The study provides new evidence that the mRNA-destabilizing protein Tristetraprolin is a modulator of HIF-1α mRNA turnover and may contribute to the control of HIF-1α and HIF-1α target gene expression during the adaptive response of endothelial cells to hypoxia.
Abstract
Endothelial cells (ECs) are the primary sensors of variations in blood oxygen concentrations. They use the hypoxia-sensitive stabilization of the hypoxia-inducible factor-1α (HIF-1α) transcription factor to engage specific transcriptional programs in response to oxygen changes. The regulation of HIF-1α expression is well documented at the protein level, but much less is known about the control of its mRNA stability. Using small interfering RNA knockdown experiments, reporter gene analyses, ribonucleoprotein immunoprecipitations, and mRNA half-life determinations, we report a new regulatory mechanism of HIF-1α expression in ECs. We demonstrate that 1) sustained hypoxia progressively decreases HIF-1α mRNA while HIF-1α protein levels rapidly peak after 3 h and then slowly decay; 2) silencing the mRNA-destabilizing protein tristetraprolin (TTP) in ECs reverses hypoxia-induced down-regulation of HIF-1α mRNA; 3) the decrease in the half-life of Luciferase-HIF-1α-3′UTR reporter transcript that is observed after prolonged hypoxia is mediated by TTP; 4) TTP binds specifically to HIF-1α 3′UTR; and 5) the most distal AU-rich elements present in HIF-1α 3′UTR (composed of two hexamers) are sufficient for TTP-mediated repression. Finally, we bring evidence that silencing TTP expression enhances hypoxia-induced increase in HIF-1α protein levels with a concomitant increase in the levels of the carbonic anhydrase enzyme CA IX, thus suggesting that TTP physiologically controls the expression of a panel of HIF-1α target genes. Altogether, these data reveal a new role for TTP in the control of gene expression during the response of endothelial cell to hypoxia.
INTRODUCTION
Hypoxia-inducible factor (HIF)-1α, the rate-limiting and oxygen-regulated subunit of the heterodimeric transcription factor HIF-1, triggers major changes in normal and cancer cells by driving the transcription of a number of genes that control glucose metabolism, cell survival, erythropoiesis, and angiogenesis ( Forsythe et al., 1996; Dewhirst et al., 2008). In particular, the transcription of vascular endothelial growth factor (VEGF) has been shown to be stimulated by hypoxia via HIF-1 ( Forsythe et al., 1996). In endothelial cells (ECs), the expressions of VEGF and of its specific receptors VEGF-R1 and VEGF-R2 are up-regulated by hypoxia through HIF-1α–dependent mechanisms ( Tang et al., 2004). More recently, Lee et al. have reported that, in addition to its major role as an EC mitogenic factor, VEGF is also implied in an autocrine loop stimulating EC survival ( Lee et al., 2007). Thus ECs have the potential to tightly regulate their responses to changes in the oxygenation of surrounding tissues. ECs lacking HIF-1α display reduced proliferation and decreased tubular network formation under hypoxia and are unable to efficiently form new capillaries in wound-healing assays ( Tang et al., 2004). In addition, loss of HIF-1α in ECs disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis ( Tang et al., 2004).
Hypoxic regulation of HIF-1α expression is well described at the protein level. In oxygenated cells, HIF-1α is hydroxylated on proline residues. This modification promotes its interaction with and ubiquitination by the Von Hippel-Lindau (VHL) tumor suppressor protein and thereby its rapid degradation through the ubiquitin-proteasome pathway. Under hypoxia, prolyl hydroxylases are inactive; HIF-1α is not recognized by the VHL protein and is consequently stabilized in the cytoplasm ( Huang et al., 1998). After heterodimerization with the constitutively expressed β subunit, HIF-1α translocates to the nucleus and activates the transcription of genes bearing a hypoxia-responsive element (HRE) in their promoter. These include a number of genes controlling angiogenesis (VEGF, EG-VEGF, adrenomedullin, Flt-1), glucose and energy metabolism (glucose transporter-1 and -3, hexokinase 1 and 2, aldolase A and C, glyceraldehyde-3-phosphate dehydrogenase), and intracellular pH regulation (carbonic anhydrases IX and XII) ( Maxwell et al., 2001). Aside from this well-established regulatory pathway, there are few emerging reports supporting additional regulation of HIF-1α expression at the transcriptional and/or posttranscriptional levels. Increased HIF-1α mRNA levels were observed in high-grade colorectal and gastric carcinomas and validated in the latter case as a poor prognosis factor ( Furlan et al., 2007; Ma et al., 2007). Several RNA-binding proteins were reported to interact with the HIF-1α mRNA 3′-untranslated region (UTR) and to control mRNA translation. These include the cytoplasmic polyadenylation element-binding proteins (CPEB-1 and -2; Hagele et al., 2009), HuR, and the polypyrimidine tract-binding protein (PTB; Schepens et al., 2005; Galban et al., 2008; Hagele et al., 2009). HIF-1α mRNA was also recently identified in an immortalized lung epithelial cell line as a novel target for the miR-17-92 microRNA (miRNA) cluster ( Taguchi et al., 2008).
The 3′UTR of HIF-1α mRNA contains many AU-rich elements (ARE). AREs are short instability elements identified in the 3′UTR of several short-lived mRNAs, including that of VEGF ( Chen and Shyu, 1995; Bakheet et al., 2003). We have previously reported that TIS11b, the double zinc-finger protein from the tristetraprolin (TTP) family, enhances VEGF mRNA decay by interacting specifically with two AREs within a short 75-base-long region of VEGF 3′UTR ( Ciais et al., 2004). The human TTP family consists of three members, TTP (TIS11/ZFP36), TIS11b (ZFP36L1/BRF1), and TIS11d (ZFP36L2/BRF2), that own the same characteristic CCCH tandem zinc-fingers and share similar mRNA-destabilizing activity in vitro ( Blackshear, 2002). The prototypical member TTP has been shown to promote decay of several labile mRNAs, including those of TNFα, GM-CSF, IL-2, and IL-10, by interacting with the basic RNA decay machinery ( Carballo et al., 1998, 2000; Ogilvie et al., 2005; Sandler and Stoecklin, 2008).
In view of its critical role in endothelial cell function, we analyzed the regulation of HIF-1α mRNA expression by the TTP family proteins in ECs under hypoxic conditions. Our results show that hypoxia-induced decrease of HIF-1α mRNA in endothelial cells is mediated by TTP. TTP, preferentially to TIS11b or TIS11d, decreases both endogenous HIF-1α and reporter gene mRNAs fused to HIF-1α 3′UTR through direct interactions with HIF-1α 3′UTR. Furthermore, we demonstrate that endothelial cells silenced for TTP expression not only display higher levels of HIF-1α protein under hypoxia but also exhibit higher levels of carbonic anhydrase IX (CA IX), a major target of HIF-1α, which is involved in the regulation of pH homeostasis. These observations point to a novel TTP-mediated regulatory pathway of HIF-1α activity, which may contribute to the control of HIF-1α physiological target expression in EC in response to hypoxia.
RESULTS
Effect of hypoxia on HIF-1α protein and mRNA levels in endothelial cells
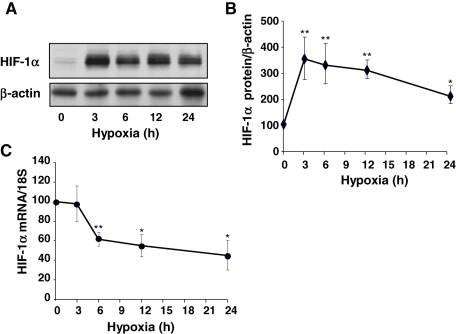
HIF-1α is the key factor for the adapted cellular response to hypoxia. It is well established that hypoxia induces the stabilization of the HIF-1α protein. However, the effect of hypoxia on HIF-1α mRNA levels has been understudied until a recent work reporting a decrease of HIF-1α mRNA stability in hypoxic adenocarcinoma cells ( Uchida et al., 2004). To evaluate the kinetics of hypoxic induction of HIF-1α protein expression in endothelial cells, SV-40-immortalized human microvascular endothelial cells (HMEC-1) were exposed to hypoxia (1.5% O2) for 3–24 h. Western blot analysis of whole-cell extracts shows that HIF-1α was almost undetectable in normoxia ( Figure 1A). Exposure of HMEC-1 cells to hypoxia resulted in a rapid and transient increase in HIF-1α protein levels peaking at 3 h of treatment ( Figure 1, A and B). Sustained hypoxia progressively decreased HIF-1α levels, which, however, remained higher than basal levels at 24 h ( Figure 1, A and B). We next evaluated HIF-1α mRNA steady-state levels in normoxic and hypoxia-treated endothelial cells. Three hours of hypoxia did not change HIF-1α mRNA levels ( Figure 1C), suggesting that hypoxia-induced increases in HIF-1α protein are likely due to translational or posttranslational changes. By contrast, prolonged hypoxia progressively decreased HIF-1α mRNA, reaching 45 ± 14% of the initial value at 24 h ( Figure 1C). These results led us to investigate whether these changes in HIF-1α mRNA during prolonged hypoxia involve one of the three TTP family members, namely TIS11b, TIS11d, or TTP.
FIGURE 1:
Time course of effects of hypoxia on HIF-1α protein and mRNA levels in HMEC-1 cells. Cells were exposed to normoxia (19% O2) or to hypoxia (1.5% O2) for various periods of time. (A) HIF-1α protein levels were determined by Western blot. A representative blot is shown. (B) Quantification of HIF-1α protein in three independent experiments. The levels of HIF-1α were normalized to β-actin protein level. (C) HIF-1α mRNA levels were determined by quantitative RT-PCR, normalized to 18S RNA levels, and plotted as a percentage of the initial value (t = 0) against time. Data are presented as means ± SE. *, **, P < 0.05 and P < 0.01 vs. 0 h.
Silencing TTP expression selectively increases endogenous HIF-1α mRNA in HMEC-1 cells during prolonged hypoxia
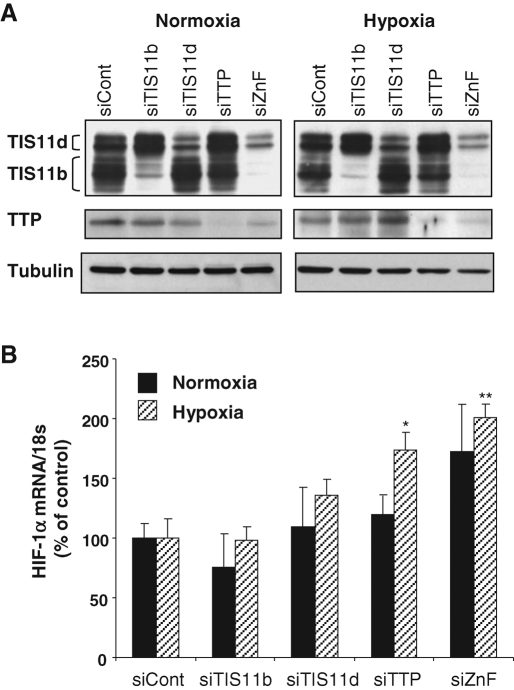
To evaluate the potential involvement of TIS11b, TIS11d, or TTP in hypoxia-induced decrease in HIF-1α mRNA, the expression of each protein was knocked down in HMEC-1 cells using either specific TIS11b, TIS11d, and TTP small interfering RNA (siRNAs) or an siRNA targeting the three family members (ZnF siRNA). HMEC-1 cells were subsequently exposed to 24 h of hypoxia. Western blot analysis of total cell extracts revealed that the expression of TIS11b, TIS11d, and TTP was strongly repressed in HMEC-1 cells transfected with their specific siRNAs or with ZnF siRNA as compared with control siRNA ( Figure 2A). Measurement of HIF-1α mRNA levels by quantitative reverse transcription PCR (RT-qPCR) shows that silencing TIS11b, TIS11d, or TTP independently or simultaneously (siZnF) had no significant effect on HIF-1α transcript in normoxic endothelial cells ( Figure 2B). Under hypoxia, down-regulation of TIS11b expression did not affect HIF-1α transcript ( Figure 2B). Although a trend to an increase in HIF-1α mRNA was consistently observed in TIS11d siRNA-treated cells, this increase did not reach statistical significance. By contrast, silencing the expression of TTP or of the three TTP family members together significantly increased HIF-1α mRNA levels to 174 ± 15% and 201 ± 11% of controls, respectively (with P < 0.05 and P < 0.01, n = 3; Figure 2B). These results suggested to us that endogenous HIF-1α mRNA is selectively targeted by TTP in endothelial cells exposed to sustained hypoxia.
FIGURE 2:
Silencing TTP expression in HMEC-1 cells selectively increases endogenous HIF-1α mRNA in response to hypoxia. HMEC-1 cells were transfected either with a negative control siRNA (siCont), or with TIS11b, TIS11d, or TTP siRNAs or with ZnF siRNA, which targets all three family members. Twenty-four hours after transfection, HMEC-1 cells were maintained in normoxia or exposed to hypoxia (1.5% O2) during an additional 24 h. (A) Western blot analysis of cell extracts showing efficient down-regulation of TIS11b, TIS11d, and TTP expression by their respective siRNAs as well as by the ZnF siRNA. β-Actin was used as a control of protein loading. (B) Total RNA was recovered and analyzed by RT-qPCR. HIF-1α mRNA values were normalized to 18S RNA values and plotted as a percentage of the siCont value. Data are presented as means ± SE of three independent experiments performed in triplicate. *, **, P < 0.05 and P < 0.01 vs. siCont, respectively.
Overexpressed TTP decreases reporter gene activity through HIF-1α 3′UTR
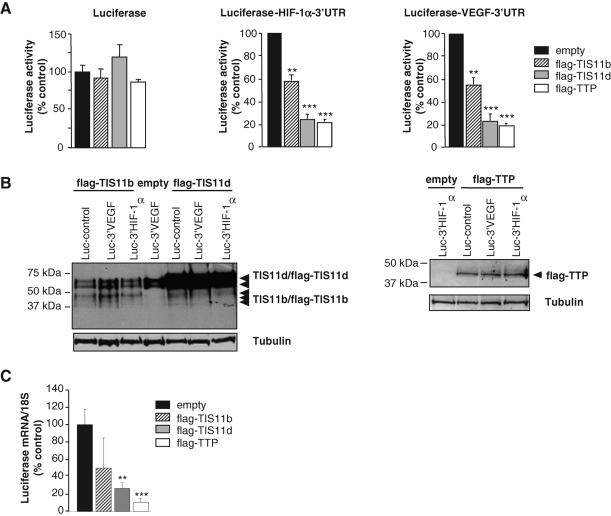
As the 3′UTR of HIF-1α mRNA contains many AREs that are potential targets for TTP family proteins, a series of experiments was designed to investigate whether HIF-1α 3′UTR could interfere with the three members of the TTP family. The full-length HIF-1α 3′UTR sequence (1175 base pairs) was cloned downstream of a luciferase reporter gene (pLuc HIF-1α-3′UTR plasmid). As a control, we elaborated a similar construct using the full-length 3′UTR of human VEGF (1923 base pairs; Ciais et al., 2004). These plasmids were cotransfected in HMEC-1 cells with expression plasmids encoding either flag-TTP, flag-TIS11b, or flag-TIS11d. As shown in Figure 3A, in the absence of any 3′UTR, luciferase activity was grossly unaffected by any TTP family member. Both TTP and TIS11d induced a 70–80% inhibition of luciferase activities, whether cloned upstream of HIF-1α- or VEGF-3′UTR, whereas TIS11b decreased luciferase activity by 40% ( Figure 3A). To determine whether this weaker repression of Luc-HIF-1α-3′UTR transcript by TIS11b was due to lower expression of TIS11b compared with TIS11d, TTP family member protein levels were checked by Western blotting. As shown in Figure 3B, the overexpression of TIS11b under the various transfection conditions (Luc, Luc-HIF-1α-3′UTR and Luc-VEGF-3′UTR) was lower than that of TIS11d, suggesting that the repression levels of luciferase activity were correlated with the levels of TTP family protein expression. We then checked the effect of TTP family members on the levels of Luc-HIF-1α-3′UTR transcripts by RT-qPCR. As seen in Figure 3C, a significant decrease of luciferase mRNA levels was observed in cells overexpressing TIS11d and TTP (26.6 ± 6.4% and 10.1 ± 1.5% of controls for TIS11d and TTP, respectively), indicating that the repression of luciferase activity by TIS11d and TTP ( Figure 3A) was correlated to a decrease in the level of Luc-HIF-1α-3′UTR transcript. As endogenous HIF-1α mRNA was selectively targeted by TTP in hypoxic endothelial cells and as TTP overexpression induced the most significant decrease in Luc-HIF-1α-3′UTR transcript levels, we then focused on TTP as a regulator of HIF-1α mRNA expression under hypoxia.
FIGURE 3:
Effect of TTP family members on HIF-1α mRNA 3′UTR-mediated luciferase activity and on Luc-HIF-1α-3′UTR mRNA levels. HMEC-1 cells were transiently transfected with 1 μg of pLuc HIF-1α-3′UTR, pLuc VEGF-3′UTR, or pLuc-control plasmids and 500 ng of either pCDNA3.1-empty, pCDNA3.1-flag-TTP, pCDNA3.1-flag-TIS11b, or pCDNA3.1-flag-TIS11d. (A) Luciferase activities were determined 24 h posttransfection as described in Materials and Methods. Results are expressed as relative light units of firefly luciferase activity over relative light units of renilla luciferase activity to compensate for variations in transfection efficiency and are represented as a percentage of luciferase activity measured in HMEC-1 cells transfected with an empty vector. Transfections were performed in triplicates and values are means ± SE of at least 5 independent experiments. **, ***, significantly different from empty plasmid with P < 0.01 and P < 0.001, respectively. (B) The accumulation of overexpressed proteins in each transfection condition shown in (A) was determined by Western blot using either anti–TIS11b/TIS11d (left panel) or anti–flag antibodies (TTP, right panel). Tubulin was used as a control of protein loading. (C) HMEC-1 cells were transiently transfected for 24 h with either pCDNA3.1-empty or pCDNA3.1-flag-TTP, pCDNA3.1-flag-TIS11b, or pCDNA3.1-flag-TIS11d, and with pLuc HIF-1α-3′UTR vector. Then, the levels of Luc-HIF-1α-3′UTR transcripts and 18S RNA were determined by RT-qPCR. Data are presented as means ± SE of three independent experiments performed in triplicate. **, ***, significantly different from empty plasmid with P < 0.01 and P < 0.001, respectively.
TTP targets HIF-1α mRNA 3′UTR
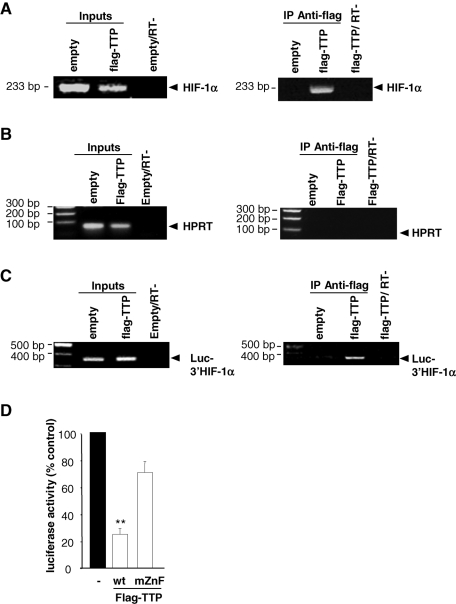
To determine whether endogenous HIF-1α mRNA is a direct target of TTP in living cells, we performed ribonucleoprotein immunoprecipitation (RNP-IP) experiments. Cross-linked RNP complexes were immunoprecipitated from HMEC-1 cells overexpressing or not flag-TTP. Figure 4A clearly shows the presence of HIF-1α mRNA in all inputs (empty plasmid and flag-TTP), whereas HIF-1α mRNA was immunoprecipitated only when flag-TTP was present. This interaction between HIF-1α mRNA and TTP was specific since HPRT mRNA was not coprecipitated with Flag-TTP ( Figure 4B). We then checked whether TTP interacts with HIF-1α mRNA 3′UTR by immunoprecipitating RNP complexes in HMEC-1 cells cotransfected with pLuc HIF-1α-3′UTR and empty or flag-TTP expression vectors. As shown in Figure 4C, a substantial amount of immunoprecipitated luciferase mRNA was detected in cells overexpressing flag-TTP, indicating that TTP targets HIF-1α mRNA 3′UTR. To confirm these observations, we mutated the two zinc finger domains of TTP that are required for TTP binding to AREs and tested the ability of this mutant to repress luciferase activity. As shown in Figure 4D, luciferase activity was strongly reduced by TTP, and this repression was mostly lost in the presence of the TTP mutant.
FIGURE 4:
TTP directly binds to HIF-1α mRNA 3′UTR in endothelial cells. Ribonucleoprotein complexes were immunoprecipitated (RNP-IP) with agarose beads-linked anti–flag antibodies from HMEC-1 cells transfected with pCDNA3.1-empty or pCDNA3.1-flag-TTP vectors as described in Materials and Methods. (A) Left panel: Input lanes show the amplification of HIF-1α mRNA in 10% of nonimmunoprecipitated lysate. Right panel: Representative RT-PCR showing enrichment of endogenous HIF-1α mRNA in RNP-IP of TTP-transfected cells and no amplification from empty plasmid-transfected cells. “RT-” represents control PCR amplification in the absence of reverse transcriptase. (B) RT-PCR analysis of HPRT mRNA in inputs and RNP-IPs from HMEC-1 lysates transfected with pCDNA3.1-empty or pCDNA3.1-flag-TTP and pLuc-HIF-1α-3′UTR vectors. Note the absence of HPRT mRNA in RNP-IPs, indicating that the interaction between HIF-1α mRNA and TTP was specific. (C) Representative RT-PCR analysis of Luc-HIF-1α-3′UTR mRNA in inputs and RNP-IPs from HMEC-1 lysates transfected with pCDNA3.1-empty or pCDNA3.1-flag-TTP and pLuc HIF-1α-3′UTR vectors. “RT-” represents control PCR amplifications in the absence of reverse transcriptase. (D) Luciferase activity was determined in extracts from HMEC-1 cells transfected with pCDNA3.1-empty plasmid, flag-wild type TTP (wt) or flag-TTP mutant (mZnF: double-zinc finger mutant). Transfections were performed in triplicates and values are means ± SE of four independent experiments. **, significantly different from TTP mutant with P < 0.01.
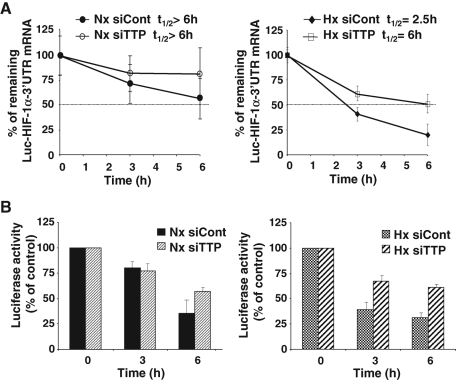
TTP controls HIF-1α mRNA stability under hypoxia
In attempts to determine whether the TTP-induced decrease in HIF-1α mRNA levels resulted from changes in HIF-1α mRNA stability, TTP-transfected HMEC-1 cells were treated with DRB (5,6-dichloro-1-β-d-ribofuranosylbenzimidazole) to block transcription. Total RNA was extracted at different time intervals and analyzed by Northern blot (data not shown). We found that HIF-1α mRNA had a very long half-life of 15 h, which is very unusual for TTP-regulated mRNAs (data not shown). Given the possible deleterious effects of DRB on cell physiology after several hours of treatment (15–24 h) and the difficulty to establish an endothelial cell line stably expressing a reporter gene, we generated a HeLa Tet-off cell line expressing a tet-repressible Luc-HIF-1α-3′UTR reporter gene as described in Materials and Methods. The role of TTP in the regulation of HIF-1α mRNA turnover on one hand, and of luciferase activity on the other hand, was assessed in normoxic or hypoxia-treated HeLa Tet-off cells transfected or not with TTP siRNA. Figure 5A shows that Luc-HIF-1α-3′UTR mRNA half-life in normoxic HeLa cells was longer than 6 h. Silencing TTP expression did not change significantly the remaining Luc-HIF-1α-3′UTR mRNA measured at the same time points under normoxia ( Figure 5A). By contrast, a marked decrease in Luc-HIF-1α-3′UTR mRNA half-life was observed in hypoxia-treated cells compared with normoxic cells (2.5 h vs. a half-life >6 h). Importantly, down-regulation of TTP expression in hypoxia-treated cells increased Luc-HIF-1α-3′UTR mRNA half-life from 2.5 to 6 h ( Figure 5A). When luciferase activity of Luc-HIF-1α-3′UTR transcript was measured in the same conditions, we observed that silencing TTP expression did not affect the reporter gene activity in normoxia ( Figure 5B). Conversely, knockdown of TTP increased luciferase activity at all time points in hypoxia-treated cells ( Figure 5B), providing evidence for a correlation between repression of luciferase activity and repression of Luc-HIF-1α-3′UTR transcript levels. Taken together, these results suggest that 1) the decrease in HIF-1α transcript levels that we observed during prolonged hypoxia is related mainly to a decrease in mRNA stability, and 2) TTP is involved in hypoxia-induced HIF-1α mRNA decay.
FIGURE 5:
TTP regulates Luc-HIF-1α-3′UTR mRNA stability in hypoxia-treated cells. (A) Half-life of Luc-HIF-1α-3′UTR transcript in HeLa cells in normoxia (Nx) and in hypoxia (Hx). HeLa-Tet off cells expressing a tet-repressible Luc-HIF-1α-3′UTR reporter gene were transfected with siTTP siRNAs. Twenty-four hours later, doxycycline (1 μg/ml) was added to block transcription. Total RNA was then recovered at the indicated time and analyzed by RT-qPCR. HIF-1α mRNA values were normalized to 18S RNA values and plotted as a percentage of the initial value against time. The data shown represent the mean ± SE of two independent experiments performed in triplicate. (B) Luciferase activity in HeLa-Tet-off cells expressing the tet-regulated Luc-HIF-1α-3′UTR reporter gene. Cells were treated as described in (A) then measurement of luciferase activity was performed as described in Materials and Methods using whole-cell extracts. Error bars indicate the SD of duplicate samples.
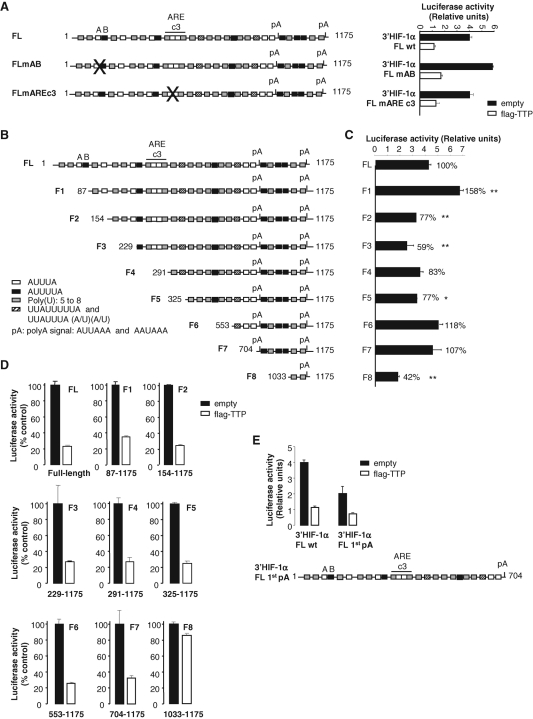
Characterization of a short fragment of HIF-1α mRNA 3′UTR that recapitulates basal activity and is sufficient for repression by TTP
The aforementioned data indicate that TTP contributes to the destabilization of HIF-1α mRNA in endothelial cells through the formation of a molecular complex with its 3′UTR. We next attempted to identify the TTP-responsive region using the heterologous reporter gene strategy. Computer analysis of the human HIF-1α 3′UTR revealed several AU-rich or U-rich elements: two nonameric motifs (UUAUUUUUA and UUAUUAUUU), seven canonical pentameric motifs (AUUUA), six hexameric motifs (AUUUUA), 16 U-rich regions as well as two poly(A) signals ( Figure 6A). According to the ARED database, the HIF-1α 3′UTR belongs to the class III AU-rich group, based on the pentamers flanked by U-rich regions located between base (b) 269 and b287 of the 3′UTR (AREc3; Figure 6A). These elements are highly conserved in mouse and rat sequences. We first mutated this cluster III (UU into CC in both pentamers). However, as shown in Figure 6A, TTP repressed the activity of wild-type and mutated constructs with the same efficiency. Consequently, we decided to modify the most 5′ ARE element present in the 3′UTR that is composed of one pentamer (A) close to one hexamer (B) ( Figure 6A). The same mutation as above was introduced in the pentamer and UUU were substituted by CCC in the hexameric motif. As seen in Figure 6A, even though we observed a 1.5-fold increase of the basal AB mutant activity, it was still repressed by TTP with the same efficiency. These results thereby demonstrate that other cis elements in the 3′UTR are crucial for repression by TTP.
FIGURE 6:
A short fragment of HIF-1α mRNA 3′UTR from b704 to b1175 carries full basal activity and TTP-mediated repression. In (A–E), HMEC-1 cells were transfected with 1 μg of pLuc plasmids in the presence or in the absence of 500 ng of pCDNA3.1 empty or pCDNA3.1-flag-TTP. Luciferase activity was measured 24 h posttransfection as described in Materials and Methods. Results are expressed as relative light units of firefly luciferase activity over relative light units of renilla luciferase activity. (A) Left panel: Schematic representation of the full-length (FL) HIF-1α mRNA 3′UTR (1175 base pairs) inserted in pLuc reporter constructs. White boxes represent pentameric motifs AUUUA, black boxes hexameric motifs AUUUUA, hatched boxes nonameric motifs (UUAUUUUUA and UUAUUAUUU), gray boxes polyU regions and pA, polyadenylation signal. FL mARE c3 and FL mAB represent FL HIF-1α 3′UTR mutated on ARE cluster 3 and HIF-1α 3′UTR mutated on the first pentamer and hexamer, respectively, that were also inserted in pLuc reporter constructs. Right panel: Luciferase activies of FL mARE c3 and FL mAB in the presence or in the absence of TTP. (B, C) Basal luciferase activities of HIF-1α 3′UTR truncations from F1 to F8. *, **, significantly different from FL with P < 0.05 and P < 0.01 (n = 3), respectively. (D) TTP repression efficiency of the eight HIF-1α 3′UTR truncations (F1 to F8). (E) TTP repression efficiency of HIF-1α 3′UTR lacking the F7 fragment (FL 1st polyA construct). Transfections were performed in triplicates and values are means ± SE. A representative experiment of three to five independent experiments is shown.
To answer this question, we initiated a deletion analysis. To keep the polyadenylation signals in all constructs, the full-length (FL) HIF-1α 3′UTR was progressively deleted from 5′ to 3′ (from F1 to F8; Figure 6B). As shown in Figure 6, C and D, the F1 construct (b87– b1175) gave the same results as the AB mutated full-length construct, confirming the presence of destabilizing elements within the 1-86b fragment. In contrast, we observed that the basal luciferase activity of fragment F2 was reduced, thus illustrating the presence of stabilizing elements within the 87-154b fragment ( Figure 6C). Further deletion between b155 and b705 did not affect the repression by TTP ( Figure 6D). The small F7 fragment (b704–b1175) bears by itself the basal activity and the repression by TTP of the full-length 3′UTR ( Figure 6, C and D). In contrast, the shortest deletion F8 (b1033–b1175), that does not contain any AU-rich element, presents a 58% reduced basal activity and is no longer repressed by TTP. It can thus be deduced that the 704–1033b fragment recapitulates the regulation of HIF-1α 3′UTR by TTP. This fragment displays 3 hexameric AU-rich motifs, a target sequence for miR-17-92 miRNA cluster (Tagushi et al., 2008) overlapping hexamer 1, one U-rich region and 2 poly(A) signals ( Figure 7A). To determine whether the 704–1033-base-long fragment was required for TTP-mediated repression of HIF-1α mRNA, we truncated the full length HIF-1α 3′UTR beyond the first polyA ( Figure 6E, FL1stpA construct), thus eliminating the F7 fragment. Figure 6E shows that Luc-FL1stpA-mediated luciferase activity was decreased by half as compared with Luc-FL luciferase activity, suggesting that this truncated transcript was less stable. Overexpression of TTP repressed the FL1stpA construct by 65 ± 1%, as compared with 75 ± 13% for FL HIF-1α 3′UTR ( Figure 6E), indicating that the remaining instability elements could somewhat take over when the F7 fragment was absent. It is worth mentioning that similar observations were made with VEGF 3′UTR ( Ciais et al., 2004). Indeed, when the TIS11b binding element within VEGF 3′UTR was deleted, luciferase activity was still repressed by overexpressed TIS11b, possibly through the remaining AU-rich elements.
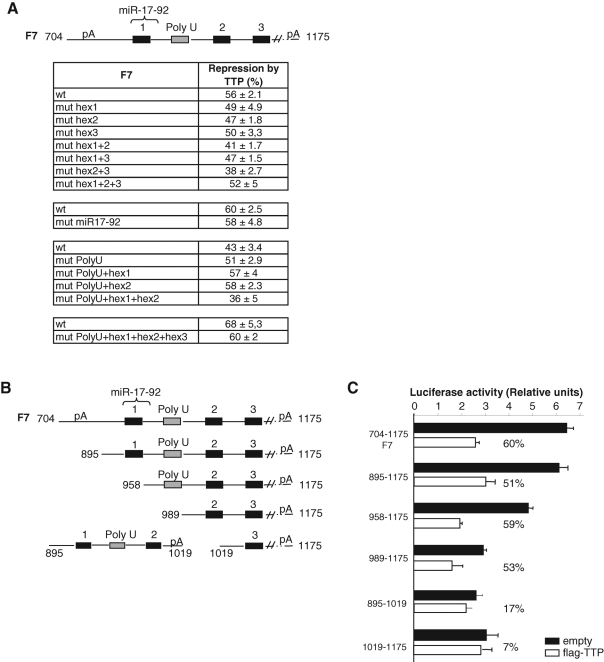
FIGURE 7:
The most distal hexamers AUUUUA within the 704- to 1033-base-long fragment are both implied in repression by TTP. (A) Upper panel: Schematic representation of the 704–1175 fragment of HIF-1α 3′UTR, which contains three hexameric motifs (black boxes), a target sequence for miR-17-92 miRNA cluster (curly bracket), and a poly U region (gray box). Lower panel: TTP-mediated repression of luciferase activity in cells transfected with the different mutants of the F7 fragment inserted in pLuc reporter vectors. Hexamer AUUUUA was mutated to AUgUgA, the poly U region UUUUUUU to UggUggU and the target sequence for miR-17-92 miRNA cluster AUGUUUGAUUUUAUGCACUUUG to AUGGUGGAUUUUAUAAACGGGG. (B) The various deletion mutants represented on the figure were inserted in pLuc reporter vectors. (C) TTP-mediated repression of luciferase activity in cells transfected with the different truncations of the 704–1175 fragment (percentages represent levels of repression of luciferase activity by TTP). Transfections were performed in triplicate and values are means ± SD. A representative experiment of two to three independent experiments is shown.
TTP-mediated repression of HIF-1α mRNA via the F7 fragment requires the two most distal AUUUUA motifs
To identify the motif within the 704–1033 sequence that is sufficient for TTP-mediated repression, we mutated the AREs present in this sequence ( Figure 7A). Single or combined mutations of each ARE were performed as well as mutations of the target sequence for the miR-17-92 miRNA cluster. Figure 7A sums up all the generated mutants and the efficiency of TTP at repressing them. None of these mutants lost its ability to be repressed by TTP. The fragment mutated on hexamers 2 and 3 was the only one to be less repressed (38% repression for hex 2+3 mutant vs. 56% for F7 wt). In addition, a shorter deletion construct (895-1175) of this TTP-responsive fragment was shown to keep the full-inhibitory response to TTP ( Figure 7, B and C). When this fragment was further shortened (fragments 958–1175 and 989–1175), the basal luciferase activity progressively decreased, whereas the repression by TTP was maintained ( Figure 7, B and C). Interestingly, when we cleaved the 895–1175 fragment into fragments 895–1019 and 1019–1175, repression by TTP was lost, indicating that the presence of both hexamers 2 and 3 is important for TTP function ( Figure 7, B and C). We concluded therefore that the 187-base-long fragment between b989 and b1175 is sufficient for repression by TTP.
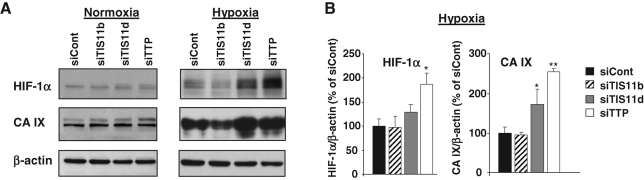
Knockdown of TTP modulates the expression of the HIF-1α downstream target CA IX under hypoxia
CA IX is a cancer-related enzyme involved in the regulation of pH homeostasis, cell proliferation, and adhesion. In human malignancy, high levels of CA IX expression are consistently seen in a high proportion of carcinomas. The existing data support the notion that CA9, due to the unique structure of its promoter, is one of the most sensitive endogenous sensors of HIF-1α activity ( Kaluz et al., 2009). Therefore, we investigated the impact of TTP, TIS11b, or TIS11d knockdown on HIF-1α protein as well as on its downstream target CA IX. HMEC-1 cells were transfected either with TTP, TIS11b, or with TIS11d siRNAs, then maintained in normoxia or exposed to hypoxia for 24 h. Figure 8 shows a Western blot analysis of whole-cell extracts for HIF-1α and CA IX expression. HIF-1α and CA IX were almost undetectable in normoxia unless the membranes were overexposed for 15 min ( Figure 1A). By contrast, exposure of HMEC-1 cells to hypoxia resulted in a substantial increase in HIF-1α protein with a concomitant increase in CA IX protein ( Figure 8A). Quantification of three independent experiments revealed that TIS11b siRNA treatment had significant effect neither on hypoxia-induced increase in HIF-1α nor on CA IX as compared with control siRNA (98 ± 23% of controls for HIF-1α and 95 ± 6% of controls for CA IX; Figure 8B). Silencing TIS11d induced no significant changes in HIF-1α protein (129 ± 17% of control siRNA; Figure 8B). Surprisingly, a moderate but significant effect of TIS11d siRNA on CA IX expression was observed (172 ± 31% of control siRNA, n = 3, P = 0.049; Figure 8B). By contrast, down-regulation of TTP using TTP siRNAs significantly potentiated hypoxia-induced increase in HIF-1α (186 ± 24% of control siRNA; Figure 8, A and B). This increase in HIF-1α was accompanied by a marked up-regulation of CA IX expression (254 ± 9% of control siRNA; Figure 8, A and B). Altogether, these results suggest that modulation of HIF-1α expression by TTP leads to modulation of the levels of the major HIF-1α downstream target CA IX and provide evidence that TTP is an inhibitor of the HIF-1α system.
FIGURE 8:
Silencing TTP expression in endothelial cells potentiates hypoxia-induced increase of both HIF-1α protein and its downstream target CA IX. HMEC-1 cells were transfected either with a control siRNA (siCont) or with TIS11b, TIS11d, or TTP siRNAs. Twenty-four hours after transfection, HMEC-1 cells were maintained in normoxia or exposed to hypoxia for an additional 24 h. (A) Expression levels of HIF-1α and CA IX proteins in normoxia and hypoxia were determined by Western blot. β-Actin was used as a control of protein loading. As the levels of HIF-1α and CA IX proteins were barely detectable in normoxic cells, the immunoblots shown (left panels) were overexposed for 15 min as compared with 2 min for those from hypoxia-treated cells (right panels). (B) Quantification of HIF-1α and CA IX proteins in three independent experiments, after normalization to β-actin protein level. *, **, significantly different from SiCont with P < 0.05 and P < 0.01, respectively.
DISCUSSION
Hypoxia occurs under physiological conditions such as embryogenesis and under pathological conditions such as tissue ischemia and cancer growth. The HIF family of transcription factors is now considered as a primary effector of the adaptative response to hypoxia ( Hickey and Simon, 2006; Brahimi-Horn and Pouyssegur, 2007). Importantly, HIF-1α conditional deletion in endothelial cells impairs tumor angiogenesis and is associated with a decrease in tumor vessel density ( Tang et al., 2004). The most striking level of regulation of HIF-1α expression is the control of protein stability. Much less is known about the control of HIF-1α mRNA levels. Here, we report that TTP protein interacts with endogenous HIF-1α mRNA 3′UTR and is involved in hypoxia-mediated decrease of HIF-1α mRNA levels in endothelial cells. In addition, TTP counteracts hypoxia-mediated increase in HIF-1α protein levels, thus contributing to a negative feedback loop aiming at preventing excessive HIF-1α protein accumulation during prolonged hypoxia. To our knowledge, this is the first study evaluating the posttranscriptional regulation of HIF-1α expression in endothelial cells.
We first investigated HIF-1α mRNA regulation under hypoxia. Our results are in agreement with those reported by Uchida et al. (2004) in A549 lung epithelial cells. Indeed, we show that, whereas HIF-1α protein expression increases during acute hypoxia (3 h) in endothelial cells, HIF-1α mRNA and, in turn, protein levels progressively decrease during prolonged hypoxia. Using an siRNA strategy, we established that TTP, preferentially to TIS11b or TIS11d, is involved in hypoxia-mediated decrease in HIF-1α mRNA levels. Interestingly, hypoxia has been shown to increase the expression of an endogenous antisense transcript (aHIF) for the 3′UTR of HIF-1α concomitantly with the decrease in HIF-1α mRNA in kidney cancer cells ( Thrash-Bingham and Tartof, 1999; Uchida et al., 2004). We also found that hypoxia markedly increases aHIF mRNA levels in HMEC-1 cells in parallel to the decrease in HIF-1α transcripts (data not shown). It has been suggested that aHIF could increase HIF-1α mRNA instability via exposing AU-rich elements in HIF-1α 3′UTR to potential destabilizing proteins ( Rossignol et al., 2002). One could speculate that, in the presence of TTP siRNA, the unmasking effect of aHIF is no longer efficient, thus leading to an increased stabilization of HIF-1α transcript.
Tristetraprolin family members are double-zinc finger proteins that bind to AREs located in the 3′UTR of several short-lived mRNAs encoding cytokines, growth factors, and inflammatory response proteins ( Baou et al., 2009). These include the mRNAs encoding proteins involved in tumorigenesis and angiogenesis, namely TNFα ( Taylor et al., 1996; Carballo et al., 1998; Twizere et al., 2003), interleukin-3 ( Stoecklin et al., 2001), cyclo-oxygenase-2 ( Sawaoka et al., 2003), c-Myc ( Marderosian et al., 2006), c-Fos ( Patino et al., 2006), cyclin D1 ( Marderosian et al., 2006), p21 ( Patino et al., 2006), and VEGF ( Ciais et al., 2004; Essafi-Benkhadir et al., 2007). Our data show that, although the three TTP family members are capable of decreasing HIF-1α-3′UTR-controlled luciferase activity in vitro, TTP appears to be the only member able to down-regulate endogenous HIF-1α mRNA under hypoxia, thus confirming that TTP and its family members are able to promote the decay of physiologically nonrelevant mRNAs when artificially overexpressed in cells. Our ribonucleoprotein immunoprecipitation experiments and the use of a TTP zinc finger mutant clearly demonstrate that TTP acts through direct binding to HIF-1α mRNA 3′UTR.
We further show that a short 187-base-long fragment within the 3′UTR (from b989 to b1175), excluding the canonical motif listed in the ARED database ( Bakheet et al., 2003), is sufficient for TTP-mediated repression. The target sequence for miR-17-92 miRNA cluster that is present in this 187 base-long TTP-response element was also dispensable for the repressive activity of TTP. This is different from the context of the TNFα 3′UTR, where it has been reported that miR-16 can pair with eight bases in the TNFα ARE and form a complex with TTP and Argonaute/eiF2C that leads to mRNA degradation ( Jing et al., 2005). Our data revealed an unexpected complexity of ARE-mediated posttranscriptional regulation of HIF-1α mRNA. The 187-base-long fragment that is repressed by TTP contains 2 hexamers AUUUUA. Mutations of these motifs into AUgUgA partially altered TTP repressive activity, whereas splitting led to a complete lost of TTP action. A hexameric AUUUUA motif has been reported to contribute to the formation of a stem–loop structure within the AT1 receptor mRNA 3′UTR and to regulate receptor mRNA stability ( Berger et al., 2005). Our results suggest that TTP-mediated repression of the short 187-base-long fragment is dictated by AUUUUA sequence motifs combined with secondary or tertiary structures within the 3′UTR. It is worth mentioning that deletion of this short fragment from HIF-1α 3′UTR ( Figure 6E) decreases, although moderately, the efficiency of TTP-mediated repression. Moreover, this deletion led to a less stable transcript, suggesting that the 187-base-long fragment also harbors some stability elements. Indeed, three polyU sequences that are potential binding sites for the mRNA-stabilizing protein HuR are located in this region.
As a long-term treatment with DRB may have some general deleterious effects on cell physiology and as we have not been successful at generating stable transfectants from endothelial cell lines, we generated a HeLa Tet-off cell line expressing a tetracycline-repressible construct containing the luciferase reporter gene cloned upstream of the HIF-1α 3′UTR. We could determine that the chimeric mRNA was a long-lived mRNA whose half-life exceeded 6 h in normoxia. It is noteworthy that Ushida et al. reported a HIF-1α mRNA half-life of 19.3 h in the A549 carcinoma cell line ( Uchida et al., 2004). Interestingly, silencing TTP expression did not affect HIF-1α mRNA half-life in normoxia. These results were in sharp contrast with those obtained in hypoxia-treated cells. Indeed, the mRNA half-life of the chimeric mRNA was decreased to 2.5 h under hypoxic conditions. In addition, silencing TTP led to a significant increase in HIF-1α mRNA half-life compared with controls (6 vs. 2.5 h), thus demonstrating a novel inhibitory role of TTP in the regulation of HIF-1α expression in hypoxic endothelial cells.
Searching for functional significance of TTP-mediated repression of HIF-1α mRNA and protein, we focused our attention on a major downstream target of HIF-1α during hypoxia, carbonic anhydrase IX. CA IX is a zinc metalloenzyme that catalyzes the reversible conversion of CO2 to bicarbonate and proton and can thus be involved in pH regulation. It has recently emerged as one of the most promising endogenous markers of cellular hypoxia. The utility of CA IX as a tumor/hypoxia biomarker, as a prognostic factor predictive of relative survival, and as a potential tumor therapeutic target have been extensively studied ( Pastorekova et al., 2006). Since identification of the Hypoxia Response Element (HRE) in the CA9 promoter, the critical role of the HIF-1 pathway in the regulation of CA9 has been supported by overwhelming evidence from numerous reports ( Kaluz et al., 2009). Our results show that silencing TTP expression enhances hypoxia-induced increase in HIF-1α protein levels with a concomitant increase in CA IX protein levels, thus confirming the tight link between these two hypoxia response genes. In addition, these original observations reveal a new role for TTP in the control of gene expression during the response to hypoxia.
High levels of HIF-1α mRNA were recently described in high-grade colorectal and gastric carcinomas ( Furlan et al., 2007; Ma et al., 2007). In colorectal cancers, HIF-1α mRNA overexpression is associated with elevated expression of VEGF and active angiogenesis and is considered as a poor prognosis predictor. This overexpression could be related to modifications of TTP expression. In agreement with this hypothesis, reduction in TTP, at both mRNA and protein levels, was described in colorectal cancer ( Young et al., 2009), as well as in lung, breast, and cervix tumors ( Brennan et al., 2009). This suppression was particularly associated with a tumorigenic phenotype ( Brennan et al., 2009). Searching for potential changes in the expression of TTP in endothelial cells submitted to prolonged hypoxia revealed that neither TTP mRNA nor TTP protein levels were significantly affected after 24 h of hypoxia (data not shown). However, changes in TTP phosphorylation and in its capacity to recruit the mRNA decay machinery might be affected under hypoxia. How precisely hypoxia affects TTP function in endothelial cells remains to be solved at the molecular level.
The role of hypoxia-induced stabilization of HIF-1α and transcriptional activation of HIF-1α target genes in cancer is clearly established in tumors. This has stimulated several therapeutic strategies aiming at interfering with HIF-1α protein expression ( Brown, 2007; Melillo, 2007; Koh et al., 2009; Semenza, 2009). These include inhibitors of HIF-1α translation such as the recent molecule KC7F2 ( Narita et al., 2009) or the compound PX-478 ( Koh et al., 2008), promoters of HIF-1α degradation such as the guanylate cyclase inhibitor YC-1 ( Li et al., 2008), or inhibitors of HIF-1α binding to DNA. On the basis of our results, we propose that TTP family proteins might represent very interesting therapeutic tools to simultaneously decrease a number of finely tuned mRNAs, including that of HIF-1α, that are known to be required for cancer progression and endothelial cell survival. Two recent reports support this hypothesis. Essafi-Benkhadir et al. reported that overexpression of TTP in a Ras-dependent tumor cell line prevents its in vivo growth and angiogenic response, when subcutaneously implanted into nude mice ( Essafi-Benkhadir et al., 2007). This effect was correlated with a decrease in VEGF mRNA and protein levels. Using a different strategy, we came to the same conclusions. We could generate a fusion polyarginine-TIS11b protein that has the ability to diffuse through the cellular plasma membranes via its protein transduction domain. We observed that intratumoral injection of this protein into subcutaneous LLC tumors resulted in massive decreases of VEGF expression, tumor angiogenesis, and tumor growth ( Planel et al., 2010). From our present study, it is conceivable that part of these anti-angiogenic and anti-tumorigenic effects are also mediated by a direct effect of exogenous TTP on HIF-1α expression.
MATERIALS AND METHODS
Cell culture
HMEC-1 cells, a human microvascular endothelial cell line immortalized by the large T antigen of SV40 ( Ades et al., 1992), were obtained from V. Abbas (Centers for Disease Control, Atlanta, GA). They were grown at 37°C in a 5% CO2–95% air atmosphere in DMEM supplemented with sodium pyruvate, 1 g/l glucose (Invitrogen, Cergy-Pontoise, France), 10% fetal calf serum (Biowest, Nuaillé, France), antibiotics (penicillin/streptomycin), and amphotericin B. The Hela Tet-off cell line expressing the Tet repressor protein fused with the VP16 transactivation domain was purchased from BD Clontech (Mountain View, CA). The cells were maintained in DMEM supplemented with 10% Tet system–approved fetal bovine serum, penicillin, streptomycin and gentamicin, and were kept under antibiotic selection with G418 (400 μg/ml). For hypoxic conditions, cells were incubated in a 1.5% O2 atmosphere for indicated periods of time.
Plasmid constructs
The plasmid pLuciferase (pLuc) derives from the previously described pLuc-V3′ ( Levy et al., 1996; Ciais et al., 2004). Two unique restriction sites HindIII and SacI were generated downstream of the firefly luciferase reporter gene to include HIF-1α (NM_001530, 1175 base pairs) or VEGF (AF_024710, 1923 base pairs) full-length 3′UTR sequences. The control vector without 3′UTR was generated by end-blunting of HindIII/SacI restriction sites and religation. The eight deletion fragments from the HIF-1α 3′UTR (F1 to F8) were generated by high-fidelity PCR with the same reverse primer: 5′-gatgcgagctcgcctggtccacagaagat-3′ containing the SacI restriction site and different forward primers described in Supplemental Table 1, containing the HindIII restriction site. The different mutants were generated by site-directed mutagenesis (QuickChange Site-directed Mutagenesis kit; Stratagene, Amsterdam, The Netherlands) with the forward primers illustrated in Supplemental Table 2. The TTP zinc finger mutant was generated by site-directed mutagenesis using two successive steps with primers described in Supplemental Table 2. The mutant of the target sequence for miR-17-92 miRNA cluster was obtained by PCR with the forward primer containing the mutation (Supplemental Table 2) and the same reverse primer described above.
To derive a tetracycline-regulated expression vector for the chimeric construct Luc-HIF-1α-3′UTR, two unique restrictions sites KpnI and BamHI were introduced upstream and downstream of Luc-HIF-1α-3′UTR cDNA, respectively, using the pLuc HIF-1α-3′UTR plasmid as a template, the forward primer 5′-GGCGCGGTACCCGAGGAGCTTGGCATTCC-3′ and the reverse primer 5′-GGCCGGGATCCCACATTCCACAGAATTAATTCG-3′. Luc-HIF-1α-3′UTR cDNA was then excised with KpnI and BamHI and inserted into the same restriction sites of pTRE-Tight vector (Clontech), which harbors a tetracycline-responsive element.
Transient transfections and luciferase assays
HMEC-1 cells were seeded at a density of 0.32 × 106 cells per 35-mm plate and transfected the next day using fugene-6 transfection reagent (Roche, Meylan, France) according to the standard protocol. Transfection efficiency was around 40% as determined by flow cytometry analysis using green fluorescent protein reporter gene (data not shown). For each experiment, 1 μg of pLuc plasmid and 500 ng of either empty, flag-TTP-, flag-TIS11b-, or flag-TIS11d-encoding pCDNA3.1 vectors were transfected. To compensate for variations in transfection efficiency, cells were cotransfected with 50 ng of the Renilla luciferase-encoding plasmid pRL-TK (Promega, Charbonnières Les Bains, France). Cells lysates were prepared 24 h after transfection and luciferase activities were measured with the Dual Luciferase System (Promega).
SDS–PAGE and immunoblotting
SDS–PAGE–resolved proteins (20–30 μg) were electrophoretically transferred onto a polyvinylidene fluoride (PVDF) membrane and analyzed by immunoblotting as previously described ( Cherradi et al., 2006). Peroxydase-conjugated anti–flag M2 (Sigma, St. Louis, MO), anti–HIF-1α antibodies (BD Biosciences, Le Pont de Claix, France) and anti–CA IX antibodies (Novus Biologicals, Littleton, CO) were used at concentrations of 2, 0.25, and 1 μg/ml, respectively. Rabbit polyclonal anti–BRF1/BRF2 antibodies recognizing both TIS11b and TIS11d were a generous gift from C. Moroni (University of Bl, Switzerland). Anti-TTP was a generous gift from G. Pagès (Institute of Developmental Biology and Cancer, Nice, France). The different membranes were stripped and reprobed with anti-tubulin (a generous gift from D. Job, Grenoble Institute of Neurosciences, France) or anti–β-actin (Sigma) to check for equal protein loading. Quantification of the Western blots was performed using ImageJ software.
RT-qPCR
Total RNA was extracted using the Macherey-Nagel kit according to the manufacturer's recommendations. An amount of 1 μg of total RNA was reverse-transcribed with the iScript System (Bio-Rad) according to the manufacturer's guidelines and random primers from Invitrogen. cDNAs were diluted in a 50-μl final volume, except in the case of RNA immunoprecipitation experiments. Quantitative PCR was performed using the GoTaq qPCR Master Mix (Promega) and 2-μl aliquots of the RT reaction. The amplification of the samples was carried out in a final volume of 20 μl using a CFX96 Real-Time System thermocycler (Bio-Rad) with the following program: initial denaturation at 95°C for 5 min followed by 40 cycles of denaturation at 95°C for 10 s and annealing at 60°C for 30 s. All PCRs were performed in duplicates. Primer sequences are described in Supplemental Table 3.
Ribonucleoprotein immunoprecipitation (RNP-IP) experiment and RT-PCR analysis
HMEC-1 cells were transfected as described above. Twenty-four hours after transfection, cells were cross-linked with 1% formaldehyde for 15 min at 37°C. After a brief PBS wash, cells were lysed and incubated overnight at 4°C with agarose-linked anti–flag M2 monoclonal antibody (Sigma) according to the manufacturer's guidelines. A 10% volume of each cell lysate was put aside as the input sample. On the next day, the affinity gels were washed three times and subjected, in parallel with the input samples, to cross-linking reversion for 45 min at 70°C in Tris-HCl (50 mM, pH 7) containing 5 mM EDTA, 10 mM DTT, and 1% SDS. RNA was purified from immunoprecipitates and input samples, treated with DNase I, and reverse transcribed with the Improm-II system from Promega. Endogenous HIF-1α mRNA was amplified with the forward primer 5′-ctactagtgccacatcatcacca-3′ and the reverse primer 5′-aagtgaaccatcatgttcca-3′, giving a PCR product of 233 base pairs. A total of 39 and 45 cycles of PCR were performed for immunoprecipitates and input samples, respectively. Luc-HIF-1α-3′UTR mRNA was detected by PCR with the forward primer 5′-gtaccgaaaggtcttaccgg-3′ hybridizing on the luciferase coding sequence and the reverse primer 5′-tgatgctactgcaatgcaatggt-3′ hybridizing on the HIF-1α 3′UTR, giving a 341–base pair PCR product with 33 and 36 cycles for immunoprecipitates and input samples, respectively. HPRT mRNA was amplified with the forward primer 5′-atg gac agg act gaa cgt ctt gc t-3′ and the reverse primer 5′-ttg agc aca cag agg gct aca atg-3′.
RNA interference
siRNAs were designed to target either a conserved sequence present in all three TTP family members (siZnF, base pairs 539–559 in TTP, NM_003407; base pairs 644–664 in TIS11b, NM_004926; base pairs 922–942 in TIS11d, NM_006887) or a specific member of the family (siTIS11b and siTIS11d) using DSIR software ( Vert et al., 2006). TTP siRNAs and control siRNAs (Negative Control 1) were purchased from Ambion (Austin, TX). The siRNA sequences are described in Supplemental Table 4. HMEC-1 cells were seeded at 2 × 106 cells per 60-mm-diameter plate and transfected the next day with 10 nM of siRNA using the RNAimax system from Invitrogen. A negative control siRNA (Negative Control 1; Ambion) that does not target any gene product was used to control for the effects of siRNA delivery.
Measurement of mRNA stability in HeLa Tet-off cells
HeLa Tet-off cells (2 × 106 cells) were transfected with pTRE-Tight-Luc-HIF-1α-3′UTR (2 μg per 60-mm-diameter plate) using Lipofectamine 2000 (Invitrogen, Cergy Pontoise, France). Twenty-four hours after the first transfection, the cells were transfected with siRNAs targeting TTP (siTTP) or scramble control siRNAs (siCont) using RNAimax system (Invitrogen) according to the manufacturer's instructions. Twenty-four hours later, 1 μg/ml of doxycycline (a tetracycline analogue) was added to stop transcription. Total RNA was isolated at different time intervals after the addition of doxycycline using the RNeasy Mini Kit from Qiagen (Courtaboeuf, France). Luc-HIF-1α 3′UTR mRNA was analyzed by quantitative RT-PCR.
Statistical analysis
Statistical analysis was carried out using GraphPad software (Prism 4, San Diego, CA). Data were analyzed using one-way analysis of variance. Results are expressed as means ± SE. A value of P < 0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
This work was supported by funds from INSERM, Sanofi-Aventis, and INCa (Cancéropole Rhône-Alpes). We are indebted to the GEFLUC and the Association pour la Recherche sur le Cancer for their financial support for the purchase of the microscopy equipment of our laboratory. D.C. is recipient of a postdoctoral grant from the Association pour la Recherche sur le Cancer. We thank C. Moroni, G. Pagès, and D. Job for their generous gift of anti–TIS11b/TIS11d, anti–TTP, and anti–tubulin antibodies, respectively. We specially thank Yves Vandenbrouck for his assistance in the design of TIS11b, TIS11d, and ZnF siRNAs using the DSIR software.
Abbreviations used:
- ARE
adenosine uridine-rich element
- TIS11b and TIS11d
TPA-inducible sequence 11b and 11d
- TTP
tristetraprolin
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0617) on July 20, 2011.
*These authors contributed equally to this work.
REFERENCES
- Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DC, Lawley TJ. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J Invest Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- Bakheet T, Williams BR, Khabar KS. ARED 2.0: an update of AU-rich element mRNA datab. Nucleic Acids Res. 2003;31:421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baou M, Jewell A, Murphy JJ. TIS11 family proteins and their roles in posttranscriptional gene regulation. J Biomed Biotechnol. 2009;2009:634520. doi: 10.1155/2009/634520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Stierkorb E, Nickenig G. The role of the AUUUUA hexamer for the posttranscriptional regulation of the AT1 receptor mRNA stability. Biochem Biophys Res Commun. 2005;330:805–812. doi: 10.1016/j.bbrc.2005.03.043. [DOI] [PubMed] [Google Scholar]
- Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn MC, Pouyssegur J. Harnessing the hypoxia-inducible factor in cancer and ischemic disease. Biochem Pharmacol. 2007;73:450–457. doi: 10.1016/j.bcp.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Brennan SE, Kuwano Y, Alkharouf N, Blackshear PJ, Gorospe M, Wilson GM. The mRNA-destabilizing protein tristetraprolin is suppressed in many cancers, altering tumorigenic phenotypes and patient prognosis. Cancer Res. 2009;69:5168–5176. doi: 10.1158/0008-5472.CAN-08-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM. Tumor hypoxia in cancer therapy. Methods Enzymol. 2007;435:297–321. doi: 10.1016/S0076-6879(07)35015-5. [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Cherradi N, Lejczak C, Desroches-Castan A, Feige JJ. Antagonistic functions of tetradecanoyl phorbol acetate-inducible-sequence 11b and HuR in the hormonal regulation of vascular endothelial growth factor messenger ribonucleic acid stability by adrenocorticotropin. Mol Endocrinol. 2006;20:916–930. doi: 10.1210/me.2005-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciais D, Cherradi N, Bailly S, Grenier E, Berra E, Pouyssegur J, Lamarre J, Feige JJ. Destabilization of vascular endothelial growth factor mRNA by the zinc-finger protein TIS11b. Oncogene. 2004;23:8673–8680. doi: 10.1038/sj.onc.1207939. [DOI] [PubMed] [Google Scholar]
- Dewhirst MW, Cao Y, Moeller B. Cycling hypoxia and free radicals regulate angiogenesis and radiotherapy response. Nat Rev Cancer. 2008;8:425–437. doi: 10.1038/nrc2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essafi-Benkhadir K, Onesto C, Stebe E, Moroni C, Pages G. Tristetraprolin inhibits Ras-dependent tumor vascularization by inducing vascular endothelial growth factor mRNA degradation. Mol Biol Cell. 2007;18:4648–4658. doi: 10.1091/mbc.E07-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlan D, Sahnane N, Carnevali I, Cerutti R, Uccella S, Bertolini V, Chiaravalli AM, Capella C. Up-regulation and stabilization of HIF-1alpha in colorectal carcinomas. Surg Oncol. 2007;16((Suppl 1)):S25–27. doi: 10.1016/j.suronc.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Galban S, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagele S, Kuhn U, Boning M, Katschinski DM. Cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 bind to the HIF-1alpha mRNA 3′-UTR and modulate HIF-1alpha protein expression. Biochem J. 2009;417:235–246. doi: 10.1042/BJ20081353. [DOI] [PubMed] [Google Scholar]
- Hickey MM, Simon MC. Regulation of angiogenesis by hypoxia and hypoxia-inducible factors. Curr Top Dev Biol. 2006;76:217–257. doi: 10.1016/S0070-2153(06)76007-0. [DOI] [PubMed] [Google Scholar]
- Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Kaluz S, Kaluzova M, Liao SY, Lerman M, Stanbridge EJ. Transcriptional control of the tumor- and hypoxia-marker carbonic anhydrase 9: A one transcription factor (HIF-1) show. Biochim Biophys Acta. 2009;1795:162–172. doi: 10.1016/j.bbcan.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh MY, Spivak-Kroizman T, Venturini S, Welsh S, Williams RR, Kirkpatrick DL, Powis G. Molecular mechanisms for the activity of PX-478, an antitumor inhibitor of the hypoxia-inducible factor-1alpha. Mol Cancer Ther. 2008;7:90–100. doi: 10.1158/1535-7163.MCT-07-0463. [DOI] [PubMed] [Google Scholar]
- Koh MY, Spivak-Kroizman TR, Powis G. Inhibiting the hypoxia response for cancer therapy: the new kid on the block. Clin Cancer Res. 2009;15:5945–5946. doi: 10.1158/1078-0432.CCR-09-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Chen TT, Barber CL, Jordan MC, Murdock J, Desai S, Ferrara N, Nagy A, Roos KP, Iruela-Arispe ML. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007;130:691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AP, Levy NS, Goldberg MA. Post-transcriptional regulation of vascular endothelial growth factor by hypoxia. J Biol Chem. 1996;271:2746–2753. doi: 10.1074/jbc.271.5.2746. [DOI] [PubMed] [Google Scholar]
- Li SH, Shin DH, Chun YS, Lee MK, Kim MS, Park JW. A novel mode of action of YC-1 in HIF inhibition: stimulation of FIH-dependent p300 dissociation from HIF-1{alpha} Mol Cancer Ther. 2008;7:3729–3738. doi: 10.1158/1535-7163.MCT-08-0074. [DOI] [PubMed] [Google Scholar]
- Ma J, Zhang L, Ru GQ, Zhao ZS, Xu WJ. Upregulation of hypoxia inducible factor 1alpha mRNA is associated with elevated vascular endothelial growth factor expression and excessive angiogenesis and predicts a poor prognosis in gastric carcinoma. World J Gastroenterol. 2007;13:1680–1686. doi: 10.3748/wjg.v13.i11.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marderosian M, Sharma A, Funk AP, Vartanian R, Masri J, Jo OD, Gera JF. Tristetraprolin regulates Cyclin D1 and c-Myc mRNA stability in response to rapamycin in an Akt-dependent manner via p38 MAPK signaling. Oncogene. 2006;25:6277–6290. doi: 10.1038/sj.onc.1209645. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Pugh CW, Ratcliffe PJ. Activation of the HIF pathway in cancer. Curr Opin Genet Dev. 2001;11:293–299. doi: 10.1016/s0959-437x(00)00193-3. [DOI] [PubMed] [Google Scholar]
- Melillo G. Hypoxia-inducible factor 1 inhibitors. Methods Enzymol. 2007;435:385–402. doi: 10.1016/S0076-6879(07)35020-9. [DOI] [PubMed] [Google Scholar]
- Narita T, Yin S, Gelin CF, Moreno CS, Yepes M, Nicolaou KC, Van Meir EG. Identification of a novel small molecule HIF-1alpha translation inhibitor. Clin Cancer Res. 2009;15:6128–6136. doi: 10.1158/1078-0432.CCR-08-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- Pastorekova S, Parkkila S, Zavada J. Tumor-associated carbonic anhydrases and their clinical significance. Adv Clin Chem. 2006;42:167–216. [PubMed] [Google Scholar]
- Patino WD, Kang JG, Matoba S, Mian OY, Gochuico BR, Hwang PM. Atherosclerotic plaque macrophage transcriptional regulators are expressed in blood and modulated by tristetraprolin. Circ Res. 2006;98:1282–1289. doi: 10.1161/01.RES.0000222284.48288.28. [DOI] [PubMed] [Google Scholar]
- Planel S, Salomon A, Jalinot P, Feige JJ, Cherradi N. A novel concept in antiangiogenic and antitumoral therapy: multitarget destabilization of short-lived mRNAs by the zinc finger protein ZFP36L1. Oncogene. 2010;29:5989–6003. doi: 10.1038/onc.2010.341. [DOI] [PubMed] [Google Scholar]
- Rossignol F, Vache C, Clottes E. Natural antisense transcripts of hypoxia-inducible factor 1alpha are detected in different normal and tumour human tissues. Gene. 2002;299:135–140. doi: 10.1016/s0378-1119(02)01049-1. [DOI] [PubMed] [Google Scholar]
- Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- Sawaoka H, Dixon DA, Oates JA, Boutaud O. Tristetraprolin binds to the 3′-untranslated region of cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell line lacks the binding site. J Biol Chem. 2003;278:13928–13935. doi: 10.1074/jbc.M300016200. [DOI] [PubMed] [Google Scholar]
- Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, Cornelis S. The polypyrimidine tract-binding protein stimulates HIF-1alpha IRES-mediated translation during hypoxia. Nucleic Acids Res. 2005;33:6884–6894. doi: 10.1093/nar/gki1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1 inhibitors for cancer therapy: from gene expression to drug discovery. Curr Pharm Des. 2009;15:3839–3843. doi: 10.2174/138161209789649402. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Stoeckle P, Lu M, Muehlemann O, Moroni C. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA. 2001;7:1578–1588. [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster. Cancer Res. 2008;68:5540–5545. doi: 10.1158/0008-5472.CAN-07-6460. [DOI] [PubMed] [Google Scholar]
- Tang N, Wang L, Esko J, Giordano FJ, Huang Y, Gerber HP, Ferrara N, Johnson RS. Loss of HIF-1alpha in endothelial cells disrupts a hypoxia-driven VEGF autocrine loop necessary for tumorigenesis. Cancer Cell. 2004;6:485–495. doi: 10.1016/j.ccr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- Twizere JC, et al. Interaction of retroviral Tax oncoproteins with tristetraprolin and regulation of tumor necrosis factor-alpha expression. J Natl Cancer Inst. 2003;95:1846–1859. doi: 10.1093/jnci/djg118. [DOI] [PubMed] [Google Scholar]
- Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J Biol Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.