A novel perinuclear nuage organelle, the piNG-body, is associated with piRNA silencing in testes of Drosophila. This body contains the known ovarian nuage proteins Vasa, Aub, AGO3, Tud, Spn-E, Bel, Squ, and Cuff, as well as AGO1.
Abstract
Proteins of the PIWI subfamily Aub and AGO3 associated with the germline-specific perinuclear granules (nuage) are involved in the silencing of retrotransposons and other selfish repetitive elements in the Drosophila genome. PIWI proteins and their 25- to 30-nt PIWI-interacting RNA (piRNAs) are considered as key participants of the piRNA pathway. Using immunostaining, we found a large, nuage-associated organelle in the testes, the piNG-body (piRNA nuage giant body), which was significantly more massive than an ordinary nuage granule. This body contains known ovarian nuage proteins, including Vasa, Aub, AGO3, Tud, Spn-E, Bel, Squ, and Cuff, as well as AGO1, the key component of the microRNA pathway. piNG-bodies emerge at the primary spermatocyte stage of spermatogenesis during the period of active transcription. Aub, Vasa, and Tud are located at the periphery of the piNG-body, whereas AGO3 is found in its core. Mutational analysis revealed that Vasa, Aub, and AGO3 were crucial for both the maintenance of the piNG-body structure and the silencing of selfish Stellate repeats. The piNG-body destruction caused by csul mutations that abolish specific posttranslational symmetrical arginine methylation of PIWI proteins is accompanied by strong derepression of Stellate genes known to be silenced via the piRNA pathway.
INTRODUCTION
In many eukaryotic species, germ cells contain specific electron-dense cytoplasmic granules. During Drosophila oogenesis, these granules form a perinuclear organelle called nuage, which is believed to be involved in the selection and translational control of mRNAs transported from the nucleus ( Findley et al., 2003; Snee and Macdonald, 2004). Recent studies accentuated the participation of nuage in the PIWI-interacting RNA (piRNA) biogenesis and piRNA-dependent silencing of transposons and some other harmful selfish elements ( Lim and Kai, 2007; Lim et al., 2009). The piRNA pathway is a genomic defense mechanism conserved in germinal tissues of many eukaryotes ( Aravin et al., 2007; Thomson and Lin, 2009). In flies, proteins of the PIWI subfamily Aubergine (Aub) and Argonaute 3 (AGO3), which associate with small, 25- to 30-nt piRNAs and possess an RNA-slicing function, are considered as the main players of the piRNA pathway ( Aravin et al., 2001; Vagin et al., 2006; Brennecke et al., 2007; Gunawardane et al., 2007; Li et al., 2009). In the ovaries, Aub and AGO3 proteins realize posttranscriptional silencing via the piRNA ping-pong amplification cycle ( Brennecke et al., 2007; Gunawardane et al., 2007). It has been shown that Aub mainly associates with the antisense piRNAs derived from specialized genomic “master” loci, whereas AGO3 preferentially associates with the sense ones ( Li et al., 2009; Nishida et al., 2007). Long sense piRNA precursors are believed leave the nucleus to be recognized by the Aub-containing antisense piRNA-induced silencing complex (piRISC), where the secondary sense piRNAs are generated by the transcript cleavage. These newly generated sense piRNAs are subsequently loaded into the AGO3-containing sense piRISC, which presumably recognizes the long antisense transcripts producing the antisense piRNAs, which in turn enter the Aub-containing silencing complex, and so on ( Brennecke et al., 2007; Gunawardane et al., 2007; Ghildiyal and Zamore, 2009). The amplification cycle appears to take place in the perinuclear RNP-containing nuage granules of nurse cells in Drosophila ovaries ( Lim and Kai, 2007; Lim et al., 2009; Li et al., 2009; Malone et al., 2009). Along with Aub and AGO3, several other proteins taking part in silencing of retrotransposons and endogenous Stellate repeats via the piRNA pathway (Maelstrom, Krimper, Spindle E, Squash, Zucchini, Cutoff, Tejas) and also in mRNA degradation (DCP1, Me31B, Pacman) were identified as components of ovarian nuage ( Harris and Macdonald, 2001; Findley et al., 2003; Snee and Macdonald, 2004; Lim and Kai, 2007; Chen et al., 2007; Pane et al., 2007; Lim et al., 2009; Patil and Kai, 2010). The main marker of nuage is Vasa protein, with the predicted function of a DEAD-box RNA-helicase ( Liang et al., 1994; Findley et al., 2003; Snee and Macdonald, 2004). It was shown that the proper perinuclear localization of Aub, Krimper, and Maelstrom in the ovaries depends on Vasa ( Lim and Kai, 2007); however, its exact role in nuage remains elusive.
More than a century ago, the chromatoid body (CB), a presumed nuage homologue, was found in the cytoplasm of mammalian male germ cells from early spermatocytes to round spermatids ( Yokota, 2008). The CB consists of an electron-dense material and seems to move around the nuclei in close contact with the nuclear envelope and pore complexes ( Parvinen and Parvinen, 1979; Ventela et al., 2003). Its main function is supposedly related to posttranscriptional mRNA sorting and processing ( Kotaja and Sassone-Corsi, 2007). An essential CB component is the mammalian homologue of Drosophila Vasa protein MVH (mouse Vasa homologue). Many other CB constituents, including principal participants of the piRNA silencing pathway, have been found ( Yokota, 2008; Kotaja et al., 2006; Vagin et al., 2009).
The presence of nuage in Drosophila melanogaster spermatocytes was supported by live-imaging experiments ( Snee and Macdonald, 2004); however, its structure and functions remain poorly explored. It was shown that the repression of repeated Stellate genes in spermatocytes is realized via the piRNA pathway ( Aravin et al., 2001; Vagin et al., 2006; Nishida et al., 2007), and several ovarian nuage proteins (Aub, AGO3, Spindle E, Zucchini, Squash, and Tejas) were found to take part in Stellate silencing ( Aravin et al., 2001; Vagin et al., 2006; Li et al., 2009; Nishida et al., 2007; Pane et al., 2007; Patil and Kai, 2010; Nagao et al., 2010).
Here we present the results of nuage analysis in the testes of D. melanogaster. We demonstrated size heterogeneity of Vasa-containing cytoplasmic particles and found a new large organelle in the primary spermatocytes, the piNG-body, enriched with Vasa, Aub, and AGO3. We detected several other ovarian nuage proteins as components of the piNG-body and determined their mutual arrangement within the body. It was shown recently that the functions and protein interactions of the PIWI proteins depend on symmetrical arginine methylation (SAM) catalyzed by Csul methyltransferase ( Kirino et al., 2009). We found that csul mutations lead to piNG-body disruption accompanied by derepression of testis-specific repeated Stellate genes.
RESULTS
Visualization of the piNG-body and determination of its protein composition
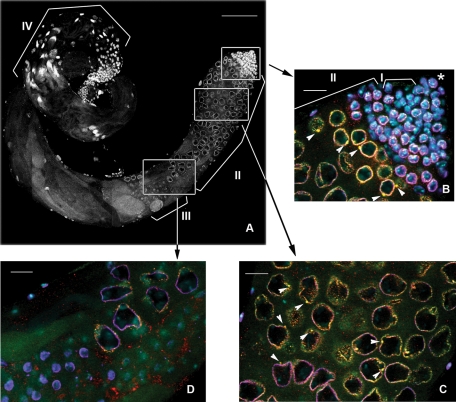
To visualize nuage in the testes of adult flies, we used antibodies against Vasa protein, a well-known component and specific marker of this structure. Immunofluorescence staining and confocal microscopy analysis of the whole-mount testis preparations from wild-type flies clearly demonstrated Vasa-stained nuage granules of at least two types located near the nucleus in the primary spermatocytes. The conventional nuage granules were scattered on the nuclear surface arranged in discontinuous rings around the nuclei on confocal slices. Among these small, dot-like particles ∼0.62 ± 0.13 μm in diameter (n = 229, where n is the number of the measured particles), we observed significantly larger structures of ∼2.38 ± 0.35 μm (n = 70), mainly one per cell ( Figure 1, B and C, and Supplemental Figure S1, A and B). In a spherical approximation, the volume of the larger granules was more than 50 times that of the smaller ones.
FIGURE 1:
Nuage granules of at least two types are detected in the perinuclear area of primary spermatocytes. Spatiotemporal pattern of nuage and piNG-bodies in the testes. (A) A full-size yw testes. White boxes indicate positions of the fragments enlarged in B–D. Scale bar, 100 μm. Brackets in A and B indicate different germinal cells: I, spermatogonial cells; II, spermatocytes; III, round spermatids; IV, elongated spermatids. Asterisk indicates the germinal proliferative center. Testes of yw flies were stained with anti-Vasa (green), anti-Aub (red), and anti-lamin (violet) antibodies; chromatin was stained with DAPI (blue). Colocalization of green (Vasa) and red (Aub) signals yields yellow color. (B) Testis apical tip. Small nuage granules (<1 μm) form discontinuous rings around the nuclei. Large nuage granules, the piNG-bodies (2.38 ± 0.35 μm), are indicated with white arrowheads. Note that not all large nuage granules can be seen on a single confocal slice. Nuage first appears in germinal stem cells and spermatogonial cells, whereas piNG-bodies are formed later in primary spermatocytes at the S2b stage (nuclear diameter of ∼6–10 μm). See also Supplemental Figure S1A for separate channel presentation. (C) The S5 stage (nuclear diameter of ∼16–20 μm). The piNG-bodies are indicated with white arrowheads. See also Supplemental Figure S1B for separate channel presentation. (D) Spermatocytes at the end of the S stage (nuclear diameter of ∼15 μm) and round spermatids (nuclear diameter of ∼4–6 μm). Nuage is preserved at the end of the S stage; however, prominent piNG-bodies are absent. No detectable nuage structure can be seen in round spermatid cells. See also Supplemental Figure S2 for separate channel presentation. Scale bars for B–D, 15 μm.
It was shown previously that Aub and AGO3 proteins colocalize with Vasa in nuage in the ovaries ( Harris and Macdonald, 2001; Snee and Macdonald, 2004; Lim and Kai, 2007; Malone et al., 2009), and their localization is interdependent ( Li et al., 2009). Piwi, the third member of the PIWI subfamily of Argonaute proteins, is known to be expressed only in the germinal proliferative center located at the testis apical tip, which contains somatic hub cells and germ stem cells ( Cox et al., 2000; Saito et al., 2006; Nishida et al., 2007). Because Aub and AGO3 are considered as the main participants of the piRNA machinery in male germinal cells ( Vagin et al., 2006; Nishida et al., 2007; Nagao et al., 2010), we studied their localization in different types of nuage granules in the testes. Using anti-Aub and anti-AGO3 antibodies, we found significant but incomplete colocalization of the Aub (67.9% ± 5.1) or AGO3 (52.0% ± 6.8) signals with the Vasa signal in the smaller perinuclear particles (Supplemental Figure S3). The Vasa signals colocalized with Aub (68.5% ± 9.2) and AGO3 (54.3% ± 13.1), respectively. In the larger structures, the Aub or AGO3 signal always colocalized with the Vasa signal ( Figures 1, B and C, and 2A). Therefore we called them piNG-bodies (piRNA nuage giant bodies).
FIGURE 2:
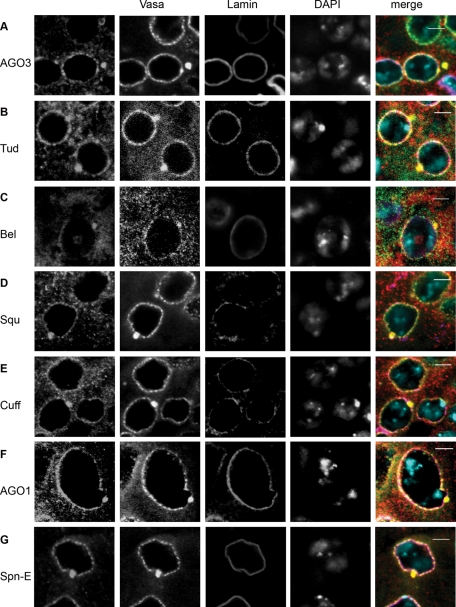
Protein components of the piNG-body and conventional nuage granules: AGO3 (A), Tud (B), Bel (C), Squ (D), Cuff (E), AGO1 (F), and Spn-E (G). Testes of yw flies were immunofluorescently stained with anti-Vasa (green) and anti-lamin (violet) antibodies; chromatin was stained with DAPI (blue). Signals from AGO3, Tud, Bel, Squ, Cuff, AGO1, and Spn-E are shown in red. Scale bars, 5 μm.
Furthermore, we performed immunofluorescence staining using antibodies against other proteins known to be associated with ovarian nuage: Tudor (Tud; Arkov et al., 2006), Spindle E (Spn-E; Lim and Kai, 2007), Squash (Squ; Pane et al., 2007), Cutoff (Cuff; Chen et al., 2007), and Belle (Bel; Johnstone et al., 2005). We found that Tud, Spn-E, Squ, and Cuff were present in nuage of spermatocytes and enriched in the piNG-bodies ( Figure 2, B, D, E, and G). A special pattern of distribution (the cytoplasm, nuage, piNG-bodies, and nucleolus-like structures within the nuclei) was shown for RNA-helicase Bel ( Figure 2C).
We also wondered whether the piNG-body was related to microRNA (miRNA) processing similar to mammalian CB, which contains Dicer ( Kotaja et al., 2006). The miRNA pathway was shown to be Dicer 1–dependent in Drosophila ( Lee et al., 2004); however, we detected no Dicer 1 signal in the piNG-body (Supplemental Figure S4A). Partial nuage-like localization of Dicer 1 was observed in spermatogonia and very early spermatocytes (Supplemental Figure S4C), whereas in mature spermatocytes, the cytoplasm was stained homogeneously (Supplemental Figure S4, A and C). Vasa intronic gene protein (VIG), a RISC component ( Caudy et al., 2002; Gracheva et al., 2009), was revealed preferentially in the nuclei but not in nuage and piNG-bodies (Supplemental Figure S4B). However, AGO1, another Argonaute protein and the principal component of the miRNA silencing complexes, was found in both the regular small nuage granules and piNG-bodies ( Figure 2F).
piNG-body formation is dynamic and associated with the meiotic prophase
The piNG-bodies were detected in primary spermatocytes, whereas the smaller, Vasa-stained nuage perinuclear granules could be seen significantly earlier, in germinal stem cells and spermatogonial cells ( Figure 1, A and B; see also Supplemental Figure S1A), where the Aub staining could also be observed in the perinuclear area, albeit less clearly. During development, spermatocytes undergo significant growth, which includes a remarkable increase in the nuclear size, up to 25 times in volume. This growth is accompanied by active transcription of constitutive and testis-specific genes, as well as by drastic chromatin reorganization and chromatid pairing events ( Cenci et al., 1994; Vazquez et al., 2002). We detected distinct piNG-bodies starting from the S2b stage (as defined by the classification of Cenci et al. [1994]; nuclear diameter of ∼6–10 μm), during the prolonged S3–S4 stages (nuclear diameter of ∼12–15 μm and chromatin separated into three clumps), and up to the S5 stage (nuclear diameter of ∼16–20 μm and completely formed discrete chromosome territories; Figure 1, B and C, and Supplemental Figure S1, A and B). At the end of the S stage (nuclear diameter of ∼15 μm), we still observed the small nuage granules but not the piNG-bodies. After meiosis, no detectable nuage structures were seen in round spermatid cells containing small spherical nuclei (4–6 μm in diameter) with highly compact chromatin ( Figure 1, A and D, and Supplemental Figure S2); however, vestigial homogeneous Vasa and Aub signals were still visible in the cytoplasm. Thus the presence of the regular nuage structures and piNG-bodies was strongly restricted to the prolonged primary spermatocyte stage.
Architecture of the piNG-body
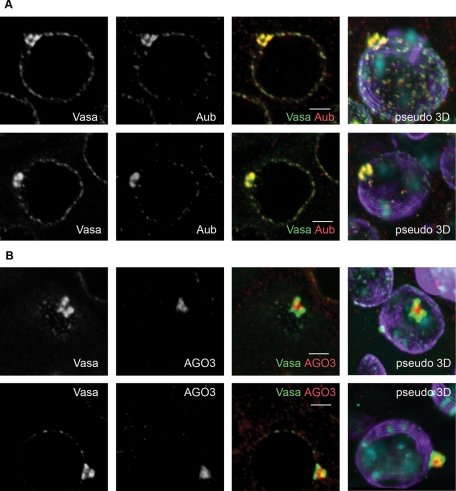
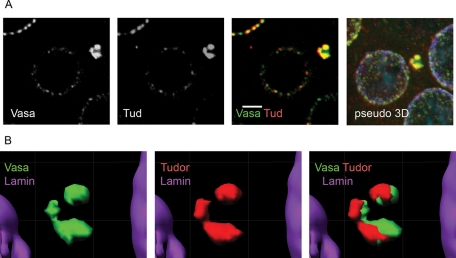
To investigate the piNG-body structure, we scanned the immunostained testes with a high-magnification lens (63×, with 4× zoom). The confocal images were examined using three-dimensional reconstruction to reveal the mutual arrangement of the piNG-body components ( Figure 3). Vasa and Aub proteins showed complete colocalization and always occupied the external parts of the piNG-body. Their signals reflected the existence of substructures of various shapes, from horseshoe to torus or pyramid, with the internal space free from the signals ( Figure 3A). At the same time, AGO3 protein was found inside the body colocalizing with Vasa only at the boundary lines ( Figure 3B and Supplemental Figure S5). Thus Aub and AGO3 occupied separate but closely located zones within the piNG-body. Similar to Aub and Vasa, Tud occupied the peripheral shell, leaving the core free from the staining. However, the Tud signal showed only partial overlap with the Vasa signal ( Figure 4, A and B). Finally, Spn-E, Squ, Cuff, and AGO1 completely colocalized with the Vasa-stained zones at the periphery of the piNG-body (Supplemental Figure S6).
FIGURE 3:
Mutual arrangement of Vasa, Aub, and AGO3 within the piNG-body. (A) A high level of Vasa (green) and Aub (red) colocalization in the piNG-bodies. Two different piNG-bodies stained with Vasa (green) and Aub (red) as well as the corresponding pseudo–three-dimensional (3D) images are shown. Note that both Vasa and Aub occupy the periphery of the piNG-body rather than the internal space. (B) AGO3 protein is found in the piNG-body core and colocalizes with Vasa only along the boundary line regions. Two different piNG-bodies stained with Vasa (green) and AGO3 (red) and the corresponding pseudo-3D images are shown. Testes of yw flies were stained with anti-Vasa (green), anti-Aub (A), or anti-AGO3 (B) (red) and anti-lamin (violet) antibodies; chromatin was stained with DAPI (blue). Colocalization of green and red signals yields yellow color. Scale bars, 3 μm. For more images of AGO3 and Vasa distribution in the piNG-body in the isosurface format see Supplemental Figure S5.
FIGURE 4:
Partial colocalization of Vasa and Tud in the piNG-body. (A) The piNG-body stained with anti-Vasa (green) and anti-Tud (red) antibodies, as well as the corresponding pseudo-3D image, are shown. Testes of yw flies were stained with anti-Vasa (green), anti-Tud (red), and anti-lamin (violet) antibodies; chromatin was stained with DAPI (blue). Colocalization of green and red signals yields yellow color. Scale bars, 3 μm. (B) An isosurface image for the piNG-body presented in A. In spite of the observed significant colocalization of Vasa and Tud signals, nonoverlapping zones are clearly seen.
Destruction of the piNG-body and derepression of Stellate genes
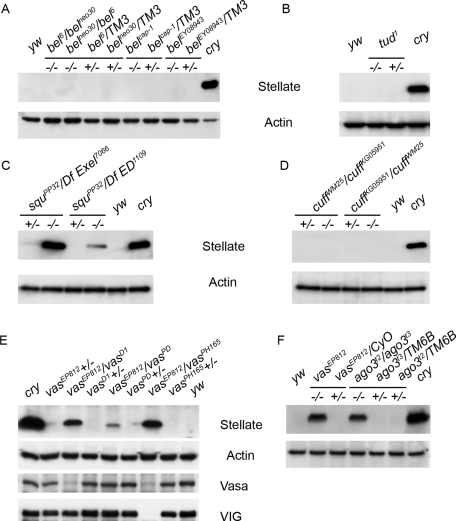
It was shown that transcripts of the X-linked Stellate tandem repeats are the main target of the piRNA silencing pathway in the testes of D. melanogaster ( Nishida et al., 2007; Nagao et al., 2010). In the absence of the Y-linked crystal locus, which encodes the Suppressor of Stellate [Su(Ste)] repeats responsible for the production of antisense piRNAs taking part in the Stellate silencing, high-level Stellate expression occurs in male germinal tissues. Stellate derepression leads to the accumulation of protein crystals and to abnormalities in meiotic chromosome condensation and segregation in spermatocytes, which cause male sterility ( Hardy et al., 1984; Livak, 1984). Transcription of Stellate genes starts in primary spermatocytes ( Aravin et al., 2004) and, according to our data, temporally coincides with the piNG-body formation ( Figure 1B). Loss-of-function aub, ago3, and spn-E mutations lead to the loss of Su(Ste) piRNAs, derepression of Stellate genes, and translation of their transcripts ( Aravin et al., 2001, 2004; Vagin et al., 2006; Li et al., 2009). Among the mutations affecting other piNG-body components (Vasa, Bel, Tud, Squ, and Cuff), only vasa and squ induced strong hyperexpression of Stellate protein ( Figure 5, C, E, and F), whereas bel, tud, and cuff did not lead to Stellate derepression ( Figure 5, A, B, and D). Often mutations in the vasa locus disturb the expression of both vasa and vig genes ( Figure 5E). However, we found that the vasEP812/vasD1 allelic combination was characterized by undisturbed expression of VIG ( Figure 5E) and clearly demonstrated Stellate derepression. These observations indicate that Vasa is responsible for the piRNA pathway disturbances and Stellate derepression in all mutant alleles.
FIGURE 5:
Stellate derepression analysis. Western blots of testis extracts probed with anti-Stellate antibodies are shown. Mutations in bel (A), tud (B), and cuff (D) cause no Stellate derepression, whereas mutations in vasa [vasEP812/vasD1, vasEP812/vasPH165 (E), and vasEP812 (F)], ago3 (F), and squ (C) lead to Stellate hyperexpression. Testes of crystal (cry) and yw flies were used as positive and negative controls, respectively. Anti-actin antibodies were used as a loading control. Anti-Vasa and anti-VIG antibodies were used for the analysis of vasa allelic combinations presented in E. Note trace Stellate derepression in the vasEP812/vasPD hypomorphic combination. vasEP812 mutant presented in F expresses neither VIG nor Vasa proteins ( Vagin et al., 2004; Gracheva et al., 2009).
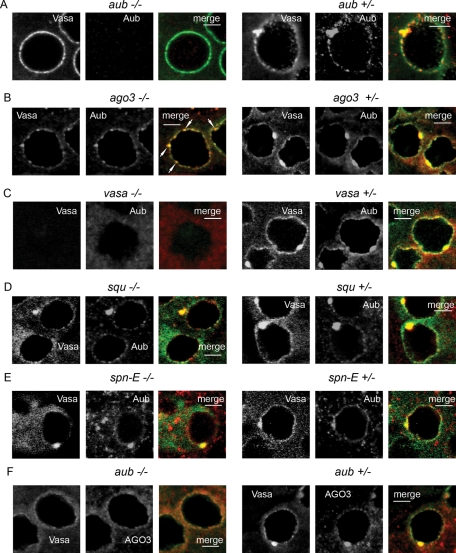
We wondered whether the mutations leading to Stellate derepression also affected piNG-body formation. In the testes of aubHN/QC42 transheterozygous flies, we observed neither the piNG-bodies nor the smaller Aub-stained nuage granules, although Vasa and AGO3 were detectable in nuage ( Figure 6, A and F). In the ago3t2/t3 testes, we also observed Aub- and Vasa-stained small nuage granules together with several discrete bright fragments of Aub/Vasa–stained dense particles (∼1 μm) randomly distributed on the nuclear surface and in the cytoplasm of spermatocytes ( Figure 6B). These particles demonstrated no internal structure typical for the piNG-body and appeared to be its precursors unable to finish the assembly process. Complete piNG-body disappearance was discovered in the vasa testes (vasEP812/D1), where Aub relocation from nuage to the cytoplasm was revealed ( Figure 6C). Thus strong Stellate derepression and piNG-body disassembly occur in case of aub, ago3, or vasa mutations. However, we clearly detected Vasa- and Aub-stained piNG-bodies in spermatocytes in spn-E and squ mutant testes, spn-E616 ( Figure 6E) and squPP32/DfExel7833 ( Figure 6D), where Stellate derepression was shown ( Figure 5C). Thus we suggest the existence of defective piNG-bodies in the case of certain mutations.
FIGURE 6:
Mutational analysis of the piNG-body. In all experiments, the testes were stained with anti-Vasa (green) and anti-Aub (red) antibodies, except for F, in which they were stained with anti-AGO3 (red) antibodies. Scale bars, 5 μm. (A, F) The aub mutation effect on the piNG-body formation. In the aubHN/aubQC42 testes, small AGO3- and Vasa-stained nuage granules are visible, but no piNG-body formation is observed. (B) The ago3 mutation effect on the piNG-body formation. No normal-sized piNG-bodies are visible; however, up to several Aub/Vasa–stained particles of ∼1 μm size are detected on the nuclear surface or in the cytoplasm of spermatocytes (white arrows). (C) No nuage is found in the testes of vasEP812/vasD1 flies (the same results were obtained with vasEP812/vasPH165 and vasEP812 mutants). (D) piNG-bodies and conventional nuage granules are distinctly seen in the squPP32/DfExel7066 transheterozygous flies. (E) piNG-bodies and conventional nuage granules are preserved in the spn-E616 homozygous flies.
Deficiency of several other piNG-body components caused no Stellate derepression ( Figure 5, A, B, and D). Analysis of bel6/belms(3)neo30 flies revealed severe testis atrophy and complete loss of germ cells prior to the spermatocyte growth stage (when Stellate transcription starts and piNG-bodies emerge in wild type; Supplemental Figure S7A). We investigated the piNG-body and conventional nuage maintenance in case of strong tud1 mutation and found no disturbance (Supplemental Figure S7B). Thereby, Tud is not crucial for the piNG-body formation. We also observed piNG-body and nuage preservation in cuffWM25/cuffKG05951 transheterozygous testes (Figure S7C).
Capsuleen affects the piNG-body assembly
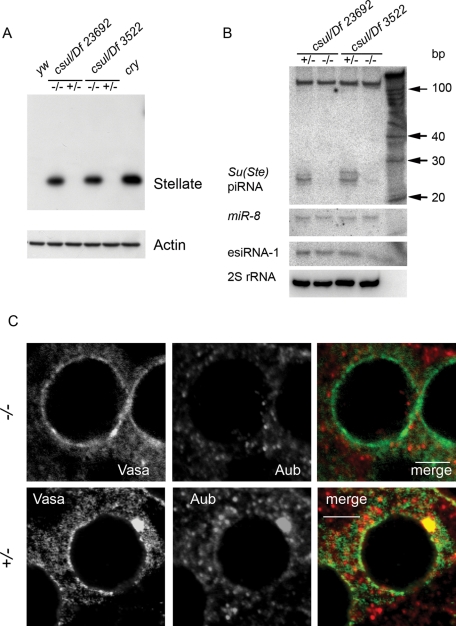
Loss-of-function capsuleen (csul) mutations lead to transposon derepression in the ovaries ( Kirino et al., 2009) and male sterility ( Gonsalvez et al., 2006). Csul, or Dart5, is an arginine methyltransferase responsible for SAM of Piwi, Aub, AGO3, and Vasa ( Kirino et al., 2009, 2010a) that has been shown to be essential for piRNA pathway functioning in the ovaries ( Kirino et al., 2009). We analyzed Stellate silencing and piNG-body formation in the testes of csulRM50/Df(2R)Jp7 and csulRM50/Df 23692 transheterozygous males and found strong Stellate derepression ( Figure 7A) along with disappearance of the piNG-body in spermatocytes ( Figure 7C). Vasa-containing small nuage granules were clearly detectable in the csul mutant testes, whereas the Aub signals were randomly distributed in the cytoplasm of spermatocytes and did not colocalize with the Vasa-stained perinuclear granular material. This finding demonstrates that piNG-body assembly depends on SAM-mediated interactions. In the csul testes, we also found a dramatic loss of antisense Su(Ste) piRNAs, whereas miRNA and endoribonuclease-prepared short interfering RNA (esiRNA) levels remained unaffected ( Figure 7B). Thus disassembly of the piNG-body was concomitant with the loss of SAM modification of Aub and AGO3 and with the piRNA pathway disruption.
FIGURE 7:
Mutation in csul prevents piNG-body assembly. (A) Western blots of testis extracts of csul mutants probed with anti-Stellate antibodies. Stellate hyperexpression is strong in both csulRM50/Df(2R)Jp7 and csulRM50/Df 23692 transheterozygous allelic combinations. Testes of cry and yw flies were used as positive and negative controls, respectively. Anti-actin antibodies were used as a loading control. (B) Northern blot analysis of small RNAs from the csul testes (see Materials and Methods for details). Antisense Su(Ste) piRNAs but neither miR-8 miRNA nor esiRNA-1 was absent owing to csul mutations. 2S rRNA was used as a loading control. (C) Immunofluorescence staining of the testes from csulRM50/Df(2R)Jp7 transheterozygous and heterozygous flies. In csul transheterozygous flies (−/−), no piNG-body formation can be observed, but Vasa-stained small nuage granules remain detectable. The Aub signals are randomly distributed in the cytoplasm of spermatocytes and do not colocalize with the Vasa-stained nuage material. Testes were stained with anti-Vasa (green) and anti-Aub (red) antibodies. Scale bars, 5 μm.
DISCUSSION
Here we studied the localization and compartmentalization of the piRNA pathway components in nuage of Drosophila male germinal tissues, the structure and protein composition of which have been poorly investigated to date. Using the immunofluorescence approach, we identified two types of granules comprising nuage. Along with the conventional small nuage particles evenly scattered on the nuclear surface in primary spermatocytes, we found a novel large granular organelle, one per cell. This organelle contained Vasa, Aub, AGO3, Tud, Spn-E, Bel, Squ, Cuff, and AGO1 proteins ( Figures 1 and 2). Because most of the identified components are known to be participants of the piRNA pathway, we called the organelle the piNG-body. piNG-bodies appeared in spermatocytes at the S2b stage and were recognizable until the S5 stage ( Figure 1, B and C, and Supplemental Figure S1, A and B). Drosophila spermatocytes are characterized by active transcription, which drops dramatically after meiotic divisions ( White-Cooper, 2010). We detected neither piNG-bodies nor other nuage granules in round spermatids ( Figure 1D and Supplemental Figure S2) and thus supposed that piNG-body formation was strongly associated with the period of active transcription in primary spermatocytes, where defects in the piRNA system caused derepression of Stellate genes and transposons ( Aravin et al., 2004; Nagao et al., 2010).
The mammalian CB is another large granular organelle involved in RNA processing in male germinal tissues. Although it appears in late pachytene spermatocytes and exists through the round spermatid stage, its formation, like that of to the piNG-body, coincides with the strong wave of transcription ( Yokota, 2008; Kotaja and Sassone-Corsi, 2007). CB is considered a nuage derivative. It contains MVH, the mammalian homologue of Drosophila Vasa protein, as well as MIWI, a piRNA pathway component, and Dicer, the nuclease necessary for the miRNAs processing ( Kotaja et al., 2006). We did not detect Dicer 1 or VIG, members of the Drosophila miRNA silencing complexes ( Lee et al., 2004; Caudy et al., 2002; Gracheva et al., 2009), within the piNG-body (Supplemental Figure S4, A and B); however, it contained AGO1 and Bel ( Figure 2, C and F). AGO1 is another principal participant of the miRNA silencing machinery, whereas Bel, a putative DEAD-box RNA-helicase, has been shown to associate with VIG, FMR, AGO1, and AGO2 and is considered to be a component of the miRNA pathway ( Zhou et al., 2008). Preferentially uniform cytoplasmic distribution of Dicer 1 in spermatocytes (Supplemental Figure S4, A and C) was similar to that observed in the nurse cells of Drosophila ovaries ( Findley et al., 2003). Thus we suggest that the piNG-body and CB can be functionally related.
The piNG-body volume is about 50 times that of a conventional nuage granule, and we succeeded in revealing its internal structure. We suggest that the observed relative positions of Aub and AGO3 proteins within the piNG-body ( Figure 3 and Supplemental Figure S5) are essential for correct functioning of the piRNA processing. Aub, which plays a role complementary to AGO3 in silencing of transposons and other repetitive selfish elements, is located at the periphery of the piNG-body ( Figure 3A), whereas AGO3 is found in the core ( Figure 3B and Supplemental Figure S5). We suppose that the observed piNG-body architecture reflects the existence of separate functions of the sense and antisense piRISCs, keeping both parts in close contact necessary for a rapid exchange of piRNA intermediates. Testis transposon piRNAs show clear signatures of the ping-pong cycle ( Nagao et al., 2010). Although this mechanism appears to be nonprevalent in Stellate silencing, both Aub and AGO3 are strongly required for the accumulation of Su(Ste) piRNAs in the testes ( Vagin et al., 2006; Li et al., 2009; Nagao et al., 2010). The concentration and compartmentalization of macromolecular complexes are believed to be essential for efficient kinetics of RNA sorting and processing ( Kurland et al., 2006).
Recent papers described two types of the nuage granules in mouse fetal gonocytes ( Aravin et al., 2009; Shoji et al., 2009). The first type, pi-bodies, contain MILI and TDRD1 proteins, whereas the second type, piP-bodies, harbor MIWI2, TDRD9, and Maelstrom, as well as known P-body proteins (DCP1, XRN1, and GW182). PIWI family proteins MILI and MIWI2 play complementary roles in transposon silencing in mice embryos, being involved in the piRNA amplification cycle ( Aravin et al., 2008). The pi- and piP-bodies are closely located in the cytoplasm of gonocytes but exist as separate particles preserving functional specialization and compartmentalization of the piRNA pathway modules. We demonstrated that a similar separation of piRISC complexes in flies could be achieved within a single organelle, the piNG-body, due to a well-ordered disposition of its components.
We observed that aub, ago3, and vasa mutations affecting Stellate silencing also impaired piNG-body formation and caused its disruption ( Figure 6, A–C and F). In contrast, the deficiency of Squ, a putative nuclease believed to be responsible for piRNA biogenesis ( Pane et al., 2007), led to Stellate hyperexpression ( Figure 5C) but caused no visible disturbances in the Vasa- and Aub-stained piNG-bodies ( Figure 6D). spn-E mutation also did not lead to visible loss of the Vasa- and Aub-stained piNG-bodies ( Figure 6E). Although the process of piNG-body formation is not understood mechanistically, we can assume a hierarchic order of protein recruitment: Vasa and Aub can be considered as basic architectural components, the incorporation of which is independent of Squ and Spn-E. Several piNG-body components (Vasa, Aub, and AGO3) are responsible for both general piNG-body assembly and Stellate silencing, whereas others (Squ and Spn-E) may determine the correct internal structure of the body, and their deficiency may result in the assembly of a defective piNG-body lacking some essential functions.
It is known that SAM modification is responsible for Aub and AGO3 recognition by Tud ( Kirino et al., 2010b; Nishida et al., 2009), and disruption of SAM modification caused Stellate derepression ( Figure 7A). Thus the absence of Stellate hyperexpression in the tud testes was unexpected. Here we found that in the piNG-body, the Tud-stained zones significantly overlapped with Vasa and Aub at the periphery rather than with AGO3 in the core ( Figure 4). However, we showed that tud1 mutation did not lead to Stellate derepression ( Figure 5B) and piNG-body disappearance (Figure S7B). In Drosophila, 25 Tudor domain–containing proteins have been described ( Siomi et al., 2010), and some of them, such as Krimper ( Lim and Kai, 2007) and Tejas ( Patil and Kai, 2010), were shown to be involved in the piRNA pathway in the ovaries and testes. It is possible that yet-unidentified proteins carrying Tudor domains also interact with the PIWI proteins in the piNG-body and their functions can be interchangeable.
We demonstrated that SAM provided by Csul arginine methyltransferase was essential for the assembly of the piNG-body but not of the conventional small nuage granules. Strong Stellate derepression associated with the loss of Su(Ste) piRNAs was observed in the csul testes ( Figure 7, A and B) lacking the piNG-body ( Figure 7C). Because SAM is required for at least Aub and AGO3 functionality ( Kirino et al., 2009, 2010b; Nishida et al., 2009), our observations highlight the functional involvement of the piNG-body in the piRNA-mediated silencing in the testes. Similar to ovarian nuage ( Anne et al., 2007), testis nuage localization of Vasa appears to be SAM independent ( Figure 7C).
Here we detected a novel spermatocyte-specific nuage-associated organelle, the piNG-body, significantly larger than the conventional nuage granule. We found that the piNG-body contained the PIWI-subfamily proteins Aub and AGO3 and demonstrated that the body disruption was accompanied by the derepression of Stellate repeats. We also showed SAM modifications of Aub and AGO3 to be essential for both piNG-body assembly and piRNA pathway functions. We suggest that the internal structure of the piNG-body provides effective compartmentalization and functioning of the piRNA pathway components, as well as cross-talk between the piRNA and miRNA pathways, since AGO1 was shown to be a component of the piNG-body.
MATERIALS AND METHODS
Fly stocks
Germinal tissues of adult D. melanogaster males raised at 23°C were used. The Df(1)w67c23(2)y (designated as yw) and Canton S lines were used as wild types in immunostaining experiments. The yw67c23;cry1BS, Yy+;+/+;P{pCaSpeR-6Ste-lacZ} line (designated as cry) carrying a deletion of the crystal locus on the Y chromosome was used as a positive control for the analysis of Stellate derepression. The aubHN/aubQC42 and ago3t2/ago3t3 transheterozygous allelic combinations and spn-E616 homozygous flies were used for the analysis of mutation effects on the piNG-body formation. The vasEP812, vasEP812/vasD1, vasEP812/vasPH165, squPP32/DfExel7066, and squPP32/Df(2L squ)ED1109 flies were used for the analysis of Stellate derepression and immunostaining. The cuffWM25/cuffKG05951 and cuffKG05951/cuffWM25 transheterozygous allelic combinations (opposite-direction crosses; the first indicated parent is the mother) and the tud1 homozygous allele also were used for Western blot analysis of Stellate derepression and immunostaining. The belms(3)neo30/bel6 and bel6/belms(3)neo30 transheterozygous allelic combinations, as well as belcap-1 and belEY08943 homozygous alleles, were used for the same purpose. The csulRM50/Df(2R)Jp7 and csulRM50/Df 23692 flies were used for Western and Northern blot analyses and immunostaining.
Immunofluorescence staining and confocal microscopy
Testes of 0- to 2-d-old adult male flies were dissected in phosphate-buffered saline (PBS) at 4°C, washed with PBT (1× PBS, 0.1% Tween 20), and fixed in 3.7% formaldehyde and PBT for 30 min at room temperature. All of the following procedures were carried out as described previously ( Egorova et al., 2009). Staining was detected by laser scanning confocal microscopy using a Carl Zeiss LSM 510 META machine (Carl Zeiss, Jena, Germany) in multichannel mode with 63× oil objective (numerical aperture 1.4); frame size 1024 × 1024 pixels; scan speed 7, with four repetitions. All images, except for magnified ones (4–5× digital zoom) recorded with the z-resolution of 0.5 μm, were taken with the z-resolution of 1 μm. The obtained pictures were imported into Imaris 5.0.1 (Bitplane, Zurich, Switzerland) for subsequent processing. Colocalization was calculated using Coloc software (Imaris 5.0.1), Advanced Coloc method.
Antibodies
The following antibodies were used for immunofluorescence staining: a mix of murine monoclonal anti–lamin Dm0 ADL67.10 and ADL84 antibodies (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), 1:500; rabbit polyclonal anti-lamin antibodies ( Osouda et al., 2005), 1:500; rabbit polyclonal anti-Aub antibodies ( Brennecke et al., 2007), 1:200; murine monoclonal anti–Aub 4D10 antibody ( Nishida et al., 2007), 1:100; rabbit polyclonal anti-AGO3 antibodies ( Brennecke et al., 2007), 1:300; rat monoclonal anti-Vasa antibody (Developmental Studies Hybridoma Bank), 1:300; rabbit polyclonal anti-Bel antibodies ( Johnstone et al., 2005), 1:250; rabbit polyclonal anti-Tud antibodies ( Arkov et al., 2006), 1:300; rabbit polyclonal anti–VIG CSH1801 antibodies ( Gracheva et al., 2009), 1:500; rabbit polyclonal anti–Dicer 1 ab4735 antibodies (Abcam, Cambridge, MA) 1:300; murine monoclonal anti-Squ antibody (Developmental Studies Hybridoma Bank), 1:50; murine monoclonal anti-Cuff antibody (Developmental Studies Hybridoma Bank), 1:50; murine monoclonal anti–AGO1 1B8 antibody ( Miyoshi et al., 2005), 1:100; murine monoclonal anti–Spn-E 3D8-E4D4 antibody ( Nishida et al., 2009), 1:100.
Alexa Fluor 488 goat anti–rat IgG, Alexa Fluor 546 goat anti–rabbit IgG, and Alexa Fluor 647 goat anti–mouse IgG (Invitrogen, Carlsbad, CA) were used as secondary reagents at a dilution of 1:1000. DAPI (4′,6-diamidino-2-phenylindole) was used for chromatin staining.
For Western blot analysis, the following antibodies were used: murine polyclonal anti-Stellate antibodies described in Egorova et al. (2009), 1:3000; murine monoclonal anti–β-actin ab8224 antibody (Abcam), 1:800. Alkaline phosphatase–conjugated anti–mouse antibodies (Sigma-Aldrich, St. Louis, MO) were used as a secondary reagent at a dilution of 1:20,000. Samples were resolved by SDS–PAGE and blotted onto Immobilon-P PVDF membrane (Sigma-Aldrich). Blots were developed using the Immun-Star AP detection system (Bio-Rad Laboratories, Hercules, CA) in accordance with the recommendations of the manufacturer.
Northern blot analysis
A total of 5 μg of RNA isolated from testes by TRIzol reagent (Invitrogen) was resolved by 20% denaturing PAGE. After that, the RNA was transferred onto the Hybond N+ membrane (Amersham-Pharmacia Biotech, GE Healthcare Bio-Sciences, Piscataway, NJ) and cross-linked by UV radiation (Stratagene, Santa Clara, CA). The subsequent prehybridization and hybridization were performed in Church buffer at 37°C for 1 h and overnight, respectively. For hybridization, 10 pmol of DNA oligonucleotides radiolabeled with [γ-32P]ATP by T4 polynucleotide kinase (New England BioLabs, Ipswich, MA) was used. The following probes were used for the detection of Su(Ste) piRNAs, 5′-GGGCTTGTTCTACGACGATGA-3′; dme-mir-8 miRNA, 5′-GACATCTTTACCTGACAGTATTA-3′; endo-siRNA-1, 5′-GGAGCGAACTTGTTGGAGTCAA-3′; and 2S rRNA, 5′-TACAACCCTCAACCATATGTAGTCCAAGCA-3′. After hybridization, the blots were washed twice with 2× 150 mM sodium chloride, 15 mM sodium citrate, and 0.05% SDS at 37°C and analyzed by the STORM PhosphorImager system (Amersham-Pharmacia).
Supplementary Material
Acknowledgments
We thank G. Hannon, A. Aravin, P. Lasko, A. Arkov, M. Siomi, and T. Kai for kindly providing antibodies and P. Zamore, J. Anne, and T. Schüpbach for fly stocks. We also thank the Bloomington Drosophila Stock Center, Indiana University, Bloomington, IN, for fly stocks. We thank A. Aravin for a helpful discussion. This work was supported by the Molecular and Cellular Biology Program, Russian Academy of Sciences, and the Russian Foundation for Basic Research (Project 10-04-00535-a). K.S.E. was supported by a grant from the Dynasty Foundation for Young Scientists.
Abbreviations used:
- DAPI
4′,6-diamidino-2-phenylindole
- PBS
phosphate-buffered saline
- piRNA
PIWI-interacting RNA
- RISC
RNA-induced silencing complex
- SAM
symmetrical arginine methylation
- Su(Ste)
Suppressor of Stellate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0168) on July 20, 2011.
REFERENCES
- Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuléen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germ line. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van deer Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov AL, Wang, Ju-YuS, Ramos A, Lehmann R. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–4062. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci G, Bonnacorsi S, Pisano C, Verni F, Gatti M. Chromatin and microtubule organization during premeiotic, meiotic and early post meiotic stages of Drosophila melanogaster spermatogenesis. J Cell Sci. 1994;107:3521–3534. doi: 10.1242/jcs.107.12.3521. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schüpbach T. cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Egorova KS, Olenkina OM, Kibanov MV, Kalmykova AI, Gvozdev VA, Olenina LV. Genetically derepressed nucleoplasmic Stellate protein in spermatocytes of D. melanogaster interacts with the catalytic subunit of protein kinase 2 and carries histone-like lysine-methylated mark. J Mol Biol. 2009;389:895–906. doi: 10.1016/j.jmb.2009.04.064. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol. 2006;16:1077–1089. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- Gracheva E, Dus M, Elgin SC. Drosophila RISC component VIG and its homolog Vig2 impact heterochromatin formation. PLoS One. 2009;4:e6182. doi: 10.1371/journal.pone.0006182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′-end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Hardy RW, Lindsley DL, Livak KJ, Lewis B, Siversten AV, Joslyn GL, Edwards J, Bonaccorsi S. Cytogenetic analysis of a segment of the Y chromosome of Drosophila melanogaster. Genetics. 1984;107:591–610. doi: 10.1093/genetics/107.4.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and is related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Deuring R, Bock R, Linder P, Fuller MT, Lasko P. Belle is a Drosophila DEAD-box protein required for viability and in the germline. Dev Biol. 2005;277:92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT is required for Ago3 and Aub stability. Nat Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z. Arginine methylation of Vasa protein is conserved across phyla. J Biol Chem. 2010a;285:8148–8154. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010b;16:70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell specific RNA-processing centre. Nat Rev Mol Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci USA. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Cillins LJ, Penny D. Genomics and the irreducible nature of eukaryotic cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li C, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:1–13. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. A unique germline organelle, Nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics. 1984;107:611–634. doi: 10.1093/genetics/107.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA. 2010;16:2503–2515. doi: 10.1261/rna.2270710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine-piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osouda S, Nakamura Y, de Saint Phalle B, McConnell M, Horigome T, Sugiyama S, Fisher PA, Furukawa K. Null mutants of Drosophila B-type lamin Dm(0) show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev Biol. 2005;284:219–232. doi: 10.1016/j.ydbio.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvinen M, Parvinen LM. Active movements of the chromatoid body. A possible transport mechanism for haploid gene products. J Cell Biol. 1979;80:621–628. doi: 10.1083/jcb.80.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM. Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci. 2004;117:2109–2120. doi: 10.1242/jcs.01059. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–58. [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Belmont AS, Sedat JW. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr Biol. 2002;12:1473–1483. doi: 10.1016/s0960-9822(02)01090-4. [DOI] [PubMed] [Google Scholar]
- Ventela S, Toppari J, Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol Biol Cell. 2003;14:2768–2780. doi: 10.1091/mbc.E02-10-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction. 2010;139:11–21. doi: 10.1530/REP-09-0083. [DOI] [PubMed] [Google Scholar]
- Yokota S. Historical survey on chromatoid body research. Acta Histochem Cytochem. 2008;41:65–82. doi: 10.1267/ahc.08010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of Argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.