perrimon 1994; Hou et al. 1996). Stained embryos were mounted in 70% glycerol and photographed by use of differential interference contrast (DIC) microscopy.
Molecular identification of upd
The limits of the upd locus were determined by Southern analysis of upd and os alleles. The distal limit of upd is based on the position of the breakpoint in Df(1)os4-51-1 at ∼370–380 kb in the Shaker complex chromosomal walk (see Fig. 2). The proximal limit was mapped by the position of breakpoints in the segmental aneuploidy, T(1;Y)v29, and Df(1)osUE69 (Ferrus et al. 1990; Eberl et al. 1992) at ∼410–415 kb. Restriction fragment length polymorphism (RFLP) analysis of recombinants between Df(1)osUE69 and porcPB16 is consistent with this mapping (N. Perrimon, D.A. Harrison, and E. Wilder, unpubl.).
DNA from λ phages M1I1, M33, E721, and EA1 were used to probe Northern blots of RNA from embryonic stages (0–1, 1–5, 5–9, and 9–13 hr). A 4-kb BamHI fragment was subcloned from the M33 phage (pUpd9-4), and labeled by random priming to probe cDNA libraries. Libraries used were a 4- to 8-hr embryonic cDNA library cloned into a plasmid (Brown and Kafatos 1988) and a 8- to 12-hr embryonic cDNA library cloned in λ phage (Zinn et al. 1988). A single clone was recovered from the phage library, whereas three clones were recovered from the plasmid library. All three plasmid clones were identical, whereas the phage clone contained a slightly longer 5′ end. Sequencing of clones was performed with the Sequenase and Thermo-Sequenase kits (Amersham/ U.S. Biochemical). Sequence from cDNAs was compared with that of the λM33 genomic clone. DNA from flies with the EMS-induced updYM55 and updYC43 alleles carried over FM7 was PCR amplified and sequenced to identify the specific lesions associated with those mutations.
Detection of glycosylation
A 761-bp SacI–EcoRI fragment of the upd cDNA clone pBS-GR51 was cloned into the SacI–EcoRI sites of pRSETB (Invitrogen) to encode a fusion of His-tagged protein with the carboxyl terminus of Upd. The fusion protein was produced and prepared as described previously (Harrison et al. 1995), and was injected into both rats and rabbits to stimulate an immunological response.
293T cells in 60-mm plates were transfected with empty or upd-containing plasmids under the control of the CMV promoter in the vector pCG (Natesan et al. 1997) by use of lipofectamine (Life Technologies) according to manufacturer’s instructions. The following day, cells were metabolically labeled by the addition of 70 μCi/plate of Express labeling mix (Dupont-NEN) for 6.5 hr. Plus tunicamycin samples were treated with 3 μg/ml tunicamycin for the same time period. Cells were lysed in RIPA buffer [150 mm NaCl, 50 mm Tris (pH 8.0), 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm EGTA] plus 0.5 mm PMSF, 20 μg/ml aprotinin solution (Sigma), 3 μg/ml leupeptin, 3 μg/ml pepstatin and spun. Upd protein was immunoprecipitated with rabbit anti-upd antibody and collected and washed after the addition of Gammabind G–Sepharose (Pharmacia). Eluted immunoprecipitates were run on 4%–20% SDS–polyacrylamide Novex gels that were fixed, treated with Amplify (Amersham), and dried prior to autoradiography.
Protein localization
Schneider cells were transfected in 60 mm plates with constructs containing upd in the pD vector (Natesan and Gilman 1995) or empty pD vector by calcium phosphate coprecipitation (Di Nocera and Dawid 1983). HA-tagged constructs contained the sequence YPYDVPDYA at the extreme amino terminus (HA–upd) or carboxyl terminus (Stat92E-HA) of the ORF. In cases in which medium was assayed for the presence of upd protein, cells were refed with Sf-900 II Serum-Free Medium (Life Technologies) the day after transfection with or without the addition of 50 μg/ml heparin (Sigma H-9399). A day later, medium was collected by spinning out cells, and concentrated in Centriprep 10,000 MWCO ultrafiltration devices (Amicon). Transfected cells were harvested 2 days after transfection by washing twice in ice-cold PBS followed by lysis in RIPA buffer plus protease inhibitors prepared as above. To prepare ECM, plates were scraped and washed with 2 m urea followed by several washes with PBS prior to harvesting with 90°C Laemmli sample buffer (Bradley and Brown 1990).
For immunoprecipitation of Upd from cell lysates, equal amounts of protein were used for all samples. Protein concentrations in medium and ECM were below detection limits, and for these samples, equivalent volumes of medium or number of plates were used for comparisons. Immunoprecipitations were performed as above with rabbit anti-Upd or mouse anti-HA monoclonal antibody (BabCO). Eluted immunoprecipitates were run on 4%–20% Novex gels and blotted to PVDF membranes (Bio-Rad). Detection of untagged Upd proteins was with rat anti-Upd antibody, and of HA-tagged proteins, with mouse anti-HA monoclonal antibody (BABCO), diluted 1/1000 followed by HRP-conjugated secondary antibodies (Bio-Rad) diluted 1/10,000 and ECL (Dupont–NEN) detection.
Analysis of Hop phosphorylation
Clone 8 cells grown to ∼80% confluence in 60 mm dishes were either cocultured with transiently transfected Schneider cells or transfected with pD-upd or empty vector by calcium phosphate coprecipitation (Di Nocera and Dawid 1983). Schneider cells used for coculture were transfected with pD-upd in 100 mm dishes, then washed with PBS and transferred to fresh plates the next day. One day later, the transfected Schneider cells were spun down, resuspended in clone 8 medium and cocultured with clone 8 cells for 3.5 hr. The nonadherent Schneider cells and medium were removed by gentle washing and RIPA lysates of these cells for immunoprecipitations were prepared as described above.
Clone 8 cells, either cocultured or transfected 2 days previously, were washed three times with ice-cold PBS containing 2 mm sodium vanadate and scraped into either Triton X-100 lysis buffer [150 mm NaCl, 10 mm Tris (pH 8.0), 10 mm EGTA, 1% Triton X-100] or RIPA lysis buffer plus protease inhibitors and 2 mm sodium vanadate. Equal amounts of total protein were used when immunoprecipitations involving the same antibodies were carried out. upd immunoprecipitation and detection was carried out as described above. Hop protein was immunoprecipitated with rabbit anti-hop antiserum (Harrison et al. 1995). Equal volumes of the resulting immunoprecipitate were loaded into two separate lanes of 8% SDS–polyacrylamide Novex gels for electrophoresis and blotting to PVDF as described above. Hop protein was detected with rat anti-hop antiserum diluted 1/1000 (Harrison et al. 1995) or monoclonal antiphosphotyrosine antibody 4G10 (UBI) diluted 1/10,000 followed by HRP-conjugated secondary antibodies diluted 1/10,000 and ECL (Dupont–NEN) detection.
Acknowledgments
We thank O. Pongs for his generous gift of λ phages, R. Nusse for Clone 8 cells, M. Frasch for antibodies, and the Bloomington Stock Center for fly lines. We thank T. Cline and L. Sefton for sharing data prior to publication. This work was supported by fellowships from the Damon Runyon/Walter Winchell Cancer Research Fund (DRG1154) and The Medical Foundation/Charles A. King Trust (DAH), by the National Science Foundation (IBN9723944 to D.A.H.), by the Howard Hughes Medical Institute (N.P.), and by the National Institutes of Health (CA45642 to M.G.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The GenBank accession no. for the upd cDNA sequence is AF072823.
Footnotes
E-MAIL DougH@pop.uky.edu; FAX (606) 257-1717.
References
- Ashburner M. Drosophila: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Baumgartner S, Martin D, Hagios C, Chiquet-Ehrismann R. Tenm, a Drosophila gene related to tenascin, is a new pair-rule gene. EMBO J. 1994;13:3728–3740. doi: 10.1002/j.1460-2075.1994.tb06682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes & Dev. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- Bradley RS, Brown AM. The proto-oncogene int-1 encodes a secreted protein associated with the extracellular matrix. EMBO J. 1990;9:1569–1575. doi: 10.1002/j.1460-2075.1990.tb08276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Casanova J, Struhl G. The torso receptor localizes as well as transduces the spatial signal specifying terminal body pattern in Drosophila. Nature. 1993;362:152–155. doi: 10.1038/362152a0. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Di Nocera PP, Dawid IB. Transient expression of genes introduced into cultured cells of Drosophila. Proc Natl Acad Sci. 1983;80:7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Eberl D, Perkins L, Engelstein M, Hilliker A, Perrimon N. Genetic and developmental analysis of polytene section 17 of the X chromosome of Drosophila melanogaster. Genetics. 1992;130:569–583. doi: 10.1093/genetics/130.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrus A, Llamazares S, de la Pompa JL, Tanouye MA, Pongs O. Genetic analysis of the Shaker gene complex of Drosophila melanogaster. Genetics. 1990;125:383–398. doi: 10.1093/genetics/125.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen P, Wieschaus E. Localized requirements for gene activity in segmentation of Drosophila embryos: Analysis of armadillo, fused, giant and unpaired mutations in mosaic embryos. Wilhelm Roux’s Arch Dev Biol. 1986;195:49–62. doi: 10.1007/BF00444041. [DOI] [PubMed] [Google Scholar]
- Goto T, Macdonald P, Maniatis T. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57:413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- Harrison D, Binari R, Nahreini T, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Perrimon N. The JAK-STAT pathway in Drosophila. Trends Genet. 1997;13:105–110. doi: 10.1016/s0168-9525(97)01006-8. [DOI] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. marelle acts downstream of the Drosophila Hop/Jak kinase and encodes a protein similar to the mammalian Stats. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Ihle JN. STATs: Signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- Ihle JN, Nosaka T, Thierfelder W, Quelle FW, Shimoda K. Jaks and Stats in cytokine signaling. Stem Cells. 1997;15:105–111. doi: 10.1002/stem.5530150814. [DOI] [PubMed] [Google Scholar]
- Ingham PW. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988;335:25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Juni N, Awasaki T, Yoshida K, Hori SH. The Om(1E) mutation in Drosophila ananassae causes compound eye overgrowth due to tom retrotransposon-driven overexpression of a novel gene. Genetics. 1996;143:1257–1270. doi: 10.1093/genetics/143.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Wilder E, Klingensmith J, Zachary K, Perrimon N. The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes & Dev. 1996;10:3116–3128. doi: 10.1101/gad.10.24.3116. [DOI] [PubMed] [Google Scholar]
- Kilchherr F, Baumgartner S, Bopp D, Frei E, Noll M. Isolation of the paired gene of Drosophila and its spatial expression during early embryogenesis. Nature. 1986;321:493–499. [Google Scholar]
- Krah-Jentgens I. “Klonierung des Shaker-Komplexes von Drosophila melanogaster und Charakterisierung einer Transkriptionseinheit aus diesem Gen-Komplex.” Ph.D. Thesis. Hamburg, Germany: Ruhr-Universitat Bochum; 1989. [Google Scholar]
- Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard OA. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- Lindsley D, Zimm G. The genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Luo H, Hanratty W, Dearolf C. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P, Villa A, Gillani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, Shea JJ O, Vezzoni P, Notarangelo LD. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–68. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- Macdonald P, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: A second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- Natesan S, Gilman M. YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol Cell Biol. 1995;15:5975–5982. doi: 10.1128/mcb.15.11.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natesan S, Rivera VM, Molinari E, Gilman M. Transcriptional squelching re-examined. Nature. 1997;390:349–350. doi: 10.1038/37019. [DOI] [PubMed] [Google Scholar]
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Nosaka T, van Deursen JM, Tripp RA, Thierfelder WE, Witthuhn BA, McMickle AP, Doherty PC, Grosveld GC, Ihle JN. Defective lymphoid development in mice lacking Jak3. Science. 1995;270:800–802. doi: 10.1126/science.270.5237.800. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Weischaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Peel D, Milner M. The expression of PS integrins in Drosophila melanogaster imaginal disc cell lines. Wilhelm Roux’s Arch Dev Biol. 1992;201:120–123. doi: 10.1007/BF00420423. [DOI] [PubMed] [Google Scholar]
- Peeters P, Raynaud SD, Cools J, Wlodarska I, Grosgeorge J, Philip P, Monpoux F, Van Rompaey L, Baens M, Van den Berghe H, Marynen P. Fusion of TEL, the ETS-variant gene 6 (ETV6), to the receptor- associated kinase JAK2 as a result of t(9;12) in a lymphoid and t(9;15;12) in a myeloid leukemia. Blood. 1997;90:2535–2540. [PubMed] [Google Scholar]
- Perrimon N, Mahowald A. l(1)hopscotch, A larval-pupal zygotic lethal with a specific maternal effect on segmentation in Drosophila. Dev Biol. 1986;118:28–41. doi: 10.1016/0012-1606(86)90070-9. [DOI] [PubMed] [Google Scholar]
- Riedl A, Jacobs-Lorena M. Determinants of Drosophila fushi tarazu mRNA instability. Mol Cell Biol. 1996;16:3047–3053. doi: 10.1128/mcb.16.6.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93:373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- Rottier PJ, Florkiewicz RZ, Shaw AS, Rose JK. An internalized amino-terminal signal sequence retains full activity in vivo but not in vitro. J Biol Chem. 1987;262:8889–8895. [PubMed] [Google Scholar]
- Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175:314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- Sprang SR, Bazan JF. Cytokine structural taxonomy and mechanisms of receptor engagement. Curr Opin Struct Biol. 1993;3:815–827. [Google Scholar]
- St Johnston D, Nüsslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- Stein D, Nüsslein-Volhard C. Multiple extracellular activities in Drosophila egg perivitelline fluid are required for establishment of embryonic dorsal-ventral polarity. Cell. 1992;68:429–440. doi: 10.1016/0092-8674(92)90181-b. [DOI] [PubMed] [Google Scholar]
- Vignais ML, Sadowski HB, Watling D, Rogers NC, Gilman M. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol Cell Biol. 1996;16:1759–1769. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C, Jurgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Part III. Zygotic loci on the X-chromosome and fourth chromosome. Wilhelm Roux’s Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Ueki K, Tobe K, Tamemoto H, Sekine N, Wada M, Honjo M, Takahashi M, Takahashi T, Hirai H, Tushima T, Akanuma Y, Fujita T, Komuro I, Yazaki Y, Kadowaki T. Tyrosine phosphorylation of the EGF receptor by the kinase Jak2 is induced by growth hormone. Nature. 1997;390:91–96. doi: 10.1038/36369. [DOI] [PubMed] [Google Scholar]
- Yan R, Luo H, Darnell JE, Jr, Dearolf CR. A JAK-STAT pathway regulates wing vein formation in Drosophila. Proc Natl Acad Sci. 1996a;93:5842–5847. doi: 10.1073/pnas.93.12.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996b;84:421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- Zhang F, Basinski MB, Beals JM, Briggs SL, Churgay LM, Clawson DK, DiMarchi RD, Furman TC, Hale JE, Hsiung HM, Schoner BE, Smith DP, Zhang XY, Wery JP, Schevitz RW. Crystal structure of the obese protein leptin-E100. Nature. 1997;387:206–209. doi: 10.1038/387206a0. [DOI] [PubMed] [Google Scholar]
- Zinn K, McAllister L, Goodman CS. Sequence analysis and neuronal expression of fasciclin I in grasshopper and Drosophila. Cell. 1988;53:577–587. doi: 10.1016/0092-8674(88)90574-0. [DOI] [PubMed] [Google Scholar]
- Zong C, Yan R, August A, Darnell JE, Hanafusa H. Unique signal transduction of Eyk: Constitutive stimulation of the JAK–STAT pathway by an oncogenic receptor-type tyrosine kinase. EMBO J. 1996;15:4515–4525. [PMC free article] [PubMed] [Google Scholar]