Ciliary delivery of polycystin-1 depends on a conserved (K/R/Q)VxPx motif. The signal enables Arf4 GTPase binding and assembly of a multimeric trafficking complex. Functional importance of Arf4 and Rab8 in ciliary trafficking is shown. The studies offer the first unifying molecular rationale for human cystic kidney diseases and retinopathies.
Abstract
Primary cilia regulate epithelial differentiation and organ function. Failure of mutant polycystins to localize to cilia abolishes flow-stimulated calcium signaling and causes autosomal dominant polycystic kidney disease. We identify a conserved amino acid sequence, KVHPSST, in the C-terminus of polycystin-1 (PC1) that serves as a ciliary-targeting signal. PC1 binds a multimeric protein complex consisting of several GTPases (Arf4, Rab6, Rab11) and the GTPase-activating protein (GAP), ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ASAP1) in the Golgi, which facilitates vesicle budding and Golgi exocytosis. A related N-terminal ciliary-targeting sequence in polycystin-2 similarly binds Arf4. Deletion of the extreme C-terminus of PC1 ablates Arf4 and ASAP1 binding and prevents ciliary localization of an integral membrane CD16.7-PC1 chimera. Interactions are confirmed for chimeric and endogenous proteins through quantitated in vitro and cell-based approaches. PC1 also complexes with Rab8; knockdown of trafficking regulators Arf4 or Rab8 functionally blocks CD16.7-PC1 trafficking to cilia. Mutations in rhodopsin disrupt a similar signal and cause retinitis pigmentosa, while Bardet-Biedl syndrome, primary open-angle glaucoma, and tumor cell invasiveness are linked to dysregulation of ASAP1 or Rab8 or its effectors. In this paper, we provide evidence for a conserved GTPase-dependent ciliary-trafficking mechanism that is shared between epithelia and neurons, and is essential in ciliary-trafficking and cell homeostasis.
INTRODUCTION
Autosomal dominant polycystic kidney disease (ADPKD) is a common form of chronic cystic kidney disease that leads to renal failure in adulthood (Wilson, 2004). ADPKD is caused by aberrant expression of mutant polycystin-1 (PC1) or polycystin-2 (PC2; Wilson, 2004). PC1 functions in cell–cell adhesion, cell–matrix adhesion, and signal transduction, and, together with PC2, functions in ion channel regulation (Wilson, 2004). In keeping with the many functions of PC1, its localization is temporally regulated and orchestrates cellular polarization. PC1 localizes to basal focal adhesions and lateral desmosomes and adherens junctions at early time points of cell polarization (Roitbak et al., 2004; Silberberg et al., 2005; Wilson, 2009). Once junctions are stabilized and cells are fully confluent, PC1 disappears from the lateral membranes and relocalizes to the apical primary cilium (Barr et al., 2001; Yoder et al., 2002a; Wilson, 2004; Silberberg et al., 2005; Xu et al., 2007). In cilia, PC1 and PC2 interact to form a receptor complex that transduces Ca2+ signals in response to renal flow and contributes to the preservation of the fully differentiated state (Qian et al., 1997; Tsiokas et al., 1997; Hanaoka et al., 2000; Gonzalez-Perret et al., 2001; Vassilev et al., 2001; Nauli et al., 2003, 2006; Xu et al., 2007). The maintenance of a functional, fully differentiated epithelium is closely linked to proper localization of ciliary proteins and ciliary signals. In ADPKD cells, PC1 fails to localize to junctional complexes and primary cilia (Roitbak et al., 2004; Xu et al., 2007). A complete understanding of polycystin biology therefore depends on the elucidation of the regulatory mechanisms governing polycystin localization.
Inherited cystic kidney diseases range from the adult onset ADPKD to the embryonic lethal Meckel-Gruber Syndrome, yet share commonalties in cyst formation, matrix alteration, and fluid production within the kidney (Hildebrandt et al., 2009; Nachury et al., 2010). Disease severity reflects the extent to which individual mutant ciliary proteins are required for development and function of one or more target organs, including kidney, liver, pancreas, and retina, among others. Ciliary signals are closely linked to the integrity of junctional and focal adhesion complexes, as well as the regulation of epithelial to mesenchymal transitions. The growing list of human diseases (including retinal degeneration, craniofacial disorders, obesity, polydactyly, and renal cystic kidney diseases) resulting from altered ciliary protein localization and function reflect profound alterations in planar cell polarity and epithelial homeostasis (Pazour, 2004; Inglis et al., 2006; Bacallao and McNeill, 2009; Hildebrandt et al., 2009; Hurd et al., 2010; Nachury et al., 2010). Commonalities between the sorting mechanisms of ciliary proteins in diverse cell types have yet to be uncovered, but may exist and help explain observed disease pathologies.
The primary cilium is a rod-shaped sensory organelle positioned on the apical surface of most eukaryotic cells; it depends on discrete mechanisms for protein delivery. Early studies on protein localization to cilia focused on intraflagellar transport (IFT). IFT is a kinesin- and dynein-dependent microtubular protein transport mechanism required for the formation and maintenance of motile and nonmotile (primary) cilia, and responsible for the transport of proteinaceous particles within cilia and flagella (Cole et al., 1998). Dissection of IFT mechanisms has provided key insights into the trafficking mechanisms utilized by nonmembrane, cilium-associated proteins, and defects in IFT machinery are linked to cystic kidney disease (Pazour et al., 2000; Yoder et al., 2002a, b; Lin et al., 2003; Follit et al., 2006; Jonassen et al., 2008; Omori et al., 2008). However, membrane proteins such as the polycystins are synthesized in the Golgi and depend on vesicular transport for plasma membrane and ciliary targeting, which is not explained by the IFT mechanisms.
Insights into the vesicular-based mechanisms of membrane protein trafficking to cilia have recently emerged. Rab GTPases are key regulators of vesicular transport; Rab GTPase-mediated ciliary-trafficking events have been described for fibrocystin, which causes autosomal recessive PKD when mutant, and Bardet-Biedl Syndrome (BBS) proteins (Nachury et al., 2007; Follit et al., 2010). Reports on BBS proteins, which form a large nonmembranous protein complex, first suggested a link between these renal ciliogenic proteins and Rab GTPase-mediated vesicular trafficking via the Rab8 GTPase exchange factor Rabin8 and the Arf-like GTPase Arl6 (Nachury et al., 2007; Jin et al., 2010). BBS is an inherited disorder with symptoms that include retinal degeneration and renal cyst formation, suggesting commonalities in ciliogenesis between renal and retinal cells. Retinal photoreceptor cells contain a modified form of the primary cilium known as the rod outer segment to which rhodopsin, a seven-transmembrane G-coupled receptor, is continuously shuttled. Studies on rhodopsin offer the best-characterized evidence of the requirement for vesicular transport to cilia and provide clues to the underlying mechanism. Rhodopsin contains a conserved C-terminal VxPx targeting sequence; this sequence interacts with Arf4, a small GTPase involved in vesicle formation (Deretic et al., 2005; Mazelova et al., 2009). Mutations in the VxPx motif disrupt the rhodopsin-Arf4 interaction and prevent rhodopsin trafficking to the rod outer segment, which results in rhodopsin mislocalization, a common occurrence in autosomal dominant retinitis pigmentosa (Deretic et al., 2005). PC2, a PC1-interacting protein and TRP calcium channel family member, shares a similar N-terminal RVxP motif, which is critical for its ciliary localization (Vassilev et al., 2001; Geng et al., 2006). Though we previously identified a PC1 ciliary localization signal within the carboxy-terminal 112 amino acids (Geng et al., 2006; Xu et al., 2007), the specific ciliary-targeting signal of PC1 remains to be elucidated. In this study, we sought to determine whether a conserved mechanism for ciliary trafficking is shared between retinal and kidney epithelial cells. We did this by defining the critical ciliary-targeting sequence in PC1, the functional associations of both polycystins with Arf4, and the novel multimeric exocytic complex that assembles with PC1 in the Golgi complex.
RESULTS
CD16.7-PC1 tail mutants do not localize to primary cilia
We previously showed that CD16.7-PC1 fusion proteins (described in Materials and Methods) encoding the membrane distal wild-type (WT) 112 amino acids of the PC1 cytoplasmic C-terminus (total length 226 amino acids; sequence shown in Figure 1A) or bearing a mutation (L152P) in the coiled-coil PC2-interaction domain both localized to primary cilia in normal primary human kidney and ADPKD cells (Vandorpe et al., 2002; Xu et al., 2007). These findings suggested that the sorting machinery of ADPKD cells is intact and that mutant polycystins most likely bear a defect in their ciliary-targeting signals. In this study, we demonstrate that the integral membrane CD16.7-PC1 fusion protein encoding the last 112 amino acids of the conserved human PC1 tail (termed CD16.7-PC1-WT) also localizes to the primary cilia (Figure 1, B–C) of immortalized normal human renal cortical tubular epithelia (RCTE) and mouse Swiss 3T3 cells (Supplemental Figure S1). Ninety-five percent of RCTE cells with cilia showed localization of CD16.7-PC1-WT to primary cilia (Figure 1B). Thus, our construct is ideally suited for further delineation of the trafficking signal.
FIGURE 1:
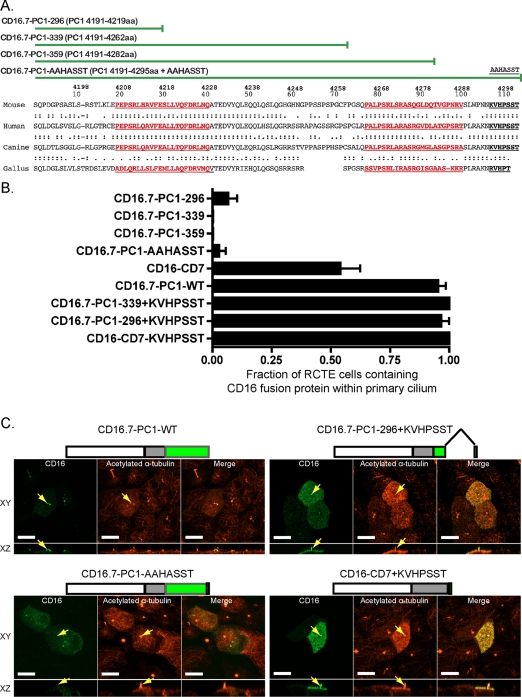
CD16.7-PC1-truncation and alanine-scanning mutants do not localize to primary cilia. (A) Alignment of the C-terminal 112 amino acids of PC1 across multiple species. Top, amino acid numbering sequence for human full-length PC1; bottom, 112 amino acids tail numbering. Red underlined regions represent conserved domains, x(1,2)-P-[GS]-x(4)-[APTV]-x(2)-[AGP]-x(3)-[ADQ]-x-[DG]-x(3)-[AQT], repeated in the PC1 C-terminus (see Materials and Methods and Table 1). Black underlined text indicates a conserved (Q/R/K)-V-x-P-x ciliary-targeting domain also found in rhodopsin and PC2. Green lines denote PC1 C-terminal truncation and alanine-scanning mutants tested in ciliary localization studies. (B) Truncation and alanine mutants identify critical KVHPSST sequence for ciliary targeting. RCTE cells were transfected 3 d postconfluence for 24 h and analyzed for CD16.7-PC1 fusion proteins expressed in the primary cilia. Analyzed were: CD16.7-PC1-WT, three different truncation mutants +/− KVHPSST sequence, alanine-scanning mutant, or a CD16 chimera bearing either the complete CD7 C-terminus (CD16-CD7) or the complete CD7 C-terminus fused to the KVHPSST sequence (CD16-CD7-KVHPSST). Graph shows the fraction of RCTE cells containing each fusion protein in the primary cilium. Quantification was performed on Z-stacks of 30–50 ciliated cells for each mutant. n = 2; one-way ANOVA, p < 0.001. (C) Representative micrographs with cartoon drawing of construct (CD16, white box; CD7, gray box; PC1, green box) show CD16.7-chimera localization detected with mAb specific for CD16 (green) and cilia labeled with acetylated α-tubulin (red). Four of eight constructs are represented. Yellow arrows indicate location of a primary cilium in each XY panel. Bars 10 μm.
Beginning with the minimal CD16.7-PC1-WT construct, we further dissected the critical sequences for ciliary delivery, and identified three regions in the C-terminal 112 amino acids of PC1 that were evolutionarily conserved (Figure 1A). We then generated C-terminal truncation mutants lacking the last 83 amino acids (CD16.7-PC1-296), 40 amino acids (CD16.7-PC1-339), or 20 amino acids (CD16.7-PC1-359), of the PC1 tail to create CD16.7-fusion proteins with respective total lengths of 296 amino acids, 339 amino acids, or 359 amino acids (Figure 1A). The minimally truncated fusion protein CD16.7-PC1-359, which lacked the last 20 amino acids (ATGPSRTPLRAKNKVHPSST) of PC1, failed to traffic to the primary cilium in 3T3 or RCTE cells, accumulated intracellularly in the perinuclear region thought to be the Golgi, and occasionally backed up into the endoplasmic reticulum (ER), which was observed as a rimming of the nuclear envelope. (quantification: Figure 1B; images: Figure S1). Similarly, CD16.7-PC1-339 and CD16.7-PC1-296 failed to reach the cilium but were found predominantly at the apical plasma membrane of RCTE cells (quantification: Figure 1B, images: Figure 1C). In 3T3 cells, CD16.7-PC1-339 accumulated intracellularly (Figure S1) and CD16.7-PC1-296 caused cell death, suggesting that different cell types may have differential sensitivities to the intracellular accumulation of mutant and poorly sorted integral membrane proteins in the incorrect membrane domain.
The results of the CD16.7-PC1 C-terminal deletion constructs suggested that the ciliary-trafficking signal was contained within the C-terminal 20 amino acids of PC1, which includes a ciliary-targeting VxPx motif found in rhodopsin and PC2 (Deretic, 2006; Geng et al., 2006; Mazelova et al., 2009). Therefore we introduced three point mutations (KVHPSST→AAHASST) within the last seven amino acids of the CD16.7-PC1-WT to determine whether the KVxPx motif served as a ciliary-targeting sequence. The alanine-scanning mutant abolished ciliary delivery, as the CD16.7-PC1-AAHASST fusion protein failed to localize to the primary cilium in 97% (34/35 cells) of transfected RCTE cells (Figure 1, B and C). In a complementary approach, we were able to rescue ciliary-targeting of the shortest CD16.7-PC1-truncation mutants [CD16.7-PC1-296, -339] by adding the KVHPSST sequence back to the C-terminus of the truncated PC1 tails (Figure 1, B and C). Analysis of another CD16 construct, which lacked the PC1 tail but contained both the transmembrane domain and full C-terminal tail of CD-7, revealed that the CD16-CD7 tail fusion protein localized to apical and basolateral plasma membrane domains as previously reported in Madin-Darby canine kidney (MDCK) cells (Haller and Alper, 1993) and was found in the primary cilium in 54% (20/37 cells) of ciliated, transfected cells (Figure 1B). Addition of KVHPSST to the CD16-CD7 fusion protein resulted in a twofold increase in delivery of the CD16-CD7+KVHPSST fusion protein to the primary cilium in 100% (40/40 cells) of ciliated RCTE cells (Figure 1, B and C), thus supporting the ciliary-targeting specificity of the KVHPSST sequence in the C-terminus of PC1.
Because soluble proteins, including GFP chimeras can follow an IFT-mediated transport pathway to cilia, our study utilized only integral membrane protein chimeras (containing the CD7 transmembrane domain), which are sorted to cilia directly from the Golgi. Thus, the data show that the trafficking signal for the vesicular transport of PC1 to the primary cilium is contained within the C-terminal seven amino acids and depends on a VxPx motif known in rhodopsin to bind Arf4 (Mazelova et al., 2009). Furthermore, the minimal ciliary-targeting motif is recognized by targeting machinery that appears to be conserved across species and ciliated cell types.
PC1 and PC2 termini interact with the small GTPase, Arf4
Arf GTPases are crucial regulators of membrane trafficking and cycle between inactive GDP-bound and active membrane-associated, GTP-bound states. Activated Arf GTPases recruit coat proteins, promote vesicle budding, and are involved in several exocytic and endocytic transport pathways (Nie and Randazzo, 2006). Arf4 in its GTP-bound, active state is unique to date in its interaction with rhodopsin, a seven transmembrane G-coupled receptor. In retinal epithelial cells, this interaction is necessary for rhodopsin trafficking from the trans-Golgi network to the rod outer segment, a specialized primary cilium. We therefore tested to determine whether the C-terminal, 20 amino acids containing the ciliary-targeting signal in PC1 might also directly bind Arf4.
In vitro binding of the extreme C-terminus of PC1 to purified Arf4 was tested first. Purified glutathione S-transferase (GST)-Arf4 was shown to be active in vitro based on binding of fluorescently labeled GTP (Figure S2A). Bead-immobilized, active, pure GST-Arf4 was loaded with nonhydrolyzable GTP-analogue preferred by Arf GTPases (GppNHp) and incubated with HeLa cell lysates overexpressing CD16.7-PC1-WT or CD16.7-PC1-truncation mutants. Six independent experiments using either GTP-γS or GTP to GST-Arf4 revealed that GST-Arf4 lacking the preferred nonhydrolyzable GppNHp was unable to bind PC1, thus underscoring the need for active Arf4 in PC1-Arf4 interactions. Subsequent immunoblot analysis of bound proteins (Figure 2A) showed interaction of purified Arf4 with CD16.7-PC1-WT. Deletion of the last 20 amino acids of the PC1 C-terminus (CD16.7-PC1-339) decreased the interaction between PC1 and GST-Arf4 nearly 40%; removal of 40 amino acids of the PC1 tail sequence severely diminished Arf4-binding activity (>80% decrease), despite similar protein input and ratio analysis. Further deletion also increased CD16.7 homooligomerization (multiple bands seen in lysate lane CD16.7-PC1-296), but the oligomers were not enriched in the Arf4-bound fraction. GST-Arf4 interactions with the alanine-scanning mutant were also decreased to levels similar to the CD16.7-PC1 truncation mutants (unpublished data). However, the add-back of the KVHPSST sequence found to be sufficient for ciliary targeting to the CD16.7-PC1 truncation mutants could not be shown to increase in vitro Arf4 binding in a statistically significant manner when compared with the truncated isoforms. These data suggest that while the KVHPSST sequence does enhance interaction with active, GTP-bound Arf4 and is required for ciliary trafficking of PC1 tail, additional residues in the extreme C-terminus of PC1 may stabilize the in vitro binding of Arf4 or further molecular mediators dependent on adjacent residues may be involved in PC1-Arf4 interactions.
FIGURE 2:
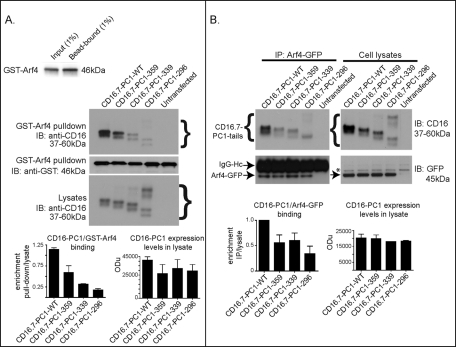
Arf4 interacts with the PC1 C-terminus. (A) In vitro interaction of PC1 and Arf4. Purified GppNHp-activated GST-Arf4 was immobilized on glutathione-Sepharose 4B (top left blot) and incubated with lysates from HeLa cells transfected with CD16.7-PC1-WT, -359, -339, or -296. Immunoblots were performed using antibodies specific for CD16 or GST. Left graph, enrichment of CD16.7-PC1 tails bound by bead-immobilized GST-Arf4 (binding normalized to amount of GST-Arf4 bound to the beads and lysate expression). Right graph, quantification in relative optical density units (OD) of the amount of CD16-PC1 protein expression in lysates. Experiments were performed in triplicate; representative panels from multiple experiments are shown. Input for immunoblots: lysate, 7%; Arf4 in vitro capture, 30%. (B) Cellular interaction of PC1 and Arf4. HeLa cells were cotransfected with Arf4-GFP and CD16.7-PC1-WT or CD16.7-PC1-truncation mutants. An antibody specific for GFP was used to immunoprecipitate Arf4-GFP and PC1 fusion proteins from HeLa lysates. Immunoblots were performed using antibodies specific for CD16 or GFP. Immunoprecipitated CD16.7-PC1 fusion proteins were normalized to corresponding immunoprecipitated Arf4-GFP and CD16-PC1 fusion protein expression in lysates between experiments. Left graph, binding of CD16.7-PC1 mutants to Arf4-GFP was calculated as a relative percentage of the CD16.7-PC-WT binding to Arf4-GFP. Differences in CD16.7-PC1 mutant protein expression levels were taken into consideration by first calculating the fraction coprecipitated with Arf4-GFP as a function of CD16.7-PC1 protein expression in the lysate. Right graph, the results were the same. Graph depicts the means ± SE of three separate experiments. Brackets indicate CD16.7-PC1 chimeric proteins; arrows point to Arf4-GFP or the heavy chain of the immunoprecipitating antibody (as labeled); * refers to nonspecific or unidentified bands. Input for immunoblots: CD16-PC1 lysates, 7%; Arf4-GFP IP blot for CD16, 22%; Arf4-GFP lysates, and IP analyzed for GFP, 1%.
Intracellular interaction between Arf4 and the PC1 tail was demonstrated by coexpressing Arf4-GFP with CD16.7-PC1-WT or CD16.7-PC1 truncation mutants in HeLa cells and performing coimmunoprecipitation assays. Immunoprecipitation of Arf4-GFP using a GFP antibody readily coprecipitated CD16.7-PC1-WT, while all the CD16.7-PC1-truncation mutant proteins exhibited a 55–70% reduction in binding (Figure 2B). Similar amounts of Arf4-GFP were immunoprecipitated in all cases. The reciprocal immunoprecipitation of CD16 fusion proteins and immunoblot analysis for coprecipitated Arf4-GFP was also conducted and showed similar trends (unpublished data). The differences in the interaction of CD16.7-PC1-WT and the CD16.7-PC1 truncation mutants with Arf4-GFP in cells mirrored the differences observed for binding to GST-Arf4 in vitro, suggesting that the C-terminal 20 amino acids ciliary-targeting region serves as an Arf4-binding site.
PC2 also localizes to the primary cilia of renal epithelial cells, where it interacts with PC1 and functions as a Ca2+-permeable cation channel (Hanaoka et al., 2000; Gonzalez-Perret et al., 2001; Vassilev et al., 2001). PC2 contains an RVxP targeting motif at the N-terminus that is required for ciliary delivery (Geng et al., 2006). To determine if PC2 also interacts with Arf4, we used GST-PC2 and FLAG-PC2 fusion proteins containing the minimal ciliary-targeting sequence to test protein interaction. Purified, bead-immobilized GST-PC2 (aa 1–15), but not GST, specifically bound purified GppNHp-bound Arf4 in vitro (Figure S2B). Deletion of the RVxP motif (GST-PC2-ΔRVqP) significantly diminished Arf4 binding, but was above the nonspecific interaction between Arf4 and the GST-only control. Use of a second FLAG-tagged PC2 (aa 1–219) further confirmed Arf4-binding specificity (Figure S2C). The composite data demonstrate that both PC1 and PC2 interact with Arf4 through a conserved domain that includes (R/K/Q)VxPx motifs.
Endogenous PC1 interacts with Arf4 and the Arf4 GTPase-activating protein, ASAP1
Endogenous PC1 resident in the Golgi and cilia was monitored using two independent polyclonal antibodies directed against a peptide epitope in the third cytoplasmic loop region of PC1 (NM002 and NM032) and remote from the C-terminal ciliary-targeting sequence (Figure 3A). For the complete antibody characterization and immunoelectron microscopy of Golgi and ciliary localizations, see the Supplemental Text and Figure S3.
FIGURE 3:
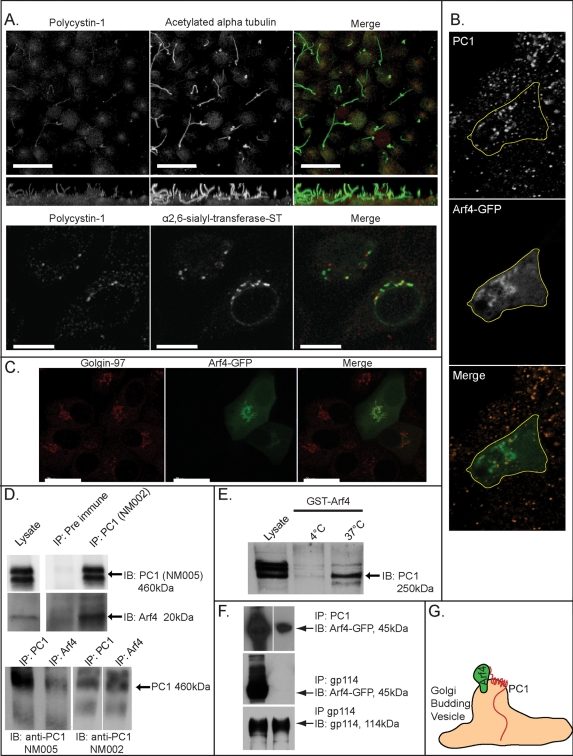
Arf4 colocalizes and forms a complex with PC1 in the Golgi. Note: Because each channel in (A) and (B) was rendered in three dimensions using separate palettes in Voxx2 software, regions of colocalization do not appear yellow. (A) A pAb against PC1 detects a pool of PC1 in the Golgi apparatus and in cilia. Top row, human RCTE cells were grown 3 d postconfluence on filter supports and labeled with rabbit pAb against PC1 (NM002, green) and mouse mAb against acetylated α-tubulin (red). Bottom row, human RCTE cells were transfected with ST tyr isoform. Cells were immunostained for PC1 with NM002 and rhodamine-conjugated secondary antibody, and for myc-tagged α2,6-sialyl-transferase with a specific mAb directed against myc and a fluorescein-conjugated secondary antibody. Scale bar: 10 μm. (B) Arf4 and PC1 colocalize in the Golgi. MDCKII cells expressing GFP-Arf4 (green) were fixed and immunostained for PC1 (NM002, ocher), and a single transfected cell within the monolayer is outlined (single confocal Z-slice). See also Supplemental Movie 1. (C) Arf4-GFP colocalizes with Golgin-97. (D) Arf4 and PC1 form a complex that can be reciprocally coprecipitated. PC1 antibody NM002 specifically immunoprecipitated PC1 and Arf4 from RCTE lysate compared with control rabbit IgG (top panels). Equivalent results were found using MDCKII cells transfected with Arf4-GFP (unpublished data). PC1 and Arf4 protein levels were enriched in PC1-specific immunoprecipitation lanes (detailed in the text) and calculated as fraction of lysate protein immunoprecipitated with PC1 antibody relative to the fraction of lysate protein precipitated with control IgG (n = 3). Endogenous Arf4 was immunoprecipitated and immunoblotted for endogenous PC1 using RCTE (or MDCKII, unpublished data) lysates and two different rabbit pAbs against PC1 (NM002 and NM005, bottom panels). (E) Arf4 and PC1 form a complex in vitro. Purified GppNHp-activated GST-Arf4 was incubated with MDCKII lysate at 4°C or 37°C. Blots were probed for PC1; relative protein migration was 250 kDa due to matrix metalloprotease cleavage in the presence of magnesium. See Figures S3F and S8. (F) Arf4-GFP interacts with endogenous PC1, but does not interact with nonciliary, apical plasma membrane protein gp114. Arf4-GFP MDCKII lysate was immunoprecipitated in parallel with antibodies specific for PC1 or gp114. Blots were probed with antibodies specific for gp114 and GFP. (G) Cartoon representation of the predicted interactions between PC1 and Arf4 in specialized Golgi exit sites.
Arf4-GFP fusion was localized predominantly to the Golgi apparatus in mammalian kidney cells (Figure 3, B and C), consistent with previous reports describing Arf4 localization in the retinal photoreceptors (Mazelova et al., 2009). Colocalization of endogenous PC1 and Arf4-GFP revealed the two proteins present together in discrete punctae closely associated with the Golgi ribbon of MDCKII cells (Figure 3B and Supplemental Movie 1), suggesting that endogenous PC1 is present with Arf4 within subdomains of the Golgi.
Coimmunoprecipitation assays revealed that NM002, but not a control rabbit immunoglobulin G (IgG), coprecipitated endogenous Arf4 (Figure 3D, top panels) as well as overexpressed Arf4-GFP in RCTE and MDCKII cells (Figure 3F, top panel). The reciprocal immunoprecipitation of endogenous Arf4 coprecipitated PC1 (Figure 3D, bottom panels; confirmation of Arf4 immunoprecipitation is shown in Figure S4). Immunoprecipitation of endogenous PC1 was independently confirmed by immunoblotting with antibodies against two different epitopes (Figure 3D, bottom panels; PC1 antibodies are described in Figure S3F). Quantification revealed enrichment of coprecipitated PC1 (5.44-fold, SE ± 0.26, n = 3) and Arf4 (5.64-fold, SE ± 0.26, n = 3) compared with control IgG. Endogenous PC1 from kidney cell lysates could bind purified, GppNHp-loaded, GST-Arf4 in vitro, and binding was enhanced at physiological temperatures, as has been reported for in vitro protein–protein interactions (Figure 3E; Dong et al., 2004). Endogenous PC1 bound Arf4-GFP, whereas a parallel control immunoprecipitation of the nonciliary, apical plasma membrane protein gp114/CEACAM did not immunoprecipitate Arf4-GFP (Figure 3F). The combined immunostaining, in vitro, and cell-based biochemical data suggest PC1 forms a complex with Arf4 in the Golgi and is segregated from apical cargo in discrete punctae, as shown by the lack of gp114-Arf4 interaction.
The activity of Arf GTPases is closely regulated by GTPase-activating proteins (GAPs), which temporally regulate GTP hydrolysis and likely participate in vesicle budding through interaction with coat proteins via a variety of scaffolding domains (Nie and Randazzo, 2006; Kahn et al., 2008; Spang et al., 2010). ASAP1 functions as an Arf4-specific GAP, participates in rhodopsin trafficking, is associated with tumor cell metastasis, and regulates vesicle coat assembly via a Bin/Amphiphysin/Rvs domain (Nie et al., 2006; Mazelova et al., 2009). As with PC1, active (GppNHp-bound) GST-Arf4 was able to bind ASAP1 in vitro, in a temperature-dependent manner (Figure 4A). Hydrolyzable GTP and GDP failed to support measurable binding (unpublished data). Coimmunoprecipitation experiments demonstrate that ASAP1 coprecipitates with PC1 or endogenous Arf4 from renal epithelial cell lysates (Figure 4, B and C; confirmation of ASAP1 immunoprecipitation is shown in Figure S4). Comparisons with rabbit control IgG precipitations confirm coprecipitation specificity with a ≥5.2-fold (SE ± 1.86, n = 3) enrichment of ASAP1 compared with control IgG (Figure 4C, top panels). Conversely, immunoprecipitation of ASAP1 was able to coprecipitate PC1, as demonstrated using two different antibodies against PC1 on immunoblot (Figure 4C, bottom panels). Like Arf4, ASAP1 could also be shown to associate with the CD16.7-PC1-WT chimeric protein in both RCTE and HeLa cells, but did not associate with the CD16.7-PC1-359 protein lacking the ciliary-targeting signal (Figure 4D). The composite data show independent lines of evidence that PC1, Arf4, and ASAP1 form a multimeric complex dependent on the C-terminal 20 amino acids of PC1.
FIGURE 4:
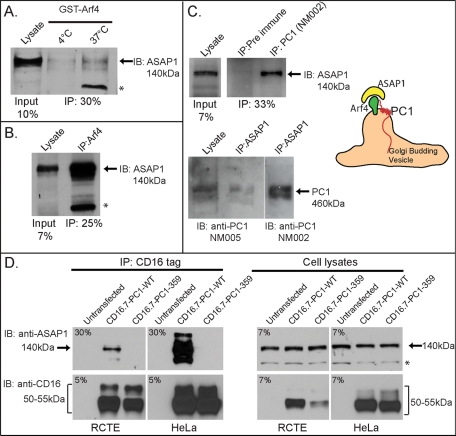
The Arf GAP, ASAP1, binds Arf4 and is part of the PC1-Arf4 complex. (A) ASAP1 interacts with Arf4 in vitro. Purified GppNHp-activated GST-Arf4 was incubated with MDCKII lysate at 4°C or 37°C. Blots were probed for ASAP1. (B) Arf4 binds ASAP1 in cells. Endogenous Arf4 was immunoprecipitated from MDCKII cell lysates. Immunoprecipitates were resolved by SDS–PAGE and immunoblotted for endogenous ASAP1. (C) PC1 binds ASAP1 in cells. Endogenous PC1 was immunoprecipitated from RCTE (and MDCK, unpublished data) cell lysates and immunoblotted for ASAP1. PC1 antibody NM002 specifically immunoprecipitated ASAP1 from RCTE lysate compared with control rabbit IgG (top panels). ASAP1 protein levels were enriched in PC1-specific immunoprecipitation lanes (detailed in the text) and calculated as fraction of lysate protein immunoprecipitated with PC1 antibody relative to the fraction of lysate protein precipitated with control IgG (n = 3). RCTE (and MDCKII, unpublished data) cell lysates were immunoprecipitated for endogenous ASAP1 and immunoblotted for PC1 using two different pAbs (NM002 and NM005). Cartoon represents predicted interaction between PC1, Arf4, and ASAP1. (D) Truncation mutants of CD16.7-PC1-WT block ASAP1 binding. mAb directed specifically against CD16 was immobilized on protein G agarose and incubated with untransfected HeLa or RCTE lysates or lysates from HeLa or RCTE cells transfected with CD16.7-PC1-WT or CD16.7-PC1-359. Samples were immunoblotted with antibodies specific for CD16 and ASAP1. Percent input shown in upper left corner of each blot. * marks nonspecific or unidentified band.
PC1 interacts with Rab GTPases
Rab GTPases are well-known regulators of membrane trafficking. Rab8 and Rab11 are required for ciliary trafficking of rhodopsin (Schwartz et al., 2007; Mazelova et al., 2009). Rab8 interacts with fibrocystin and the BBS proteins and is involved in ciliogenesis (Nachury et al., 2007; Schwartz et al., 2007; Mazelova et al., 2009). Endogenous PC1 from confluent MDCKII lysates immunoprecipitated Rab6, Rab8, and Rab11 using a covalent NM002-affinity resin (Figure 5A, left blots). Control resin lacking the PC1 antibody did not reveal coprecipitated GTPases; thus, neither PC1 nor the Rab proteins bound nonspecifically to the beads (Figure 5, A and C). Identical results were obtained using RCTE cell lysates and a TrueBlot secondary antibody that exhibits minimal reactivity with denatured antibody light chains (Figure 5A, right blots). Quantification of immunoprecipitation and comparisons with preimmune rabbit serum precipitation demonstrate the specific coprecipitation of Rab6, Rab8, and Rab11 with endogenous PC1 (Figure 5A, graph). Reciprocal immunoprecipitations using antibodies directed against Rab8 or Rab11 further confirmed that these GTPases exist in a complex with PC1 using two different PC1 antibodies (Figure 5B; confirmation of immunoprecipitation of Rab8 and Rab11 proteins by the relevant antibodies is shown in Figure S4).
FIGURE 5:
PC1 interacts specifically with Rab6, Rab8, and Rab11. (A) PC1 interacts with Rab GTPases in cells using two different methods. Left blots, endogenous PC1 was immunoprecipitated from postconfluent MDCKII cell lysates using covalently conjugated NM002-affinity resin. Coisolated proteins were resolved by SDS–PAGE and analyzed by immunoblotting for PC1, Rab6, Rab8, and Rab11. Uncoated (bead) resin served as a nonspecific binding control. Right blots, endogenous PC1 was immunoprecipitated from RCTE cell lysates; proteins were immunoblotted for individual Rab GTPases as indicated and detected using TrueBlot secondary antibody. Immunoprecipitations performed with preimmune rabbit serum served as a control. Graph represents quantification of multiple experiments represented by right-hand blots. n ≥ 3 experiments, *p ≤ 0.05, **p ≤ 0.005. (B) Rab8 and Rab11 antibodies reciprocally immunoprecipitated PC1 detected with two different anti-PC1 antibodies (NM002 and NM005). (C) PC1 does not interact with endocytic GTPases, Rab5 and Rab7. Endogenous PC1 was immunoprecipitated from postconfluent MDCKII cell lysates using covalently conjugated NM002-affinity resin. Coisolated proteins were resolved by SDS–PAGE and analyzed by immunoblotting for PC1, Rab5, and Rab7. (D) Truncation mutants of CD16.7-PC1-WT reduce Rab8 and Rab11 binding. mAb directed specifically against CD16 was immobilized on protein G agarose and incubated with untransfected HeLa or RCTE lysates or lysates from HeLa or RCTE cells transfected with CD16.7-PC1-WT or CD16.7-PC1-359. Samples were immunoblotted with antibodies specific for Rab8 and Rab11 and detected using TrueBlot secondary antibody. Confirmation of CD16 immunoprecipitation was conducted as in Figure 4 (unpublished data). Background in untransfected lane may be due to combination of nonspecific binding to protein G agarose and comigrating antibody light chains, detection of which cannot be completely eliminated for some antibodies, even with the TrueBlot system. Graphs show quantification of representative experiments. n = 2, *p ≤ 0.05, ** p ≤ 0.005. (E) Confocal microscopy of cultured NK cells shows colocalization (arrowheads) of Rab11 (red), PC1 (green), and ASAP1 (blue) in select punctae on the Golgi ribbon identified by PC1 staining. Scale bar: 20 μm. See also Figure S5 and Supplemental Movie 2.
The specificity of the Rab GTPase interactions with PC1 was further confirmed by immunoblotting PC1 immunoprecipitates for endocytic Rab GTPases. Neither Rab5 nor Rab7 were part of the PC1-Rab GTPase complex (Figure 5C; confirmation of immunoprecipitation of Rab5 and Rab7 proteins by the relevant antibodies is shown in Figure S4). This finding was consistent with published literature that Rab5 is not involved in ciliogenesis nor Smoothened delivery to primary cilia, and the fact that Rab7 functions in late endocytosis (Feng et al., 1995; Nachury et al., 2007; Boehlke et al., 2010). Finally, immunoprecipitation of CD16.7-PC1-WT from either RCTE or HeLa cells resulted in coprecipitation of Rab8 nearly twofold above the background levels seen using untransfected cell lysates, while CD16.7-PC1-359 did not bind Rab8 above background (Figure 5D). Background in the untransfected lane may be due to combination of nonspecific binding to protein G agarose and comigrating antibody light chains, detection of which cannot be completely eliminated for some antibodies, even with the TrueBlot system. Immunoprecipitation of CD16.7-PC1-WT resulted in 1.3-fold coprecipitation of Rab11 above the background levels seen using untransfected cell lysates, while CD16.7-PC1-359 did not bind Rab11 above background (Figure 5D). Quantitatively, in multiple trials using two different cell types and appropriate quantitative measures, the wild-type fusion protein consistently bound Rab8 and Rab11 at a level that was measurably above the background and the truncated CD16.7-PC1-359 protein binding, and statistically significant at p ≤ 0.05 or better. Thus, using four different biochemical approaches, we found a select subset of Rab GTPases consistently complexed to the PC1 C-terminus containing the membrane distal 20 amino acids.
Immunolocalization of Rab11, PC1, and ASAP1 via confocal imaging revealed all three proteins in discrete punctae (Figure 5E). The subcellular localization patterns of Rab11, PC1, and ASAP1 uniquely overlapped at discrete tips on Golgi tubules in ∼10% of cells per section, but not at other cellular locations where PC1 was present (Figure S5 and Supplemental Movie 2), showing the proteins may transiently colocalize and interact at what are suggested to be Golgi exit sites.
PC1 trafficking to cilia depends on expression of Arf4 and Rab8
The data shown so far define a KVHPSST amino acid targeting signal contained within a 20 amino acid C-terminal domain that is recognized by a multimeric protein complex with known functions in exocytosis. Expression of dominant negative Rab11 (S25N) or Arf4 I46D, which exhibits impaired ASAP1-mediated GTP hydrolysis (Mazelova et al., 2009), resulted in an increased intracellular accumulation of endogenous PC1 (Figure S6). Small hairpin RNAs (shRNAs) were therefore used to deplete Arf4 or Rab8a in order to obtain a quantitative measure for the functional involvement of Arf4 or Rab8a in the ciliary trafficking of PC1. Ciliary delivery of CD16.7-PC1-WT was analyzed and quantified morphologically (Figure 6). The cilia of RCTE cells are very long (as shown in Figure 2) and are readily identified in careful slice-by slice analyses of XY-, XZ-, and YZ-projections of samples labeled for acetylated α-tubulin. The cilia identified in the images were present in >5 Z-slices (e.g., >1.4 μm in length) and clearly projected above the apical plane. For clarity, we identify cilia only on the ciliated CD16-transfected cells with arrows on XZ planes. Further evidence of ciliary localization is provided in Supplemental Movies 3–5 (shRNA knockdown and CD16/acetylated α-tubulin labeling). For Rab8a, we used an shRNA closely matched to the small interfering RNA (siRNA) sequence previously used by others to effectively deplete Rab8a mRNA and protein (Schuck et al., 2004; Hattula et al., 2006) resulting in 25–40% reduction (n = 2; SE ± 14%) in expression in RCTE cells (Figure 6). Because the monoclonal anti-Rab8 antibody recognizes Rab8a and Rab8b, we suspect that the less-than-expected reduction of total Rab8 may be due to Rab8b expression. Five different shRNAs against Arf4 were first tested in HeLa cells, and two that were found capable of decreasing Arf4 protein expression up to 70% after 48 h were used for all further experiments (Figure S7). Arf4271123 and Arf462890 shRNAs used in combination (to minimize off-target effects) in RCTE cells reduced Arf4 expression by 50–70% (Figure 6; n = 2; SE ± 19%). For analyses of shRNA effects on ciliary trafficking, RCTE cells were plated on filter supports, transfected with shRNA plasmids, and incubated for 72 h for maximal GTPase depletion, at which time the cells were retransfected to express CD16.7-PC1-WT as a ciliary transport marker. When plated at 90% confluence, RCTE cells produce primary cilia within 12 h of plating. Therefore, under the experimental conditions used for measuring ciliary transport, inhibition of ciliary formation was not observed (541 cells counted, 76% ciliated untransfected, 70% ciliated glyceraldehyde-3-phosphate dehydrogenase [GAPDH] shRNA, 80% ciliated Rab8 shRNA, 72% ciliated Arf4 shRNA), since ciliation would have occurred well before the peak of GTPase depletion at 72 h posttransfection. To monitor ciliary transport, cells were immunostained for acetylated α-tubulin to identify cilia and for CD16 to identify PC1 fusion protein, and shRNA-expressing cells were identified based on GFP expression (encoded by the same plasmid vector as the shRNA; Figure 6 and Supplemental Movies 3–5). In two independent experiments, CD16.7-PC1-WT protein was detected in cilia of 80–87% of ciliated control cells without shRNA transfection or ciliated cells expressing GAPDH shRNA (Figure 6). In contrast, cells expressing Arf4 or Rab8 shRNA exhibited a >60% reduction in the number of cells exhibiting detectable ciliary expression of the integral membrane protein CD16.7-PC1-WT. Statistical significance was evaluated as described in Materials and Methods, and based on counting over 1300 cells (>250 cells/shRNA condition, including those counted to evaluate impact on ciliogenesis), using representative confocal images consisting of 20–25 slices per Z-stack collection for quantification. Thus we demonstrate that Arf4 and Rab8 are functionally required for the transport of PC1 to the primary cilium.
FIGURE 6:
ShRNA-mediated depletion of Arf4 or Rab8 impairs ciliary trafficking. RCTE cells were sequentially transfected with pGIPZ-GFP shRNA plasmids (GAPDH, Arf4, or Rab8) for 72 h, followed by transfection and expression of CD16.7-PC1-WT for an additional 20 h. Samples were labeled for CD16 and acetylated α-tubulin. Micrographs, confocal stacks of samples were collected and individual cells (CD16.7-PC1-WT transfected only, or CD16.7-PC1-WT + shRNA GFP [labeled as GAPDH, Arf4, or Rab8 shRNA]) were analyzed for cilia (Cy5), shRNA transfection (GFP coexpression), and CD16.7-PC1-WT (Alexa 546) expression within cilia. Graph, p values represent comparison of counts of dually transfected cells relative to the counts of cells transfected with CD16.7-PC1-WT alone (labeled as “None”). Comparisons of Arf4 or Rab8 dually transfected cells to GAPDH dually transfected cells yielded the same results as comparisons to CD16.7-PC1-WT “None” control. Statistical analyses based on > 250 cells per shRNA condition. (Immunoblot) For biochemical studies, RCTE cells were sequentially transfected with pGIPZ-GFP shRNA plasmids (GAPDH, Arf4, or Rab8) for 72 h. ShRNA-transfected cells were isolated by flow-sorting based on GFP coexpression and average shRNA-mediated reduction in Rab8 and Arf4 was quantified by immunoblot analyses (n = 2).
DISCUSSION
Defective ciliary protein targeting and signaling causes altered cellular differentiation and cell cycle regulation. ADPKD gene mutations perturb temporal and spatial localization of polycystins, causing these proteins to be absent from cilia. To date, a detailed assessment of the ciliary-targeting mechanisms for the polycystins has not been reported. In this paper, we show a conserved (K/R/Q)VxPx ciliary-targeting motif, previously identified in rhodopsin and PC2 (Deretic et al., 2005; Geng et al., 2006), is necessary to traffic PC1 to primary cilia. The last 20 amino acids of PC1 promotes direct association between the C-terminus of PC1 and active, GTP-bound Arf4. Truncation of the PC1 tail and deletion of KVHPSST negatively impacts binding of membrane-trafficking components Arf4, ASAP1, and Rab GTPases (Rab8 and Rab11). Consequently, chimeric PC1 proteins bearing the truncated signal fail to reach cilia and accumulate intracellularly in the Golgi region and ER or apical plasma membrane. While this work was under review, PC2 transport to cilia was reported to bypass the trans-Golgi, and instead exit from cis-Golgi for ciliary targeting (Hoffmeister et al., 2011). Our demonstration that Arf4 is colocalized with the trans-Golgi marker Golgin-97, and that the known N-terminal ciliary-targeting motif (RVxP) of PC2 interacts with Arf4, but not when mutant, suggests that there must also be a Golgi-mediated pathway for PC2 to the cilia. Arf4 or Rab8 knockdown significantly impairs PC1 delivery to cilia. Together, these data define a conserved domain in the termini of both polycystins with functional importance in the assembly of a ciliary-trafficking complex (Figure 7).
FIGURE 7:
PC1 ciliary-trafficking pathway depends on a conserved K/R/QVxPx sequence and Arf4, Rab6, Rab11, Rab8, and ASAP1-trafficking machinery, and is distinct from the endocytic-trafficking route regulated by Rab5 and Rab7.
The known activities of the ciliary-trafficking constituents predict an ordered sequence of events beginning with cargo recognition in the Golgi and followed by vesicle budding, translocation, docking, and vesicle fusion. Arf GTPases function as adapters to link cargo proteins with cytosolic coat proteins to promote vesicle budding (Kahn, 2009; Spang et al., 2010). Our demonstration that Arf4 is central to the recognition of the polycystins and transport of PC1 from the Golgi to cilia using shRNA knockdown has important implications for the transport mechanism. ASAP1 regulates GTPase hydrolysis, induces membrane curvature, and scaffolds Rab GTPase effectors (Nie et al., 2006; Inoue et al., 2008). The GAP activity of ASAP1 is tightly modulated through conformational changes induced by differential protein–protein interactions (Jian et al., 2009). Our finding that expression of an Arf4 mutant that fails to bind ASAP1 collapses the Golgi, taken together with the fact that ASAP1 stimulates GTP-hydrolysis by Arf4 (Mazelova et al., 2009), suggests that the nucleotide-binding and hydrolysis cycle of Arf4 and its regulation by ASAP1 are critical in Golgi function. The formation of an Arf4/ASAP1/PC1 complex in the Golgi suggests that Arf4 binds PC1 and recruits ASAP1 to the Golgi, where it contributes to the further assembly and regulation of the cilia-directed exocytosis complex at specialized Golgi exit sites. We envision that vesicle budding and recruitment of Rab proteins precludes ASAP1-triggered Arf-GTP hydrolysis until budding is complete and subsequent hydrolysis may allow Arf4 recycling in preparation for subsequent docking and fusion events.
We demonstrate that among the proteins recruited by the Arf4/ASAP1 complex are Rab8 and Rab11a, which have well-established functions in post-Golgi trafficking and cytoskeletal-mediated plasma membrane targeting, and emerging roles in ciliogenesis (Huber et al., 1993; Chen et al., 1998; Lapierre et al., 2001; Nachury et al., 2007; Schwartz et al., 2007; Yoshimura et al., 2007; Knodler et al., 2010). Our findings tie in well with the previously demonstrated interactions between the Rab8 guanine exchange factor (GEF), Rabin8, and Rab11a that occur at the base of cilia and suggest that Rab8 activity is required for centrosomal vesicle docking (Knodler et al., 2010; Westlake et al., 2011). Rab11 is well known to be associated with epithelial apical recycling endosomes, which are utilized for the exocytosis of a subset of newly synthesized basolateral proteins en route to the plasma membrane, in addition to being a sorting station for rerouting endocytosed proteins (Ullrich et al., 1996; Chen et al., 1998; Sheff et al., 1999; Orzech et al., 2000; Chen and Wandinger-Ness, 2001). Rab6 participates in vesicle tethering in the Golgi and cooperates with Rab11 (Miserey-Lenkei et al., 2007; Hayes et al., 2009). Using biochemical analyses, we show that PC1 interacts with Rab6, Rab8, and Rab11, but not Rab5 and Rab7, indicating that ciliary-targeted PC1 is not processed through the early endocytic pathway. These data are consistent with the rhodopsin-trafficking model, and demonstrate that trafficking mechanisms consisting of Rab GTPases (Rab6, Rab11, Rab8), Arf4 GTPase, and Arf GAP ASAP1 are conserved between retinal photoreceptor cells and renal epithelial cells. We predict that additional molecules, such as Rabin8 and FIP3, may contribute to PC1 trafficking. Recent studies revealed that transport protein particle II complex components (C3, C9, and C10) and Rab11 are required for vesicular trafficking of Rabin8 to the centrosome, where Rabin8 subsequently recruits and activates Rab8 (Knodler et al., 2010; Westlake et al., 2011). Our data, taken in the context of published literature, suggest that PC1 participates in the GTPase vesicle ciliary relay, starting with Rab6 and Arf4 in the Golgi, and then trafficks with Rab11 to the ciliary base, where Rab8 participates with various effectors to facilitate ciliary transport.
The commonalities between the basolateral and ciliary-trafficking machinery are also of interest from the perspective of the temporal and spatial regulation of PC1 localization during epithelial polarization (Wilson, 2001; Silberberg et al., 2005). PC1 disappears from junctional complexes and relocalizes to cilia in fully polarized cells (Silberberg et al., 2005; Xu et al., 2007). Rab8 and Rab11 are regulators of Golgi to basolateral plasma membrane trafficking in epithelia and the dendrites of neurons (Huber et al., 1995; Chen et al., 1998; Chen and Wandinger-Ness, 2001; Ang et al., 2003), yet localize to cilia in fully differentiated epithelia (Mazelova et al., 2009; Knodler et al., 2010; Nachury et al., 2010). The analogy is also exemplified in Caenorhabditis elegans, where the primary cilium is located at the end of a neuronal dendrite, and here Rab8 also plays a role in ciliary trafficking (Kaplan et al., 2010). In primary ADPKD cells, Rab8 localization and basolateral, but not apical, trafficking are perturbed with concomitant Golgi accumulation of cargo (Charron et al., 2000a, 2000b). Changes in localization depend on the differentiation state of renal epithelia, and suggest the need for a shunting mechanism that switches the sorting of proteins like the polycystins from basolateral to ciliary targeting; this process is likely to involve centrosome positioning, the cytoskeleton, exocyst components, and specific adaptors that are activated upon cell differentiation (Bacallao et al., 1989; Folsch et al., 2003; Rogers et al., 2004; Ishikawa et al., 2005; Marshall, 2008; Zuo et al., 2009). Primary cilia have ciliary “necklaces” at their bases that serve as a docking sites and filters (Pedersen et al., 2008; Jin et al., 2010). The exclusion of apical membrane protein gp114 from the trafficking complex offers further evidence for a model wherein ciliary membrane protein trafficking from the Golgi is largely distinct from the apical pathway driven by lipid rafts (Simons and Wandinger-Ness, 1990; Wandinger-Ness et al., 1990), though there may be some exceptions, as exemplified by Smoothened and somatostatin receptor 3 (Milenkovic et al., 2009; Jin et al., 2010). Interestingly, the CD16-CD7 chimera bearing the full-length CD7 C-terminus was promiscuous in its membrane localization, but demonstrated at least a twofold increase (minimal estimate, since we used an all-or-none measure for ciliary localization) in ciliary targeting upon addition of the KVHPSST. This suggests that some proteins may be devoid of specific inclusion or exclusion mechanisms, and that minimal signals can increase pathway selectivity. The PC1 ciliary transport pathway shares commonalities with the basolateral-sorting pathway that is dependent on the recognition of specific signals in cargo proteins (Schuck and Simons, 2004). Connections to the recycling endosome could offer a mechanism for PC1 rerouting from the basolateral membrane. However, only a small pool of PC1 likely transits from basolateral membrane to cilia, since PC1 was not associated with early endosomal Rab5 or late endosomal Rab7. Through trafficking machinery components with dual roles in basolateral and ciliary trafficking, the cilium may remain intimately connected to events occurring at the noncontiguous basolateral membrane domain.
As the ciliary-trafficking mechanisms emerge, it is evident there are multiple transport machineries and pathways, which may interface and cross-talk. For example, Smoothened interacts with Rab23, but utilizes a lateral diffusion mechanism to enter cilia (Milenkovic et al., 2009; Boehlke et al., 2010). Cross-talk between IFT and vesicular-mediated trafficking is evidenced by IFT20, which localizes to the Golgi with PC2, and associates with Rab8 (Scheffers et al., 2002; Follit et al., 2006). These observations highlight the complexity of ciliary trafficking, and solidify the close link between defects in IFT and GTPase-regulated vesicular trafficking and disease.
We identified a (K/R/Q)VxPx ciliary-targeting signal at the PC1 C-terminus, and established a functional requirement for Arf4 and Rab8 in the delivery of PC1 to cilia. The terminal signals and machinery utilized by polycystins bear strong homology to the signal and trafficking mechanism of rhodopsin (Deretic et al., 2005; Deretic, 2006; Mazelova et al., 2009). Conservation of ciliary-trafficking mechanisms in diverse cell types provides a compelling molecular rationale for ciliopathies that affect multiple target organs. The close coupling between cargo and transport machinery illustrates how mutations in ciliary proteins may preclude assembly of the macromolecular-trafficking complex or trapping of trafficking machinery and result in Golgi retention or cell death. Our results constitute a critical step in identifying key contributors to cystogenesis that represent potential targets for therapy.
MATERIALS AND METHODS
Cell lines and reagents
Madin-Darby canine kidney II (MDCKII; Ward et al., 2004), primary normal human kidney 1 (NK1; Roitbak et al., 2004), RCTE (Nauli et al., 2006), 9-12 PKD (Nauli et al., 2006) Swiss 3T3, and HeLa cells (Huber et al., 1993) were cultured as described in the cited references. All transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Normal rabbit IgG control consisted of pooled preimmune sera from two rabbits (Covance, Princeton, NJ). Primary antibodies were: mouse mAb directed against myc epitope (a gift from Karen Colley, University of Illinois College of Medicine, Chicago, IL); mouse mAb directed against acetylated α-tubulin (Sigma-Aldrich, St. Louis, MO); rabbit pAb directed against GFP (Molecular Probes, Invitrogen, Eugene, OR); rabbit pAb directed against α-ß-tubulin (Cell Signaling Technology, Danvers, MA); mouse mAb directed against Rab8, Rab11 (immunoprecipitation and immunoblots), and ASAP1 (BD BioSciences, San Jose, CA); rabbit pAb against Rab6, rabbit pAb directed against Arf4, mouse mAb against CD16, and goat pAb directed against Rab11 (immunofluorescence; Santa Cruz Biotechnology, Santa Cruz, CA); mAb directed against gp135 (Ojakian and Schwimmer, 1988); mAb directed against gp114 (provided by Karl Matlin, University of Chicago, Chicago, IL); rabbit pAb directed against Arf4 (generated using purified Arf4 as an antigen; Deretic et al., 2005); mouse mAb directed against Rab5 (Qiu et al., 1994); rabbit pAb directed against Rab7 (Feng et al., 1995); rabbit pAb directed against PC1 NM005 (Roitbak et al., 2004); and NM002 and NM032 (described in the Supplemental Text). Fluorophore-conjugated secondary antibodies raised in goat were from Jackson Laboratories or in donkey from Invitrogen (Alexa Fluor dyes). Xenopus WT-Arf4-GFP (Mazelova et al., 2009) was excised and spliced into mammalian expression vector pcDNA3.1 for these studies. All reagents were from Sigma, unless otherwise noted.
Sequence analyses
Alignment of the C-terminal ∼112 amino acid sequences of human, mouse, canine, and gallus PC1 (illustrated in Figure 1A) revealed two areas with a high degree of sequence conservation (human PC1 aa 4208–4229 and aa 4266–4289). ExPASy PRATT pattern searches (http://web.expasy.org/pratt; Jonassen et al., 1995) were used to comparatively evaluate multiple ciliary proteins in various combinations for conserved patterns, consistently identifying related regions in both polycystins and fibrocystin. The sequences highlighted in red in Figure 1 were derived from a comparison of: hPC2 N-terminal sequence (MVNSSRVQPQQPGDAKRPPAPRAPDPGRLMA), hPC1 C-terminal sequence 1 (EPEPSRLQAVFEALLTQFDRLNQATEDVYQLEQQLHSLQ GRRS), hPC1 C-terminal sequence 2 (PALPSRLARASRGVDLATGPSRTPLRAKNK VHPSST), and hFC C-terminus (KRSKSRKTKPEEIPESQTNNQNIHIHISSKRRESQGPKKEDTVVGEDMRMKVMLGKVNQCPHQLMNGVSRRKVSRHIVREEEAAVPAPGTTGITSHGHICAPGAPAQQVYLQETGNWKEGQEQLLRYQLAGQNQLLLLCPDFRQERQQLPGQSRLSKQSGSLGLSQEKKASCGATEAFCLHSVHPETIQEQL). The predicted pattern P-x(1,2)-P-[GS]-x(4)-[APTV]-x(2)-[AGP]-x(3)-[ADQ]-x-[DG]-x(3)-[AQT] is expected to yield ∼5.3 random matches in 100,000 sequences (50,000,000 residues; Nicodeme, 2001). Inclusion of soluble ciliary protein sequences failed, yielding only highly degenerate motifs with a high probability of random occurrence. When the predicted motif was used in a protein database search, the motif was reidentified in the sequences of interest found in Table 1. We noted an abundant ∼250 kDa cleavage fragment of polycystin-1 when conducting in vitro GTPase binding experiments, which require magnesium to stabilize bound nucleotide. The 250 kDa polycystin-1 fragment was absent in EDTA-containing buffers. Sequence analyses identified a series of potential matrix metalloprotease sites in the ectoplasmic domain near the G-protein–coupled receptor proteolytic cleavage site. The sites were identified based on similarity to consensus metalloprotease cleavage sites (Caescu et al., 2009). The 250 kDa fragment consists of the remaining transmembrane segments and C-terminus and is recognized by NM002 and NM005 antibodies (Figure S8).
TABLE 1:
Conserved sequences identified in PC1, PC2, fibrocystin, and progressive rod-cone degeneration protein homologue.
| Protein | Residues | Homology |
|---|---|---|
| PC1: Homo sapiens (human) P98161, -2, -3 | 4210–4231 | Pe.PSrlqaVfeAlltQfDrlnQ |
| 4199–4220 | ||
| 4209–4230 | ||
| 4268–4290 | PalPSrlarAsrGvdlAtGpsrT | |
| 4257–4279 | ||
| 4267–4289 | ||
| PC2: Homo sapiens (human) Q13563 | 9–31 | PqqPGdakrPpaPrapDpGrlmA |
| Fibrocystin: Homo sapiens (human) Q8TCZ9 | 3968–3989 | Pa.PGttgiTshGhicApGapaQ |
| Progressive rod-cone degeneration protein homologue: Mus musculus (mouse) | 24–45 | Pe.PSrvdgTvvGsgsDtDlqsT |
Immunofluorescence staining
The myc-tagged trans-Golgi marker α2,6-sialyl-transferase (ST tyr isoform; Fenteany and Colley, 2005) was expressed in MDCKII and human RCTE or GFP-N-acetylgalactosaminyltransferase-2 was expressed in MDCKII cells (Organelle Lights Golgi-GFP, Invitrogen). Cells were fixed with 3% paraformaldehyde (PFA) 24 h posttransfection, and processed for immunostaining.
GFP-Arf4 was localized by fixing cells and mounting and viewing them directly or after immunostaining for other markers as described in the paragraph below.
For ciliary-trafficking and immunolocalization experiments, RCTE cells were grown on filter supports to 3 d postconfluence and fixed with 3% PFA. Cells were processed using Triton-X 100 (Roitbak et al., 2004) or 1% SDS (Xu et al., 2007), and primary and secondary antibody incubations were performed in a humidified chamber at 37°C. Primary NK1 cells immunostained for Rab11, PC1, and ASAP1 were fixed with 3% PFA or by the pH shift-fix method (Bacallao et al., 2006), and labeled as previously described (Roitbak et al., 2004). Cells costained with antibodies to α-ß-tubulin and to CD16 were first labeled for α-ß-tubulin and washed, then labeled with anti-CD16, and then washed with phosphate-buffered saline (PBS) and labeled with fluorescently tagged antibodies. Ciliary colocalization of CD16.7-PC1 fusion proteins with acetylated α-tubulin or α-ß-tubulin was defined as exhibiting colocalization of the two signals across at least two pixels within the cilium as imaged in the confocal Z-stack data.
For shRNA cell-based assays, cells were labeled with mAb anti-CD16 and anti–mouse Alexa Fluor 555 secondary, and then fixed for 20 min at room temperature with 3% PFA, repermeabilized, and labeled with mAb antiacetylated α-tubulin, followed by anti–mouse Alexa Fluor 647.
Confocal immunofluorescence images were collected using a Zeiss LSM510 laser-scanning confocal microscope (Carl Zeiss, Thornwood, NY) with 40×, numerical aperture (NA) 1.3 or 63×, NA 1.4 oil immersion objectives. LSM 510 Image Acquisition software (Carl Zeiss) was used to acquire images. Specimens were mounted in Mowiol or 1,4-diazabicyclo[2.2.2]octane/glycerol (Bacallao et al., 2006) mounting medium. Confocal Z-stacks were processed with the Zeiss Image Browser or Voxx2 (provided freely for noncommercial use by the Indiana Center for Biological Microscopy, Indianapolis, IN, www.nephrology.iupui.edu/imaging/voxx/index.htm), and assembled using Photoshop and Illustrator (Adobe, San Jose, CA). Movies were generated using the movie maker feature in Voxx2 software, and converted to .avi and .mpg file formats using QuickTime Player (Apple, Cupertino, CA) and TMPGEnc (Pegasys, Tokyo, Japan), respectively.
Immunoelectron microscopy
Samples were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer, dehydrated through a graded series of ethyl alcohols, and embedded in Unicryl (Vector Labs, Burlingame, CA). Thin sections (70–90 nm) were mounted on Formvar/carbon-coated nickel grids. After drying, the grids were floated on drops of 0.05 M glycine for 15 min to quench the aldehydes, then rinsed with PBS. Grids were blocked and permeabilized in 2% BSA, 0.1% cold water fish gelatin, and 0.1% Tween in PBS, and placed overnight at 4°C in incubation buffer (0.1% BSA-c [Aurion, Hatfield, PA], 0.05% Tween in PBS) containing primary antibody. Grids were rinsed with incubation buffer and then floated on drops of the incubation buffer containing secondary antibody with attached 10-nm gold particles overnight at 4°C. After rinsing, grids were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer for 5 min. After rinses in distilled water, grids were dried and stained for contrast with uranyl acetate and lead citrate. Samples were viewed with a Tecnai Bio Twin transmission electron microscope (FEI, Hillsboro, OR).
CD16.7-PC1 mutagenesis
The CD16.7-PC1-WT fusion protein contains an NH2-terminal ectodomain of human CD16, a single transmembrane span from human CD7, and the last 112 amino acids of human PC1 (Vandorpe et al., 2002). Site-directed mutagenesis was performed using a PCR approach. CD16.7-PC1-WT served as a template, and three antisense oligonucleotides (359 cdPC1r 5′-TCGAGCGGCCGCCCTACAGGTCCACACCCCGACT-3′; 339 cdPC1r 5′-TCGAGCGGCCGCCCTAGGATGGGCCACGGGAAGA-3′; 296 cdPC1r 5′-TCGAGCGGCCGCCCTACTCGAACACGGCTTGGA-3′) were designed to truncate the PC1 tail. Each antisense oligonucleotide was designed with a NotI recognition site and a four-nucleotide overhang to facilitate cleavage of the PCR product. Each antisense primer was combined with a sense primer (CD-PC1-F 5′-CCACACTCAAAGATAGCG-3′), which corresponded to a region within the CD16 sequence, upstream of the BamHI site. PCR reactions were carried out using the Phusion High-Fidelity PCR kit and GC buffer pack, according to manufacturer's instructions (Finnzymes, OY, Finland). PCR products and CD16.7-PC1-WT were digested with BamHI and NotI in sequential reactions. PCR products were separated on 1% agarose gels and extracted using the QIAquick Gel Extraction kit (Qiagen, Valencia, CA). Digested PCR products were ligated with digested CD16.7-PC1-WT vector using T4 ligase (New England Biolabs, Ipswich, MA). Each vector was fully sequenced by the University of New Mexico Health Sciences Center (UNM HSC) DNA Research Services (http://hsc.unm.edu/som/programs/cidi/dna.shtml). The following constructs: CD16.7-PC1-AAHASST, CD16.7-CD7 C-terminus+KVHPSST, and CD16.7-PC1-296 and -339 truncation mutants with KVHPSST added were prepared under contract with Genscript (Piscataway, NJ). All constructs were verified with sequence analysis.
In vitro GST-Arf4 binding
GST-Arf4 was prepared and purified as previously described (Mazelova et al., 2009). GST-Arf4 activation measurements using flow cytometry were performed as previously described for Rab and Rho GTPases (Schwartz et al., 2008). GST-Arf4 was bound to glutathione-Sepharose beads (GE Healthcare, Piscataway, NJ) to saturation at 4°C (4 h) or 37°C (1 h) on a rotator. Beads were washed with HPS buffer (30 mM HEPES, pH 7.4, 20 mM NaCl, 100 mM KCl) containing 1 mM ethylenediaminetetraacetic acid (EDTA) and centrifuged to remove unbound protein. Beads were incubated in GTPase exchange buffer (20 mM HEPES, pH 7.4, 25 mM NaCl, 2 mM EDTA) with freshly prepared 100 mM GppNHp (2 mM final concentration) for 20 min in a 37°C water bath. After incubation, MgCl2 was added to a final concentration of 10 mM. One-day postconfluent MDCKII cells (on 10-cm dishes) or HeLa cells transfected for 24 h with CD16.7-PC1 constructs (60-mm dishes) were lysed with 1 ml of buffer containing 20 mM HEPES (pH 7.4), 20 mM NaCl, 100 mM KCl, 2.5 mM MgCl2, 1% Triton-X 100, and protease inhibitors. Lysates were precleared with glutathione-Sepharose beads for 1 h at 4°C. Precleared lysates were added to active GST-Arf4 beads, and incubated on a rotator at 4°C (4 h) or 37°C (1 h). Beads were centrifuged and washed three times with HPS containing 2.5 mM MgCl2. Bound proteins were eluted with twofold concentrated SDS sample buffer at 70°C for 15 min. Eluates were resolved with SDS–PAGE and immunoblotted with NM002 or with antibodies specific for CD16, GST, and ASAP1. ImageJ 1.42i gel analysis tools (http://rsb.info.nih.gov/ij/download.html) and Quantity One software (Bio-Rad, Hercules, CA) were used to quantify band intensities. Statistical analyses and graphs were generated using GraphPad Prism, version 4.0a for Macintosh (GraphPad Software, San Diego, CA, www.graphpad.com).
Generation of GST-PC2 (aa 1–15) and the GST pulldown of Arf4
A nucleotide sequence corresponding to the N-terminal of human PC2 (aa 1–15) was generated by PCR using a sense primer (5′-cggtttcgggatccatggtgaactccagtcgcgtgcagcctcagcagcc-3′) and an antisense primer (5′-agagagcggaattcttaggcgtccccgggctgctgaggctgcac-3′) synthesized by the UNM DNA Research Services. Oligonucleotides corresponding to the N-terminal sequence of PC2 were inserted in the EcoRI and BamHI sites of the pGEX-KG vector (ATCC). GST-PC2 and GST-Arf4 were expressed and purified as is in the preceding section. Arf4 was cleaved from GST by incubation with thrombin for 17 h at 22°C, cleaved GST and uncleaved GST-Arf4 were removed by glutathione absorption, and the pure Arf4 was used for the GST-PC2 binding assays. GST or GST-PC2 (5 μg) were incubated with 5 μg Arf4 in PBS containing 0.05% NP-40, 1.5 mM phenylmethylsulfonyl fluoride (PMSF) and 0.16 mg/ml bovine serum albumin (BSA) for 1.5 h at 4°C, and then incubated with glutathione-Sepharose 4B beads for 1h at 4°C. Bound Arf4 was detected with affinity-purified rabbit anti-Arf4 (1:5000). Site-directed mutagenesis of GST-PC2 was performed using the QuikChange II site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene, La Jolla, CA). Removal of RVQP (aa 6–9) and RVQPQQ (aa 6–11) gave similar results.
Generation of C-terminal FLAG-tagged PC2 and PC2 immunoprecipitation
The oligonucleotide sequence corresponding to the N-terminal of PC2 (aa 1–219) was generated by PCR using full-length human PC2 in pcDNA3 (a kind gift from Christopher Ward, Mayo Clinic, Rochester, MN) with a sense primer (5′-agagaggcaagcttatggtgaactccagtcgc-3′) and an antisense primer (5′-gaacggcagatctacttttaaggtatttctctcggttagtg-3′). The PCR product was digested with HindIII and BglII and inserted in the HindIII and BglII sites of pFLAG-CTC (Sigma-Aldrich). FLAG-tagged protein was expressed using lipofection of HeLa cells. For immunoprecipitation experiments, cell lysates containing FLAG-tagged PC2, or the FLAG-tagged control protein BAP, were incubated with 15 μl bed volume of anti-FLAG-M2 agarose (Sigma-Aldrich) for 2h at 4°C, and washed three times with the reaction buffer (50 mM HEPES, 150 mM NaCl, 5 mM MgCl2, 0.1% Triton X-100, 0.1% BSA, 1 mM PMSF). Each sample was incubated with 5 μg purified Arf4 in 500 μl of reaction buffer overnight at 4°C. Agarose was washed eight times with reaction buffer, and bound proteins were resolved by SDS–PAGE and immunoblotted with rabbit anti-Arf4 and rabbit anti-FLAG antibody. The RVQPQQ deletion mutant was generated using a two-step PCR approach. The first reaction was performed using PC2 in pFLAG-CTC as a template and primers (sense 5′-GCTTATGGTGAACTCCAGTCCCGGGGACG and antisense 5′-GAACGGCAGATCTACTTTTAAGGTATTTCTCTC). The second reaction was performed using the first PCR product as a template and primers (sense primer 5′-AGAGAGGCAAGCTTATGGTGAACTCCAGTC and antisense primer 5′- GAACGGCAGATCTACTTTTAAGGTATTTCTCTC). The second PCR product was inserted into pFLAG-CTC vector at HindIII and BglII sites.
Coimmunoprecipitation assays
Coimmunoprecipitations assays with immunoblots were performed with protein A (rabbit pAb primary antibodies) or protein G (mouse mAb primary antibodies) agarose (Upstate, Lake Placid, NY) as previously described (Roitbak et al., 2004), using MDCKII and RCTE cell lysates collected at 1–5 d postconfluency. Preimmune rabbit serum was used for control immunoprecipitations. Immunoprecipitations using MDCKII lysates with immunoblots for Rab5, Rab6, Rab7, Rab8, and Rab11 were performed using the ProFound Mammalian Co-immunoprecipitation kit to avoid light chain detection (Pierce, Rockford, IL) according to the manufacturer's instructions. Immunoprecipitations using RCTE lysates for CD16, Rab5, Rab6, Rab7, Rab8, and Rab11 were performed using Rabbit or Mouse IgG TrueBlot (eBioscience, San Diego, CA) systems according to the manufacturer's instruction. Quantification of immunoprecipitation was performed using a BioRad ChemiDoc XRS+ imaging system and Quantity One software, and enrichment was calculated as a fraction of lysate protein immunoprecipitated with PC1 antibody relative to the fraction of lysate protein precipitated with control IgG. Approximately 5% of the total lysate volume was used per immunoblot, and 25% of the total lysate was used per immunoprecipitation sample.
Preparation of NM002 and NM032 peptide antibody and affinity purification
A peptide corresponding to aa 3633–3645 of human PC1, and with an N-terminal cysteine (CKRLHPDEDDTLVE) was synthesized (Suzanna Horvath, California Institute of Technology, Pasadena, CA). The peptide was conjugated to keyhole limpet hemocyanin using benzoquinone and was used to immunize rabbits NM002 and NM032 (Covance, Denver, PA). For affinity purification, the same peptide was conjugated to Sulfalink gel (#20401; Pierce) according to the manufacturer's instructions (1 mg peptide/1 ml resin bed volume). Ten milliliters NM002 serum was incubated with 5 ml resin and washed to remove unbound immunoglobulin and serum proteins. Bound immunoglobulin was eluted with 0.1 M glycine (pH 2.5), immediately neutralized with Tris base, and used for immunoblotting and immunostaining experiments at a concentration of 10 μg/ml.
siRNA-mediated PC1 depletion and quantification of loss of NM002 reactivity
A PC1 siRNA sequence complementary to nucleotides 584–605 within the leucine-rich repeat (LRR) domain, demonstrated previously to ablate PCI expression (Xu et al., 2007), was synthesized from sense and antisense oligodeoxynucleotides (PC-17 sense 5′-AAACAGTCACACTCAAACGGGCCTGTCTC-3′; PC-17 antisense 5′-AACCCGTTTGAGTGTGACTGTCCTGTCTC-3′; Integrated DNA Technologies, Coralville, IA) according to the manufacturer's instructions. GAPDH control siRNA was purchased from Ambion (control template set 4800; Austin, TX). Ambion's Silencer siRNA construction kit was used to generate siRNA according to the manufacturer's instructions. Cy3 labeling of siRNA was performed using the Ambion labeling kit according to manufacturer's instructions (immunofluorescence evaluation only). Human pancreatic adenocarcinoma (HPAC) cells were transfected with PC1 siRNA for 48 h (100 pmol per well of a six-well plate) using Lipofectamine 2000 per the manufacturer's instructions. Knockdown efficacy, evaluated by immunoblot using NM005 anti-PC1 antibody or monoclonal anti-GAPDH antibody (Ambion), showed a fivefold decrease in protein levels (Xu et al., 2007). The effect of PC1 depletion on NM002 staining was quantified using confocal micrographs and Slidebook software (Intelligent Imaging Innovations, Denver, CO). NM002 staining was quantified in 30 Cy3-labeled PC1 siRNA-positive cells and reported relative to NM002 staining in 30 Cy3-labeled GAPDH siRNA controls with SDs.
ShRNA-mediated depletion of Arf4 and Rab8 and statistical analyses
Arf4 shRNA (OligoIDs: V3LHS-410096, V2LMM-62890, V2LHS-271123, V3LHS-371138, V2LHS-92212) and Rab8 shRNA (OligoID: V3LHS-359728) clones were obtained in pGIPZ-GFP vectors (Thermo Scientific Open Biosystems, Huntsville, AL) and transfected into HeLa and RCTE cells using Lipofectamine 2000. The Rab8 shRNA (AAGAGACAAGTTTCCAAGGAA) exhibited 100% identity over 17 of 21 nucleotides with the siRNA sequence used by others to effectively deplete Rab8 mRNA and protein (Schuck et al., 2004; Hattula et al., 2006). The sequence is unique to Rab8a and not present in Rab8b despite the significant amino acid conservation. ShRNA plasmids were transfected into HeLa cells and expressed for 48 h to determine percent knockdown compared with untransfected and cells transfected with GAPDH control. Based on transient transfection results, V2LHS-271123 and V2LMM-62890 Arf4 shRNA vectors were pooled and used for RCTE cilia-trafficking studies (Supplemental Figure 7). Percent knockdown in RCTE cells was determined by first flow-sorting the shRNA-transfected cells coexpressing GFP and immunoblotting for the target protein. Arf4 and Rab8 protein expression levels in shRNA and untransfected lysate lanes were normalized to actin, then expression levels of shRNA-treated lanes were compared with untransfected controls. For immunofluorescence trafficking assays, RCTE cells were plated on filter supports and transfected with shRNA plasmids using Lipofectamine 2000. Cells were allowed to express shRNA products for 72 h. RCTE cells were then transfected with CD16.7-PC1-WT plasmid, and grown for an additional 20 h. Cell labeling and image collection methods are described in the Immunofluorescence staining section. Labeled cells were imaged using a battlement pattern to ensure capture of all dually transfected cells.
The proportion of cells with CD16.7-PC1-WT in the cilium was estimated for each plasmid as a weighted average of sample-specific proportions, with weights equal to the number of ciliated cells per sample. To assess differences in proportions across plasmids, we used a logistic regression analogue of analysis of variance (ANOVA), allowing for overdispersion, which may be present due to within-group variation in proportions. F-tests were used to test for differences among plasmids and for pair-wise comparisons of plasmids. Separate analyses were performed for distinct experiments. To test whether data from different experiments could be pooled, we added experiment and experiment-by-group interactions to the ANOVA models. and tested the significance of these added effects in the combined data set. There were no significant differences among separate experiments, or interaction between experiments and groups. All analyses were performed using SAS 9.2 (SAS Institute, www.sas.com). Statistical significance was defined as p < 0.05.
Supplementary Material
Acknowledgments
Studies were supported by National Institute of Diabetes and Digestive and Kidney Diseases R01 DK50141 to A.W.N., R01 EY12421 to D.D., R01 DK68581to V.H.G., and R01 DK57662 to S.L.A. H.H.W. was supported by a Research Fellowship from the National Kidney Foundation. We gratefully acknowledge Elsa Romero and Samantha Schwartz for expert technical assistance and Janet Kelly and Lauren Thal for administrative support (all at the University of New Mexico HSC, Albuquerque, NM). We thank Jason Byars (University of New Mexico HSC, Albuquerque, NM) for technical assistance with image conversion and Voxx2 software and Caroline Miller (Indiana University, Indianapolis, IN) for processing and collection of electron micrographs. We thank Scott Ness for helpful discussions, Genevieve Phillips for quantification of siRNA depletion of immunofluorescence staining, Sophia Endischee for immunoblot analysis of NM0032, and Stephanie Jerman for critical review of the manuscript (all at the University of New Mexico HSC, Albuquerque, NM). DNA and protein concentrations were quantified using instrumentation provided by the Keck-UNM Genomics Resource (KUGR) Facility (http://hsc.unm.edu/som/micro/Genomics). The electron microscopy studies were performed at the Indiana University School of Medicine EM Center, thanks to the generous support of that facility by the Polycystic Kidney Disease Foundation. Immunofluorescence images in this paper were generated in the University of New Mexico Cancer Center Fluorescence Microscopy Facility, supported as detailed on the webpage: http://hsc.unm.edu/crtc/microscopy/Facility.html.
Abbreviations used:
- ADPKD
autosomal dominant polycystic kidney disease
- ASAP1
ArfGAP with SH3 domain ankyrin repeat and PH domain 1
- BBS
Bardet-Biedl syndrome
- BSA
bovine serum albumin
- EDTA
ethylenediaminetetraacetic acid
- GAP
GTPase-activating protein
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GEF
guanine exchange factor
- GST
glutathione S-transferase
- HPAC
human pancreatic adenocarcinoma cells
- IFT
intraflagellar transport
- IgG
immunoglobulin G
- LRR
leucine-rich repeat
- MDCK
Madin-Darby canine kidney
- NK1
normal human kidney 1 cells
- PBS
phosphate-buffered saline
- PC1
polycystin-1
- PC2
polycystin-2
- PFA
paraformaldehyde
- PMSF
phenylmethylsulfonyl fluoride
- RCTE
renal cortical tubular epithelia cells
- shRNA
small hairpin RNA
- siRNA
small interfering RNA
- ST tyr isoform
myc-tagged trans-Golgi marker α2, 6-sialyl-transferase
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-01-0082) on July 20, 2011.
The authors declare no conflict of interest.
REFERENCES
- Ang AL, Folsch H, Koivisto UM, Pypaert M, Mellman I. The Rab8 GTPase selectively regulates AP-1B-dependent basolateral transport in polarized Madin-Darby canine kidney cells. J Cell Biol. 2003;163:339–350. doi: 10.1083/jcb.200307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacallao R, Antony C, Dotti C, Karsenti E, Stelzer EH, Simons K. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J Cell Biol. 1989;109:2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacallao RL, McNeill H. Cystic kidney diseases and planar cell polarity signaling. Clin Genet. 2009;75:107–117. doi: 10.1111/j.1399-0004.2008.01148.x. [DOI] [PubMed] [Google Scholar]
- Bacallao R, Sohrab S, Phillips CL. Guiding principles of specimen preservation for confocal fluorescence microscopy. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. New York: Springer; 2006. pp. 368–379. [Google Scholar]
- Barr MM, DeModena J, Braun D, Nguyen CQ, Hall DH, Sternberg PW. The Caenorhabditis elegans autosomal dominant polycystic kidney disease gene homologs lov-1 and pkd-2 act in the same pathway. Curr Biol. 2001;11:1341–1346. doi: 10.1016/s0960-9822(01)00423-7. [DOI] [PubMed] [Google Scholar]
- Boehlke C, Bashkurov M, Buescher A, Krick T, John AK, Nitschke R, Walz G, Kuehn EW. Differential role of Rab proteins in ciliary trafficking: Rab23 regulates Smoothened levels. J Cell Sci. 2010;123:1460–1467. doi: 10.1242/jcs.058883. [DOI] [PubMed] [Google Scholar]
- Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J. 2009;424:79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron AJ, Bacallao RL, Wandinger-Ness A. ADPKD: a human disease altering Golgi function and basolateral exocytosis in renal epithelia. Traffic. 2000a;1:675–686. doi: 10.1034/j.1600-0854.2000.010811.x. [DOI] [PubMed] [Google Scholar]
- Charron AJ, Nakamura S, Bacallao R, Wandinger-Ness A. Compromised cytoarchitecture and polarized trafficking in autosomal dominant polycystic kidney disease cells. J Cell Biol. 2000b;149:111–124. doi: 10.1083/jcb.149.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Feng Y, Chen D, Wandinger-Ness A. Rab11 is required for trans-Golgi network-to-plasma membrane transport and a preferential target for GDP dissociation inhibitor. Mol Biol Cell. 1998;9:3241–3257. doi: 10.1091/mbc.9.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Wandinger-Ness A. Expression and functional analyses of Rab8 and Rab11a in exocytic transport from trans-Golgi network. Methods Enzymol. 2001;329:165–175. doi: 10.1016/s0076-6879(01)29077-6. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D. A role for rhodopsin in a signal transduction cascade that regulates membrane trafficking and photoreceptor polarity. Vision Res. 2006;46:4427–4433. doi: 10.1016/j.visres.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc Natl Acad Sci USA. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Chen W, Welford A, Wandinger-Ness A. The proteasome alpha-subunit XAPC7 interacts specifically with Rab7 and late endosomes. J Biol Chem. 2004;279:21334–21342. doi: 10.1074/jbc.M401022200. [DOI] [PubMed] [Google Scholar]
- Feng Y, Press B, Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenteany FH, Colley KJ. Multiple signals are required for alpha2,6-sialyltransferase (ST6Gal I) oligomerization and Golgi localization. J Biol Chem. 2005;280:5423–5429. doi: 10.1074/jbc.M412396200. [DOI] [PubMed] [Google Scholar]
- Follit JA, Li L, Vucica Y, Pazour GJ. The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J Cell Biol. 2010;188:21–28. doi: 10.1083/jcb.200910096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsch H, Pypaert M, Maday S, Pelletier L, Mellman I. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J Cell Biol. 2003;163:351–362. doi: 10.1083/jcb.200309020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L, Okuhara D, Yu Z, Tian X, Cai Y, Shibazaki S, Somlo S. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J Cell Sci. 2006;119:1383–1395. doi: 10.1242/jcs.02818. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perret S, Kim K, Ibarra C, Damiano AE, Zotta E, Batelli M, Harris PC, Reisin IL, Arnaout MA, Cantiello HF. Polycystin-2, the protein mutated in autosomal dominant polycystic kidney disease (ADPKD), is a Ca2+-permeable nonselective cation channel. Proc Natl Acad Sci USA. 2001;98:1182–1187. doi: 10.1073/pnas.98.3.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller C, Alper SL. Nonpolarized surface distribution and delivery of human CD7 in polarized MDCK cells. Am J Physiol. 1993;265:C1069–C1079. doi: 10.1152/ajpcell.1993.265.4.C1069. [DOI] [PubMed] [Google Scholar]
- Hanaoka K, Qian F, Boletta A, Bhunia AK, Piontek K, Tsiokas L, Sukhatme VP, Guggino WB, Germino GG. Co-assembly of polycystin-1 and -2 produces unique cation-permeable currents. Nature. 2000;408:990–994. doi: 10.1038/35050128. [DOI] [PubMed] [Google Scholar]
- Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpaa K, Laakkonen P, Peranen J. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J Cell Sci. 2006;119:4866–4877. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- Hayes GL, Brown FC, Haas AK, Nottingham RM, Barr FA, Pfeffer SR. Multiple Rab GTPase binding sites in GCC185 suggest a model for vesicle tethering at the trans-Golgi. Mol Biol Cell. 2009;20:209–217. doi: 10.1091/mbc.E08-07-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Attanasio M, Otto E. Nephronophthisis: disease mechanisms of a ciliopathy. J Am Soc Nephrol. 2009;20:23–35. doi: 10.1681/ASN.2008050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeister H, Babinger K, Gurster S, Cedzich A, Meese C, Schadendorf K, Osten L, de Vries U, Rascle A, Witzgall R. Polycystin-2 takes different routes to the somatic and ciliary plasma membrane. J Cell Biol. 2011;192:631–645. doi: 10.1083/jcb.201007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber LA, Dupree P, Dotti CG. A deficiency of the small GTPase rab8 inhibits membrane traffic in developing neurons. Mol Cell Biol. 1995;15:918–924. doi: 10.1128/mcb.15.2.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J Cell Biol. 1993;123:35–45. doi: 10.1083/jcb.123.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd T, Zhou W, Jenkins P, Liu CJ, Swaroop A, Khanna H, Martens J, Hildebrandt F, Margolis B. The retinitis pigmentosa protein RP2 interacts with polycystin 2 and regulates cilia-mediated vertebrate development. Hum Mol Genet. 2010;19:4330–4344. doi: 10.1093/hmg/ddq355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet. 2006;22:491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Inoue H, Ha VL, Prekeris R, Randazzo PA. Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol Biol Cell. 2008;19:4224–4237. doi: 10.1091/mbc.E08-03-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- Jian X, Brown P, Schuck P, Gruschus JM, Balbo A, Hinshaw JE, Randazzo PA. Autoinhibition of Arf GTPase-activating protein activity by the BAR domain in ASAP1. J Biol Chem. 2009;284:1652–1663. doi: 10.1074/jbc.M804218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen I, Collins JF, Higgins DG. Finding flexible patterns in unaligned protein sequences. Protein Sci. 1995;4:1587–1595. doi: 10.1002/pro.5560040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen JA, San Agustin J, Follit JA, Pazour GJ. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol. 2008;183:377–384. doi: 10.1083/jcb.200808137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA. Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Lett. 2009;583:3872–3879. doi: 10.1016/j.febslet.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, et al. Consensus nomenclature for the human ArfGAP domain-containing proteins. J Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan OI, et al. The AP-1 clathrin adaptor facilitates cilium formation and functions with RAB-8 in C. elegans ciliary membrane transport. J Cell Sci. 2010;123:3966–3977. doi: 10.1242/jcs.073908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler A, Feng S, Zhang J, Zhang X, Das A, Peranen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LA, Kumar R, Hales CM, Navarre J, Bhartur SG, Burnette JO, Provance DWJ, Mercer JA, Bahler M, Goldenring JR. Myosin Vb is associated with plasma membrane recycling systems. Mol Biol Cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Hiesberger T, Cordes K, Sinclair AM, Goldstein LS, Somlo S, Igarashi P. Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc Natl Acad Sci USA. 2003;100:5286–5291. doi: 10.1073/pnas.0836980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. Basal bodies platforms for building cilia. Curr Top Dev Biol. 2008;85:1–22. doi: 10.1016/S0070-2153(08)00801-6. [DOI] [PubMed] [Google Scholar]
- Mazelova J, Astuto-Gribble L, Inoue H, Tam BM, Schonteich E, Prekeris R, Moritz OL, Randazzo PA, Deretic D. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28:183–192. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic L, Scott MP, Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserey-Lenkei S, et al. Rab6-interacting protein 1 links Rab6 and Rab11 function. Traffic. 2007;8:1385–1403. doi: 10.1111/j.1600-0854.2007.00612.x. [DOI] [PubMed] [Google Scholar]
- Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier. Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauli SM, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- Nauli SM, Rossetti S, Kolb RJ, Alenghat FJ, Consugar MB, Harris PC, Ingber DE, Loghman-Adham M, Zhou J. Loss of polycystin-1 in human cyst-lining epithelia leads to ciliary dysfunction. J Am Soc Nephrol. 2006;17:1015–1025. doi: 10.1681/ASN.2005080830. [DOI] [PubMed] [Google Scholar]
- Nicodeme P. Fast approximate motif statistics. J Comput Biol. 2001;8:235–248. doi: 10.1089/10665270152530836. [DOI] [PubMed] [Google Scholar]
- Nie Z, et al. A BAR domain in the N terminus of the Arf GAP ASAP1 affects membrane structure and trafficking of epidermal growth factor receptor. Curr Biol. 2006;16:130–139. doi: 10.1016/j.cub.2005.11.069. [DOI] [PubMed] [Google Scholar]
- Nie Z, Randazzo PA. Arf GAPs and membrane traffic. J Cell Sci. 2006;119:1203–1211. doi: 10.1242/jcs.02924. [DOI] [PubMed] [Google Scholar]
- Ojakian GK, Schwimmer R. The polarized distribution of an apical cell surface glycoprotein is maintained by interactions with the cytoskeleton of Madin-Darby canine kidney cells. J Cell Biol. 1988;107:2377–2387. doi: 10.1083/jcb.107.6.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori Y, Zhao C, Saras A, Mukhopadhyay S, Kim W, Furukawa T, Sengupta P, Veraksa A, Malicki J. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat Cell Biol. 2008;10:437–444. doi: 10.1038/ncb1706. [DOI] [PubMed] [Google Scholar]
- Orzech E, Cohen S, Weiss A, Aroeti B. Interactions between the exocytic and endocytic pathways in polarized Madin-Darby canine kidney cells. J Biol Chem. 2000;275:15207–15219. doi: 10.1074/jbc.275.20.15207. [DOI] [PubMed] [Google Scholar]
- Pazour GJ. Intraflagellar transport and cilia-dependent renal disease: the ciliary hypothesis of polycystic kidney disease. J Am Soc Nephrol. 2004;15:2528–2536. doi: 10.1097/01.ASN.0000141055.57643.E0. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Veland IR, Schroder JM, Christensen ST. Assembly of primary cilia. Dev Dyn. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- Qian F, Germino FJ, Cai Y, Zhang X, Somlo S, Germino GG. PKD1 interacts with PKD2 through a probable coiled-coil domain. Nat Genet. 1997;16:179–183. doi: 10.1038/ng0697-179. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Xu X, Wandinger-Ness A, Dalke DP, Pierce SK. Separation of subcellular compartments containing distinct functional forms of MHC class II. J Cell Biol. 1994;125:595–605. doi: 10.1083/jcb.125.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KK, Wilson PD, Snyder RW, Zhang X, Guo W, Burrow CR, Lipschutz JH. The exocyst localizes to the primary cilium in MDCK cells. Biochem Biophys Res Commun. 2004;319:138–143. doi: 10.1016/j.bbrc.2004.04.165. [DOI] [PubMed] [Google Scholar]
- Roitbak T, Ward CJ, Harris PC, Bacallao R, Ness SA, Wandinger-Ness A. A polycystin-1 multiprotein complex is disrupted in polycystic kidney disease cells. Mol Biol Cell. 2004;15:1334–1346. doi: 10.1091/mbc.E03-05-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers MS, Le H, van der Bent P, Leonhard W, Prins F, Spruit L, Breuning MH, de Heer E, Peters DJ. Distinct subcellular expression of endogenous polycystin-2 in the plasma membrane and Golgi apparatus of MDCK cells. Hum Mol Genet. 2002;11:59–67. doi: 10.1093/hmg/11.1.59. [DOI] [PubMed] [Google Scholar]
- Schuck S, Manninen A, Honsho M, Fullekrug J, Simons K. Generation of single and double knockdowns in polarized epithelial cells by retrovirus-mediated RNA interference. Proc Natl Acad Sci USA. 2004;101:4912–4917. doi: 10.1073/pnas.0401285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Simons K. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J Cell Sci. 2004;117:5955–5964. doi: 10.1242/jcs.01596. [DOI] [PubMed] [Google Scholar]
- Schwartz SL, Cao C, Pylypenko O, Rak A, Wandinger-Ness A. Rab GTPases at a glance. J Cell Sci. 2007;120:3905–3910. doi: 10.1242/jcs.015909. [DOI] [PubMed] [Google Scholar]
- Schwartz SL, Tessema M, Buranda T, Pylypenko O, Rak A, Simons PC, Surviladze Z, Sklar LA, Wandinger-Ness A. Flow cytometry for real-time measurement of guanine nucleotide binding and exchange by Ras-like GTPases. Anal Biochem. 2008;381:258–266. doi: 10.1016/j.ab.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff DR, Daro EA, Hull M, Mellman I. The receptor recycling pathway contains two distinct populations of early endosomes with different sorting functions. J Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberberg M, Charron AJ, Bacallao R, Wandinger-Ness A. Mispolarization of desmosomal proteins and altered intercellular adhesion in autosomal dominant polycystic kidney disease. Am J Physiol Renal Physiol. 2005;288:F1153–F1163. doi: 10.1152/ajprenal.00008.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Wandinger-Ness A. Polarized sorting in epithelia. Cell. 1990;62:207–210. doi: 10.1016/0092-8674(90)90357-k. [DOI] [PubMed] [Google Scholar]
- Spang A, Shiba Y, Randazzo PA. Arf GAPs: gatekeepers of vesicle generation. FEBS Lett. 2010;584:2646–2651. doi: 10.1016/j.febslet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiokas L, Kim E, Arnould T, Sukhatme VP, Walz G. Homo- and heterodimeric interactions between the gene products of PKD1 and PKD2. Proc Natl Acad Sci USA. 1997;94:6965–6970. doi: 10.1073/pnas.94.13.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandorpe DH, Wilhelm S, Jiang L, Ibraghimov-Beskrovnaya O, Chernova MN, Stuart-Tilley AK, Alper SL. Cation channel regulation by COOH-terminal cytoplasmic tail of polycystin-1: mutational and functional analysis. Physiol Genomics. 2002;8:87–98. doi: 10.1152/physiolgenomics.00092.2001. [DOI] [PubMed] [Google Scholar]
- Vassilev PM, et al. Polycystin-2 is a novel cation channel implicated in defective intracellular Ca(2+) homeostasis in polycystic kidney disease. Biochem Biophys Res Commun. 2001;282:341–350. doi: 10.1006/bbrc.2001.4554. [DOI] [PubMed] [Google Scholar]
- Wandinger-Ness A, Bennett MK, Antony C, Simons K. Distinct transport vesicles mediate the delivery of plasma membrane proteins to the apical and basolateral domains of MDCK cells. J Cell Biol. 1990;111:987–1000. doi: 10.1083/jcb.111.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward HH, Wang J, Phillips C. Analysis of multiple Invs transcripts in mouse and MDCK cells. Genomics. 2004;84:991–1001. doi: 10.1016/j.ygeno.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Westlake CJ, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci USA. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PD. Polycystin: new aspects of structure, function, and regulation. J Am Soc Nephrol. 2001;12:834–845. doi: 10.1681/ASN.V124834. [DOI] [PubMed] [Google Scholar]
- Wilson PD. Polycystic kidney disease. N Engl J Med. 2004;350:151–164. doi: 10.1056/NEJMra022161. [DOI] [PubMed] [Google Scholar]
- Wilson PD. Mouse models of polycystic kidney disease. Curr Top Dev Biol. 2009;84:311–350. doi: 10.1016/S0070-2153(08)00606-6. [DOI] [PubMed] [Google Scholar]
- Xu C, Rossetti S, Jiang L, Harris PC, Brown-Glaberman U, Wandinger-Ness A, Bacallao R, Alper SL. Human ADPKD primary cyst epithelial cells with a novel, single codon deletion in the PKD1 gene exhibit defective ciliary polycystin localization and loss of flow-induced Ca2+ signaling. Am J Physiol Renal Physiol. 2007;292:F930–F945. doi: 10.1152/ajprenal.00285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder BK, Hou X, Guay-Woodford LM. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J Am Soc Nephrol. 2002a;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. [DOI] [PubMed] [Google Scholar]
- Yoder BK, Tousson A, Millican L, Wu JH, Bugg CE Jr, Schafer JA, Balkovetz DF. Polaris, a protein disrupted in orpk mutant mice, is required for assembly of renal cilium. Am J Physiol Renal Physiol. 2002b;282:F541–F552. doi: 10.1152/ajprenal.00273.2001. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Guo W, Lipschutz JH. The exocyst protein Sec10 is necessary for primary ciliogenesis and cystogenesis in vitro. Mol Biol Cell. 2009;20:2522–2529. doi: 10.1091/mbc.E08-07-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.