A whole-genome RNAi screen identified phy-1 as a novel interaction partner of the Caenorhabditis elegans gene ten-1. It is shown that the catalytic subunit of prolyl 4-hydroxylase, which is coded for by phy-1, is important for type IV collagen secretion and that the transmembrane protein TEN-1 links the epidermis to muscle cells through the basement membrane.
Abstract
Teneurins are a family of phylogenetically conserved proteins implicated in pattern formation and morphogenesis. The sole orthologue in Caenorhabditis elegans, ten-1, is important for hypodermal cell migration, neuronal migration, path finding and fasciculation, gonad development, and basement membrane integrity of some tissues. However, the mechanisms of TEN-1 action remain to be elucidated. Using a genome-wide RNA interference approach, we identified phy-1 as a novel interaction partner of ten-1. phy-1 codes for the catalytic domain of collagen prolyl 4-hydroxylase. Loss of phy-1 significantly enhanced the embryonic lethality of ten-1 null mutants. Double-mutant embryos arrested during late elongation with epidermal defects, disruption of basement membranes, and detachment of body wall muscles. We found that deletion of phy-1 caused aggregation of collagen IV in body wall muscles in elongated embryos and triggered the loss of tissue integrity in ten-1 mutants. In addition, phy-1 and ten-1 each genetically interact with genes encoding collagen IV. These findings support a functional mechanism in which loss of ten-1, together with a reduction of assembled and secreted basement membrane collagen IV protein, leads to detachment of the epidermis from muscle cells during late elongation of the embryo when mechanical stress is generated by muscle contractions.
INTRODUCTION
Morphogenesis, together with cell growth and cellular differentiation, is one of the fundamental processes of development. The coordination of morphogenetic movements involves interactions among the extracellular matrix, the cell surface, and the cytoskeleton (Chin-Sang and Chisholm, 2000). The epidermis is the largest organ in Caenorhabditis elegans, and its structure defines the shape and the size of the animal. During embryogenesis an ovoid ball of cells changes into a worm-shaped larva, driven by the migration, fusion, and elongation of epidermal cells (Simske and Hardin, 2001). Disruption of any of these processes leads to arrested embryos. Whereas early embryonic elongation depends on circumferentially oriented actin in the epidermal cells (Priess and Hirsh, 1986), processes beyond the twofold stage require proper connections among the epidermis, basement membrane (BM), and underlying body wall muscles (Francis and Waterston, 1991; Williams and Waterston, 1994). Once elongation is complete, hypodermal cells secrete the extracellular cuticle to hold the hypodermal cells in their final shape (Priess and Hirsh, 1986).
Teneurins are a family of phylogenetically conserved proteins described in Drosophila (Baumgartner et al., 1994; Levine et al., 1994; Fascetti and Baumgartner, 2002; Rakovitsky et al., 2007), zebrafish (Mieda et al., 1999), chicken (Minet et al., 1999; Tucker et al., 2001), mouse (Oohashi et al., 1999; Ben-Zur et al., 2000; Zhou et al., 2003), and C. elegans (Drabikowski et al., 2005). Teneurins are expressed during pattern formation and morphogenesis. They encode type II transmembrane proteins with a conserved domain structure consisting of an intracellular domain containing several conserved proline-rich regions, a single transmembrane domain, and an extracellular domain, which encompasses the major part of the protein. The extracellular domain consists of eight consecutive epidermal growth factor–like repeats, an extended region of conserved cysteines, and a stretch of YD repeats toward the C-terminus. The predicted mass of teneurin monomers is ∼300 kDa (Feng et al., 2002).
A single gene, named ten-1, encodes the sole orthologue of teneurins in C. elegans. Gene expression is controlled by two alternative promoters, ten-1a and ten-1b, resulting in two transcript versions differing in the length of the intracellular domain. Promoter green fluorescent protein (GFP) fusion proteins show distinct expression patterns: the upstream promoter ten-1a is predominantly active in mesoderm, whereas the downstream promoter ten-1b is predominantly active in the ectoderm (Drabikowski et al., 2005). TEN-1 is important for epidermal morphogenesis, gonad migration, neuronal path finding, and BM integrity of several tissues (Drabikowski et al., 2005; Trzebiatowska et al., 2008). Two deletion alleles of ten-1 were characterized as genetic null alleles: ok641 and tm651 (Trzebiatowska et al., 2008). Both mutants display pleiotropic phenotypes, including embryonic lethality, larval arrest, sterility, protruding vulva, or bursting through the vulva. Often germ cells are leaking from the developing gonad into the body cavity because of the rupture in the gonadal BM during the second larval stage. Attempts to investigate the function of ten-1 led to the discovery of genetic interactions of ten-1 with the BM-associated genes related to dystroglycan (dgn-1), integrin (ina-1), laminin (epi-1), and nidogen (nid-1). These experiments suggested that ten-1 acts in a parallel pathway with a partly redundant function to dystroglycan and/or integrin receptors (Trzebiatowska et al., 2008). Several studies on mutations in genes encoding BM molecules illustrate the importance of these components in morphogenesis. In C. elegans, a thin BM lines the pseudocoelomic cavity and separates the body wall muscle cells from the hypodermis and nervous system (White et al., 1976). A similar BM surrounds the intestine and gonad, whereas a thicker BM surrounds the pharynx (Albertson and Thomson, 1976; White, 1988).
In C. elegans, type IV collagen is expressed by the body wall muscle cells (Graham et al., 1997). Two genes have been identified to encode BM collagen IV, emb-9 and let-2. Before the proteins are assembled and deposited in the BM they undergo several steps of modifications in the endoplasmic reticulum. In vertebrates an essential role has been identified for the prolyl 4-hydroxylase (P4H). Proline hydroxylation of procollagen is important for the folding of the collagen chains into stable trimers at physiological temperatures (Fessler and Fessler, 1978). The enzyme P4H consists of an enzymatic subunit and a protein disulfide isomerase subunit. The loss of the enzymatic subunit causes embryonic lethality in the mouse due to the loss of BM integrity (Holster et al., 2007). Genes encoding the subunits of the P4H are phylogenetically conserved. Four genes have been identified to encode the enzymatic subunit of the C. elegans P4H: phy-1 (also known as dpy-18), phy-2, phy-3, and phy-4 (Winter and Page, 2000; Myllyharju et al., 2002; Riihimaa et al., 2002; Keskiaho et al., 2008). Epistasis analyses show that phy-1 in complex with phy-2 is essential for the survival of the C. elegans (Friedman et al., 2000). phy-1 mutations alone result in a mild dumpy phenotype, whereas animals lacking phy-2 alone are superficially wild type. This indicates that phy-1 codes for the most important subunit for the function of P4H at normal physiological conditions. P4H in C. elegans has been implicated in the modification of cuticle collagens but not in the maturation of BM collagen.
In this study, we characterize a novel genetic interaction between ten-1 and phy-1 and investigate ten-1 function during late embryonic elongation in a phy-1 deletion background. The characterization of the genetic interaction between ten-1 and phy-1 indicates a further link between TEN-1 and the extracellular matrix involving BM collagen IV. Furthermore, we contribute new insights into the function of phy-1 in C. elegans.
RESULTS
Loss of P4H function in ten-1 null mutants results in embryonic lethality
We performed a genome-wide RNA interference (RNAi) screen to identify novel genetic interaction partners of ten-1 (unpublished data). This screen led to the identification of phy-3 as an interaction partner of ten-1. Knockdown of phy-3 by RNAi in a ten-1 deletion background resulted in enhanced embryonic and larval lethality, as well as an overall reduced brood size in comparison to an empty vector control. Knockdown of phy-3 in a wild-type background did not lead to any obvious effect. phy-3 belongs to a family of genes coding for catalytic subunits of the collagen-modifying enzyme prolyl 4-hydroxylase. Four isoforms have been identified in C. elegans: phy-1, phy-2, phy-3, and phy-4. The mRNA sequences of all genes are very similar. To investigate whether the decrease of phy-3 mRNA level caused off-target effects, we performed quantitative real-time PCR analysis during rescreening of this candidate. We found that the RNAi for phy-3 also affects the expression levels of phy-1 and phy-2 (Supplemental Figure S1). To determine whether a single gene or a combination of them caused the enhancement of the ten-1 mutant phenotype, we generated double- and triple-knockout mutants using the null alleles phy-1(ok162), phy-2(ok177), and phy-3(ok199). We found that only the deletion of phy-1 results in a significant increase (20%) in embryonic lethality in a ten-1 mutant background (Table 1). Deletion of phy-3 in a ten-1 phy-1 mutant background did not increase any of the analyzed phenotypes (Supplemental Table S2). We also examined ten-1 phy-1 double-mutant animals for sterility, protruding vulva, and bursting-through-the-vulva phenotypes but could not find any differences in comparison with the ten-1 single mutant (unpublished data). Furthermore, ten-1 phy-1 mutant animals were dumpy to the same extent as the phy-1(ok162) single mutant itself. To show that the genetic interaction of ten-1 and phy-1 is allele independent, we repeated the double-mutant analysis for the second ten-1 deletion allele, tm651. Indeed, we confirmed the increase in embryonic lethality when phy-1 function is depleted (Table 1). Thus the genetic interaction between ten-1 and phy-1 is true for two independent alleles of ten-1, ok641, and tm651.
TABLE 1:
ten-1 genetically interacts with phy-1.
| Worm strain | Genotype | n | Total laid eggs (95% CI) | Embryonic lethal, % (95% CI) | Larval arrest, % (95% CI) |
|---|---|---|---|---|---|
| N2 | Wild type | 26 | 269 (246–289) | 0.5 (0.4–0.7) | 0.3 (0.2–0.5) |
| RU90 | ten-1(ok641) | 16 | 249 (213–284) | 3.8 (3.2–4.4)** | 30.0 (29.3–32.2) |
| RU98 | ten-1(tm651) | 18 | 227 (193–260) | 3.9 (3.3–4.5)** | 21.3 (19.5–22.0) |
| RU170 | phy-1(ok162) | 13 | 254 (233–274) | 1.0 (0.7–1.4) | 0.3 (0.2–0.6) |
| RU171 | ten-1(ok641) phy-1(ok162) | 15 | 160 (137–182) | 20.2 (19.1–22.3)***1 | 40.9 (38.7–42.6) |
| RU197 | ten-1(tm651) phy-1(ok162) | 20 | 264 (239–288) | 17.2 (15.9–17.9)***1 | 35.1 (34.9–37.5) |
| RU194 | kdEx132(phy-1p::phy-1; unc-122::gfp) | 7 | 260 (220–299) | 1.3 (0.8–1.9) | 1.5 (1.0–2.2) |
| RU196 | ten-1(ok641) phy-1(ok166); kdEx132 | 21 | 168 (151–184) | 4.5 (3.7–5.0)***2 | 25.7 (25–27.9) |
| RU191 | kdEx131(myo-3p::phy-1::gfp; unc-122::gfp) | 5 | 234 (209–258) | 0.2 (0–0.6) | 0 (0–0.3) |
| RU195 | ten-1(ok641) phy-1(ok162); kdEx131 | 13 | 163 (147–178) | 5.4 (4.6–6.5)***2 | 34.3 (31.9–35.9) |
| RU198 | kdEx133(eff-1p::phy-1::gfp; unc-122::gfp) | 15 | 170 (158–181) | 2.6 (2.0–3.3) | 0.8 (0.5–1.2) |
| RU199 | ten-1(ok641) phy-1(ok162); kdEx133 | 8 | 205 (159–250) | 18.2 (16.9–20.7), n.s. | 26.6 (24.1–28.4) |
Mean percentage and 95% confidence interval (CI) of wild-type and mutant worms and rescue lines of ten-1(ok641) phy-1(ok162) double mutant analyzed for embryonic lethality and larval arrest. n, number of animals tested for brood size.
**p < 0.003 compared with N2.
***1p < 10−7 compared with RU90 or RU98, respectively.
***2p < 10−7 compared with RU171.
n.s., p > 0.8 compared with RU171.
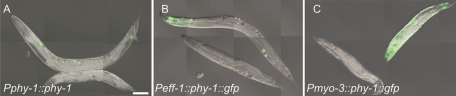
To show that the phenotype of the ten-1(ok641) phy-1(ok162) double mutant is specific for the loss of phy-1, we expressed the phy-1 cDNA under its endogenous promoter in the double mutant. The construct rescued the dumpy phenotype, as well as the increased embryonic lethality (Figure 1A and Table 1). Thus our analysis identified phy-1 as a novel genetic interaction partner of ten-1, specifically affecting embryonic lethality.
FIGURE 1:
Tissue-specific rescue of the dumpy phenotype of ten-1(ok641) phy-1(ok162) double mutants. Transgenic and nontransgenic animals of the same population of each rescue line are shown. The dumpy phenotype is rescued by expression of phy-1 under the control of its endogenous promoter (Pphy-1::phy-1) (A) (coinjection marker unc-122::gfp is expressed only in the coelomocytes) or by an epidermal specific promoter (Peff-1::phy-1::gfp) (B). Expression driven by a muscle-specific promoter (Pmyo-3::phy-1::gfp) does not rescue the dumpy phenotype (C) but rescues the embryonic lethality (see Table 1). Scale bar, 100 μm.
phy-1 is predominantly expressed in the epidermis, where it functions in the hydroxylation of cuticle collagens (Hill et al., 2000). We tested whether the loss of phy-1 function in the epidermal cells is responsible for the increase in embryonic lethality in ten-1(ok641) deletion mutants. Expression of phy-1 under the control of the epidermis-specific eff-1 promoter did rescue the dumpy phenotype but not the embryonic lethality of the double mutant (Figure 1B and Table 1). In contrast, expression of phy-1 under the control of the muscle-specific promoter myo-3 was able to rescue the embryonic lethality but not the dumpy phenotype (Figure 1C and Table 1). Thus the embryonic lethality of the ten-1(ok641) phy-1(ok162) double mutant is due to the loss of a previously unidentified function of phy-1 in the body wall muscles of C. elegans.
ten-1 phy-1 mutants arrest during late elongation with morphological defects
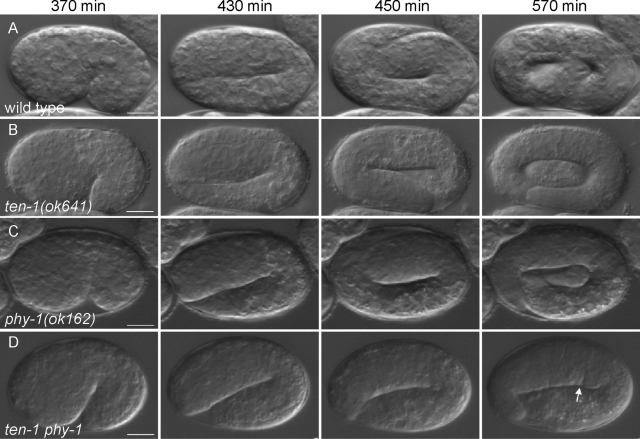
To investigate the nature of the embryonic lethality in the ten-1 phy-1 double mutant, we analyzed embryos using time-lapse Nomarski microscopy. We found that ten-1 phy-1 double-mutant embryos display defects in epidermal elongation (Figure 2). Wild-type embryos cultured at 20°C begin to elongate ∼350 min after first cleavage and complete elongation by ∼600 min. At the 1.75-fold stage, muscle contractions begin and embryos start to twitch. By the twofold stage, embryos roll vigorously. Synthesis and deposition of the larval cuticle start by ∼690 min (Figure 2A and Supplemental Movie S1). The embryos hatch at ∼800 min. phy-1(ok162) mutant embryos were indistinguishable from wild type in their embryonic elongation (Figure 2C and Supplemental Movie S3). Embryonic elongation following epidermal enclosure was found to be mostly normal in the ten-1(ok641) deletion mutant (Figure 2B and Supplemental Movie S2). About 20% of the ten-1 phy-1 double-mutant embryos were embryonic lethal due to arrest at various stages of late elongation. Double-mutant embryos displayed normal elongation prior to the twofold stage. Using time-lapse analysis, we recorded embryos arresting at the 2- or 2.5-fold stage (Figure 2D). Embryos were twitching and occasionally rolling. Nevertheless, movements seemed to be distinct from wild-type movements at the same stage of elongation (Supplemental Movies S4 and S5). The development continued, as indicated by cuticle formation and development of the pharynx. None of the recorded embryos displaying the elongation defects hatched.
FIGURE 2:
ten-1 phy-1 double-mutant embryos arrest late in elongation. Nomarski images of representative embryos undergoing elongation of (A) wild-type, (B) ten-1(ok641), (C) phy-1(ok162), and (D) ten-1 phy-1 double-mutant strains are shown at the time points indicated. Elongation and morphology of both single mutants are comparable to those of the wild-type embryo. Double mutants stop elongation at the 2- or 2.5-fold stage. Embryos look swollen and develop constrictions in the epidermis (arrow). Scale bar, 10 μm.
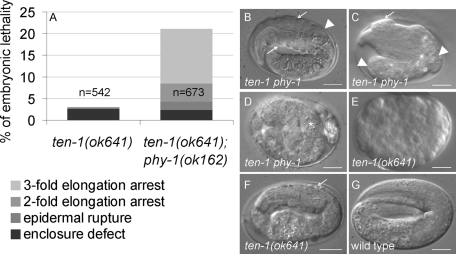
Embryos showed morphological defects of different severities (Figure 3). Morphology was analyzed ∼15 h after eggs were laid. By this time control animals have all hatched. About 4% of the embryos had visible body shape defects and bulge formations (Figure 3C). About 2% of the embryos ruptured during elongation (Figure 3D). However, 13% of the double-mutant embryos elongated to a threefold-like stage prior to arrest with constrictions in the epidermis or with other mild morphological defects (Figure 3B). A thicker epidermis at the anterior side of the animal was often observed. The pharynx appeared bent in the anterior part and misplaced from its central position in comparison to a wild-type embryo at a threefold stage (Figure 3, B and G). Double-mutant embryos, arresting at the threefold stage, showed rolling movements. We never observed that arrested embryos at the threefold stage had defects in cuticle formation, since embryos kept their shape and never shrank. Double mutants and ten-1(ok641) single-mutant embryos showed a similar percentage of embryonic arrest during epidermal enclosure (Figure 3A). Only a minor percentage of ten-1(ok641) single-mutant embryos showed late elongation defects, usually accompanied by severe morphological defects and epidermal rupture (Figure 3F). In summary, phy-1 deletion intensified morphological defects of ten-1 mutants, causing late embryonic elongation defects in ten-1 phy-1 double mutants.
FIGURE 3:
Loss of phy-1 function in the ten-1 mutant background enhances embryonic lethality and morphological defects of late embryos. (A) Distribution of embryonic lethal phenotypes of ten-1(ok641) and ten-1(ok641) phy-1(ok162). The graph represents a summary of three independent experiments of each strain. N, the number of all embryos analyzed. (B–F) Representative images of arrested embryos with most common morphological defects. (B) Approximately 13% of the double-mutant embryos arrest at a threefold stage, showing constrictions in the epidermis (arrows) and misplacement of the pharynx from its central position (arrowhead). (C) Approximately 6% of the double-mutant embryos arrest before the threefold stage. These embryos develop anterior blisters (arrow) and strong deformations of the posterior part (arrowhead). (D) Some embryos rupture during elongation (the asterisk marks the developed pharynx). (E) ten-1(ok641) single mutants arrest mainly during embryonic enclosure. Equivalent arrested embryos can be found in the double mutant (unpublished data). (F) Occasionally, the ten-1(ok641) single mutant arrests during embryonic elongation, showing constrictions (arrow) and deformations of the epidermis. (G) A threefold wild-type embryo does not show any of the described phenotypes. Scale bar, 10 μm.
ten-1 phy-1 embryos have defects in epidermal development and body wall muscles
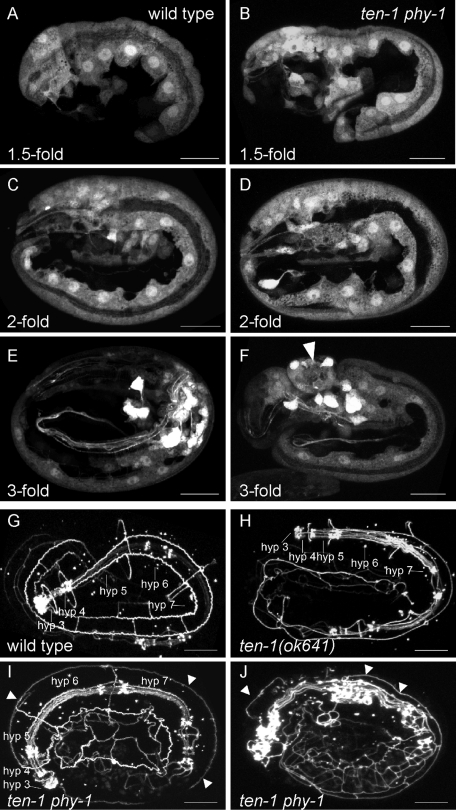
Elongation of embryos reflects the elongation of epidermal cells along the anterior–posterior axis (Priess and Hirsh, 1986). We investigated this process by expressing the ten-1b promoter translational fusion to GFP as a reporter in ten-1 phy-1 double-mutant animals and all control strains. As previously shown, this promoter GFP fusion protein is expressed in neurons and in epidermal cells, excluding seam cells (Drabikowski et al., 2005). We analyzed randomly picked embryos of various stages during late elongation of wild-type and mutant animals (Figure 4). We found that at the 1.5-fold stage epidermal cells of the double-mutant embryos were already misshapen and mislocalized (Figure 4B). Syncytia of epidermal cells of embryos at the twofold stage were misplaced (Figure 4D). Ventral epidermal cells still showed cell protrusions similar to migrating cells during ventral enclosure. Threefold embryos of the double mutant showed protrusion encompassing epidermal cells and neurons (Figure 4F).
FIGURE 4:
ten-1 phy-1 embryos display defects during epidermal development. (A–F) Expression of gfp under the ten-1b promoter in the epidermis and neurons in wild type (left) in comparison to ten-1 phy-1 double-mutant embryos (right) at indicated stages of embryonic elongation. Confocal images of the lateral view of the embryos are shown. (A, B) Hypodermal cells are misshapen and delayed in migration in 1.5-fold double-mutant embryos. (C, D) Mutant embryos at the twofold stage show misplacement of the epidermal cell layer. (E, F) Mutant threefold embryos exhibit bulges in the anterior part containing epidermal cells and neurons (arrowhead). Corresponding images of threefold embryos of ten-1(ok641) and phy-1(ok162) single mutants were indistinguishable from wild type and are presented in Supplemental Figure S2. (G, H) ten-1 phy-1 embryos show mislocalization of the epidermal junction marker AJM-1. Confocal images of embryos expressing the apical epidermal junction marker AJM-1 fused to GFP are shown. (G) Threefold wild-type embryo showing syncytia of fused hypodermal cells as indicated. (H) Threefold embryo of the ten-1(ok641) single mutant. Hypodermal cells are fused, comparable to wild type. Note the minor disorganization of the lateral epidermal cells. (I, J) Arrested ten-1 phy-1 embryos showing major disorganization of epidermal cell junctions. Mislocalization of AJM-1 on the apical side of dorsal epidermal cells is indicated by arrowheads. (J) Note defects of the pharyngeal cells. The corresponding image of a threefold embryo of the phy-1(ok162) single mutant is indistinguishable from wild type and is presented in Supplemental Figure S2. Scale bar, 10 μm.
Soon after their birth, major epidermal cells start to express and localize AJM-1, an apical epidermal junction protein. Knockdown of ten-1 by RNAi caused defects in hypodermal morphogenesis (Drabikowski et al., 2005). We used AJM-1 fused to GFP to analyze extent to which epidermal cell fusion was affected in arrested ten-1 phy-1 double-mutant embryos and the control strains (Figure 4). A wild-type embryo at a threefold stage showed localization of AJM-1 only at cell–cell junctions. Seam cells can be distinguished from the main hypodermal syncytia hyp 3 to hyp 7 (Figure 4G). The ten-1(ok641) single mutant had correctly localized AJM-1, although the overall appearance seemed mildly disorganized (Figure 4H). AJM-1 localization in phy-1(ok162) single mutants was indistinguishable from wild type (Supplemental Figure S2C). Arrested ten-1 phy-1 double-mutant embryos showed severely disorganized AJM-1 expression that was often mislocalized at basal as well as apical positions in the epidermis (Figure 4, I and J).
We addressed the question of whether the primary reason for the disorganization of AJM-1 could be a fusion defect of epidermal cells. The fusogen eff-1 is known to be essential for fusion of epidermal cells, and mutations in this gene cause embryonic and postembryonic cell fusion defects (Mohler et al., 2002). EFF-1 and TEN-1 are both transmembrane proteins and have overlapping expression patterns in epidermis, pharynx, uterus, and some head neurons. Knocking down eff-1 by RNAi in the ten-1(ok641) mutant did not enhance the epidermal defect that results in embryonic lethality in the ten-1 mutant animals (Supplemental Table S3). Thus ten-1 and eff-1 do not act in parallel or synergize in the process of epidermal cell fusion. In summary, arrested ten-1 phy-1 double mutants showed severe morphological defects that accumulate during elongation and could be the result of compromised cell migration rather than fusion defects of epidermal cells.
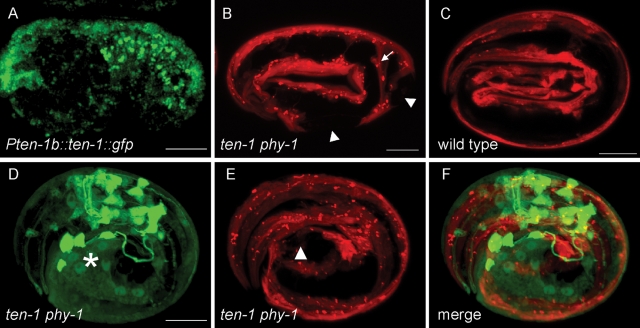
During late embryogenesis, body wall muscles were shown to be important for proper elongation (Hresko et al., 1994). The fact that double–mutant embryos arresting around the twofold stage had difficulties with twitching and rolling (cf. Supplemental Movies S4 and S5) prompted us to examine the structure of the body wall muscles. We visualized the body wall muscle structure of arrested ten-1 phy-1 embryos by expressing a myo-3p::mCherry marker (Figure 5). Wild-type embryos at a threefold stage showed continuous muscle strands along the body (Figure 5C). Arrested double-mutant embryos had gaps in the muscle strands resulting from muscles that detached from the epidermis (Figure 5B). We investigated whether there is a correlation between the defects in the epidermis and in muscles. To show that TEN-1 is indeed expressed in the epidermis, we expressed ten-1 fused to gfp under the ten-1b promoter. We found expression in the dorsal epidermal cells during embryogenesis and in neuronal precursor cells (Figure 5A). During larval development and in the adult worm TEN-1 was mainly expressed in the nerve ring, the ventral nerve cord, and some neurons in the tail (unpublished data). Thus the expression correlated well with the expression of the ten-1b promoter gfp fusion (Drabikowski et al., 2005). By crossings we combined the myo-3p::mCherry muscle marker and the pten-1b::gfp epidermal marker in the ten-1 phy-1 double mutant. Indeed, we found that in arrested embryos regions with defective epidermal cells overlap with disruptions of the muscle strands, which suggests that there is a compromised connection between these tissues (Figure 5, D–F).
FIGURE 5:
Epidermal defects correlate with defects in body wall muscles in arrested ten-1 phy-1 embryos. (A) TEN-1 is expressed in dorsal epidermal cells during embryogenesis. (B, C) Confocal images of embryos expressing mCherry under the muscle-specific myo-3 promoter. (B) Double-mutant embryo arrested at the 2.5-fold stage. Muscle strands are disrupted (arrowhead) and detached from the epidermal cells (arrow). (C) Wild-type embryos at the threefold stage show continuous strands of body wall muscles from head to tail. (D–F) Three-dimensional representation combining the epidermal (green) and muscle (red) markers in an arrested ten-1 phy-1 embryo. (D) Expression of pten-1b::gfp. The major epidermal defect is indicated by an asterisk. (E) Expression of pmyo-3::mCherry. The disrupted muscle strand is indicated by an arrowhead. (F) Merged image showing that the epidermal and muscle defects are in close proximity. Scale bar, 10 μm.
The ten-1 phy-1 mutant shows basement membrane defects in arrested embryos
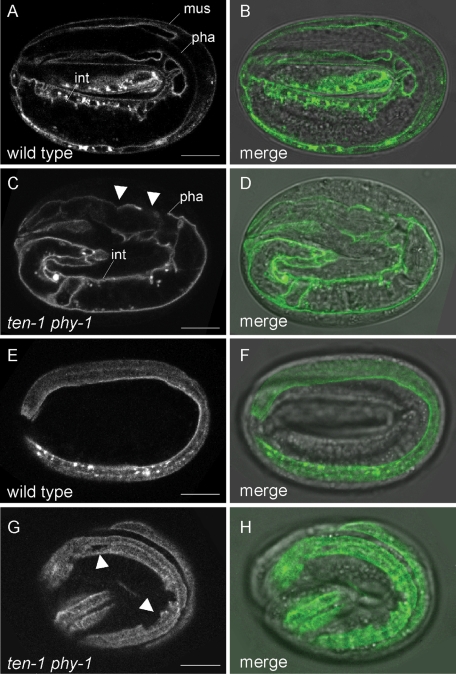
The body wall muscles and epidermis are separated by a BM. Loss of components connecting the body wall muscles, BM, and epidermis leads to arrest in elongation and paralysis of the embryos (Hresko et al., 1999; Bercher et al., 2001; Mackinnon et al., 2002; Bosher et al., 2003; Ding et al., 2008; Woo et al., 2004). We investigated whether a loss of ten-1 in the phy-1 mutant has an effect on BM integrity. To visualize BMs, we used the LAM-1::GFP reporter. Frequently, we found disorganized BM surrounding the pharynx (Figure 6, C and D).We found that arrested ten-1 phy-1 double-mutant embryos showed disrupted BMs between the muscle and the hypodermis (Figure 6, G and H). Defects in intestinal BM and gonadal BM could not be observed at these stages of development. These defects were not found in wild-type or phy-1 or ten-1 single-mutant embryos (Figure S3).
FIGURE 6:
Basement membrane integrity is affected in arrested ten-1 phy-1 mutant embryos. Localization of LAM-1 fused to GFP in a single focal plane (left) and merged with the corresponding differential interference contrast image (right). (A, B) Laminin is localized to BMs of the pharynx (pha), intestine (int), and developing gonad (not in focus) of a threefold wild-type embryo. (C, D) Disruption and disorganization of the pharyngeal BM in arrested ten-1 phy-1 embryos. Laminin is generally localized correctly. (E, F) Localization of laminin to the BM between the hypodermis and body wall muscles in a threefold wild-type embryo. One muscle strand is in focus. (G, H) The BM between body wall muscles and hypodermis shows breaks in double-mutant embryos (arrowheads). Corresponding images of threefold embryos of ten-1(ok641) and phy-1(ok162) single mutants are presented in Supplemental Figure S3. Scale bar 10, μm.
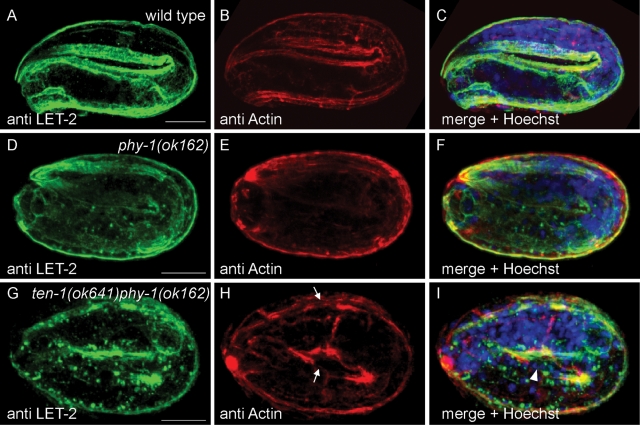
Collagen IV is essential to stabilize the BMs, and loss of collagen IV leads to embryonic lethality due to late elongation arrest and detachment of body wall muscles from the epidermis (Gupta et al., 1997). We stained phy-1(ok162) single mutants for EMB-9 and LET-2, the two C. elegans collagen IV chains (Figure 7 and Supplemental Figure S4). In wild-type C. elegans, collagen IV is expressed by body wall muscles and first appears as bright clusters of protein in the endoplasmic reticulum at the twofold stage. Most of the protein is assembled and secreted into the BMs surrounding the intestine, pharynx, developing gonad, and between body wall muscles and epidermis at the threefold stage (Figure 7A and Supplemental Figure S4A). In contrast, we found that in the phy-1(ok162) mutant intracellular aggregates of protein were still present in muscles of threefold embryos stained with anti–collagen IV antibodies (Figure 7, D and F, and Supplemental Figure S4, D and F). In addition, the BM staining did not appear as smooth in the mutant as in the wild type. Sometimes staining appeared to be weaker in some parts of the BM, although we never observed a disruption or deformation of BMs. Note that we could not observe any general defects with the laminin GFP reporter either (Supplemental Figure S3).
FIGURE 7:
Loss of phy-1 function causes aggregation of collagen IV in body wall muscles. Immunostaining with antibodies against LET-2 (green) and actin (red). DNA is stained with Hoechst dye (blue). (B, E, H) Longitudinal actin bundles correspond to the position of body wall muscles. (A, C) LET-2 is almost completely secreted into the BM in wild-type embryos at the threefold stage. The focus is on the BM separating muscles from epidermal cells. No aggregates of LET-2 can be detected in muscles. (D, F) phy-1(ok162) single mutants at the threefold stage frequently show intracellular aggregates of LET-2 as spots in close proximity to muscle nuclei. (G, I) In arrested ten-1 phy-1 double-mutant embryos, cluster formation of LET-2 protein is enhanced. (H) Actin bundles appear disorganized and are disrupted in some places (arrow). Similar results were obtained with anti EMB-9 antibody and are represented in Supplemental Figure S4. Scale bar, 10 μm.
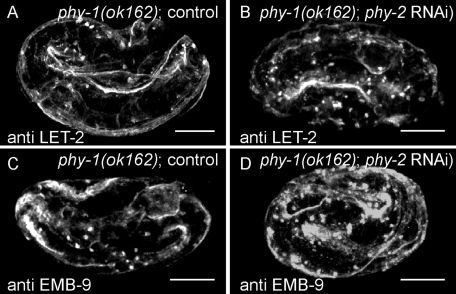
phy-1 and phy-2 are components of the enzymatic subunit of prolyl 4-hydroxylase important for the posttranslational modification of collagens. Loss of both phy genes causes embryonic lethality (Friedman et al., 2000). We knocked down phy-2 by RNAi in phy-1(ok162) deletion mutants and stained arrested embryos for collagen IV; we found an increase in protein aggregates accumulating intracellularly. Only a small fraction of collagen IV was secreted into the BM, which seemed disorganized (Figure 8). We also observed aggregates of collagen IV in the ten-1 phy-1 double-mutant embryos arrested around the 2- to 2.5-fold stage (Figure 7, G and I, and Supplemental Figure S4I) to an increased extent compared with the phy-1 single mutant. Nevertheless, a large fraction of the collagen IV was secreted and correctly localized. We observed a weakening of basement membranes at places where actin bundles of muscles were disrupted. In addition, we found a loss of integrity of the pharyngeal BM but not the one surrounding the intestine or the developing gonad. These findings are in agreement with the ones made with the laminin GFP reporter described earlier. In summary, ten-1 phy-1 double mutants showed defects in the pharyngeal BM and the BM outlining the pseudocoelomic cavity. Loss of phy-1 causes intracellular accumulation of collagen IV protein and weakens BMs in the double mutant.
FIGURE 8:
Knockdown of phy-2 in phy-1(ok162) mutants increases intracellular aggregation of collagen IV. Confocal images of phy-1(ok162) mutant embryos on control RNAi (A, C) or phy-2 RNAi (B, D) stained with anti–LET-2 or anti–EMB-9 antibodies. (A, C) Control embryos at the threefold stage show few collagen IV aggregates. Collagen IV protein localizes to BMs. (B, D) Elongation arrested embryos show an increase in collagen IV aggregates, and the muscle BM appears disorganized. Scale bar, 10 μm.
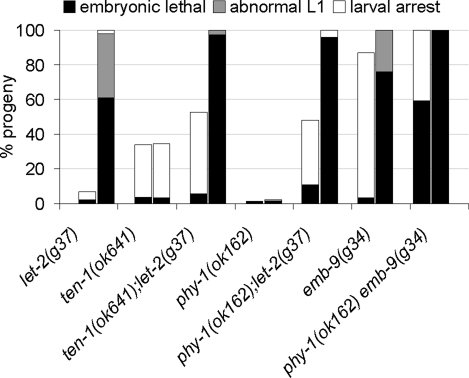
Finally, we investigated whether ten-1 and/or phy-1 interact genetically with the collagen IV genes themselves. We used the hypomorphic temperature-sensitive alleles emb-9(g34) and let-2(g37) and analyzed double mutants for embryonic lethality and larval arrest at 20 and 25°C (Figure 9). The loss of let-2 in the ten-1 mutant increased the number of larval-arrested animals by ∼20% compared with ten-1 single mutants at 20°C. Because only 2% of let-2(g37) worms are larval arrested at 20°C, the phenotype of the double mutants indicates a functional interaction rather than just an additive effect. At 25°C embryonic lethality is more pronounced in ten-1(ok641);let-2(g37) double mutants than in the let-2 or ten-1 single mutants. Combining the collagen IV mutant alleles with the phy-1 deletion allele increased embryonic lethality and larval arrest significantly at 20 and 25°C. Because phy-1 was not implicated in the function of BMs, we checked for genetic interactions with other important BM genes (Table 2). We found that phy-1 also genetically interacted with the BM components laminin epi-1 and collagen XVIII cle-1. The reduction of laminin caused enhanced embryonic lethality in phy-1 single mutants. Interference with collagen XVIII expression caused an increase in embryonic lethality and worms bursting through the vulva. This could be an indication for a role of phy-1 in the modification of collagen XVIII. In contrast, we did not find a genetic interaction between phy-1 and integrin ina-1 combining phy-1(ok162) mutant with a hypomorphic allele of ina-1, gm144.
FIGURE 9:
ten-1 and phy-1 genetically interact with the collagen IV genes. The mean percentages of progeny of homozygous single and double mutants that arrest during embryogenesis (black), hatch as severely deformed L1 (gray), or arrest as larvae (white) are indicated. Each allele was tested at 20°C (left bars) and 25°C (right bars). The loss of ten-1 increased larval lethality of the let-2 mutant at 20°C by ∼20% (p < 0.001) in comparison to the ten-1(ok641) single mutant alone. In addition, the combination of the phy-1 and let-2 mutants resulted in increased larval (p < 10−7 compared with single mutants) and embryonic lethality (p < 10−5 compared with single mutants) at 20°C. The embryonic lethality of ten-1;let-2 or phy-1;let-2 double mutants is more pronounced at 25°C than with the let-2(g37) single mutant. The loss of phy-1 function in the emb-9(g34) mutant resulted in increased embryonic lethality at 20 and 25°C (p < 10−7). ten-1(ok641) and phy-1(ok162) single mutants did not show a temperature-sensitive phenotype. At least 400 progeny were examined for each allele at each temperature.
TABLE 2:
phy-1 genetically interacts with the basement membrane components epi-1 and cle-1.
| Genotype | Embryonic lethal, % (95% CI) | Larval arrest, % (95% CI) | Sterile and/or vulva defects, % (95% CI) | Fertile adults, % (95% CI) | n |
|---|---|---|---|---|---|
| Wild type; L4440 (RNAi) | 0.9 (0.2–1.8) | 0.5 (0.1–1.6) | 0 (0–0.5) | 98.6 (97.3–99.5) | 537 |
| phy-1(ok162); L4440 (RNAi) | 1.9 (0.7–3.4) | 1.5 (0.4–2.7) | 1.4 (0.4–2.7) | 95.3 (93.6–97.6) | 418 |
| Wild type; epi-1 (RNAi) | 5.5 (3.8–7.0) | 30.5 (28.3–34.9) | 64.7 (60.6–67.4) | 0 (0–0.3) | 799 |
| phy-1(ok162); epi-1 (RNAi) | 24.3 (21.3–27.2)***1 | 38.1 (34.7–41.3), n.s. | 37.4 (34.7–41.1) | 0 (0–0.4) | 843 |
| Wild type; cle-1 (RNAi) | 0.8 (0.2–1.6) | 1.1 (0.4–2.0) | 0 (0–0.3) | 98.1 (96.9–99.0) | 774 |
| phy-1(ok162); cle-1 (RNAi) | 7.0 (5.4–9.4)* | 0 (0–0.4) | 10.9 (8.0–12.7)***2 | 82.2 (79.6–85.4) | 681 |
| ina-1(gm144) | 13.5 (9.2–16.9) | 37.3 (27.4–38.1) | 28.6 (27.4–38.1) | 19.6 (16.1–25.5) | 307 |
| phy-1(ok162) ina-1(gm144) | 11.2 (9.3–13.9) | 39.3 (36.7–43.8) | 31.8 (28.9–35.6) | 19.9 (16.0–21.6) | 776 |
Mean percentage and 95% confidence interval (CI) of wild-type and mutant worms analyzed for embryonic lethality, larval arrest, sterility and vulva defects (include protruding and bursting-at-the-vulva phenotype), and fertile adults. n, number of individuals analyzed.
*p < 0.05 compared with wild type; cle-1 (RNAi).
***1p < 0.001 compared with wild type; epi-1 (RNAi).
***2p < 0.001 compared with wild type; cle-1 (RNAi).
n.s., p > 0.2 compared with wild type; epi-1 (RNAi).
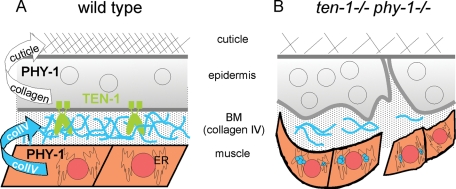
In summary, our data suggest that phy-1 and ten-1 act in parallel in influencing the connection between epidermis and muscle during embryonic development. Proposed roles of TEN-1 and PHY-1 are summarized in Figure 10. In this model, TEN-1 acts as an epidermal receptor for the BM collagens modified by PHY-1 and secreted by the muscle cells.
FIGURE 10:
Model for the functional interaction between TEN-1 and PHY-1. (A) In wild-type C. elegans TEN-1 is expressed in the epidermis and presumably localized basally binding to BM collagen IV. PHY-1 functions in the epidermis and in the muscle to modify procollagens, which are then incorporated into the extracellular matrix. (B) Loss of phy-1 function in muscle cells results in intracellular accumulation of incompletely processed procollagen and less secreted collagen IV into the BM. Loss of TEN-1 causes developmental defects in the epidermis and reduced binding to the BM. The combined effect of the missing BM receptor with less collagen further weakens the connection between the epidermis and body wall muscles. This leads to detachment of epidermis and muscle cells during late elongation of the embryo when mechanical stress is generated by muscle contractions.
DISCUSSION
Loss of phy-1 function enhances ten-1 embryonic lethality
We found that phy-1 loss of function significantly increases embryonic lethality of ten-1 mutant animals and that this interaction is allele independent. We analyzed double-mutant embryos by time-lapse microscopy and found that ten-1 phy-1 embryos arrest during late elongation. The loss of phy-1 enhances the late embryonic defects that are rarely found in ten-1 single mutants. The phy-1(ok162) mutant is indistinguishable from wild-type C. elegans in terms of embryonic and larval lethality. Therefore the phenotype of the double mutant is not just an additive effect but reflects a mechanistic connection between TEN-1 and PHY-1. We characterized the arrested embryos using several GFP fusion markers and found defects in the migration and fusion of epidermal cells. A recent study showed late embryonic defects of the ten-1(ok641) mutant (Morck et al., 2010). However, embryos with gross epidermal defects are rarely found in ten-1(ok641) mutants, and our analysis shows that embryos predominantly arrest during enclosure. Only 0.36% of ten-1(ok641) mutant animals arrest around the twofold stage or disrupt during elongation.
A proper connection between body wall muscles, epidermal cells, and the separating BM is crucial for successful late elongation (Zhang and Labouesse, 2010). The ten-1 phy-1 double mutant displayed detachment of body wall muscles and misplacement of epidermal cells during development. These observations suggest a compromised connection between these tissues. We found that TEN-1 is expressed in epidermal cells during embryogenesis, but the subcellular localization of TEN-1 within the epidermis remains to be determined. Of interest, mutants defective in the basal components of the epidermal attachment structures like myotactin, let-805, or the spectraplakin isoform vab-10A display similar phenotypes as we found in the ten-1 phy-1 double mutant (Hresko et al., 1999; Bosher et al., 2003). In addition, ten-1 mutants show low penetrance of embryonic lethality due to late elongation arrest, indicating a redundant role of ten-1 with other genes during embryonic elongation. Thus loss of phy-1 function seems to affect a process that supports proper ten-1 function.
Does PHY-1 modify basement membrane collagens?
To form stable helices, procollagens need to be posttranslationally modified. phy-1 codes for the enzymatic subunit of C. elegans P4H and has been shown to be important for modification of cuticle collagens (Hill et al., 2000). phy-1 and phy-2 double mutants are embryonic lethal, and embryos burst around the time of cuticle secretion (Winter and Page, 2000). Nevertheless, other observations of double-mutant embryos elongating only to a twofold-like stage prior to their explosion suggest an additional role for the enzyme complex (Friedman et al., 2000). Staining phy-1(ok162) mutant embryos with anti–collagen IV antibodies revealed accumulations of protein aggregates around the nuclei of body wall muscle cells in fully elongated embryos. We did not observe similar aggregates in wild-type C. elegans. Additional reduction in phy-2 by RNAi increased intracellular collagen IV accumulation in arrested embryos compared with phy-1 mutant embryos. Thus insufficient secretion of collagen IV protein into the BM contributes to the embryonic lethality in phy-1;phy-2 loss-of-function mutants. Furthermore, we found that phy-1 genetically interacts with emb-9 and let-2. These findings indicate a functional connection between PHY-1 and collagen IV.
Our observations linking phy-1 with collagen IV are supported by ten-1 phy-1 double-mutant rescue experiments. Embryonic lethality could be rescued by transient overexpression of phy-1 in body wall muscles. This argues for an important function of PHY-1 in the muscle cells, which express collagen IV. Our results indicate that in C. elegans P4H is responsible for the modification of type IV procollagen, and this is in agreement with similar results obtained in vertebrates (Holster et al., 2007).
TEN-1 and its role in extracellular matrix assembly/maintenance
There is increasing evidence that TEN-1 functions in the assembly or maintenance of the extracellular matrix in C. elegans. Previously, genetic interactions of ten-1 with the BM receptors integrin ina-1 and dystroglycan dgn-1 were reported. In addition, loss of ten-1 function sensitizes animals toward an enhanced phenotype when combined with mutations in the BM components laminin EPI-1 and nidogen NID-1 (Trzebiatowska et al., 2008). However, there is specificity of the interaction between ten-1 and BM components. Mutations in cle-1 or unc-52 did not enhance embryonic lethality, larval arrest, or sterility of the ten-1(ok641) mutant (Trzebiatowska et al., 2008). This does not preclude that unc-52 interacts genetically with ten-1 in other processes, such as axon positioning, as observed by Morck et al. (2010) using a different unc-52 allele than the one used in our study.
In this study we found that a compromised BM lacking collagen IV enhances ten-1 mutant lethality. The morphological appearance of ten-1 phy-1 double mutants partially resembles defects seen in collagen IV mutants. Collagen IV mutants arrest at the twofold or threefold stage due to detachment of body wall muscles from the epidermis (Gupta et al., 1997). Accumulations of collagen IV protein in ten-1 phy-1 mutant animals appear frequently as two strong spots close to the body wall muscle cell nuclei (Figure 7 and Supplemental Figure S4). Similar staining has been reported when emb-9 mutants were stained for LET-2 protein, indicating that LET-2, the alpha2(IV) chain, is not secreted in the absence of full-length EMB-9, the alpha1(IV) chain (Gupta et al., 1997). We postulate that collagen IV is insufficiently modified by PHY-1, resulting in intracellular accumulation of protein and subsequent instability of basement membrane.
We found defects in the BM surrounding the pharynx and between body wall muscles and epidermal cells. During elongation, these sites are exposed to mechanical forces by muscle contraction. Fibrous organelles, the structural homologues of vertebrate hemidesmosomes, provide mechanical stability with the onset of muscle contraction during elongation. However, whether collagen IV affects fibrous organelle formation is not clear. In addition, receptors responsible for collagen IV binding are not yet defined in C. elegans. Of interest, we found that loss of ten-1 enhances lethality of the collagen IV mutant let-2(g37), indicating that TEN-1 might be a receptor for type IV collagen and/or guide its assembly in the BM. In summary, our results provide evidence that TEN-1 has a role in BM formation or stabilization required for mechanical stability during the process of late elongation in C. elegans.
Conservation in higher organisms
Our data provide evidence that TEN-1 links epidermal cells via BM proteins, such as collagen IV, to muscle structures. In Drosophila, the Ten-a protein localizes to muscle attachment structures during embryogenesis (Fascetti and Baumgartner, 2002). Muscle attachment in Drosophila depends on PS integrin (Prokop et al., 1998), and loss of integrin leads to detachment of epidermal and muscle cells from the ECM. Nevertheless, in the absence of PS integrin, the connection between microtubules and the epidermis is retained, suggesting involvement of additional receptors (Brown, 2000).
In chicken, teneurin-2 colocalizes with laminin in the optic cup and ventricular endocardium (Tucker et al., 2001). A recent study investigating the expression of teneurin-4 in the avian embryo describes colocalization of teneurin-4 with laminin in the BM surrounding the endoderm, as well as nests of cells in the mesenchyme (Kenzelmann-Broz et al., 2010). Thus teneurins might be important for the organization and/or stabilization of basement membranes in vertebrates as well. Recently, another report described a coexpression of the mouse teneurin isoform Odz3 with collagen I and II in distinct regions of the fibrous layer and in the proliferating layer of mandibular condylar cartilage (Murakami et al., 2010).
The coexpression and colocalization of teneurins with collagens and basement membranes and the action of TEN-1 in conjunction with PHY-1 point to a function of TEN-1 in morphogenetic events requiring the proper deposition of collagenous extracellular matrices and adequate attachment of cells to these extracellular matrices.
MATERIALS AND METHODS
General methods and C. elegans strains
C. elegans strains were cultured at 20°C as described in Brenner (1974). Wild type refers to the C. elegans variety Bristol strain N2. In addition, the following strains were used: JK2729 [phy-1(ok162)], JK2757 [phy-2(ok177)], TP7 [phy-3(ok199)], RU90 [ten-1(ok641)], RU98 [ten-1(tm651)], GG34 [emb-9(g34)], GG37 [let-2(g37)], and NG144 [ina-1(gm144)]. Strains were at least four times outcrossed prior to analysis. The following GFP marker strains were used: RU9 [kdEx31(ten-1b::gfp)], IM253 [urEx131(lam-1::gfp)], SU93 [(ajm-1::gfp;rol-6); him-5], and GW397 [gwIs28(myo-3p::mCherry; unc-119+); gwIs39] (Meister et al. 2010). Double and triple mutants were maintained as RU171 [ten-1(ok641) phy-1(ok162)], RU197 [ten-1(tm-651) phy-1(ok162)], RU179 [ten-1(ok641); phy-2(ok177)], RU168 [ten-1(ok641); phy-3(ok199)], RU177 [ten-1(ok641) phy-1(ok162); phy-3(ok199)], RU182 [ten-1(ok641); phy-2(ok177); phy-3(ok199)], RU181 [phy-2(ok177); phy-3(ok199)], RU202 [ten-1(ok641); let-2(g37)], RU203 [phy-1(ok162); let-2(g37)], RU206 [phy-1(ok162) emb-9(g34)], and RU205[phy-1(ok162) ina-1(gm144)] strains. Primers used for genotyping are listed in Supplemental Table S4.
Constructs and plasmids
The phy-1 rescue construct under the endogenous phy-1 promoter was generated as follows: phy-1 3′ untranslated region (UTR) was amplified from wild-type genomic DNA using 5′gtatccaacaaatggatccacg3` and 5`TAGCAGCCGACACTAAACAG3′. The open reading frame of phy-1 was amplified from cDNA and fused to its 3′UTR by PCR using 5′AGAAATATACCGGTAATCCTTGAC3′ and 5′TATGGCGCCTAATTTATTCACGG CAAAGAAAAAGGCAG3′. The resulting PCR fragment was cloned into pPD117.01 (A. Fire) into AgeI and NarI sites. Three kilobases of the phy-1 5′UTR were amplified by PCR using 5′TTTGCATGCTCAACTCTAGCAAAATGGCAC3′ and 5′GGATTACCGGTATATTTCTTTCAG3′. The PCR fragment was cloned into SphI and AgeI sites.
To generate the body wall muscle–specific rescue construct under the myo-3 promoter, phy-1 cDNA was amplified by PCR using 5′TTTCTAGAATGCGCCTGGCACTCCTTGTAC3′ and 5′TTACCGGTAGGGTCTCCCAGACGTC3′. The PCR fragment was cloned in pPD136.64 (A. Fire) into XbaI and AgeI sites. To express phy-1 under an epidermal promoter, the myo-3 promoter sequence of the aforementioned construct was replaced by 3 kb of the eff-1 5′UTR, which was amplified by PCR from genomic DNA using 5′ATGAATAAGCTTAATATCTGGCTCAG3′ and 5′TTTTCTAGAGCTTCTGAAAATAAAATGATACG3′. The PCR fragment was cloned into HindIII and XbaI sites.
To generate the ten-1 expression construct, the entire genomic DNA containing the ten-1b promoter and open reading frame was amplified from the cosmid F36A3 by PCR using 5′TTTCCGCGGTTCTAAAGGAAATGTGAAGCATG3′ and 5′TTTGCCGGCCCTTCAGATTTTCGGAACTTCC3′. The PCR fragment was cloned into the pPD117.01 (A. Fire) into SacII and NgoMIV sites, resulting in a translational fusion to gfp C-terminally.
Transgenic animals
Transgenic animals were generated by microinjection of DNA into the distal arms of gonads as described (Mello et al., 1991). phy-1 under its own promoter was injected into N2 at 5 ng/μl plus 10 ng/μl of unc-122::gfp injection marker (made by P. Sengupta) and 85 ng/μl plasmid p3T (MoBiTe). Injection of the full rescue construct resulted in three independent lines, one of which is RU192 [kdEx132(Pphy-1::phy-1::phy-1_3′UTR, unc-122::gfp)]. The muscle-specific rescue construct was injected into N2 at a concentration of 5 ng/μl plus 95 ng/μl plasmid p3T (MoBiTe). One line could be obtained and was named RU191 [kdEx131(Pmyo-3::phy-1::gfp)]. The epidermal-specific construct was injected into N2 at 5 ng/μl plus 10 ng/μl of unc-122::gfp injection marker and 85 ng/μl plasmid p3T (MoBiTe). Injection resulted in two independent lines, one of which is RU198 [kdEx133(Peff-1::phy-1::gfp, unc-122::gfp)]. All plasmids were linearized with AhdI or FspI prior to injection. For rescue of the ten-1(ok641) phy-1(ok162) double mutant, hermaphrodites were crossed with transgenic males carrying the extrachromosomal arrays. For a summary of results see Table 1.
Transgenic strains carrying ten-1::gfp expression construct were obtained by microparticle bombardment. The unc-119(ed3) III strain (DP38) was cobombarded with the ten-1::gfp expression construct and the unc-119 rescuing construct. Three lines were isolated, one of which is RU204 [kdIs134(Pten-1b::ten-1::gfp); unc-119+].
Phenotypic analysis
L4 hermaphrodites were placed on separate plates and allowed to lay eggs. The mother was transferred to new plates twice a day. Animals were analyzed for brood size and the progeny for embryonic lethality and larval lethality.
For time-lapse microscopy embryos were placed on 2% agarose pad in egg buffer (Shakes and Epstein, 1995). Recording was performed for 4–6 h at 20°C in multiple focal planes using Nomarski optics. Images were acquired with a Z1 microscope (Zeiss, Jena, Germany) and an Axio-Cam MRM camera (Zeiss). Movies were assembled using the ImageJ software.
For feeding mutants with epi-1 RNAi (Ahringer Library, NYU Langone Medical Center, New York, NY) or cle-1 or eff-1 RNAi (ORFeome feeding library v1.1, Open Biosystems Products, Huntsville, AL), L4 animals were placed on RNAi plates for 48 h at 15°C. Young adults were singled on fresh RNAi plates and allowed to lay eggs for 24 h at 20°C. The progeny were analyzed for embryonic lethality and larval lethality and adults for sterility and vulva defects. For statistics, 95% confidence intervals were calculated using the “exact method” (Clopper and Pearson, 1934). The p values were determined by the Tukey's multiple comparison test. All statistical analyses were performed using the program R.
Immunofluorescence staining
The protocol for staining of embryos was adapted from Graham et al. (1997). Samples were blocked with 10% normal goat serum (NGS) (Invitrogen, Carlsbad, CA) in phosphate-buffered saline (PBS) containing 0.5% TWEEN-20 (PBS-T). Primary antibodies against EMB-9 (NW1910; gift of J. M. Kramer), LET-2 (NW68; gift of J. M. Kramer), and actin (MAB 1501; Chemicon, Temecula, CA) were added in PBS-T–NGS overnight at 4°C. Embryos were washed with PBS-T and incubated with fluorescein-conjugated goat anti-rabbit (Alexa 488 from Invitrogen) or goat anti-mouse (Alexa 543 from Invitrogen) secondary antibody. Embryos were washed in PBS-T containing Hoechst, followed by PBS alone.
Microscopy
Confocal images were acquired with a Zeiss Axiovert 200M equipped with LSM510. Optical sections were collected at 0.7-μm intervals and combined using the maximum projection function. For publication, confocal images were annotated using ImageJ. For confocal imaging of embryos expressing reporters, embryos of the appropriate ages were collected in egg buffer (Shakes and Epstein, 1995). The embryos were treated with NaOCL (1:10 diluted in water) for ∼2 min. The embryos were washed in egg buffer, treated with chitinase (2.5 U in egg buffer) for ∼3 min, and washed again in egg buffer only. Embryos were transferred to a 2% agarose pad containing 10 mM sodium azide. Images of immobilized embryos were acquired immediately after treatment.
Supplementary Material
Acknowledgments
We thank James M. Kramer for the generous gift of antibodies and Andrew Fire for GFP expression vectors. We are grateful to Jacqueline Ferralli for technical assistance. We thank Joy Alcedo and Richard P. Tucker for critical comments on the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minneapolis, Minneapolis, MN), which is funded by the National Center for Research Resources, National Institutes of Health. This work was supported by the Novartis Research Foundation.
Abbreviations used:
- BM
basement membrane
- GFP
green fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0853) on July 27, 2011.
REFERENCES
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Baumgartner S, Martin D, Hagios C, Chiquet-Ehrismann R. Tenm, a Drosophila gene related to tenascin, is a new pair-rule gene. EMBO J. 1994;13:3728–3740. doi: 10.1002/j.1460-2075.1994.tb06682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zur T, Feige E, Motro B, Wides R. The mammalian Odz gene family: homologs of a Drosophila pair-rule gene with expression implying distinct yet overlapping developmental roles. Dev Biol. 2000;217:107–120. doi: 10.1006/dbio.1999.9532. [DOI] [PubMed] [Google Scholar]
- Bercher M, Wahl J, Vogel BE, Lu C, Hedgecock EM, Hall DH, Plenefisch JD. mua-3, a gene required for mechanical tissue integrity in Caenorhabditis elegans, encodes a novel transmembrane protein of ephithelial attachment complexes. J Cell Biol. 2001;154:415–426. doi: 10.1083/jcb.200103035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosher JM, Hahn BS, Legouis R, Sookhareea S, Weimer RM, Gansmuller A, Chisholm AD, Rose AM, Bessereau JL, Labouesse M. The Caenorhabditis elegans vab-10 spectraplakin isoforms protect the epidermis against internal and external forces. J Cell Biol. 2003;161:757–768. doi: 10.1083/jcb.200302151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NH. Cell-cell adhesion via the ECM: integrin genetics in fly and worm. Matrix Biol. 2000;19:191–201. doi: 10.1016/s0945-053x(00)00064-0. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-Sang ID, Chisholm AD. Form of the worm: genetics of epidermal morphogenesis in C. elegans. Trends Genet. 2000;16:544–551. doi: 10.1016/s0168-9525(00)02143-0. [DOI] [PubMed] [Google Scholar]
- Clopper C, Pearson S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- Ding M, King RS, Berry EC, Wang Y, Hardin J, Chisholm AD. The cell signaling adaptor protein EPS-8 is essential for C. elegans epidermal elongation and interacts with the ankyrin repeat protein VAB-19. PLoS One. 2008;3:e3346. doi: 10.1371/journal.pone.0003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabikowski K, Trzebiatowska A, Chiquet-Ehrismann R. ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev Biol. 2005;282:27–38. doi: 10.1016/j.ydbio.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Fascetti N, Baumgartner S. Expression of Drosophila Ten-a, a dimeric receptor during embryonic development. Mech Dev. 2002;114:197–200. doi: 10.1016/s0925-4773(02)00055-2. [DOI] [PubMed] [Google Scholar]
- Feng K, Zhou XH, Oohashi T, Morgelin M, Lustig A, Hirakawa S, Ninomiya Y, Engel J, Rauch U, Fassler R. All four members of the Ten-m/Odz family of transmembrane proteins form dimers. J Biol Chem. 2002;277:26128–26135. doi: 10.1074/jbc.M203722200. [DOI] [PubMed] [Google Scholar]
- Fessler JH, Fessler LI. Biosynthesis of procollagen. Annu Rev Biochem. 1978;47:129–162. doi: 10.1146/annurev.bi.47.070178.001021. [DOI] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465–479. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Higgin JJ, Moulder G, Barstead R, Raines RT, Kimble J. Prolyl 4-hydroxylase is required for viability and morphogenesis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;97:4736–4741. doi: 10.1073/pnas.97.9.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, Johnson JJ, Wang S, Sibley MH, Gupta MC, Kramer JM. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J Cell Biol. 1997;137:1171–1183. doi: 10.1083/jcb.137.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta MC, Graham PL, Kramer JM. Characterization of alpha1(IV) collagen mutations in Caenorhabditis elegans and the effects of alpha1 and alpha2(IV) mutations on type IV collagen distribution. J Cell Biol. 1997;137:1185–1196. doi: 10.1083/jcb.137.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KL, Harfe BD, Dobbins CA, L'Hernault SW. dpy-18 encodes an alpha-subunit of prolyl-4-hydroxylase in Caenorhabditis elegans. Genetics. 2000;155:1139–1148. doi: 10.1093/genetics/155.3.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivirikko KI, Myllyharju J. Loss of assembly of the main basement membrane collagen, type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J Biol Chem. 2007;282:2512–2519. doi: 10.1074/jbc.M606608200. [DOI] [PubMed] [Google Scholar]
- Hresko MC, Schriefer LA, Shrimankar P, Waterston RH. Myotactin, a novel hypodermal protein involved in muscle-cell adhesion in Caenorhabditis elegans. J Cell Biol. 1999;146:659–672. doi: 10.1083/jcb.146.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko MC, Williams BD, Waterston RH. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J Cell Biol. 1994;124:491–506. doi: 10.1083/jcb.124.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann-Broz D, Tucker RP, Leachman NT, Chiquet-Ehrismann R. The expression of teneurin-4 in the avian embryo: potential roles in patterning of the limb and nervous system. Int J Dev Biol. 2010;54:1509–1516. doi: 10.1387/ijdb.103139dk. [DOI] [PubMed] [Google Scholar]
- Keskiaho K, Kukkola L, Page AP, Winter AD, Vuoristo J, Sormunen R, Nissi R, Riihimaa P, Myllyharju J. Characterization of a novel Caenorhabditis elegans prolyl 4-hydroxylase with a unique substrate specificity and restricted expression in the pharynx and excretory duct. J Biol Chem. 2008;283:10679–10689. doi: 10.1074/jbc.M800972200. [DOI] [PubMed] [Google Scholar]
- Levine A, Bashan-Ahrend A, Budai-Hadrian O, Gartenberg D, Menasherow S, Wides R. Odd Oz: a novel Drosophila pair rule gene. Cell. 1994;77:587–598. doi: 10.1016/0092-8674(94)90220-8. [DOI] [PubMed] [Google Scholar]
- Mackinnon AC, Qadota H, Norman KR, Moerman DG, Williams BD. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol. 2002;14:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010;24:766–782. doi: 10.1101/gad.559610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Kikuchi Y, Hirate Y, Aoki M, Okamoto H. Compartmentalized expression of zebrafish ten-m3 and ten-m4, homologues of the Drosophila ten(m)/odd Oz gene, in the central nervous system. Mech Dev. 1999;87:223–227. doi: 10.1016/s0925-4773(99)00155-0. [DOI] [PubMed] [Google Scholar]
- Minet AD, Rubin BP, Tucker RP, Baumgartner S, Chiquet-Ehrismann R. Teneurin-1, a vertebrate homologue of the Drosophila pair-rule gene ten-m, is a neuronal protein with a novel type of heparin-binding domain. J Cell Sci. 1999;112:2019–2032. doi: 10.1242/jcs.112.12.2019. [DOI] [PubMed] [Google Scholar]
- Mohler WA, Shemer G, del Campo JJ, Valansi C, Opoku-Serebuoh E, Scranton V, Assaf N, White JG, Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- Morck C, Vivekanand V, Jafari G, Pilon M. C. elegans ten-1 is synthetic lethal with mutations in cytoskeleton regulators, and enhances many axon guidance defective mutants. BMC Dev Biol. 2010;10:55. doi: 10.1186/1471-213X-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Fukunaga T, Takeshita N, Hiratsuka K, Abiko Y, Yamashiro T, Takano-Yamamoto T. Expression of Ten-m/Odz3 in the fibrous layer of mandibular condylar cartilage during postnatal growth in mice. J Anat. 2010;217:236–244. doi: 10.1111/j.1469-7580.2010.01267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J, Kukkola L, Winter AD, Page AP. The exoskeleton collagens in Caenorhabditis elegans are modified by prolyl 4-hydroxylases with unique combinations of subunits. J Biol Chem. 2002;277:29187–29196. doi: 10.1074/jbc.M203824200. [DOI] [PubMed] [Google Scholar]
- Oohashi T, Zhou XH, Feng K, Richter B, Morgelin M, Perez MT, Su WD, Chiquet-Ehrismann R, Rauch U, Fassler R. Mouse ten-m/Odz is a new family of dimeric type II transmembrane proteins expressed in many tissues. J Cell Biol. 1999;145:563–577. doi: 10.1083/jcb.145.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Hirsh DI. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- Prokop A, Martin-Bermudo MD, Bate M, Brown NH. Absence of PS integrins or laminin A affects extracellular adhesion, but not intracellular assembly, of hemiadherens and neuromuscular junctions in Drosophila embryos. Dev Biol. 1998;196:58–76. doi: 10.1006/dbio.1997.8830. [DOI] [PubMed] [Google Scholar]
- Rakovitsky N, Buganim Y, Swissa T, Kinel-Tahan Y, Brenner S, Cohen MA, Levine A, Wides R. Drosophila Ten-a is a maternal pair-rule and patterning gene. Mech Dev. 2007;124:911–924. doi: 10.1016/j.mod.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Riihimaa P, Nissi R, Page AP, Winter AD, Keskiaho K, Kivirikko KI, Myllyharju J. Egg shell collagen formation in Caenorhabditis elegans involves a novel prolyl 4-hydroxylase expressed in spermatheca and embryos and possessing many unique properties. J Biol Chem. 2002;277:18238–18243. doi: 10.1074/jbc.M200895200. [DOI] [PubMed] [Google Scholar]
- Shakes DC, Epstein HF. Caenorhabditis elegans: Modern Biological Analysis of an Organism. San Diego, CA: Academic Press; 1995. [Google Scholar]
- Simske JS, Hardin J. Getting into shape: epidermal morphogenesis in Caenorhabditis elegans embryos. Bioessays. 2001;23:12–23. doi: 10.1002/1521-1878(200101)23:1<12::AID-BIES1003>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Trzebiatowska A, Topf U, Sauder U, Drabikowski K, Chiquet-Ehrismann R. Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol Biol Cell. 2008;19:3898–3908. doi: 10.1091/mbc.E08-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RP, Chiquet-Ehrismann R, Chevron MP, Martin D, Hall RJ, Rubin BP. Teneurin-2 is expressed in tissues that regulate limb and somite pattern formation and is induced in vitro and in situ by FGF8. Dev Dyn. 2001;220:27–39. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1084>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- White JG. The anatomy. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos Trans Royal Soc Lond. 1976;275:327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter AD, Page AP. Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans. Mol Cell Biol. 2000;20:4084–4093. doi: 10.1128/mcb.20.11.4084-4093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo WM, Goncharov A, Jin Y, Chisholm AD. Intermediate filaments are riquired for C. elegans epidermal elongation. Dev Biol. 2004;267:216–229. doi: 10.1016/j.ydbio.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Zhang H, Labouesse M. The making of hemidesmosome structures in vivo. Dev Dyn. 2010;239:1465–1476. doi: 10.1002/dvdy.22255. [DOI] [PubMed] [Google Scholar]
- Zhou XH, Brandau O, Feng K, Oohashi T, Ninomiya Y, Rauch U, Fassler R. The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr Patterns. 2003;3:397–405. doi: 10.1016/s1567-133x(03)00087-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.