Abstract
The Drosophila nucleosome remodeling factor (NURF) is a protein complex consisting of four polypeptides that facilitates the perturbation of chromatin structure in vitro in an ATP-dependent manner. The 140-kD NURF subunit, imitation switch (ISWI), is related to the SWI2/SNF2 ATPase. Another subunit, NURF-55, is a 55-kD WD repeat protein homologous to the human retinoblastoma-associated protein RbAp48. Here, we report the cloning and characterization of the smallest (38 kD) component of NURF. NURF-38 is strikingly homologous to known inorganic pyrophosphatases. Both recombinant NURF-38 alone and the purified NURF complex are shown to have inorganic pyrophosphatase activity. Inhibition of the pyrophosphatase activity of NURF with sodium fluoride has no significant effect on chromatin remodeling, indicating that these two activities may be biochemically uncoupled. Our results suggest that NURF-38 may serve a structural or regulatory role in the complex. Alternatively, because accumulation of unhydrolyzed pyrophosphate during nucleotide incorporation inhibits polymerization, NURF may also have been adapted to deliver pyrophosphatase to chromatin to assist in replication or transcription by efficient removal of the inhibitory metabolite.
Keywords: Chromatin, ISWI, NURF, pyrophosphatase, remodeling
There is now abundant evidence for a role of chromatin structure in the regulation of transcription, and indications that the organization of chromatin can also influence other chromosome activities such as recombination, replication, and DNA repair. It has been known that the wrapping of DNA on the surface of nucleosomes, the fundamental units of chromatin structure, has repressive effects on the transcriptional process (for review, see Grunstein 1990; Felsenfeld 1992; Owen-Hughes and Workman 1994; Paranjape et al. 1994; Kornberg and Lorch 1995; Pruss et al. 1995; van Holde et al. 1995; Fletcher and Hansen 1996; Koshland and Strunnikov 1996; Hartzog and Winston 1997; Ramakrishnan 1997). It is only recently, however, that advances have been made in identifying the underlying mechanisms by which nucleosomes are altered to permit the initiation of transcription. Such mechanisms include the intrinsic resistance of certain DNAs to curvature, post-translational modification of nucleosomal histone proteins, and ATP-dependent remodeling of nucleosome structure (for review, see Becker 1994; Felsenfeld 1996; Kingston et al. 1996; Tsukiyama and Wu 1997; Wu 1997).
Recent work has led the discovery of several activities that apparently use the hydrolysis of ATP in the remodeling of chromatin templates. The yeast SWI/SNF complex facilitates the binding of GAL4 derivatives to nucleosomal DNA (Owen-Hughes et al. 1996) and possesses the ability to cause a local perturbation of chromatin structure in an ATP-dependent manner (for review, see Henikoff 1993; Carlson and Laurent 1994; Peterson and Tamkun 1995; Peterson 1996). The SWI2/SNF2 gene itself was identified in genetic screens as a positive regulator of a small class of yeast genes, and encodes a protein homologous to DNA-stimulated ATPases/DNA helicases (Laurent et al. 1993; Cairns et al. 1994; Côté et al. 1994). SWI2/SNF2 homologs exist in both flies (brahma) and humans (BRG1, HBRM) and these are also present in large multisubunit protein complexes (Tamkun et al. 1992; Khavari et al. 1993; Muchardt and Yaniv 1993; Elfring et al. 1994; Dingwall et al. 1995; Tsukiyama et al. 1995). RSC (remodeling the structure of chromatin) is another yeast protein complex related to and with similar properties as SWI/SNF (Cairns et al. 1996). These complexes are different, however, in that RSC is necessary for cell viability and appears to be more abundant than SWI/SNF.
Biochemical studies in our laboratory have identified an ATP-dependent chromatin remodeling complex present in Drosophila embryo extracts, designated the nucleosome remodeling factor (NURF), which facilitates a local, transcription factor-mediated perturbation of chromatin structure within a nucleosomal array (Tsukiyama et al. 1994; Tsukiyama and Wu 1995). NURF is composed of four subunits with molecular masses of 215, 140, 55, and 38 kD assembled in a native complex of ∼500 kD (Tsukiyama and Wu 1995). The 140-kD subunit has been identified as imitation switch (ISWI), a member of the SWI2/SNF2 family that is characterized by its highly conserved ATPase domain (Tsukiyama et al. 1995). The hydrolysis of ATP during the remodeling of chromatin is likely to be mediated by ISWI. Unlike the SWI/SNF complex, the ATPase activity of NURF is stimulated by the presence of nucleosomes rather than by free DNA (Tsukiyama and Wu 1995), suggesting that NURF recognizes both the protein and DNA components of the nucleosome. The interaction between NURF and nucleosomes appears to involve the flexible histone tails that protrude from the surface of the nucleosome (Georgel et al. 1997). ISWI has been shown recently to be present in at least two other protein complexes from Drosophila that affect chromatin structure in a related but distinct manner—ACF (ATP-utilizing chromatin assembly and remodeling factor; Ito et al. 1997) and CHRAC (chromatin accessibility complex; Varga-Weisz et al. 1995, 1997). The presence of ISWI as a common subunit indicates that this ATPase may serve as the energy-transducing component of chromatin-remodeling machines.
The 55-kD subunit of NURF was found to be identical to the 55-kD subunit of dCAF-1, a complex that facilitates the assembly of chromatin in vitro (Tyler et al. 1996; Martínez-Balbás et al. 1998). A human counterpart of NURF-55, RbAp48, was originally identified as a retinoblastoma binding protein in vitro (Qian et al. 1993), and is present in several distinct complexes that affect histone metabolism—the human CAF-1 complex (Verrault et al. 1996) and mammalian histone deacetylase complexes (Taunton et al. 1996; Hassig et al. 1997; Zhang et al. 1997). RbAp46 (a smaller variant of RbAp48), was demonstrated to be a subunit of the human Hat1 histone acetyltransferase (HAT) complex, and was shown to enhance the ability of the Hat1 complex to bind its histone substrates, thereby increasing its activity (Verreault et al. 1998). The counterparts of NURF-55 in yeast, Hat2p and MSI1p, are members of a HAT complex (Parthun et al. 1996) and the yeast CAF-1 complex (Kaufman et al. 1997), respectively. Overall, the results suggest that this conserved subfamily of WD repeat proteins may assist targeting of protein complexes to chromatin.
To investigate further how the NURF complex facilitates the perturbation of chromatin structure, we set out to identify the remaining components of the complex. We report here the identification and characterization of the smallest NURF subunit, NURF-38. Our studies reveal that the NURF-38 protein is an inorganic pyrophosphatase (PPase), an enzyme essential for driving critical biosynthetic reactions including transcription, replication, and DNA repair. Although this activity appears to be dispensable for the remodeling of plasmid chromatin in vitro, the retention of PPase activity in the NURF complex suggests that NURF may have a broader function in the cell nucleus than was indicated previously.
Results
The NURF-38 gene encodes a putative inorganic PPase
NURF 38 is the smallest component among the four major polypeptides that copurify at or near stoichiometric levels with nucleosome remodeling activity (Tsukiyama and Wu 1995). We obtained peptide sequences for the NURF-38 protein by in-gel endopeptidase digestion and Edman degradation sequencing as described previously (Tsukiyama and Wu 1995; Martínez-Balbás et al. 1998). The sequences of three peptides were used to design primers for a PCR-based screen of a 0- to 12-hr Drosophila melanogaster embryonic cDNA library. Two primer pairs generated a 235-bp and a 711-bp fragment, respectively; sequencing of the cloned fragments revealed that the smaller fragment was contained within the larger.
Using the 711-bp fragment as a probe, we obtained two independent genomic clones and four independent cDNA clones. One of the cDNA clones was presumed to contain the entire open reading frame (ORF) based on the presence of a stop codon in the 5′-transcribed, untranslated region and a poly(A) stretch at the 3′ terminus. Figure 1A shows the organization of the NURF-38 gene as deduced from the genomic and cDNA sequences. We determined that NURF-38 is a single-copy gene that resides in subdivision 60 C on the right arm of chromosome 2 (data not shown).
Figure 1.
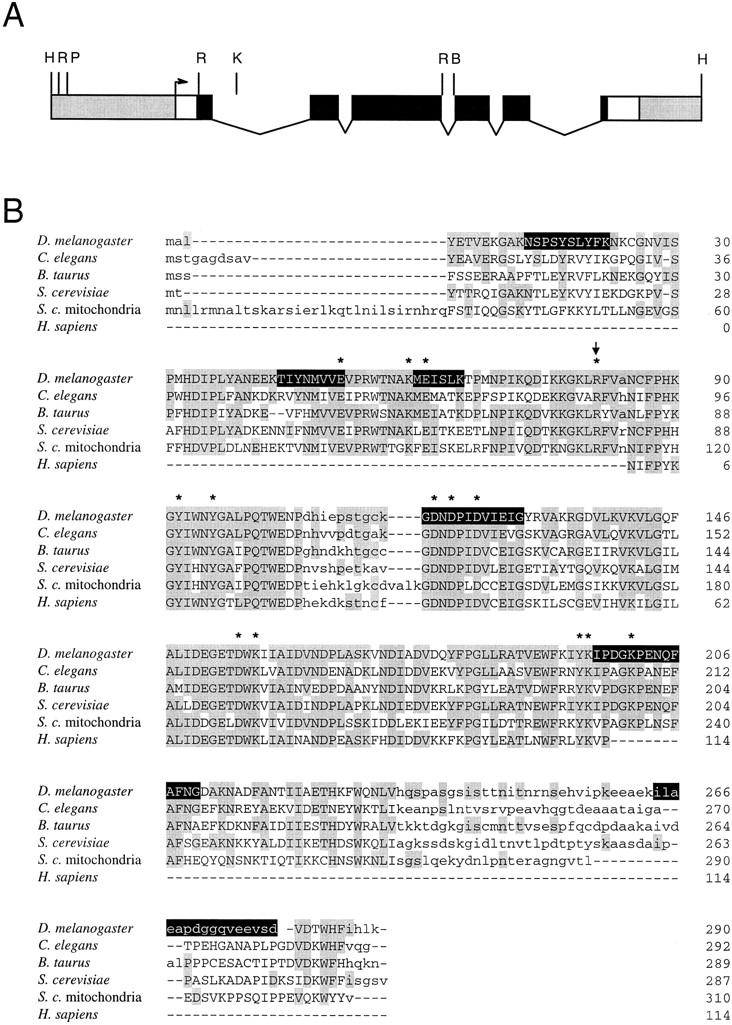
Cloning and identification of the NURF-38 protein. (A) Structure of the NURF-38 gene. Untranscribed sequences are shaded. The presumptive transcription start site is shown as an arrow. (Open boxes) Transcribed, untranslated sequences; (solid boxes) the coding region of the gene; (thin lines) introns. (B) BglII; (H) HindIII; (K) KpnI; (P) PstI; (R) EcoRI. (B) The predicted primary sequence of NURF-38 is shown aligned with eukaryotic inorganic pyrophosphatases. Uppercase letters designate conserved blocks; amino acids identical to those in the Drosophila protein are shaded. Invariant residues, known by crystallographic studies to be in the active site of the S. cerevisiae enzyme, are marked by asterisks. Arginine 81 is also marked with an arrow (see text). Amino acid sequences obtained by endopeptidase digestion of the protein and Edman degradation of purified peptides are highlighted in black.
All peptides for which we obtained amino acid sequence data were contained within the predicted protein sequence (containing 290 residues with a calculated molecular weight of 32.7 kD), including those not used in primer design (Fig. 1B). A BLAST search (Altschul et al. 1990) of existing databases revealed that NURF-38 had extensive amino acid sequence identity with inorganic PPases (Fig. 1B). Overall, this identity can be as high as 53.9%; when the central portion of the PPase sequence is examined, the identity increases to ∼70% for yeast, Drosophila, nematode, and mammalian proteins. Most striking is the absolute conservation of the 14 amino acid residues determined by crystallographic studies to be in the active site of the Saccharomyces cerevisiae enzyme (Cooperman et al. 1992 and references therein). These include an invariant arginine (arginine-78 in S. cerevisiae, arginine-81 in D. melanogaster) also shown by chemical modification to be necessary for catalysis (Bond et al. 1980).
NURF-38 is a subunit of the NURF complex
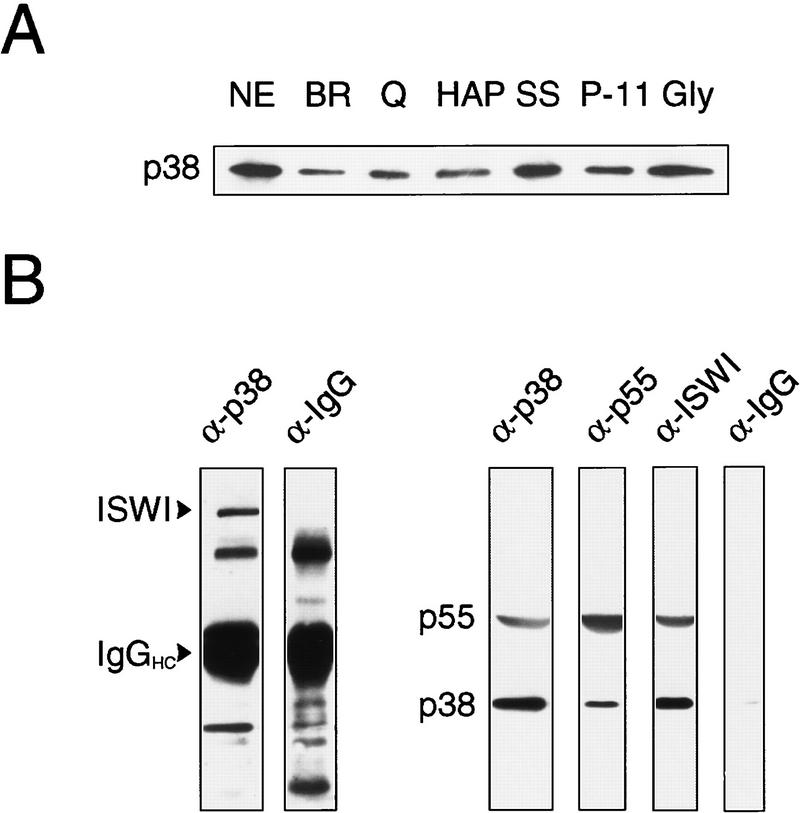
To evaluate whether NURF-38 is an integral subunit of the NURF complex, we assayed for the presence of NURF-38 protein in the peak fractions of NURF activity from each step of the purification protocol. As revealed by Western blotting using antibodies raised against bacterially expressed NURF-38 protein, approximately equivalent amounts of NURF-38 can be found in each peak fraction (Fig. 2A). We also performed co-immunoprecipitation experiments to demonstrate independently physical interactions among the NURF polypeptides. Affinity-purified antibodies against two NURF subunits identified previously, ISWI and NURF-55, were capable of immunoprecipitating NURF-38 from a crude NURF-containing fraction, whereas a mock precipitation using purified chicken IgG failed to show immunoprecipitation (Fig. 2B). Conversely, affinity-purified antibodies against NURF-38 were able to immunoprecipitate both NURF-55 and ISWI. Similar results were obtained in experiments performed with unfractionated embryo extracts (data not shown).
Figure 2.
NURF-38 is a subunit of NURF. (A) NURF-38 copurifies with active NURF fractions. The Western blot shows samples of chromatographic fractions containing approximately equivalent NURF activity (the peak fractions of the representative chromatographic columns). (NE) Nuclear extract; (BR) DEAE cellulose/Bio-Rex 70; (Q) Q Sepharose; (HAP) hydroxylapatite; (SS) single-stranded DNA cellulose; (P11) cellulose phosphate P-11; (Gly) glycerol gradient centrifugation. The blot was probed with rabbit polyclonal anti-NURF-38 antibodies. Antibody specificity was demonstrated by the ability of recombinant NURF-38 to block antibody activity. (B) NURF-38 co-immunoprecipitates with NURF-55 and ISWI. 50 μl of a crude NURF-containing fraction (material that had passed through the first step in the purification protocol, a tandem Bio-Rex 70/DEAE cellulose column) were incubated with protein A–Sepharose CL-4B beads coupled to rabbit anti-NURF-38, rabbit anti-NURF-55, rabbit anti-ISWI, or rabbit anti-chicken IgG control antibodies. Immunoprecipitates and supernatants (10 μl equivalents) were analyzed by SDS-PAGE and Western blotting. Bead-bound material is shown. Antibodies used for each precipitation are indicated above the blots. For Western blotting, ISWI was detected with antibodies raised in rabbit, therefore the background bands present on the blot. The most prominent of these corresponds to the immunoglobulin heavy chain and is denoted IgGHC. NURF-38 and NURF-55 were detected together with antibodies raised in rat and so no background bands are evident.
Importantly, immunodepletion of NURF-38 protein from the crude NURF-containing fraction also led to depletion of the in vitro chromatin remodeling activity of NURF. As shown in Figure 3A, consecutive passages of the NURF-containing fraction over protein-A beads coupled to anti-NURF-38 antibodies successively removed NURF-38, ISWI, and NURF-55 subunits from the extract. By the third passage, no NURF-38 was detectable in the supernatant. The remaining ISWI and NURF-55 are likely to be attributable either to the presence of either free protein or to protein associated in complexes other than NURF (i.e., ACF, CHRAC, HAT, and histone deacetylase complex (HDAC); Varga-Weisz et al. 1995, 1997; Taunton et al. 1996; Ito et al. 1997; Hassig et al. 1997; Zhang et al. 1997; Verrault et al. 1998). Analysis of supernatants after the third and fourth passages with anti-NURF-38 beads revealed greatly reduced nucleosome remodeling activity when compared with the robust activity of mock IgG-depleted samples (Fig. 3B, promoter probe), as shown by the reduction of the amount of submononucleosomal fragments generated by chromatin remodeling. Therefore, the physical removal of NURF-38 results in a concurrent loss of chromatin remodeling activity.
Figure 3.
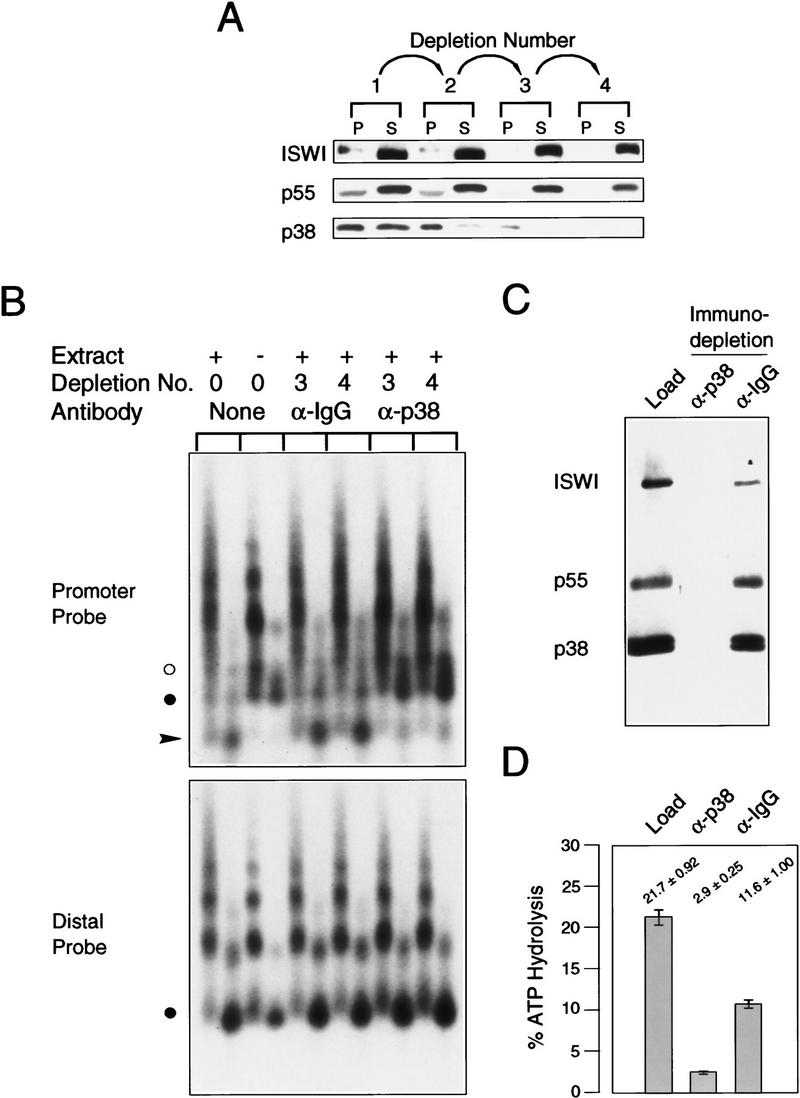
Anti-NURF-38 antibodies can deplete NURF activity. (A) Antibodies against NURF-38 can immunodeplete NURF from a partially purified embryo extract. Affinity-purified anti-NURF-38 or anti-chicken IgG control antibody was coupled to Protein A–Sepharose CL-4B beads. Serial incubations were performed and the contents of each passage are shown. (P) Pellet fraction; (S) supernatant. Numbers under arrows indicate the number of passages over the antibody-coupled beads. (B) GAGA transcription factor-mediated nucleosome disruption of reconstituted hsp70 plasmid chromatin, using partially purified embryo extract and the supernatants from the depletions in A. Southern blot showing nucleosome disruption as assayed by micrococcal nuclease digestion (3- and 15-min digestion points were taken for each chromatin sample). The blot was hybridized with radiolabeled oligonucleotides corresponding to either the hsp70 promoter (−113 to −142) or downstream sequences (+1803 to +1832) as described (Tsukiyama et al. 1994). (α-IgG) Control antibody depleted material; (α-p38) supernatant after incubation with anti-NURF-38 antibodies. Supernatant fractions used in the assays are indicated above the blot. (•) The position of the mononucleosomal band. NURF activity is gauged by the appearance of subnucleosomal bands (arrowhead) at the expense of the mononucleosomal fragment. (○) A digestion product caused by binding of GAGA factor. (C) Depletion of NURF from a purified fraction. A P-11 fraction containing NURF activity (see Materials and Methods) was depleted of NURF with antibodies raised against NURF-38. The starting material is compared with the supernatants obtained after incubation with antibodies raised against NURF-38 or a control antibody. Ten microliters of each sample was loaded. NURF-38 is often seen as a doublet, which may be attributable to a phosphorylation event. Sf9 cells infected with recombinant baculovirus bearing a NURF-38-encoding cDNA also show two bands when extracts are prepared. (D) Nucleosome-stimulated ATPase activity of the starting material and the supernatants obtained in C were tested. The control anti-IgG beads show some diminishment of ATPase activity, but the anti-NURF-38 beads show greater depletion of ATPase activity.
To demonstrate further interactions among these subunits, we performed an immunodepletion experiment with a more purified NURF-containing fraction. In this case, NURF purified to the penultimate step (P-11 fraction, material that is free of other ISWI- or p55-containing protein complexes; Tsukiyama et al. 1995) was subjected to a single pass over beads coupled to either anti-NURF-38 or to control IgGs. Figure 3C shows that NURF-38, NURF-55, and ISWI can be removed effectively from the sample by incubation with bead-coupled anti-NURF-38 antibodies. Conversely, the control antibodies failed to deplete these proteins from the fraction. The NURF-depleted supernatant also showed a substantial loss of nucleosome-stimulated ATPase when compared with the material incubated with a control antibody (Fig. 3D).
In addition, biochemical experiments designed to assess the relative strength of the interactions among the NURF subunits showed that NURF-55 remained substantially bound to NURF-38, even after incubation with up to 4 m urea (data not shown), suggesting that these proteins are tightly associated. Together, these data provide further support that NURF-38 is an integral component of the complex.
The NURF-38 protein has inorganic PPase activity
PPases are highly conserved enzymes that hydrolyze inorganic pyrophosphate to inorganic phosphate. These enzymes are required for a broad spectrum of biosynthetic pathways, including oligosaccharide and fatty acid synthesis, tRNA charging/amino acid activation, and polynucleotide synthesis (Kornberg 1962). In the cell nucleus, pyrophosphate is generated as a metabolite of replication, transcription, and DNA repair. During nucleic acid polymerization, the incorporation of a nucleoside in the growing chain with the concomitant liberation of pyrophosphate is a readily reversible reaction. The hydrolysis of pyrophosphate by PPase eliminates metabolite inhibition, therefore shifting the reaction equilibrium in favor of polymerization (Kornberg 1962).
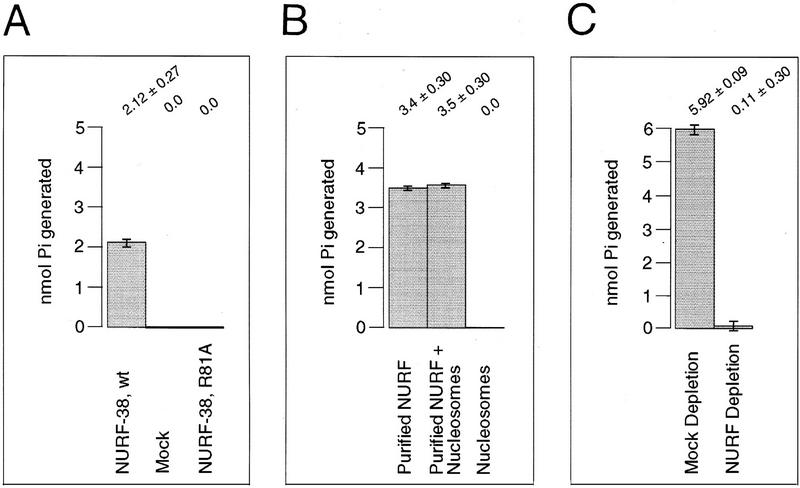
To determine whether NURF-38 possessed PPase activity, we expressed the polypeptide as a soluble polyhistidine fusion protein and purified it to >95% homogeneity by nickel–NTA affinity chromatography under nondenaturing conditions. As measured by a colorimetric assay specific for inorganic phosphate (Shatton et al. 1983), the recombinant NURF-38 protein was capable of catalyzing the hydrolysis of inorganic pyrophosphate to monophosphate. Conversely, a mock purification performed with bacteria transformed with the empty vector showed no detectable PPase activity. Importantly, we found that substitution of arginine-81 with alanine (arginine 81 corresponds to arginine 78, a residue essential for catalysis by the S. cerevisiae enzyme) led to a drastic reduction of PPase activity (Fig. 4A) without causing a detectable change in protein stability or solubility. We therefore conclude that NURF-38 is a PPase.
Figure 4.
Inorganic pyrophosphatase activity of recombinant NURF-38 and purified NURF complex. (A) PPase activity of recombinant NURF-38. Fifty nanograms of recombinant NURF-38, purified to >95% purity as judged by SDS-PAGE and Coomassie staining, were analyzed. (Mock) Mock purification made from cells transformed with empty vector. Activity is given as picomoles of inorganic phosphate generated in the course of the 10-min assay. (B) PPase activity of purified NURF in the presence and absence of nucleosomes. Approximately 0.3 ng of NURF, purified to the glycerol gradient step and free of extraneous proteins as judged by silver staining, was used in these experiments. Activity is given as in A. The assay was conducted in 50 μl, and the concentration of nucleosomes was >10 ng/μl (see Materials and Methods). (C) Depletion of PPase activity from a crude NURF-containing fraction. The same extract and conditions used in Fig. 3, A and B, were employed.
Purified NURF possesses inorganic PPase activity
We next investigated whether the native NURF complex also had inorganic PPase activity. As shown in Figure 4B, NURF highly purified by a seven-step procedure and visibly free of contaminating proteins (Tsukiyama and Wu 1995) was also found to have relatively strong PPase activity. Interestingly, we found that the PPase activity of purified NURF (specific activity ∼1.13 × 10−4 mole Pi/mg protein/min) is of the same order of magnitude as the activity of a commercially available yeast enzyme. By comparison, the specific PPase activity of bacterially expressed NURF-38 is significantly lower; we suspect this low activity may be a consequence of the bacterial expression. The high PPase activity of the native NURF complex was neither inhibited nor enhanced by the presence of reconstituted nucleosomes in the reaction (Fig. 4B). In addition, immunodepletion of NURF from a crude NURF-containing fraction with anti-NURF-38 antibodies (see Fig. 3A) resulted in quantitative depletion of total PPase activity (Fig. 4C). Taken together, our results indicate that NURF-38 in the native NURF complex retains a highly active PPase activity.
The PPase and chromatin remodeling activities of NURF can be biochemically uncoupled
The presence of both the ISWI ATPase and the NURF-38 PPase components in the NURF complex raised the interesting question of whether ATP hydrolysis by ISWI occurred at the α–β or the β–γ phosphodiester bond, releasing inorganic pyrophosphate or phosphate, respectively. To address this issue, we analyzed the products of ATP hydrolysis on stimulation of the ATPase activity of purified NURF by nucleosomes (Tsukiyama and Wu 1995). As shown by thin-layer chromatography and autoradiography, [α-32P]ATP was hydrolyzed to [α-32P]ADP, with no detectable hydrolysis to [α-32P]AMP (Fig. 5). These results indicate that the hydrolysis of ATP by ISWI occurs at the β–γ position, thereby liberating inorganic phosphate and not pyrophosphate.
Figure 5.
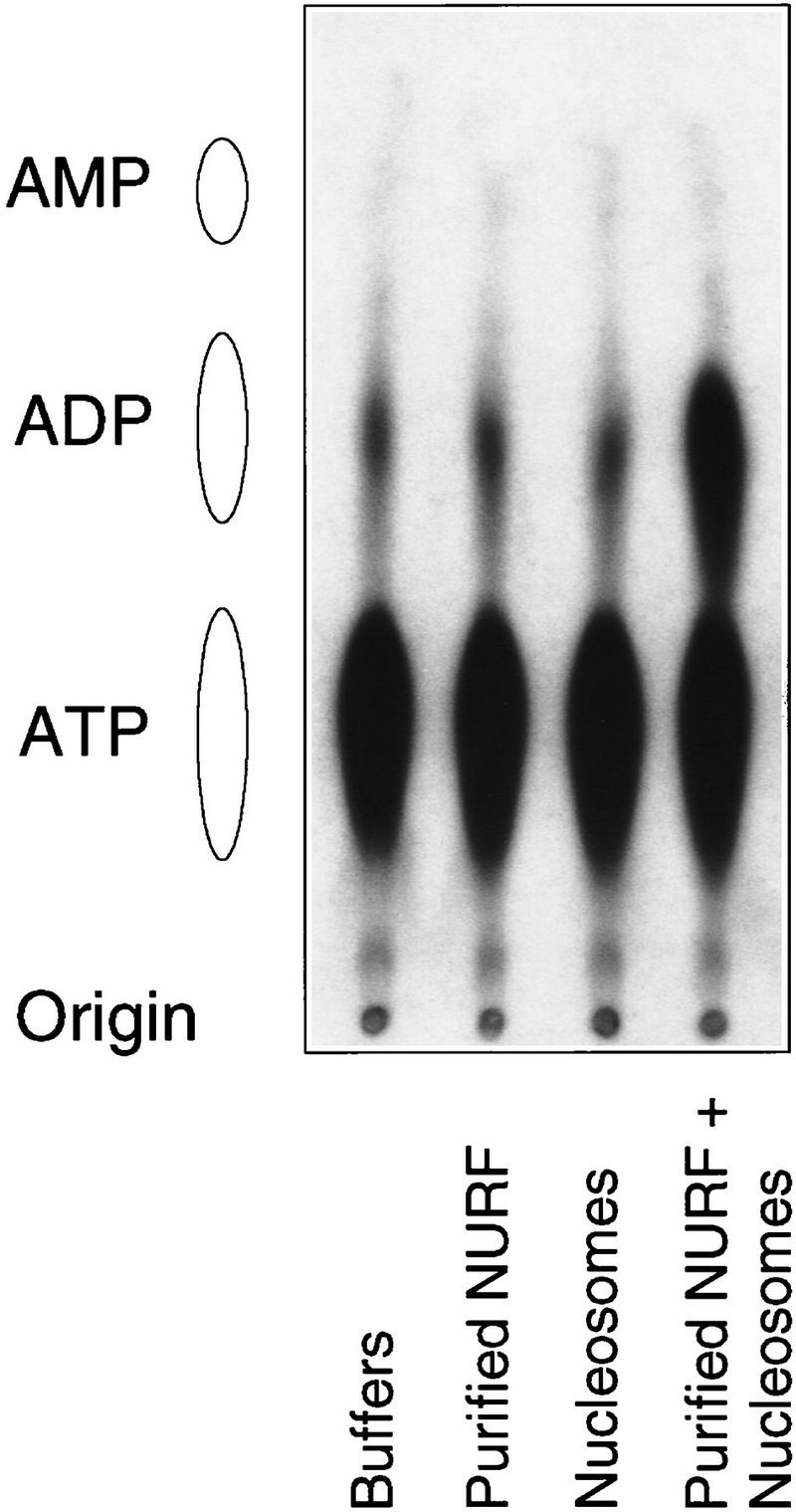
The ATPase activity of NURF in vitro does not liberate inorganic pyrophosphate. [α-32P]ATP was used as a substrate for the ATPase assay using purified NURF and nucleosomes assembled by salt dialysis. Nucleotides were separated by thin layer chromatography. The origin and the positions of marker nucleotides are shown at left.
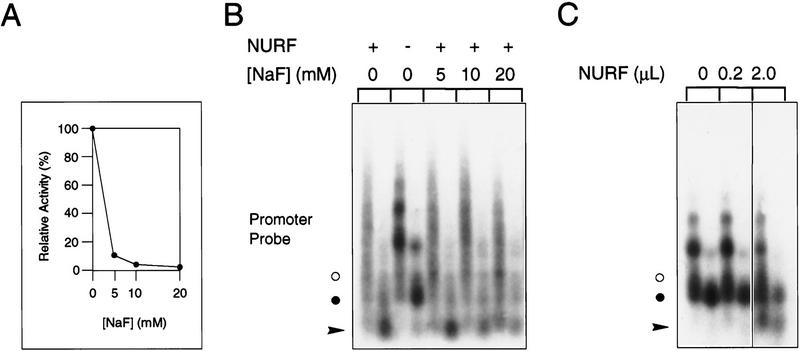
To assess further whether the PPase activity of NURF was required for the perturbation of chromatin structure in vitro, we employed sodium fluoride, an inhibitor of PPase activity (Baykov et al. 1976). At concentrations of up to 20 mm, sodium fluoride effectively reduced PPase activity in the chromatin remodeling reaction by 90%–95% (Fig. 6A). The inclusion of these levels of inhibitor had no substantial effect on the ability of NURF to facilitate GAGA factor-dependent nucleosome remodeling at the hsp70 promoter (Fig. 6B). This result suggests that nucleosome remodeling may be uncoupled from the bulk of the PPase activity of NURF. Although the few percent residual activity in the presence of fluoride cannot be excluded from having a role in the remodeling reaction, we believe this to be unlikely as the amount of NURF was not in excess. Ten percent of the input NURF in the standard remodeling reaction failed to generate detectable remodeling of chromatin (Fig. 6C).
Figure 6.
The PPase activity of NURF is not required for chromatin remodeling. (A) Titration of the inhibition of the PPase activity of purified NURF and its chromatin template in the presence of increasing concentrations of sodium fluoride. (B) Remodeling of plasmid chromatin templates by NURF in the presence of increasing amounts of sodium fluoride. The probes and methodology are the same as in Fig. 3B. (•) The position of the mononucleosomal band; (arrowhead) the submononucleosomal fragments; (○) a GAGA-specific band. (C) Control experiment demonstrating that 10% of the NURF activity used in the assays shown in B is insufficient to support remodeling of plasmid chromatin. The amount of NURF used in each sample is indicated at the top. Other symbols as in B. Two microliters corresponds to the amount of NURF employed in the experiment shown in B.
Distribution and subcellular localization of NURF-38
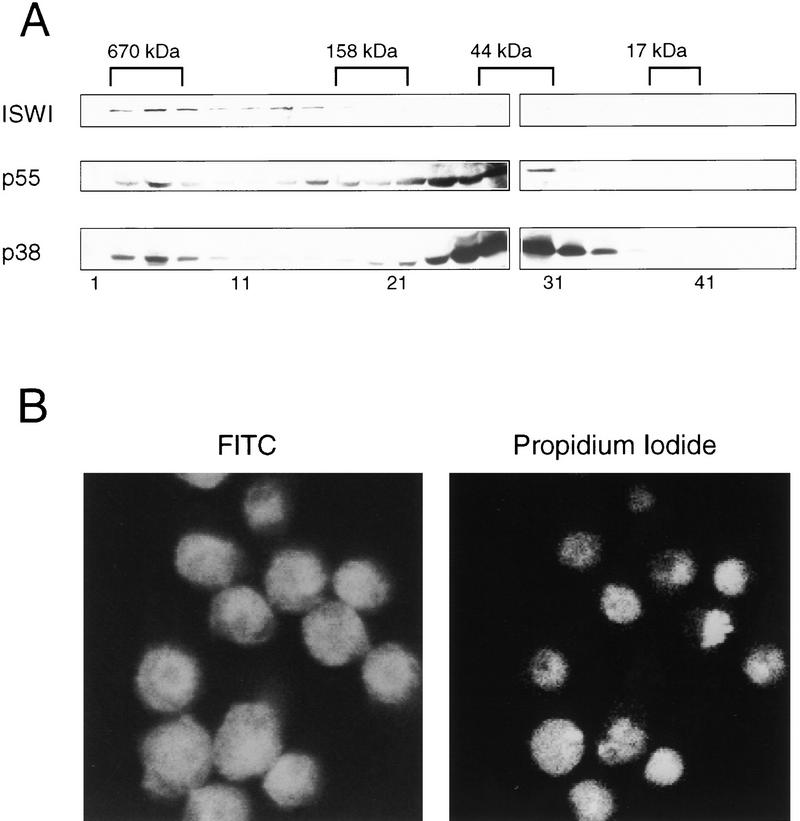
Soluble eukaryotic PPases have been shown to exist as homodimers in solution (for review, see Cooperman et al. 1992). To assess the native state of NURF-38, we analyzed its distribution in a crude Drosophila embryo extract by gel filtration and Western blotting. As shown in Figure 7A, most of the NURF-38 protein migrated in later fractions with an apparent molecular weight of ∼40,000 as compared with globular protein standards, consistent with a monomer or compact dimer. A small percentage of NURF-38, however, was clearly visible in the 600,000 molecular weight range, consistent with its inclusion in the NURF complex. These results indicate that there are at least two distinct populations of NURF-38 in Drosophila embryo extracts.
Figure 7.
NURF-38 exists in two forms and is present in both the nuclear and cytoplasmic compartments of the cell. (A) Superose 12 gel filtration chromatographic analysis of crude Drosophila embryo extract. Fifty microliters of extract was analyzed on a Pharmacia SMART system. Twenty-microliter fractions were analyzed by Western blotting using anti-NURF-38, anti-NURF-55, and anti-ISWI antibodies. The relative positions of molecular mass markers are noted above the blot; the fraction numbers are below. (B) Immunofluorescent staining and confocal analysis of Schneider line 2 cells with affinity-purified anti-NURF-38 antibodies. Specificity of the antibody reagent was demonstrated by the ability of the recombinant protein to block staining.
Whereas one form of PPase appears to be associated with the mitochondrial membrane and another has been found in cytoplasmic fractions of cell extracts (Lundin et al. 1991), the nuclear localization of these enzymes has not, to our knowledge, been investigated. As shown by indirect immunofluorescence and confocal microscopy (Fig. 7B), NURF-38 PPase can be found in both the cytoplasm and the nucleus. This is consistent with its expected requirement for efficient removal of high levels of pyrophosphate that are generated during replication and transcription in the nuclear compartment (Cooperman et al. 1992).
Discussion
The Drosophila NURF consists of four distinct polypeptides of which two, ISWI and Drosophila RbAp48/CAF-1p50, have been identified previously and are components of several macromolecular complexes involved in nucleosome dynamics or histone metabolism. In this report, we have identified and characterized the smallest, 38-kD subunit of NURF, and have provided immunological evidence that NURF-38 is an integral subunit of the complex. Sequence analysis of NURF-38 and cloning of the corresponding cDNA revealed substantial and extensive sequence identity with inorganic PPases. Accordingly, both recombinant NURF-38 protein and the purified NURF complex were found to possess inorganic PPase activity.
Inhibition of the PPase activity of NURF by sodium fluoride, which does not substantially affect nucleosome remodeling, shows that this new enzymatic activity of NURF may be uncoupled from chromatin remodeling in vitro. Moreover, the ATPase activity of NURF, essential for its nucleosome remodeling activity, results in the liberation of inorganic phosphate (not pyrophosphate) as for all studied ATPases. Taken together, these results suggest that the PPase activity of the NURF complex has no significant connection with chromatin remodeling per se. In this context, it is interesting that another ISWI-containing chromatin-remodeling complex, Drosophila CHRAC, has an associated topoisomerase II component whose known activity is also uninvolved in the remodeling function (Varga-Weisz et al. 1997).
How then might NURF-38 function as part of the NURF complex? The integral association of NURF-38 as part of a larger NURF complex with general affinity to chromatin (Mizuguchi et al. 1997, Martínez-Balbás et al. 1998) may provide a vehicle to deliver PPase activity into the proximity of the DNA replication and transcription machinery generating the pyrophosphate metabolite. Because the incorporation of nucleotides into a growing nucleic acid chain (and the concomitant liberation of pyrophosphate) is a readily reversible reaction, the hydrolysis of the inorganic pyrophosphate to free phosphate drives the reaction in favor of incorporation (Kornberg 1962). Therefore, the inclusion of NURF-38/PPase in the NURF complex should facilitate DNA and RNA synthesis in chromatin by preventing metabolite inhibition. In this respect, it is of interest that the mitochondrial PPase catalytic subunit is associated with other, as-yet-unidentified components that anchor the PPase in the mitochondrial membrane (Lundin et al. 1991, 1992).
An alternative possibility not mutually exclusive from the above is that the NURF-38 protein may be employed as a structural or regulatory component of the complex. It will be possible to test this directly when reconstitution of the NURF complex becomes feasible. A third possibility is that chromatin remodeling is sensitive to the level of pyrophosphate metabolite that is generated in the cell nucleus, and requires PPase to relieve inhibition of the remodeling reaction. This possibility appears unlikely, however, as we have been unable to observe sensitivity of the chromatin remodeling reaction to inorganic pyrophosphate at levels up to 30 mm where the action of NURF PPase was insufficient to reduce the high IPP concentration significantly.
Recently, mutations in Drosophila NURF-140/ISWI have been found to have a late larval-lethal phenotype with effects on transcription that are consistent with a role for nucleosome remodeling in vivo (J. Tamkun, pers. comm.). The presence of ISWI in three distinct remodeling complexes (NURF, ACF, and CHRAC), however, precludes a simple interpretation for the precise function of ISWI in the NURF complex. Likewise, the genetics of NURF-55 is complicated by the inclusion of the encoded protein in multiple and distinct enzymes involved in histone metabolism. Drosophila NURF-38 mutants have not yet been generated. Such mutants are expected to have pleiotropic effects, probably with a lethal phenotype, given the fundamental role of PPase in so many key biosynthetic pathways. Mutants of the cytoplasmic PPase in S. cerevisiae are lethal, and deletions of the gene encoding the mitochondrial protein result in the inability of mitochondria to replicate their genomes during cell division (Lundin et al. 1991). It will be of interest to identify mutations in Drosophila NURF-38 that affect its association with the NURF complex without impairment of the PPase activity. The isolation or design of such mutations and the analysis of mutant phenotypes in transgenic flies or mosaics lacking endogenous NURF-38 poses a special challenge for the analysis of the function of the NURF complex in vivo.
Materials and methods
Cloning of the NURF-38 gene
Peptide data for NURF-38 was obtained by in-gel endopeptidase digestion of NURF-38, HPLC purification of resultant peptides, and Edman degradation sequencing as described previously for NURF-55 (Martínez-Balbás et al. 1998). Three peptides were chosen as templates for primer construction and PCR-based screening of an embryonic cDNA library and primers were synthesized in both orientations using the Drosophila codon bias. For peptide 155, the sequence TIYNMVV was used and primers ACCATCTACAACATGGTGGTG (155-1a, sense orientation) and CACCACCATGTTGTAGATGGT (155-1b, antisense orientation) were constructed; for peptide 168, the sequence EAPDGGQVEE was used and primers GAGGCCCCCGACGGCGGCCAGGTGGAGGAG (168-1a) and CTCCTCCACCTGGCCGCCGTCGGGGGCCTC (168-1c) were synthesized; and for peptide 159, sequence DNDPID was used and primers GACAACGACCCCATCGAC (159-1a) and GTCGATGGGGTCGTTGTC (159-1b) were made.
The PCR reactions were prepared in 100-μl batches in all possible pair-wise combinations (excluding those primer pairs in which the primers were complements of each other) using 0.5 units Taq DNA polymerase (Promega), and containing 2.5 mm MgCl2, 100 pmoles of each primer, 0.5 mm dNTPs, and ∼500 ng of library DNA (Novagen). Aliquots (10 μl) were then used in ten reactions run in parallel in a gradient-heat block thermocycler using the following program: 94°C for 2 min, 1 cycle; 94°C for 30 sec, 44°C–66°C (∼2°C increments) 1 min, 72°C 1 min, 30 cycles; 72°C 1 min, 1 cycle. A fragment of 235 bp was generated with primers 155-1a/159-1b, and a 711-bp fragment with 155-1a/168-1b. These fragments were TA-cloned into pT7BlueT (Novagen) as per the manufacturer’s protocol. The clones were then sequenced in their entirety. The larger clone, after translation, revealed the presence of sequences corresponding to peptides 159, 164, and 138. We concluded that the clones were encoded by the NURF-38 gene and the larger clone was used as per the manufacturer’s instructions to directly probe the library to obtain full-length clones. Four independent positives were cloned, one of which contained the full-length cDNA, including the 5′-transcribed untranslated region (with an in-frame stop codon) and a complete poly(A) tail.
Preparation of recombinant NURF-38
The full-length cDNA (clone pEXloxNURF-38 4.1) was cut with EcoRI and XhoI and cloned into the same sites of the inducible pET28b vector (Novagen). This resulted in a gene encoding a recombinant protein containing an amino-terminal His6 tag but lacking the 12 amino-terminal residues of the wild-type NURF-38. The protein was expressed by transforming Escherichia coli BL21 cells and growing a 500-ml culture at 37°C to an OD600 of 0.6 in Luria broth (LB) supplemented with 50 μg/ml kanamycin, at which time the culture was induced with 0.5 mm isopropyl-β-d-thiogalactoside (IPTG) for 1 hr. The cells were collected by spinning at 7000 rpm in a Beckman JA-10 rotor for 5 min at 4°C, washed once in ice-cold PBS, and collected again by spinning at 15,000 rpm in a Beckman JA-20 rotor for 15 min at 4°C. The cells were disrupted by sonication in ice-cold HEGN-0.3 [25 mm HEPES–KOH at pH 7.6, 1 mm EDTA, 10% (vol/vol) glycerol, 0.02% (vol/vol) NP-40, 0.3 m KCl, 1 mm DTT, 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride/hydrochloride (AEBSF)], and the soluble fusion protein collected by spinning the lysate at 15,000 rpm in a Beckman JA-20 rotor for 10 min at 4°C to pellet the debris. The clarified supernatant was then added to a 1-ml bed volume of nickel–NTA agarose beads (Qiagen) in the presence of 25 mm imidazole (Sigma), and incubated at 4°C for 1 hr with gentle rotation. The protein-coupled beads were then washed three times each with 10 ml of HEGN-0.3 containing 25 mm imidazole for 30 min at 4°C, and the protein eluted with 1 ml of HEGN-0.3 containing 100 mm imidazole for 30 min at 4°C. The resulting protein was found to be >95% pure as judged by SDS-PAGE and Coomassie staining. Concentrations were estimated against a BSA standard (Bio-Rad) loaded on the same gel. Mock purifications were prepared the same way except empty pET28b vector was used. Faint background bands present in both preparations were used to standardize them to each other and equal volumes were used in all experiments using a mock control. Protein preparations were aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Preparation of anti-NURF-38 polyclonal immune sera
Antibodies were raised in both rabbits and rats by Berkeley Antibody Company (Richmond, CA) according to that company’s recommendations, using the recombinant His6–NURF-38 protein described above as antigen. The first inoculation was performed with protein purified by SDS-PAGE and used denatured, directly from the acrylamide gel. All subsequent inoculations were performed with soluble protein stored in elution buffer (see above).
Affinity purification of anti-NURF-38, anti-NURF-55, and anti-ISWI
Affinity-purified antibodies raised in rabbits against NURF-55 and ISWI were prepared from whole immune sera essentially as described (Martínez-Balbás et al. 1998) using His6-tagged antigens coupled to nickel–NTA agarose beads (Qiagen); however, instead of PBS, the antibodies were dialyzed against antibody storage buffer (20 mm Tris-HCl at pH 7.4, 28 mm NaCl), and stored in same supplemented with 1 mg/ml of BSA. Antibodies against NURF-38 were prepared in the same fashion, using the His6-fusion protein described above. The bed volume was 1 ml (containing ∼2 mg/ml antigen), and 2 ml of serum was used per preparation.
For some experiments, controls containing depleted antibody (i.e., affinity-purified antibody pre-adsorbed to a His6–NURF-38 antigen column) were employed to demonstrate the specificity of the antibody. These were prepared as follows: 200 μl of affinity purified anti-NURF-38 (∼0.2 mg/ml) was incubated with 100 μl bed volume of antigen resin (see above) containing ∼2 mg/ml recombinant NURF or with an equal volume of a mock preparation control at 4°C for 1 hr. The beads were then pelleted and the supernatant collected.
SDS-PAGE and Western blotting
Proteins were analyzed by SDS-PAGE using 10% acrylamide separating gels and Western blotting using nitrocellulose membranes (0.22-μm pore size, Schleicher & Schuell) and chemiluminescent detection methodology (ECL, Amersham). Primary antibodies raised in rat against NURF-38 and NURF-55 were used at 1:1000 dilution (sera), whereas rabbit anti-ISWI antibodies were used at 1:250 (serum) or 1:500 (affinity purified). HRP-conjugated secondary antibodies raised in donkey (Amersham) were used at 1:10,000 dilution (anti-rat IgG) or 1:20,000 (anti-rabbit IgG).
Gel filtration chromatography
Drosophila embryo S-150 extract was prepared as described previously (Becker and Wu 1992). Extract (50 μl) was loaded onto a Superose-12 column (Pharmacia) and analyzed using the SMART system (Pharmacia). The elution was performed with modified embryo extraction buffer [10 mm HEPES–KOH at pH 7.6, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm EGTA, 5% (vol/vol) glycerol, 1 mm DTT, 1 mm AEBSF] and 20-μl fractions were collected. Odd-numbered fractions were loaded in their entirety on a 10% SDS-polyacrylamide gel and analyzed by Western blot as described above.
Indirect immunofluorescence
Drosophila Schneider line 2 cells were cultured in 1-ml slide flasks (Nunc). Cells were fixed in 4% formaldehyde in PBS for 20 min at room temperature, followed by treatment with methanol for 10 min. After blocking with 3% (vol/vol) BSA in PBS, 0.1% Tween-20 for 30 min, the slides were incubated with 0.5 ml of a 1:100 dilution of either affinity-purified rabbit anti-NURF-38 or a 1:100 dilution of antigen-blocked antibody preparation (see above) in blocking buffer and incubated for 2 hr at room temperature. After washing twice for 10 min at room temperature with PBS, 0.05% (vol/vol) Tween-20, each sample was incubated with 0.5 ml of a 1:250 dilution in blocking buffer of fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Cappel) for 1 hr at room temperature. The cells were washed twice as above and treated with 0.5 ml of 0.4 mg/ml RNase A (Boehringer Mannheim) in PBS for 1 hr at 37°C, washed twice as above, and stained with 50 μg/ml propidium iodide (Sigma) for 30 min at room temperature. The samples were analyzed on a Zeiss Axiovert confocal microscope using a 63× objective with oil immersion as per the manufacturer’ instructions. Polytene squashes were performed as described previously (Martínez-Balbás et al. 1998), using a 1:100 dilution of affinity-purified anti-NURF-38 as the primary antibody. FITC-conjugated donkey anti-rabbit secondary antibody (Cappel) was used at 1:200 dilution.
NURF purification
NURF purification was prepared as described (Tsukiyama and Wu 1995). Partially purified NURF used in immunoprecipitation and immunodepletion studies was material used after the first step in the purification of the crude embryo extract, with the sole modification being the reversal of order of the tandem DEAE cellulose and Bio-Rex 70 columns.
Database searches and sequence analyses
The National Center for Biotechnology Information web site (www.ncbi.nlm.nih.gov) was used to perform searches of databases employing the BLAST program (Altschul et al. 1990). All eukaryotic inorganic PPase were examined and their scores reviewed, indicating a positive identification for NURF-38 as an inorganic pyrophosphatase. For alignment of these proteins, we employed the Macaw sequence analysis software package (version 2.0.5, Win 32I, NCBI) and alignments were made allowing gaps. More refined alignments and calculations of percent identity along the entire length of the protein were performed with the Align program at the GeneStream Homepage (http://dot.imgen.bcm.tmc.edu).
Inorganic PPase assays
Assays were performed as described by Shatton et al. (1983), except that the reaction buffer used was embryo extraction buffer (see above). All blank and standard samples included the appropriate buffer (see above) as a control for protein samples. For samples including antibodies, protein was pre-incubated with antibody on ice for 1 hr before analysis. For the experiments involving nucleosomes, the assay was performed in 50 μl total volume, keeping the nucleosome concentration >10 ng/μl to ensure their stability. After the assay was performed, the sample volume was increased to 0.5 ml volume with color reagent/buffer mix and the OD660 measured. Experiments with purified NURF were performed in the presence of 1 μg/ml BSA as carrier.
Immunoprecipitation assays and immunodepletions
A slurry of protein A–Sepharose CL-4B (Pharmacia) (40 μl) was incubated for 1 hr at 4°C with 25 μg of affinity-purified anti-NURF-38, anti-NURF-55, or anti-ISWI antibodies (see above) or 25 μg of purified rabbit anti-chicken IgG (Jackson Immunochemicals) in interaction buffer [IB: 50 mm Tris-HCl at pH 7.5, 120 mm KCl, 0.1% (vol/vol) NP-40, 0.1 mm EDTA, 3 mm MgCl2, 1 mm AEBSF]. The beads were then pelleted, washed three times in IB at 4°C with rotation, and incubated with 40 μl of a partially purified embryo extract (DEAE flow-through, see above) for 1 hr at 4°C with rotation. The beads were then pelleted, washed three times in IB as above, and boiled in 40 μl of SDS-PAGE sample buffer. An SDS-PAGE sample buffer (40 μl, 2×) was added to the supernatant samples that were then boiled. Equivalents (20 μl) were loaded on a 10% SDS-polyacrylamide gel and analyzed by Western blot as described above.
Immunodepletions of native NURF from crude fractions were performed as follows: 200 μg of affinity-purified anti-NURF-38 antibody or control rabbit anti-chicken IgG was coupled to 100-μl bed volume of protein A–Sepharose beads, as above. The beads were then aliquoted into four separate tubes in equal amounts. Partially purified extract (50 μl) was added to the beads in the first tube and incubated for 1 hr at 4°C with gentle rotation. The resulting supernatant (10 μl) was set aside for Western analysis, and the remaining 40 μl were added to the second tube and processed as above. Two more incubations were performed similarly, four incubations were performed in total. Supernatant (2.5 μl) was used in chromatin disruption assays (see below). For immunodepletions using more purified NURF, 10 μl of a NURF-containing P-11 fraction (in HEGN-0.3) was diluted 1:2 in HEGN containing no KCl to bring the concentration of KCl to 150 mm. Insulin was added to 0.2 mg/ml and AEBSF to 1 mm. The immunoprecipitations were performed as above, with 20-μl bed volume of antibody-coupled beads and 20 μl of diluted NURF. Resulting supernatant (1 μl) was then assayed for ATPase activity, described below, and 10-μl equivalents were used for Western blot analysis.
Chromatin assembly and disruption assays
Assembly, Sarkosyl-treatment, purification of plasmid chromatin, and GAGA-mediated disruption were performed as described (Tsukiyama and Wu 1995).
ATPase assays and nucleosome assembly
Nucleosome assembly was carried out as described (Georgel et al. 1997). All assays were performed such that the concentration of nucleosomes was above 10 ng/μl to ensure their stability. ATPase assays were performed as described (Georgel et al. 1997) except [α-32P]ATP was used to monitor hydrolysis. The products of the ATPase reaction were separated by TLC (polyethyleneimine cellulose on polyester, Sigma) using 0.75 m KH2PO4. The hydrolysis products were measured by analyzing the labeled ADP generated by autoradiography.
Acknowledgments
We thank C. Klee for performing microsequence analysis on NURF peptides; R. Kobayashi for protocols on in-gel proteolytic cleavage and HPLC of peptides; J. Tamkun for anti-ISWI antibodies and discussions of unpublished data; M. Mortin and T. Jones for assistance with in situ localization of the NURF-38 gene; P. Wagner for discussions of ATPases and advice on ATPase assays; and M. Martínez-Balbás, G. Marchler, G. Mizuguchi, and members of our laboratory for valuable assistance and suggestions. This work was supported in part by the Intramural Research Program of the National Cancer Institute. D.A.G. is a Leukemia Society of America Postdoctoral Fellow. R.S. and V.O. are supported by postdoctoral fellowships from the Human Frontier Science Program Organization. The sequences for NURF-38 have been submitted to GenBank under accession nos. AF085600 (genomic) and AF085601 (cDNA).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL carlwu@helix.nih.gov; FAX (301) 435-3697.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baykov AA, Artjukov AA, Avaeva SM. Fluoride inhibition of inorganic pyrophosphatase. I. Kinetic studies in a Mg2+-PPi system using a new continuous enzyme assay. Biochim Biophys Acta. 1976;429:982–992. doi: 10.1016/0005-2744(76)90343-0. [DOI] [PubMed] [Google Scholar]
- Becker PB. The establishment of active promoters in chromatin. BioEssays. 1994;16:541–547. doi: 10.1002/bies.950160807. [DOI] [PubMed] [Google Scholar]
- Becker PB, Wu C. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond MW, Chiu NY, Cooperman BS. Identification of an arginine important for enzymatic activity within the covalent structure of yeast inorganic pyrophosphatase. Biochemistry. 1980;19:94–102. doi: 10.1021/bi00542a015. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multi-subunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Carlson M, Laurent BC. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Cooperman BS, Baykov AA, Lahti R. Evolutionary conservation of the active site of soluble inorganic pyrophosphatase. Trends Biochem Sci. 1992;17:262–266. doi: 10.1016/0968-0004(92)90406-y. [DOI] [PubMed] [Google Scholar]
- Côté J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Dingwall AK, Beek SK, McCallum CM, Tamkun JW, Kalpana GV, Goff SP, Scott MP. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992;335:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- ————— Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- Fletcher TM, Hansen JC. The nucleosomal array: Structure/function relationships. Crit Rev Eukaryot Gene Expr. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]
- Georgel PT, Tsukiyama T, Wu C. Role of histone tails in nucleosome remodeling by Drosophila NURF. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Winston F. Nucleosomes and transcription, recent lessons from genetics. Curr Opin Genet Dev. 1997;7:192–198. doi: 10.1016/s0959-437x(97)80128-1. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Transcriptional activator components and poxvirus DNA-dependent ATPases comprise a single family. Trends Biochem Sci. 1993;18:291–292. doi: 10.1016/0968-0004(93)90037-n. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes & Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes & Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- Kornberg A. On the metabolic significance of phosphorylytic and pyrophosphorylytic reactions. In: Kasha H, Pullman P, editors. Horizons in biochemistry. New York, NY: Academic Press; 1962. pp. 251–264. [Google Scholar]
- Kornberg RD, Lorch Y. Interplay between chromatin structure and transcription. Curr Opin Cell Biol. 1995;7:371–375. doi: 10.1016/0955-0674(95)80092-1. [DOI] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes & Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Lundin M, Baltscheffsky H, Ronne H. Yeast PPA2 gene encodes a mitochondrial inorganic pyrophosphatase that is essential for mitochondrial function. J Biol Chem. 1991;266:12168–12172. [PubMed] [Google Scholar]
- Lundin M, Deopujari SW, Lichko L, da Silva LP, Baltscheffsky H. Characterization of a mitochondrial inorganic pyrophosphatase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1992;1098:217–223. doi: 10.1016/s0005-2728(05)80339-1. [DOI] [PubMed] [Google Scholar]
- Martínez-Balbás MA, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of Nucleosome Remodeling factor NURF in transcriptional activation of chromatin. Mol Cell. 1997;1:141–150. doi: 10.1016/s1097-2765(00)80015-5. [DOI] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen-Hughes T, Utley RT, Côté J, Peterson CL, Workman JL. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T, Workman JL. Experimental analysis of chromatin function in transcription control. Crit Rev Euk Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- Paranjape SM, Kamakaka RT, Kadonaga JT. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast, links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Peterson CL. Multiple switches to turn on chromatin? Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Tamkun JW. The SWI-SNF complex, a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- Pruss D, Hayes JJ, Wolffe AP. Nucleosomal anatomy—where are the histones? BioEssays. 1995;17:161–170. doi: 10.1002/bies.950170211. [DOI] [PubMed] [Google Scholar]
- Qian Y-W, Wang Y-C, Hollingsworth RE, Jones D, Jr, Ling N, Lee EY-H. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu Rev Biophys Biomol Struct. 1997;26:83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- Shatton JB, Ward C, Williams A, Weinhouse S. A microcolorimetric assay of inorganic pyrophosphatase. Anal Biochem. 1983;130:114–119. doi: 10.1016/0003-2697(83)90657-7. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma, a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- ————— Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kD subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Bulger M, Kamakaka RT, Yashi R, Kadonaga JT. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K, Zlatanova J, Arents G, Moudrianakis E. Elements of chromatin structure, histones, nucleosome, and fibres. In: Elgin SCR, editor. Chromatin structure and gene expression. Oxford, UK: Oxford University Press; 1995. pp. 1–26. [Google Scholar]
- Varga-Weisz PD, Blank TA, Becker PB. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J. 1995;14:2209–2216. doi: 10.1002/j.1460-2075.1995.tb07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Verrault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- ————— Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]