This study identifies zwint-1 as a novel substrate for AurB during mitosis. Phosphorylation is required for outer kinetochore assembly during prometaphase. However, zwint-1 dephosphorylation is required at metaphase for checkpoint silencing.
Abstract
Aurora B (AurB) is a mitotic kinase responsible for multiple aspects of mitotic progression, including assembly of the outer kinetochore. Cytoplasmic dynein is an abundant kinetochore protein whose recruitment to kinetochores requires phosphorylation. To assess whether AurB regulates recruitment of dynein to kinetochores, we inhibited AurB using ZM447439 or a kinase-dead AurB construct. Inhibition of AurB reduced accumulation of dynein at kinetochores substantially; however, this reflected a loss of dynein-associated proteins rather than a defect in dynein phosphorylation. We determined that AurB inhibition affected recruitment of the ROD, ZW10, zwilch (RZZ) complex to kinetochores but not zwint-1 or more-proximal kinetochore proteins. AurB phosphorylated zwint-1 but not ZW10 in vitro, and three novel phosphorylation sites were identified by tandem mass spectrometry analysis. Expression of a triple-Ala zwint-1 mutant blocked kinetochore assembly of RZZ-dependent proteins and induced defects in chromosome movement during prometaphase. Expression of a triple-Glu zwint-1 mutant rendered cells resistant to AurB inhibition during prometaphase. However, cells expressing the triple-Glu mutant failed to satisfy the spindle assembly checkpoint (SAC) at metaphase because poleward streaming of dynein/dynactin/RZZ was inhibited. These studies identify zwint-1 as a novel AurB substrate required for kinetochore assembly and for proper SAC silencing at metaphase.
INTRODUCTION
Kinetochores in higher eukaryotes assemble a hierarchy of proteins responsible for interacting with microtubules, mediating chromosome movement, monitoring chromosome alignment, and executing chromosome segregation. The molecular details of these processes are only partially deciphered, with many unresolved questions. The complexities of microtubule-binding proteins at kinetochores and contributions of each, for example, are poorly understood.
Phosphorylation is a widely recognized mechanism of regulation during mitosis (Musacchio and Salmon, 2007), and multiple mitotic kinases have been identified at the kinetochore, including BubR1, Mps-1, Plk-1, and Aurora B (AurB). Functional targets for these kinases include kinetochore attachment, chromosome biorientation, tension sensing, chromosome movement, and kinetochore assembly (Mao et al., 2003; DeLuca et al., 2006; Emanuele et al., 2008; Guimaraes et al., 2008; Vader and Lens, 2008; Kang and Yu, 2009; Santaguida et al., 2010). AurB is a highly conserved serine/threonine kinase believed to play important roles in kinetochore assembly and function (Carmena et al., 2009). AurB phosphorylates histone H3 on the inner plate (Hsu et al., 2000), HEC1 on the outer plate (DeLuca et al., 2006; Welburn et al., 2010), and MCAK at metaphase (Zhang et al., 2007). AurB is also believed to regulate the dynamics of the ROD, ZW10, zwilch (RZZ) complex (Famulski and Chan, 2007). Recent work highlights the antagonistic roles of protein phosphatase 1 (PP1) and AurB in kinetochore assembly (Emanuele et al., 2008).
Kinetochore dynein plays a crucial role in checkpoint silencing at metaphase (King et al., 2000; Howell et al., 2001; Wojcik et al., 2001; Whyte et al., 2008). A mitotic phosphorylation site was identified recently that is required for targeting of dynein to kinetochores (Whyte et al., 2008); however, the kinase responsible for this phosphorylation was not identified. Several features of AurB suggest that this kinase is closely linked to phosphorylated dynein. PP1γ was identified as a dynein phosphatase (Whyte et al., 2008), and PP1 is known to dephosphorylate substrates for AurB (Hsu et al., 2000; Emanuele et al., 2008). AurB phosphorylation regulates proteins involved in dynein–driven streaming, including the RZZ complex (Famulski and Chan, 2007). The activated form of AurB localizes to the layers of the kinetochore responsible for recruiting dynein (Deluca et al., 2011).
In this study, we investigated the role of AurB in recruiting kinetochore dynein. Although AurB was required for recruitment of phospho-dynein to kinetochores, dynein was not identified as the target for AurB phosphorylation. Using ZM447439 treatment or kinase-dead (KD) AurB constructs, we identified a boundary between zwint-1 and the RZZ complex as a target for AurB regulation. Tandem mass spectrometry (MS/MS) analysis revealed three novel phosphorylation sites at the C-terminus of zwint-1 responsible for RZZ recruitment. Analysis of phosphorylation-site mutants indicates that triple phosphorylation is sufficient for recruitment of the RZZ complex. At metaphase, however, dephosphorylation of these sites is required for dynein-driven streaming of checkpoint proteins. Together these studies identify zwint-1 as a novel AurB substrate central to the regulation of the spindle assembly checkpoint (SAC).
RESULTS
Impact of Aurora B inhibition on kinetochore dynein
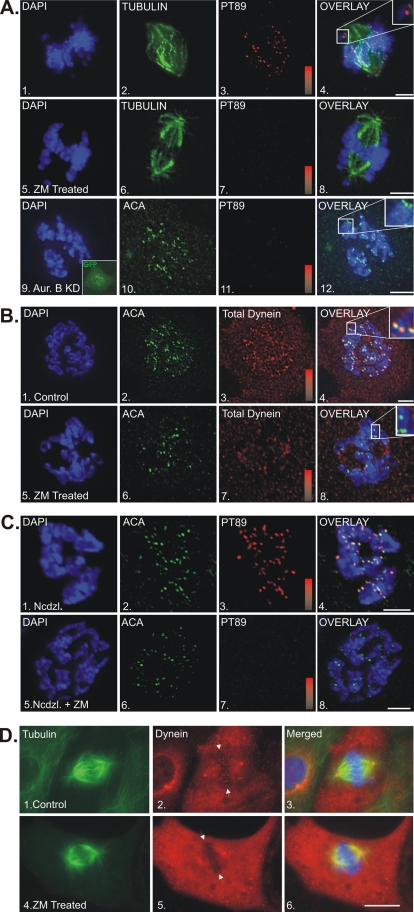
In previous work, a mitotic phosphorylation site was identified in the dynein intermediate chains (ICs) that is required for recruitment of dynein to kinetochores (Whyte et al., 2008). PP1 was identified as the dynein phosphatase (Whyte et al., 2008), raising the possibility that AurB is a dynein kinase (Hsu et al., 2000; Emanuele et al., 2008). We tested this model by inhibiting AurB and measuring levels of phospho-dynein at kinetochores using a phospho-epitope-antibody (pT89). Inhibition of AurB with ZM447439 (ZM) (Ditchfield et al., 2003) induced a dramatic loss of pT89 staining at kinetochores despite the formation of normal mitotic spindles and normal entry into mitosis (Figure 1A). To confirm AurB inhibition as the cause of this effect, we expressed a dominant-negative AurB KD construct as an alternative (Murata-Hori and Wang, 2002). Expression of this construct reduced recruitment of pT89 dynein at kinetochores to background levels even though mitotic entry proceeded normally (Figure 1A). These findings suggest that AurB activity is required for accumulation of phospho-dynein at kinetochores.
FIGURE 1:
Impact of Aurora B inhibition of recruitment of dynein to kinetochores. (A) NRK2 cells stained for chromatin (DAPI, blue), pT89 dynein (red), and tubulin (green) or ACA (green) in controls (1–4), ZM-treated cells (5–8), or cells expressing KD AurB (9–12). Intensity gradients represent a range of 0–13,000 for pT89 staining. Average pixel intensity was 11,870 ± 825 for controls and 374 ± 132 for ZM-treated cells (p < 0.0001). Bar, 5 μm. (B) NRK2 cells stained for chromatin (DAPI, blue), ACA (green), and total dynein (red) in controls (1–4) and ZM-treated cells (5–8). Overlays reveal colocalization of dynein with ACA in controls but not ZM-treated cells (insets). Intensity gradients represent a range of 0–13,000 for dynein staining. Average pixel intensity was 11780 ± 985 for controls and 1325 ± 477 for ZM-treated cells (p < 0.0001). Bar, 5 μm. (C) NRK2 cells stained for chromatin (DAPI, blue), ACA (green), and phospho-dynein (red) in cells treated with nocodazole (1–4) or nocodazole plus ZM (5–8). Intensity gradients represent a range of 0–1300 for pT89 staining. Average pixel intensity was 1277 ± 435 for controls and 654 ± 101 for ZM-treated cells. Bar, 5 μm. (D) BSC-1 cells expressing GFP-tubulin were subjected to cooling after chromosome alignment followed by rewarming to allow spindle assembly and stained for total dynein in controls (1–4) and ZM-treated cells (5–8). Dynein accumulates at metaphase kinetochores (arrows) in controls but not ZM-treated cells.
Both phospho-dynein and dephospho-dynein are detected on prometaphase kinetochores (Whyte et al., 2008), so we tested whether both populations required AurB activity. AurB inhibition with ZM led to a total loss of dynein at kinetochores but not spindle poles (Figure 1B). These findings suggest that AurB activity is required for accumulation of all kinetochore dynein during prometaphase.
The level of kinetochore dynein could be affected by reducing initial recruitment or by stimulating release through interactions with microtubules. To distinguish between these mechanisms, we used nocodazole treatment, which allows recruitment but not microtubule-dependent release (Whyte et al., 2008). Cells treated with nocodazole alone displayed robust phospho-dynein at kinetochores, whereas cells treated with nocodazole and ZM displayed a loss of phospho-dynein at kinetochores (Figure 1C). Finally, we used reduced temperature to depolymerize microtubules at metaphase, followed by rapid warming to assess the recruitment of dynein to kinetochores that had lost microtubule attachment after alignment (Khodjakov et al., 2000). Phospho-dynein was recruited to kinetochores during rewarming in controls but was reduced to background levels in cells treated with ZM (Figure 1D). These assays confirm that AurB is required for the recruitment of dynein to kinetochores.
As a more direct measure of dynein phosphorylation, we assayed AurB activity using in vitro kinase assays (Supplemental Figure 1A). Neither wild-type ICs nor T89A/T89D mutant ICs were phosphorylated to any measurable levels in these assays. Recognizing the limitations of in vitro kinase assays, we also performed Western blot analysis of cell extracts. Comparing control extracts to ZM-treated extracts, we measured levels of total dynein (V3) and phospho-dynein (pT89). Total dynein was detected in both extracts (Supplemental Figure 1B). However, phospho-dynein was detected in ZM-treated but not control extracts. This difference reflects the cosedimentation of phospho-dynein with chromosomes during the preparation of centrifuged mitotic extracts in control samples and the failure of phospho-dynein to cosediment with chromosomes in ZM-treated extracts. Because phospho-dynein was still detectable after ZM treatment, these assays suggest that AurB is not a dynein kinase. However, AurB is required for recruitment of phospho-dynein to kinetochores during prometaphase.
Determining the boundary between AurB-dependent and AurB-independent proteins
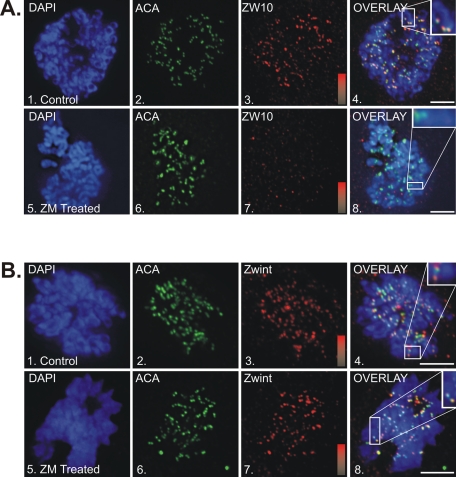
Although our analysis did not identify AurB as a dynein kinase, AurB inhibition had a profound effect on recruitment of dynein to kinetochores during prometaphase. One possibility is that AurB regulates the platform of dynein-associated proteins at kinetochores (Famulski and Chan, 2007; Ditchfield et al., 2003). We measured the impact of AurB inhibition on accumulation of dynactin, spindly, MAD2, BubR1 (see Supplemental Figure 2), and ZW10 (Figure 2A) and determined that all were depleted substantially from kinetochores. Whereas previous studies revealed a requirement for AurB in the recruitment/retention of ZW10, ROD, Mad2, Bub1, and BubR1 (Famulski and Chan, 2007; Ditchfield et al., 2003), these new studies confirm and extend the previous work to include dynein and dynein-associated proteins. Collectively, this body of work indicates that AurB is required for the recruitment of the RZZ complex and RZZ-dependent proteins to kinetochores during prometaphase.
FIGURE 2:
Impact of Aurora B inhibition on RZZ proteins at kinetochores. (A) HeLa cells stained for chromatin (DAPI, blue), ACA (green), and ZW10 (red) in controls (1–4) and ZM-treated cells (5–8). Overlays reveal colocalization of ZW10 with ACA in controls but not ZM-treated cells (insets). Intensity gradients represent a range of 0–12,000 for ZW10 staining. Average pixel intensity was 10,380 ± 619 for controls and 751 ± 152 for ZM-treated cells (p < 0.0001). Bar, 5 μm. (B) HeLa cells stained for chromatin (DAPI, blue), ACA (green), and zwint-1 (red) in controls (1–4) and ZM-treated cells (5–8). Overlays reveal colocalization of zwint-1 with ACA in both controls and ZM-treated cells (insets). Intensity gradients represent a range of 0–15,000 for zwint-1 staining. Average pixel intensity was 12,510 ± 531 for controls and 12,050 ± 441 for ZM-treated cells (p = 0.508). Bar, 5 μm.
To identify the boundary between AurB-dependent and AurB-independent proteins at kinetochores, we measured the impact of AurB inhibition on proteins implicated in RZZ recruitment. Zwint-1 is a ZW10-binding protein important for RZZ recruitment (Starr et al., 2000; Famulski et al., 2008; Vos et al., 2011), and both HEC1 (Lin et al., 2006) and KNL1 (Cheeseman et al., 2008) have been proposed to recruit zwint-1. Using this hierarchy, we observed that zwint-1 (Figure 2B), HEC1 (Supplemental Figure 2E), and KNL1 (unpublished data) were not affected by AurB inhibition. Together these experiments identify a novel boundary between zwint-1 and RZZ that is regulated by AurB and required for recruitment of dynein to kinetochores during prometaphase.
A novel substrate for Aurora B
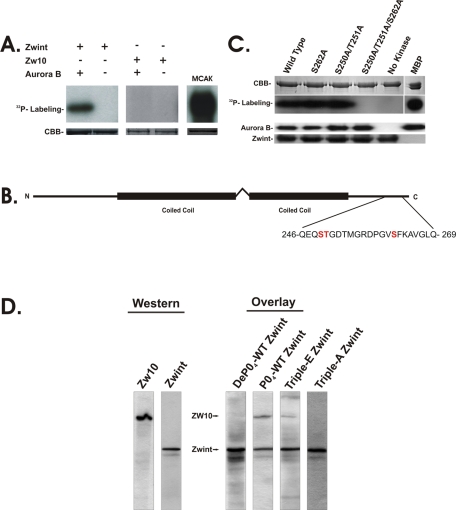
Because zwint-1 binds directly to ZW10 (Starr et al., 2000), we tested which of these proteins was phosphorylated by AurB. In vitro kinase assays with recombinant zwint-1 and ZW10 revealed phosphorylation of zwint-1 but not ZW10 (Figure 3A). This suggests that AurB phosphorylation of zwint-1 could be the basis of regulated recruitment of RZZ to kinetochores.
FIGURE 3:
Phosphorylation of zwint-1 but not ZW10 by AurB. (A) In vitro kinase assays of zwint-1 (full-length), ZW10 (full-length), and MCAK by purified AurB. CBB staining (SDS–PAGE) confirms equal loading, and 32P-labeling reflects phosphorylation of zwint-1 and MCAK but not ZW10. (B) Line diagram of human zwint-1, summarizing MS/MS mapping of AurB phosphorylation sites in human zwint-1 (full length) after trypsin/EndoGluC digestion. Compared to the two coiled-coil domains in zwint-1, phosphorylation sites were identified at S250, T251, and S262 near the C-terminus. (C) In vitro kinase assays of single, double, and triple zwint-1 mutants with purified AurB. CBB staining (SDS–PAGE) confirms equal loading of substrate, whereas Western blots detecting AurB and zwint-1 confirm the protein composition. The 32P-labeling indicates phosphorylation of wild-type and single- and double-mutant zwint-1 but not triple-mutant zwint-1. Myelin-binding protein (MBP) was used as a positive control. (D) Blot overlay assays with recombinant zwint-1 proteins probing total mitotic HeLa cell extracts. Western blots indicate mobility of ZW10 and zwint-1, whereas overlay samples indicate binding of recombinant zwint-1, phosphorylated zwint-1, triple-E zwint-1, and triple-A zwint-1. Phosphorylated zwint-1 and triple-E zwint-1 bound to ZW10, but neither dephospho–zwint-1 nor triple-A zwint-1 bound. Native zwint-1 is detected in the samples as part of the assay.
To confirm AurB phosphorylation of zwint-1, we completed MS/MS analysis of phosphorylated zwint-1 and mapped three novel phosphorylation sites near the C-terminus (S250, T251, S262) combining tryptic and endo-GluC proteolytic digests (Figure 3B and Supplemental Figure 3, A and B). In vitro kinase assays were used to assess phosphorylation at each site. Whereas single mutants [S250A (unpublished data), T251A (unpublished data), and S262A] and a double mutant (S250A/T251A) retained measurable AurB phosphorylation in kinase assays, a triple mutant (S250A/T251A/S262A) of zwint-1 displayed loss of AurB phosphorylation (Figure 3C). These studies identify zwint-1 as a novel AurB substrate and reveal the sites of phosphorylation near the C-terminus.
To determine the functional impact of zwint-1 phosphorylation, we performed blot overlay assays and identified phosphorylation-dependent binding partners (Figure 3D). Recombinant zwint-1 (dephosphorylated) failed to bind proteins in mitotic extracts, although endogenous zwint-1 is detected as part of the assay. When recombinant zwint-1 was phosphorylated in vitro by AurB, the assays revealed binding to a band comigrating with ZW10. This interaction was confirmed with dot blot assays of recombinant ZW10 (Supplemental Figure 3, C and D). We also tested the ability of a triple-E mutant (S250E/T251E/S262E) of zwint-1 to bind ZW10 in these assays. Similar to phospho–zwint-1, triple-E mutant zwint-1 bound ZW10 in mitotic extracts (Figure 3D). Consistent with the results with recombinant zwint-1, triple-A mutant (S250A/T251A/S262A) failed to bind ZW10 (Figure 3D). Together these assays reveal three novel AurB phosphorylation sites in zwint-1 that mediate a phosphorylation-dependent interaction with ZW10.
Impact of zwint-1 phosphorylation-site mutants on kinetochore assembly
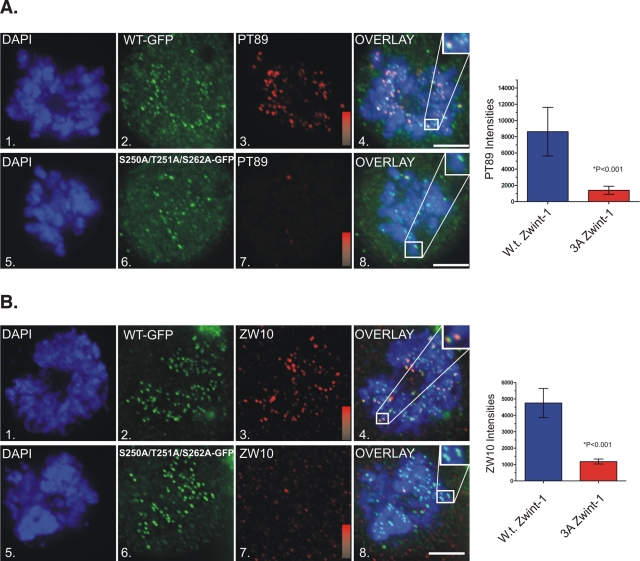
To test the requirement for triple phosphorylation of zwint-1 in cells, we expressed wild-type and triple-A zwint-1 mutant constructs in cells and assessed recruitment of dynein and ZW10. Both the wild-type and triple-A mutant constructs accumulated at kinetochores to similar levels (Figure 4). Although recruitment of phospho-dynein was normal in cells expressing wild-type zwint-1, recruitment of phospho-dynein was reduced significantly in cells expressing the triple-A mutant (Figure 4A). Similar defects were observed for ZW10 (Figure 4B). We also analyzed single and double zwint-1 mutants in these assays, but the impacts on dynein and ZW10 recruitment were incomplete, with intermediate levels of both proteins retained at kinetochores (unpublished data). Because the outcomes with the triple-A mutant zwint-1 mimic the effects of ZM treatment closely, these findings support a requirement for AurB phosphorylation of zwint-1 in kinetochore assembly.
FIGURE 4:
Impact of triple-A mutant zwint-1 expression on kinetochore dynein and ZW10. (A) NRK2 cells stained for chromatin (DAPI, blue), zwint-1-GFP (green), and pT89-dynein (red) in cells expressing wild-type zwint-1 (1–4) or triple-A mutant zwint-1 (5–8). Overlays reveal colocalization of pT89 dynein with wild-type zwint-1 but a loss of pT89 dynein in cells expressing the triple-A mutant (insets). Intensity gradients represent a range of 0–12,000 for pT89 dynein staining. Statistical analysis of pT89 signal intensity in cells expressing wild-type zwint-1 (blue) and triple-A mutant zwint-1 (red). PT89 signal is reduced significantly in cells expressing the triple-A mutant (p < 0.0001). Bar, 5 μm. (B) HeLa cells stained for chromatin (DAPI, blue), zwint-1-GFP (green), and ZW10 (red) in cells expressing wild-type zwint-1 (1–4) or triple-A mutant zwint-1 (5–8). Overlays reveal colocalization of ZW10 with wild-type zwint-1 but a loss of ZW10 in cells expressing the triple-A mutant (insets). Intensity gradients represent a range of 0–6000 for ZW10 staining. Statistical analysis of ZW10 signal intensity in cells expressing wild-type zwint-1 (blue) and triple-A mutant zwint-1 (red). ZW10 signal is reduced significantly in cells expressing the triple-A mutant (p < 0.0001). Bar, 5 μm.
Impact of zwint-1 phosphorylation-site mutants on chromosome movement
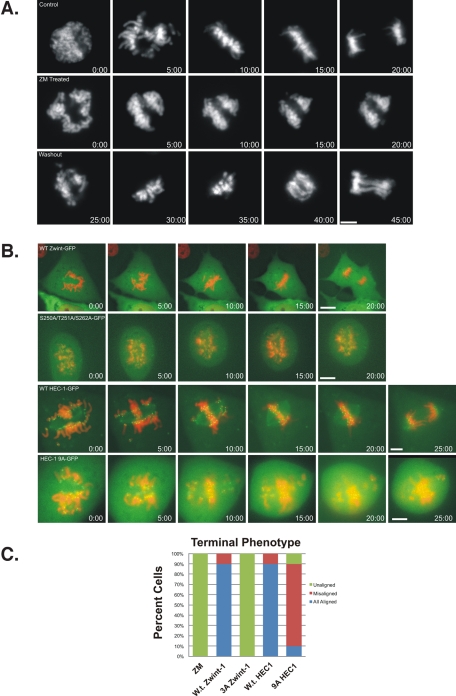
The triple-A zwint-1 mutant mimicked ZM treatment in disrupting kinetochore assembly; however, other AurB substrates (e.g., HEC1) are known to play roles in microtubule attachment and chromosome movement during prometaphase. To assess whether loss of zwint-1 phosphorylation is sufficient to explain ZM-induced defects during prometaphase, we compared cells treated with ZM to cells expressing zwint-1 or HEC1 phosphorylation-site mutants using live-cell imaging (Figure 5). To establish the details of the ZM phenotype, we performed time-lapse imaging and followed chromosome movements. These assays revealed chromosome immobilization after initial association with the mitotic spindle (Figure 5A, Supplemental Figure 4, A and B, and Supplemental Videos S1–S4). Chromosomes failed to congress during ZM treatment and retained this “locked” phenotype for the duration of time-lapse sequences (Figure 5C). Interestingly, if ZM-washout was performed after cells had achieved this “locked” phenotype, chromosomes did progress to anaphase onset (Figure 5A and Supplemental Video 4). This suggests that the AurB inhibition induces defects in dynamic chromosome movement and is reversible.
FIGURE 5:
Comparison of zwint-1 triple-A mutant to AurB inhibition. (A) Time-lapse imaging of NRK2 cells expressing mCherry-H2B. Control cells display normal mitotic progression. Cells treated with ZM (middle) display prometaphase arrest with “locked” chromosomes. ZM-treated cells with subsequent washout (bottom) reinitiate chromosome alignment and progress to anaphase onset. Bar, 5 μm. (B) Time-lapse imaging of NRK2 cells expressing mCherry-H2B and EGFP-Zwint-1 or EGFP-HEC1. Cells expressing wild-type zwint-1 EGFP (top) display normal mitotic progression. Cells expressing triple-A mutant zwint-1 EGFP (second from top) display prometaphase arrest with “locked” chromosomes. Cells expressing with-type HEC1 EGFP (third from top) display normal mitotic progression. Cells expressing HEC1 9A EGFP (bottom) display movement but errors in alignment. Bar, 5 μm. (C) Graphical representation of chromosome movement phenotypes for each condition.
To assess the contribution of known AurB substrates to this “locked” phenotype, we compared ZM treatment to expression of zwint-1 or HEC1 mutants (Guimaraes et al., 2008). The triple-A zwint-1 construct accumulated at kinetochores similar to the wild-type construct and induced a “locked” phenotype in which chromosomes stopped moving after initial binding to the mitotic spindle (Figure 5, B and C, and Supplemental Videos S5 and S6). In contrast, cells expressing the 9A HEC1 construct displayed delays in chromosome alignment and a failure to align all chromosomes. However, chromosomes continued to move in these cells, and the majority of chromosomes aligned at the metaphase plate (Figure 5, B and C, and Supplemental Videos S7 and S8). Although the zwint-1 mutant mimics ZM treatment closely during prometaphase, this represents a subset of the defects induced by AurB inhibition. It will be important to determine the contributions of other AurB substrates in future studies.
Zwint-1 phosphomimetics overcome AurB inhibition
As another method to test whether zwint-1 is a primary target for AurB regulation during prometaphase, we prepared a triple-E mutant (S250E/T251E/S262E) zwint-1 construct for expression in mammalian cells. This construct accumulated at kinetochores similar to the wild-type construct (Figure 6). To assess whether this construct mimicked phospho–zwint-1 in kinetochore assembly, we inhibited AurB in cells expressing the triple-E mutant and measured accumulation of dynein and RZZ (Figure 6). Despite a ZM-induced defect in recruitment of both dynein and ZW10 in cells expressing wild-type zwint-1, cells expressing the triple-E zwint-1 construct were resistant to AurB inhibition and displayed accumulation of both dynein and ZW10 at kinetochores (Figure 6, A and B). Single and double zwint-1 phosphomimetics were also assessed; however, they displayed incomplete resistance to AurB inhibition (unpublished data). These findings suggest that triple-E mutant zwint-1 is sufficient to rescue kinetochore assembly of the dynein-binding platform during AurB inhibition.
FIGURE 6:
Impact of triple-E mutant expression on kinetochore dynein and ZW10. (A) ZM-treated NRK2 cells (during prometaphase) expressing wild-type (1–4) or triple-E mutant (5–8) EGFP-zwint-1 (green) and stained for chromatin (DAPI, blue) and pT89-dynein (red). Intensity gradients represent a range of 0–12,000 for pT89 dynein staining. Bar, 5 μm. (B) ZM-treated HeLa cells (during prometaphase) stained for chromatin (DAPI, blue), zwint-1-GFP (green), and ZW10 (red) in cells expressing wild-type (1–4) or triple-E mutant (5–8) EGFP-zwint-1 (green). Intensity gradients represent a range of 0–6000 for pT89 dynein staining. Bar, 5 μm.
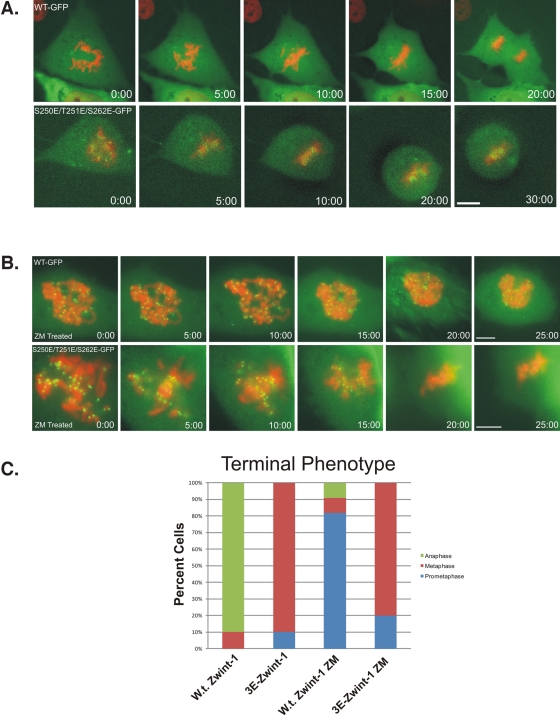
Because the triple-E zwint-1 mutant overcame AurB inhibition of kinetochore assembly, we also measured the impact on chromosome movement (Figure 7). There was no significant difference in the time from nuclear envelope breakdown to metaphase between cells expressing wild-type and those expressing triple-E mutant zwint-1 (16.4 ± 1.13 min vs. 18.4 ± 0.98 min). Image sequences of cells expressing the triple-E mutant displayed normal chromosome alignment similar to cells expressing the wild-type construct (Figure 7, A–C, and Supplemental Video S9). Of interest, however, cells expressing the triple-E mutant spent an extended time at metaphase compared with cells expressing wild-type zwint-1 (18.2 ± 4.4 min vs. 5.3 ± 1.8 min; p = 1.58 × 10−6). The time spent at metaphase for cells expressing triple-E zwint-1 is likely an underestimate because these cells displayed phototoxicity before the full duration of delay could be measured. For this reason, we scored the stage of mitosis when imaging was stopped (Figure 7C). Examples of anaphase onset were not observed for cells expressing triple-E zwint-1.
FIGURE 7:
Analysis of metaphase–anaphase transition in cells expressing triple-E mutant zwint-1. (A) NRK2 cells expressing mCherry-H2B and wild-type zwint-1 EGFP (top) or triple-E mutant zwint-1 (bottom) were subjected to time-lapse imaging. Bar, 5 μm. (B) ZM-treated NRK2 cells expressing mCherry-H2B and wild-type zwint-1 EGFP (top) or triple-E mutant zwint-1 (bottom) were subjected to time-lapse imaging. Bar, 5 μm. (C) Graphical depiction of the mitotic progression events for each condition. Colors indicate the mitotic stage at the end of each imaging experiment.
We also tested whether the triple-E mutant construct could rescue the defects in chromosome movement induced by ZM treatment. Whereas cells expressing wild-type zwint-1 displayed the “locked” phenotype described earlier, cells expressing the triple-E mutant progressed to metaphase chromosome alignment (Figure 7, B and C, and Supplemental Videos S10 and S11). Together, these experiments indicate that the triple-E zwint-1 protein mimics phospho–zwint-1 in kinetochore assembly and in mediating dynamic chromosome movement during prometaphase. Despite alignment, however, the triple-E construct delays the transition from metaphase to anaphase.
Zwint-1 dephosphorylation is required at metaphase
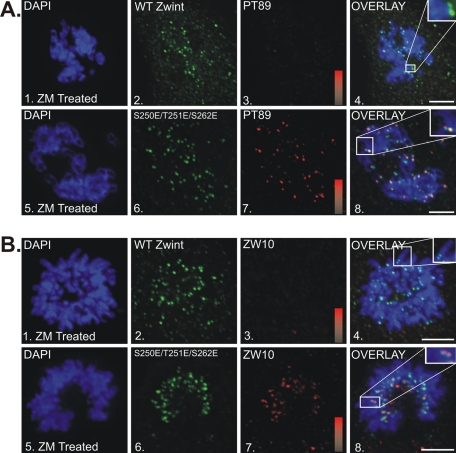
Previous work on kinetochore dynein revealed a sequence of events at metaphase including dynein dephosphorylation and dynein-mediated poleward transport of checkpoint proteins that was required for silencing the SAC in response to chromosome alignment (Whyte et al., 2008). Defects in any one of these events could explain the defects observed in cells expressing the triple-E zwint-1 construct. To determine which aspects of SAC silencing were defective, we monitored dynein dephosphorylation and poleward transport (Figure 8). Using the pT89 phospho-antibody to measure dynein dephosphorylation, we observed typical loss of phospho-dynein from metaphase kinetochores in cells expressing either wild-type or triple-E zwint-1 constructs (Figure 8A). This indicates that stretch-activated dynein dephosphorylation (Whyte et al., 2008) is retained in cells expressing triple-E zwint-1.
FIGURE 8:
Impact of triple-E mutant expression on metaphase–anaphase transition. (A) NRK2 cells (at metaphase) stained for chromatin (DAPI, blue) and pT89 dynein (red) after expression of wild-type zwint-1–EGFP (1–4) or triple-E mutant zwint-1–EGFP (5–8). Intensity gradients correspond to intensity values of 0-6000 for pT89 dynein. Bar, 5 μm. (B) NRK2 cells (at metaphase) stained for chromatin (DAPI, blue) and kinetochore proteins (red) after expression of wild-type zwint-1–EGFP (1–4) or triple-E–mutant zwint-1-EGFP (5–24). Expression of triple-E mutant zwint-1 induced metaphase kinetochore retention of total dynein (5–8), ZW10 (9–12), dynactin (13–16), MAD2 (17–20), and BubR1 (21–24). Locations of spindle poles (yellow arrows) and kinetochores (white arrows) are indicated. Intensity gradients correspond to intensity values of 0–12,000 (total dynein), 0–10,000 (dynactin and ZW10), 0–13,000 (MAD2), and 0–11,000 (BubR1). Bar, 5 μm.
We also measured levels of proteins that normally “stream” from kinetochores to spindle poles at metaphase. In contrast to cells expressing wild-type zwint-1, kinetochores of cells expressing triple-E zwint-1 retained dynein, dynactin, ZW10, MAD2, and BubR1 at kinetochores despite chromosome alignment and evidence of kinetochore stretch (Figure 8B and Supplemental Figure 5, A–D). The retention of dynein and dynactin, in particular, reflects an inability to transport checkpoint proteins away from the kinetochores. These experiments reveal that although triple-E zwint-1 is capable of mimicking the role of phosphorylated zwint-1 in mediating kinetochore assembly, it is not capable of mimicking zwint-1 dephosphorylation in response to chromosome alignment. This suggests that zwint-1 dephosphorylation is required to release streaming proteins at metaphase.
DISCUSSION
Our analysis of AurB and kinetochore dynein provides improved insight into the roles of AurB in kinetochore assembly and the consequences of AurB inhibition. In addition to proposed roles in regulating the microtubule-binding activities of HEC1 and MCAK, AurB phosphorylates zwint-1 during prometaphase. Consistent with previous studies that highlighted the requirement for AurB activity in the recruitment of the RZZ complex (Famulski and Chan, 2007), the present work defines the molecular basis of this regulation. AurB-mediated phosphorylation of zwint-1 also defines the boundary between stable kinetochore proteins and those that stream to mediate checkpoint silencing at metaphase. This represents a fundamental advance in our understanding of spindle assembly checkpoint regulation during mitosis.
Aurora B regulates dynein recruitment indirectly
AurB inhibition was achieved using either ZM447439 or expression of a KD AurB construct. Because both approaches had the same effect on assembly of the dynein-binding platform, these experiments add further support for the importance of AurB in the recruitment of the RZZ complex and RZZ-dependent proteins to kinetochores during prometaphase. Although AurB inhibition reduced accumulation of dynein at kinetochores, in vitro and in vivo assays reveal that AurB is not the kinase responsible for generating the pT89 phospho-epitope. Using immunofluorescence microscopy analysis of a hierarchy of kinetochore proteins, we determined that AurB is required for recruitment of the RZZ complex to kinetochores and for recruitment of RZZ-dependent proteins, including dynactin and dynein.
The phenotypes observed after AurB inhibition also shed light on potential roles for RZZ-dependent proteins during prometaphase. Dynein has been proposed to mediate initial microtubule capture and to drive early aspects of chromosome movement. Dynactin is also believed to play a role in both of these processes; however, the molecular details of this coordination are not understood. Other microtubule-binding proteins (e.g., spindly, CLIP-170) interact directly or indirectly with the RZZ complex and could contribute to microtubule binding. Of interest, this entire set of microtubule-binding proteins is distal to the RZZ complex and lost after AurB inhibition. Despite this loss, initial association of chromosomes with the mitotic spindle appears functional. This highlights the contributions of more-proximal microtubule-binding proteins (e.g., HEC1) in initial interactions between chromosomes and microtubules or proteins recruited independent of the RZZ complex. An alternative interpretation is that more-proximal microtubule-binding proteins become dominant during AurB inhibition as a result of the atypical composition of kinetochores under these conditions. Consistent with this possibility, the “locked” phenotype we observe after AurB inhibition is abnormal and might reflect an incomplete kinetochore structure. Of interest in this regard, dynein is believed to release initial, lateral microtubule attachments under normal conditions (Varma et al., 2008), and the loss of dynein could contribute to the “locked” phenotype. Future studies will be needed to assess the type of microtubule attachment in cells expressing the triple-A mutant zwint-1 and to compare this to ZM treatment.
Zwint-1 is a novel AurB substrate during mitosis
AurB was implicated previously in regulating MCAK and HEC1; however neither of these proteins is clearly linked to the recruitment of dynein to kinetochores. Focusing on this aspect of kinetochore assembly, we monitored a linear sequence of protein–protein interactions implicated in dynein recruitment. The simplest interpretation of our imaging studies is that recruitment of the RZZ complex was affected by AurB inhibition, but more-proximal layers of the kinetochore are not affected. Consistent with this hypothesis, zwint-1 is recognized as the primary ZW10-binding protein (Starr et al., 2000; Famulski et al., 2008; Vos et al., 2011), and we identified zwint-1 as an AurB substrate (Figure 3).
Two aspects of our MS/MS mapping are intriguing. The identification of three phosphorylation sites in close proximity was not expected in comparison to a single phosphorylation required for dynein recruitment (pT89) (Whyte et al., 2008). However, multiple phosphorylation sites have been proposed in both HEC1 (Guimaraes et al., 2008; Deluca et al., 2011) and MCAK (Zhang et al., 2007), suggesting that this mechanism might be shared with other AurB substrates. Second, the specific residues of phosphorylation are not absolutely conserved among mammals (Supplemental Figure S3B). Compared to rodent sequences, human zwint-1 contains an extended C-terminal tail harboring the phosphorylation sites we mapped. Mouse and rat zwint-1 lack this tail but contain a series of potential C-terminal phosphorylation sites missing in the human sequence. The C-terminus of zwint-1 was shown to contain the ZW10-binding domain (Vos et al., 2011), consistent with C-terminal phosphorylation playing a role in ZW10 binding. Future work will be needed to test for analogous phosphorylation of zwint-1 in other species.
Proper regulation of zwint-1 is required for checkpoint silencing
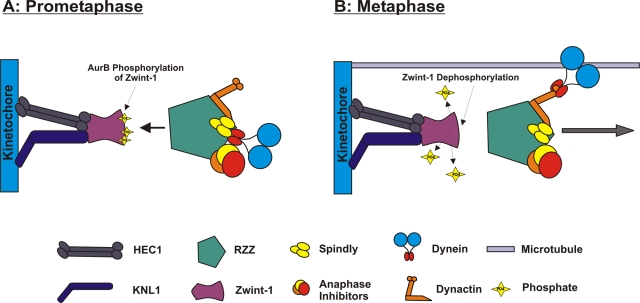
These studies suggest that phosphorylation-dependent recruitment of the RZZ complex by zwint-1 is important for several reasons. During prometaphase, it drives the sequential assembly of the multilayered kinetochore through the activity of a mitotic kinase (Figure 9). At metaphase, however, this mechanism defines a boundary between stable kinetochore proteins and those that stream poleward in response to chromosome alignment (Figure 9). The latter is dynein-driven process that requires dephosphorylation of dynein by PP1γ (Whyte et al., 2008). Mechanistically, it was unclear whether the release of RZZ and other checkpoint proteins simply reflected the strength of dynein as a motor or other events were coordinated by phosphatase activity at metaphase. Our work suggests that streaming of the RZZ complex at metaphase involves activation of dynein (Whyte et al., 2008) and release of the RZZ complex by zwint-1 dephosphorylation (Figure 9). Consistent with this model, the triple-E zwint-1 mutant mediates assembly of the RZZ platform but does not release RZZ at metaphase despite normal dephosphorylation of dynein. Together, these findings define a new aspect of the checkpoint-silencing process that is regulated by AurB.
FIGURE 9:
Model for zwint-1 phosphorylation by AurB. (A) During prometaphase, AurB phosphorylation of zwint-1 is required for recruitment of ZW10-, pT89 dynein-, and RZZ-dependent proteins to kinetochores. This is defective after AurB inhibition or after expression of the triple-A zwint-1 mutant. Triple-E mutant zwint-1 mimics phospho–zwint-1 in RZZ recruitment, even after AurB inhibition. (B) At metaphase, dephosphorylation of zwint-1 is required for poleward streaming of dynein and checkpoint proteins to silence the spindle assembly checkpoint. This process is defective in cells expressing the triple-E mutant despite typical dephosphorylation of pT89 dynein in response to chromosome alignment.
MATERIALS AND METHODS
Cell culture, drug treatments, and transfection
NRK2 and HeLa cells were cultured in 5% CO2 and 95% air in a Narco 4200 cell culture incubator at 37°C. NRK2 cells were cultured using Nutrient Mixture F12 Ham Kaighn's Modified (F12K) media (Sigma-Aldrich, St. Louis, MO). HeLa cells were cultured using DMEM (Sigma-Aldrich). Media were supplemented with 10% fetal bovine serum and 1% l-glutamine with penicillin/streptomycin. NRK2 and/or HeLa cells were treated with 2 μM ZM447439 (Tocris Bioscience, Bristol, United Kingdom) for 2 h. Plasmids were transiently transfected into NRK2 cells using nucleoporation (Lonza, Basel, Switzerland) or into HeLa using Fugene HD (Promega, Madison, WI). Aurora B kinase-dead mutant–green fluorescent protein (GFP) construct was a gift from Y. Wang (University of Massachusetts Medical School, Worcester, MA). HEC1 constructs were provided by J. DeLuca (Colorado State University, Fort Collins, CO).
Western blot analysis
After protein transfer, membranes were blocked with 5% milk or 5% PhosphoBLOCKER (Cell Biolabs, San Diego, CA) for phospho-specific antibodies in TBST (50 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Tween 20) for 1 h at room temperature (RT) with gentle agitation. Primary antibodies were used at the indicated dilutions: anti-pT89, 1:1000 (Whyte et al., 2008); anti–total dynein (V3), 1:2000 (Vaughan et al., 2001); anti-tubulin (DM1A), 1:1000 (Santa Cruz Biotechnology, Santa Cruz, CA); glutathione S transferase–horseradish peroxidase (HRP), 1:1000 (Abcam, Cambridge, MA); and polyclonal anti–hZwint-1, 1:1000 (Abcam). Membranes were incubated with primary antibodies diluted in blocking buffer for 1 h at RT with gentle agitation. After four 5-min washes with TBST, membranes were incubated with HRP-conjugated secondary antibodies and diluted at 1:10,000 in blocking buffer for 1 h at RT with gentle agitation. After four 5-min washes with TBST, membranes were incubated with LumiGLO chemiluminescent substrate (KPL, Gaithersburg, MD) for 1 min at RT. Chemiluminescence was recorded on autoradiographic film and digitalized using an HP 1370 scanner and PhotoShop (Adobe, San Jose, CA).
Recombinant protein purification and mutagenesis
Full-length pDEST47 Zwint-1 (G. Chan, University of Alberta, Alberta, CA) was cloned into pET14B. PET14B plasmids containing full-length Zwint-1 or full-length ZW10 (provided by R. Vallee, Columbia University, New York, NY) were expressed in Rosetta cells (Novagen, EMD4Biosciences, Gibbstown, NJ) and purified by nickel affinity chromatography using His-Bind resin (Novagen) as previously described (Vaughan and Vallee, 1995). Purified proteins were dialyzed against 1× TBS buffer and cleaved with thrombin (Novagen) to remove HIS tags. Protein concentration was determined by standard Bradford assay using Coomassie Plus Protein Assay reagent (Pierce, Rockford, IL). Mutants were generated by site-directed mutagenesis (Phusion; Finnzymes, Lafayette, CO), using full-length pET14B Zwint-1 or pDest47 Zwint-1 as a template. Primers used were as follows: S262A, forward primer, AGAGACCCTGGTGTGGCCTTCAAGGCTGTTGG; and S250A/T251A, reverse primer, CCCCATGGTGTCTCCTGCACCCTGCTCCTGGGG. Enhanced GFP-tagged versions of wild-type and mutant zwint-1 for mammalian expression were prepared using standard methods.
In vitro kinase assays
Recombinant Aurora B kinase (Cell Signaling Technology, Danvers, MA) was incubated with recombinant wild-type Zwint-1, ZW10, MCAK (a gift from C. Walczak, Indiana University, Bloomington, IN), MBP (New England Biolabs, Ipswich, MA), or zwint-1 mutants. Reactions were performed with the buffer provided (Cell Signaling Technology), ATP, and γ32P-ATP at 30°C for 30 min. Samples were boiled for 5 min, separated by SDS–PAGE, and stained with Coomassie brilliant blue for 15 min. After destaining, gels were exposed to autoradiograph film and processed to analyze the level of 32P incorporation.
Liquid chromatography/MS/MS analysis
Full length, wild-type Zwint was phosphorylated by AurB using the manufacturer-supplied buffer, ATP, at 30°C for 30 min and subjected to digestion with trypsin or Endoproteinase GluC (New England Biolabs), followed by liquid chromatography/MS/MS of the resulting peptides on a nanoACQUITY UPLC (Waters, Milford, MA) coupled to an amaZon ion trap mass spectrometer (Bruker Daltonics, Billerica, MA) in the Mass Spectrometry and Proteomics Facility at the University of Notre Dame. MS data searches of the Swiss-Prot database were performed using the Mascot search engine.
Blot overlay assays
Blot overlay assays were performed as previously described (Vaughan and Vallee, 1995). Cell extracts were fractionated by SDS–PAGE and transferred to polyvinylidene fluoride membranes. Membranes were incubated in 5% milk or 5% PhosphoBLOCKER overnight at 4°C with gentle agitation. Recombinant protein was diluted in blocking buffer at concentrations from 0.25 to 5.0 μg/ml and incubated with membranes for 2 h at room temperature. To visualize binding partners, membranes were washed four times for 5 min in TBST, incubated with primary antibody against the probe protein in blocking buffer for 1 h at room temperature, washed four times for 5 min in TBST, incubated with HRP-conjugated secondary antibodies in blocking buffer for 1 h at room temperature, washed four times for 5 min in TBST, and processed for enhanced chemiluminescence.
Immunofluorescence microscopy
NRK2 or HeLa cells were fixed in methanol at −20°C or 4% paraformaldehyde (PFA). PFA-fixed cells were extracted with 0.05% Triton X-100 in TBS (50 mM Tris, pH 7.4, 150 mM NaCl). Cells were rehydrated in TBS for 30 min and blocked for 1 h at RT with 3% bovine serum albumin (BSA) in TBS. Primary antibodies were used at the following dilutions: pT89, 1:600 (Whyte et al., 2008); V3, 1:200 (Vaughan et al., 2001); D'art, 1:300 (Echeverri et al., 1996); anti-centromere antibody, 1:50 (Antibodies, Davis, CA); ZW10, 1:200 (Famulski and Chan, 2007); polyclonal Zwint, 1:200 (Abcam, Cambridge, MA); HEC1, 1:300 (Imgenex, San Diego, CA); Spindly, 1:200 (Novus, Littleton, CO); DM1A, 1:100 (Cell Signaling); BubR1, 1:100 (BD, Franklin Lakes, NJ); and Mad2, 1:200 (BD). Cells were incubated with primary antibodies diluted in 3% BSA/TBS for 1 h at RT. Cells were washed four times for 5 min with TBS and incubated with Cy2-, Cy3-, or Cy5-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted in 3% BSA/TBS at 1:300 for 1 h at RT. Cells were washed four times for 5 min with TBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI) diluted 1:20,000 in TBS for 5 min. Coverslips were mounted using ProLong Gold antifade reagent (Invitrogen, Carlsbad, CA).
Images were acquired on a microscope (Axiovert 200MOT; Carl Zeiss, Jena, Germany) operated with MetaMorph software (MDS Analytical Technologies, Sunnyvale, CA) and a charge-coupled device (CCD) camera (CoolSNAP HQ; Roper Scientific, Sarasota, FL) or DeltaVision deconvolution workstation (Applied Precision, Issaquah, WA). Images were collected at RT using a 63× 1.4–numerical aperture (NA) oil immersion objective. Data were collected as z-series and subjected to digital deconvolution with Metamorph or Delta Vision software. Using matched illumination and antibody conditions, we used raw image intensities to determine intensity scales for each experiment. These raw data were maintained through image processing in Photoshop and Coreldraw (Corel, Mountain View, CA) and presented in a pseudo-color scale to convey relative image intensities.
Live-cell imaging
NRK2 cells expressing mCherry-H2B, enhanced GFP–tubulin, and zwint-1 constructs or HEC1 constructs were cultured in complete media, supplemented with 12 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer (HyClone, Dubuque, IA), at 37°C in the absence of CO2 for 24 h prior to imaging. Cells were treated with inhibitors as previously described. Images were acquired and deconvolved on a microscope (Axiovert 200MOT) operated with Metamorph software and a camera (Cascade 1k; Roper Scientific). Images were collected at 37°C using a 63× 1.4-N.A. objective. Data were collected as time-lapse image sequences.
Chilling/rewarming assays
Time-lapse images of BSC-1–GFP–tubulin cells (Hornick et al., 2011) were captured using a Leica DM RXA2 microscope (Leica Microsystems, Bannockburn, IL) with a Plan Apo 63×, 1.3-NA, 37°C glycerol immersion objective enclosed in a custom-made Plexiglas box maintained at 37°C and a Hamamatsu ORCA-ER CCD camera (Hamamatsu Photonics, Hamamatsu, Japan). When the chromosomes were judged to have aligned, either vehicle (dimethyl sulfoxide) or ZM was added to the custom-built imaging chamber (Hornick et al., 2011), and then the chamber was chilled (−20°C for 5 min, followed by 35 min at 4°C; Khodjakov et al., 2000). The chamber was rewarmed on the microscope for time-lapse imaging, and a nosepiece diamond scribe was used to mark the cell position. After methanol fixation, cells were stained using anti-dynein antibodies (V3).
Statistical analysis
Statistical analysis of intensity measurements and live-cell analysis was performed using GraphPad 5 (GraphPad Software, La Jolla, CA). Average kinetochore intensities were analyzed by averaging the peak intensities of the 10 brightest kinetochores in deconvolved images of 10 or seven different transfected cells. The p values of comparisons between control and experimental measurements were determined with a two-tailed t test, assuming unequal variance. Confidence levels were chosen at p ≤ 0.05.
Supplementary Material
Acknowledgments
We thank R. Vallee (Columbia University) for providing constructs encoding ZW10, C. Walczak (Indiana University) for providing purified MCAK fragments, YuLi Wang (Carnegie Mellon University, Pittsburgh, PA) for providing a kinase-dead AurB construct, and J. DeLuca (Colorado State University) for providing HEC1 constructs. Bruker Daltonics and Dionex Corporation (Sunnyvale, CA) are gratefully acknowledged for providing an extended in-house demonstration of an amaZon ETD ion trap mass spectrometer and an Ultimate 3000 2D nanoHPLC system, respectively, to the Notre Dame Mass Spectrometry and Proteomics Facility.
Abbreviation used:
- AurB
aurora B
- RZZ,
ROD, ZW10, zwilch
- SAC
spindle assembly checkpoint
- ZM
ZM447439
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0213) on July 20, 2011.
REFERENCES
- Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol Biol Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Deluca KF, Lens SM, Deluca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Lan W, Jwa M, Miller SA, Chan CS, Stukenberg PT. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski JK, Chan GK. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr Biol. 2007;17:2143–2149. doi: 10.1016/j.cub.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Famulski JK, Vos L, Sun X, Chan G. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J Cell Biol. 2008;180:507–520. doi: 10.1083/jcb.200708021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick JE, Mader CC, Tribble EK, Bagne CC, Vaughan KT, Shaw SL, Hinchcliffe EH. Amphiastral mitotic spindle assembly in vertebrate cells lacking centrosomes. Curr Biol. 2011;21:598–605. doi: 10.1016/j.cub.2011.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Kang J, Yu H. Kinase signaling in the spindle checkpoint. J Biol Chem. 2009;284:15359–15363. doi: 10.1074/jbc.R900005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10(2):59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- King JM, Hays TS, Nicklas RB. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J Cell Biol. 2000;151:739–748. doi: 10.1083/jcb.151.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Chen Y, Wu G, Lee WH. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene. 2006;25:6901–6914. doi: 10.1038/sj.onc.1209687. [DOI] [PubMed] [Google Scholar]
- Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- Murata-Hori M, Wang YL. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr Biol. 2002;12:894–899. doi: 10.1016/s0960-9822(02)00848-5. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Saffery R, Li Z, Simpson AE, Choo KH, Yen TJ, Goldberg ML. HZwint-1, a novel human kinetochore component that interacts with HZW10. J Cell Sci. 2000;113:1939–1950. doi: 10.1242/jcs.113.11.1939. [DOI] [PubMed] [Google Scholar]
- Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Varma D, Monzo P, Stehman SA, Vallee RB. Direct role of dynein motor in stable kinetochore-microtubule attachment, orientation, and alignment. J Cell Biol. 2008;182:1045–1054. doi: 10.1083/jcb.200710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Leszyk JD, Vaughan KT. Cytoplasmic dynein intermediate chain phosphorylation regulates binding to dynactin. J Biol Chem. 2001;276:26171–26179. doi: 10.1074/jbc.M102649200. [DOI] [PubMed] [Google Scholar]
- Vos LJ, Famulski JK, Chan GK. hZwint-1 bridges the inner and outer kinetochore: identification of the kinetochore localization domain and the hZw10 interaction domain. Biochem J. 2011;436:157–168. doi: 10.1042/BJ20110137. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR 3rd, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J, et al. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J Cell Biol. 2008;183:819–834. doi: 10.1083/jcb.200804114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik E, Basto R, Serr M, Scaerou F, Karess R, Hays T. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.