The microtubule motor Eg5 is well known for its functions during mitosis. It is shown that during interphase, Eg5 associates with ribosomes and is required for efficient protein synthesis.
Abstract
The kinesin-related molecular motor Eg5 plays roles in cell division, promoting spindle assembly. We show that during interphase Eg5 is associated with ribosomes and is required for optimal nascent polypeptide synthesis. When Eg5 was inhibited, ribosomes no longer bound to microtubules in vitro, ribosome transit rates slowed, and polysomes accumulated in intact cells, suggesting defects in elongation or termination during polypeptide synthesis. These results demonstrate that the molecular motor Eg5 associates with ribosomes and enhances the efficiency of translation.
INTRODUCTION
Even though translation can be reproduced in cell-free extracts, it is likely that additional regulatory and structural mechanisms influence polypeptide synthesis in intact cells. One mechanism that might serve to enhance translation in cells is the association of the translational machinery with the linear cytoskeletal filaments of the cytoplasm. These structural elements may support directionality, cellular localization, or efficiency of translation compared with cell-free systems. An association of various translational components with the cytoskeleton was observed previously; these components include mRNA and polyribosomes, as well as various translation initiation and elongation factors ( Jansen, 1999). In addition, ribosomes and polysomes have also been shown to functionally associate with both actin and microtubules in many eukaryotic cell types ( Lenk et al., 1977; Fulton et al., 1980; Moon et al., 1983; Ramaekers et al., 1983; Hesketh and Pryme, 1988). In sea urchin embryos, an association of ribosomes with microtubules occurs via a short stalk ( Suprenant et al., 1989) reminiscent of cargo association to microtubules by molecular motors. Consistent with this interpretation, ribosomes were shown to migrate along microtubules in hemipteran oocytes ( Macgregor and Stebbings, 1970), the kinesin motor KIF4 is required for anterograde transport of ribosomes in rat neurons ( Bisbal et al., 2009), and KIF3A is found in periaxoplasmic ribosomal plaques ( Sotelo-Silveira et al., 2004). In addition, both kinesin and dynein molecular motors are known to function in mRNA transport and localization ( Suprenant, 1993; Jansen, 1999) and participate in the formation of mRNA-silenced stress granules and P-bodies ( Loschi et al., 2009). However, a role for molecular motors in translation has not yet been demonstrated.
Mitotic motor Eg5 (kinesin 5, KIF11) is a plus-end directed homotetrameric microtubule motor and is best known for its functions during mitosis in microtubule cross-linking, antiparallel microtubule sliding, bipolar spindle formation, and, in nonmammalian systems, microtubule poleward flux ( Sawin et al., 1992; Kashina et al., 1997; Sharp et al., 1999; Miyamoto et al., 2004; Shirasu-Hiza et al., 2004; Kapitein et al., 2005; van den Wildenberg et al., 2008). However, recently, in neurons, Eg5 was found to be expressed in postmitotic neurons and to function during interphase ( Ferhat et al., 1998). In developing neurons Eg5 acts as an inhibitory factor of axonal growth, as in its absence axons grow longer, more rapidly, retract less often, and grow past signaling cues that usually redirect growth in another direction ( Haque et al., 2004; Myers and Baas, 2007; Nadar et al., 2008). In this work, we investigate whether Eg5 might also function in other cellular processes during interphase in mammalian cells.
We used both knockdown and small-molecule inhibition of Eg5 in mammalian cells to investigate the role of Eg5 in protein synthesis. First, 35S methionine and cysteine (Met/Cys) incorporation assays were used to identify how the loss of Eg5 affects global polypeptide synthesis. Second, through sucrose gradient sedimentation and immunoprecipitation assays we determined whether Eg5 associates with ribosomes and whether Eg5 is required for the association of 80S ribosomes with microtubules. Finally, sucrose gradient fractionations and polysome profiles were completed to identify the steps of translation in which Eg5 functions. The results from this study demonstrate that Eg5 has an additional role in the cell during interphase, increasing the efficiency of protein translation.
RESULTS
Eg5 is essential for normal levels of polypeptide synthesis
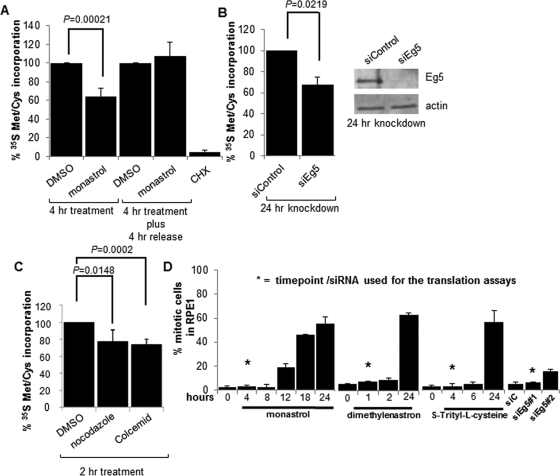
To investigate a role for the Eg5 motor in translation, protein synthesis was assayed by measuring 35S Met/Cys incorporation into nascent polypeptides. RPE1 cells (human retinal pigmented epithelial cells immortalized with human telomerase reverse transcriptase [hTERT]) were exposed to an acute 4-h treatment of monastrol, a specific inhibitor of the ATPase activity of Eg5 ( Mayer et al., 1999; Maliga et al., 2002), prior to a 30-min incubation with 35S Met/Cys. Labeled cells were lysed, and whole-cell lysates (WCLs) were trichloroacetic acid (TCA) precipitated and subjected to scintillation counting. 35S Met/Cys incorporation assays revealed a significant ∼40% reduction in protein synthesis after Eg5 inhibition ( Figure 1A). When monastrol was washed away and replaced with fresh medium, translation returned to normal levels, showing that the inhibition of protein synthesis was specific, reversible, and not due to cell death ( Figure 1A). Cycloheximide (CHX), an inhibitor of protein synthesis, was used as a positive control for this assay. To confirm the reduction in translation after Eg5 inhibition, Eg5 was knocked down by a small interfering RNA (siRNA) for 24 h, and a similar ∼40% reduction in protein synthesis was observed ( Figure 1B). In addition, to demonstrate that the decrease in protein synthesis was due to Eg5 inhibition and not due to off-target effects, the 35S Met/Cys incorporation assay was repeated after a 24-h knockdown of Eg5, a 4-h monastrol treatment, or simultaneous treatment of both. In each of these treatments, a significant ∼40% decrease in protein synthesis was observed (Supplemental Figure S1A). The fact that the phenotypes after simultaneous treatment of Eg5 knockdown and monastrol treatment were not additive suggests that the Eg5 siRNA and monastrol were inhibiting the same target. Furthermore, the reduction in translation after Eg5 inhibition was not cell line specific, as a decrease in protein synthesis was observed in multiple cell lines (RPE1, HFF-hTERT, NIH-3T3, and U2OS), demonstrating the generality of this phenotype (Supplemental Figure S1B).
FIGURE 1:
Eg5 inhibition or microtubule disruption causes a defect in protein synthesis. Quantitation of 35S Met/Cys incorporation assays in WCLs from RPE1 cells as indicated (A) 4 h after a 130 μM monastrol or DMSO (solvent control) treatment or a 4-h monastrol or DMSO treatment followed by a 4-h release into the same medium without monastrol or DMSO, (B) 24 h before or after Eg5 knockdown (siEg5#1), or (C) a 2-h treatment with 12 μM nocodazole or 0.002 mg/ml Colcemid (microtubule disruptors) or the DMSO solvent control. CHX treatment in A was used as a positive control. In B, a representative immunoblot is shown to demonstrate Eg5 was knocked down 24 h after the addition of siEg5#1 siRNA; actin is used as the loading control. Results are shown as means ± SD and are representative of at least three independent experiments; p values are derived from Student's t test (null hypothesis). (D) Quantification of mitotic indices by DAPI staining after a 130 μM monastrol treatment, 3 μM dimethylenastron, or 1.5 μM S-trityl-l-cysteine for the indicated times or Eg5 knockdown by siEg5#1 or siEg5#2 siRNA for 24 h. DMSO was used as the control for the small-molecule inhibitor treatments, and siControl was used as the control for the knockdown experiments. Asterisks represent the time point or the siRNA at which all translation experiments were completed (except where indicated). There is no significant increase in mitotic indices between the time points labeled with asterisks and controls (0 h or siControl; p > 0.1). Longer treatments of Eg5 inhibition are shown and did lead to a mitotic arrest, but these times were not used in the translation experiments; instead, they are shown to demonstrate that the Eg5 inhibitors were active at the concentration used. In each experiment, a minimum of 300 cells were counted and at least three independent experiments were completed. Results are shown as means ± SD.
To further demonstrate the specificity of Eg5 inhibition toward protein synthesis, two additional small-molecule inhibitors of Eg5—dimethylenastron and S-trityl-l-cysteine—a structurally different inhibitor from monastrol, were used ( DeBonis et al., 2004; Gartner et al., 2005). RPE1 cells treated with dimethylenastron for as briefly as 1 h, or with S-trityl-l-cysteine for 4 h, followed by a 35S Met/Cys incorporation assay, again revealed an ∼40% decrease in protein synthesis (Supplemental Figure S1C), similar to the effects on translation by monastrol or siRNA to Eg5. Therefore these data indicate that Eg5 and its ATPase activity are required for efficient protein synthesis, although these results alone do not distinguish whether Eg5 directly or indirectly effects polypeptide synthesis.
Protein synthesis occurs in both the cytosolic and the endoplasmic reticulum (ER)–associated compartments. Therefore we investigated whether Eg5 may be preferentially required for protein synthesis associated with either of these compartments. RPE1 cells were treated either with or without monastrol prior to a 35S Met/Cys incorporation assay. RPE1 cells were then separated into cytosolic or ER fractions, and nascent protein synthesis was measured. Confirmation of cell fractionation into cytosolic and ER fractions was completed by immunoblot analysis using calnexin as an ER marker, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a cytosolic marker, and actin as a loading control (Supplemental Figure S2A). Four hours after monastrol treatment, a significant ∼60% reduction in protein synthesis was observed for both the cytosolic and ER fractions (Supplemental Figure S2A). It should be noted that this assay does not address whether Eg5 is specifically required for synthesis of cytosolic or secretory proteins, as translation of cytosolic and secretory proteins could occur in either compartment ( Lerner et al., 2003). To further confirm the specificity of Eg5 inhibition on protein synthesis, the assay was repeated with two different siRNA oligonucleotides (siEg5#1 and siEg5#2) directed against Eg5 at nonoverlapping regions of the nucleotide sequence. Twenty-four hours after Eg5 knockdown, a 30–40% decrease in protein synthesis occurring in both compartments was observed (Supplemental Figure S2, B and C). Thus these assays suggest that Eg5 is required for translation of proteins associated with both compartments. In summary, a total of five different specific agents of Eg5 inhibition were used in these 35S Met/Cys incorporation assays, and each was observed to cause a similar and significant reduction in protein synthesis of the cell ( Figure 1 and Supplemental Figures S1 and S2).
As kinesin motors typically use microtubules to complete their functions, we investigated whether microtubules were important for protein synthesis in mammalian cells, similar to their importance in translation in other model systems ( Bernstam et al., 1980). RPE1 cells were treated with nocodazole or Colcemid for 2 h to depolymerize microtubules prior to completing a 35S Met/Cys incorporation assay. After microtubule depolymerization, an ∼20–25% reduction in protein synthesis was observed in WCLs ( Figure 1C; also see Discussion). These data are consistent with a general requirement for microtubule-based motors in translation.
Decrease in translation after Eg5 inhibition is not due to mitotic arrest or cell death
Translation is inhibited when cells enter mitosis ( Sivan et al., 2007), and prolonged inhibition of Eg5 can cause mitotic arrest ( Mayer et al., 1999; Kapoor et al., 2000). Therefore we took care to be certain that the translational phenotype observed after loss of Eg5 was not due to mitotic arrest. In all experiments using Eg5 inhibitors, short, 1- to 4-h exposures, depending on the inhibitor, were used that did not significantly increase the mitotic index. The mitotic index was quantitated by three different assays—4′,6-diamidino-2-phenylindole (DAPI) staining, phosphorylated histone-H3 immunofluorescence, and flow cytometry. Mitotic cells remained at <5% of the total cell population in both the control and the acutely inhibited populations by fluorescence microscopy ( Figure 1D and Supplemental Figure S3A; conditions used in the translation assays are marked with an asterisk). In addition, the two Eg5 siRNAs produced little (siEg5#2) or no increase (siEg5#1) in RPE1 mitotic cells at the 24-h time point chosen for the translational analysis ( Figure 1D; for all siRNA experiments, except Figure 1D and Supplemental Figure S2C, siEg5#1 was used, which gave no change in the mitotic index at 24 h). Finally, cell cycle analysis was conducted by flow cytometry; 24 h after monastrol treatment an ∼62% increase of cells in G2/M was observed (Supplemental Figure S3B). However, after a 4-h monastrol treatment—the time at which we conducted the translation assays—only a 5.5% increase of cells in G2/M was observed. Our interpretation of these controls is that the 0–5.5% increase of cells in G2 or M phase cannot explain the consistent ∼40% reduction in protein synthesis observed when Eg5 is inhibited. Therefore these data demonstrate that the translational phenotypes described in this article after loss of Eg5 were not a result of mitotic arrest.
We also tested whether the decrease in translation was due to cell death, as prolonged treatment of monastrol leads to mitotic arrest and eventually apoptosis ( Chin and Herbst, 2006). When cells were treated with monastrol, caspase 3–associated apoptotic cell death was not observed, even up to 16 h after treatment (Supplemental Figure S3C). A 16-h staurosporine treatment was used as a positive control to demonstrate caspase-3 cleavage in RPE1 cells. In addition, viability assays were conducted using a Promega Cell Proliferation Kit, a colorimetric assay measuring the number of viable cells by bioreduction of a tetrazolium compound by NADH or NADPH occurring only in metabolically active cells. Through the use of this assay, neither of the siRNAs to Eg5 caused a significant decrease in the metabolic activity of the cells as compared with the siControl (Supplemental Figure S3D). Although a 4-h monastrol treatment did cause a partial inhibition of metabolic activity, it was reversible after monastrol was removed, indicating that the decrease in metabolic activity was not due to cell death (Supplemental Figure S3E). These results confirm that a reduction of protein synthesis was observed before any indication of cell death could be documented.
Eg5 associates with ribosomes
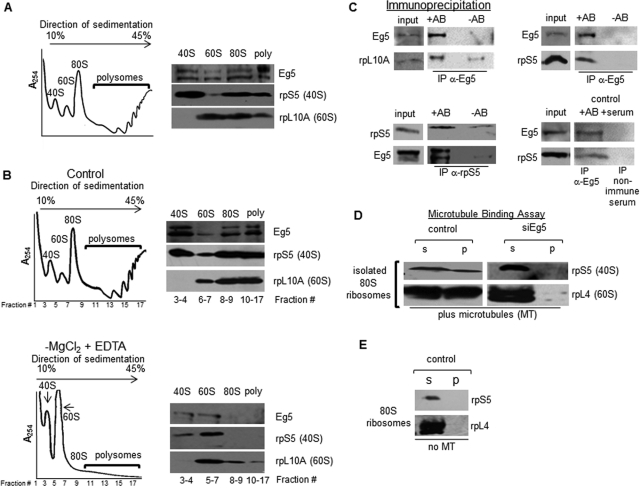
Given that we demonstrated that Eg5 is essential for normal levels of polypeptide synthesis, we wanted to determine whether Eg5 could associate with mature ribosomes. The eukaryotic 40S and 60S ribosomal subunits assemble on mRNA to form the 80S ribosome. The association of multiple 80S ribosomes on a single mRNA constitutes a translating polysome complex. RPE1 cellular lysates were laid on a 10–45% continuous sucrose gradient, centrifuged, and fractionated with constant monitoring at an absorbance of 254 nm. Four peaks were resolved in the polysome profile, representing, from left to right, the 40S ribosomal subunits, the 60S ribosomal subunits, the 80S ribosomes, and the polysomes ( Figure 2A). Fractions representing each subunit, ribosomes, or polysomes were pooled together and concentrated prior to SDS–PAGE analysis (Materials and Methods). To confirm isolation of ribosomes, immunoblot analysis of these fractions was completed with two ribosomal markers—rpS5, a component of the 40S subunit; and rpL10A, a component of the 60S subunit. As expected, rpS5 was primarily found in the 40S, the 80S, and the polysome fractions, whereas rpL10A was found in the 60S, the 80S, and the polysome fractions. Immunoblotting of the identical fractions for Eg5 revealed that Eg5 was found to cofractionate with the 40S and 60S subunits, the 80S ribosomes, and the polysomes. Note that Eg5 can be seen as doublet in these samples, most likely indicating the presence of an Eg5 proteolytic fragment. Both bands are derived from Eg5, because they are observed with different affinity-purified antibody sources and both bands are lost when Eg5 is knocked down by siRNA (Supplemental Figure S4).
FIGURE 2:
Eg5 associates with ribosomes and links ribosomes to microtubules. (A, B) Polysome profiling of mature ribosomes from RPE1 cells. WCLs were placed on a 10–45% continuous sucrose gradient, centrifuged for 2.5 h at 27,000 rpm in a Sorvall AH629 rotor, and fractionated with constant monitoring at an absorbance of 254 nm. (A) Immunoblots are representative of at least six independent experiments; the relative loads for each of the fractions are as follows: 40S and 60S subunits, 50%; 80S ribosomes, 33%; polysomes, 16.7%. Poly, polysomes. (B) Polysome profiling of control RPE1 cells (top) or in the absence of magnesium chloride (MgCl2; bottom). The positions of the 80S ribosomes and polysomes in the control gradient were used to determine the fraction numbers that would be collected as 80S ribosomes or polysomes in the −MgCl2, +EDTA sample. The relative loads for each of the fractions are as follows: 40S, 60S subunits, and 80S ribosomes, 50%; polysomes ∼40%. Each of these experiments was completed at least five times. (C) Top, immunoprecipitation of Eg5 in RPE1 cells was conducted and immunoprecipitates were probed for the presence of the 60S ribosomal subunits (rpL10A; left), or for the presence of the 40S ribosomal subunits (rpS5, right). Bottom, reciprocal IP of rpS5 was conducted and immunoprecipitates were probed for the presence of Eg5 (left; separate exposures are shown for input and IP lanes). IP of nonimmune serum was used as a negative control (right); IP of Eg5 was completed in parallel to confirm the association of Eg5 with rpS5. For each sample, 50% of the IP was loaded on the gel; input lane for Eg5 and control IP represents 10% of the total and for rpS5 IP represents 15% of the total. Results shown are representative of at least three independent experiments. −AB, no antibody added; +AB, Eg5/rpS5 antibody added. (D) Sucrose gradient–isolated 80S ribosome fractions from RPE1 WCLs were added to preformed taxol-stabilized microtubules for 45 min, followed by a 30-min centrifugation at 10,000 × g. Immunoblots in the presence of Eg5 (control) are representative of at least five independent experiments and in the absence of Eg5 (siEg5) are representative of at least three independent assays. Antibodies to rpS5 and rpL4 were used to demonstrate the 80S ribosome binding to the pelleted microtubules, and each of these antibodies represents an independent microtubule-binding assay. siEg5#1 siRNA for 24 h was used to knock down Eg5 in this experiment. P, microtubule pellet; S, supernatant. Assays completed in the presence (D; plus microtubules) or (E) in the absence (no MT) of polymerized microtubules.
To confirm that Eg5 cosedimented with 80S ribosomes and polysomes rather than another cosedimenting species, we completed sucrose gradient fractionations in the absence of magnesium. It was previously shown that ribosome integrity depends on magnesium, and in its absence, ribosomes dissociate into 40S and 60S ribosomal subunits ( Bradford and Sullivan, 1981; Wang et al., 2008). When sucrose gradients were performed in the absence of magnesium, Eg5 was no longer detected at the positions in the sucrose gradient corresponding to the 80S ribosomes and polysomes as compared with the control (fraction numbers 8–17; 80S ribosomes, polysomes). As expected, Eg5 was now only observed associated with the 40S and 60S ribosomal subunits ( Figure 2B). These results strongly suggest that Eg5 associates with 80S ribosomes and polysomes.
To confirm an association of Eg5 with ribosomes, we immunoprecipitated Eg5 and tested for the presence of ribosomal markers ( Figure 2C). The 60S ribosomal subunit marker rpL10A was present in the pellet at low levels without any Eg5 antibody added (−AB), but substantially more rpL10A was pelleted from equal amounts of cell lysate when Eg5 was immunoprecipitated (+AB). We repeated this analysis with rpS5, a marker for the 40S ribosomal subunits. rpS5 was present in the pellet only when Eg5 was immunoprecipitated ( Figure 2C, +AB) and not without Eg5 antibody addition (−AB). For the reciprocal experiment, we immunoprecipitated rpS5. In the absence of antibody, there were only trace amounts of the ribosomal complex observed; however, in the presence of the rpS5 antibody, Eg5 was found in the pellet ( Figure 2C). In each of the immunoprecipitations, the level of ribosomal protein and Eg5 increased substantially with the addition of antibody as compared with background levels of the controls (−AB), demonstrating that the precipitate was not due to nonspecific binding to the beads. To further confirm the specificity of the Eg5 pulldown, immunoprecipitation of nonimmune serum was completed in parallel with the Eg5 immunoprecipitation. Again, rpS5 was pelleted only when Eg5 antibodies were present and was not pelleted when nonimmune serum was used for the immunoprecipitation ( Figure 2C). These data support an association of Eg5 and ribosomes in cells. However, we do not yet know whether this association is direct or indirect.
Ribosomes associate with microtubules through Eg5
As discussed in the Introduction, ribosomes have been shown to associate with microtubules in numerous systems ( Suprenant et al., 1989; Hamill et al., 1994; Han et al., 2006). Therefore we tested whether Eg5 promoted the association of ribosomes with microtubules in mammalian cells. To test this, in vitro microtubule-binding assays were performed with 80S ribosomes isolated from sucrose gradient fractionation. These assays were performed by assembling microtubules using purified tubulin, adding sucrose gradient–isolated 80S ribosome fractions, and incubating them together in a binding reaction. Following centrifugation, non–microtubule-bound proteins are found in the supernatant (S), whereas microtubules and microtubule-bound proteins are observed in the pellet (P). When 80S ribosome fractions were incubated with in vitro–assembled microtubules and centrifuged, the 80S ribosomes were found to partially associate with microtubules, as demonstrated by ribosomal markers rpS5 and rpL4 ( Figure 2D, control, P). If microtubules were omitted from the binding reaction, 80S ribosomes did not associate with the pellet, demonstrating the specificity of this assay ( Figure 2E, P, no MT). These observations confirm that mammalian ribosomes can associate with microtubules, as observed previously in sea urchin embryos ( Suprenant et al., 1989). To investigate whether ribosome association with microtubules required Eg5, Eg5 was knocked down for 24 h, and 80S ribosomes were isolated by sucrose gradient fractionation and incubated in a binding reaction with assembled microtubules. In the absence of Eg5, the association of the 80S ribosomes with the microtubule pellet was lost ( Figure 2D, siEg5, P), indicating that Eg5 is required to maintain ribosome association with microtubules in these in vitro microtubule-binding assays. Although only a fraction of the ribosomes associated with the microtubule pellet, if the ribosomes are a motor-associated cargo, we expect the association to be transient, and not all of the ribosomes may bind to microtubules at any single moment in time.
Eg5 functions during the postinitiation stage of translation
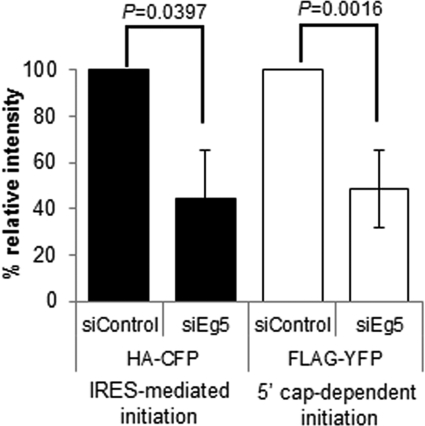
Translation can be separated into three steps: initiation, elongation, and termination. To examine why Eg5 activity is important for protein synthesis, we began by evaluating whether Eg5 is preferentially required for 5′ cap-dependent (canonical) or cap-independent translation initiation. U2OS cells, chosen for their high transfection efficiency, were transiently transfected with a bicistronic protein expression plasmid allowing both 5′ cap-dependent and the viral encephalomyocarditis internal ribosome entry site (IRES)–mediated translation initiation to be monitored simultaneously ( Nie and Htun, 2006). Translation of the yellow fluorescent protein (YFP) marker is controlled via the traditional 5′ cap-dependent pathway, whereas translation of cyan fluorescent protein (CFP) occurs via an IRES. After knockdown of Eg5, expression of both markers was significantly reduced by roughly equal amounts, 55% (CFP) and 52% (YFP). These data indicate that Eg5 functions during both the 5′ cap-dependent and the IRES-mediated translation ( Figure 3). This can occur if both types of initiation require Eg5 or if Eg5 functions at the later steps of translation in elongation or termination.
FIGURE 3:
Eg5 is required for IRES-mediated and 5′ cap-dependent translation initiation. U2OS cells were transfected with Eg5 siRNA (siEg5#1) for a total of 36 h. Eg5 was knocked down in U2OS cells for 12 h prior to cells being transfected with a bicistronic expression plasmid for an additional 24 h. Bar graph represents quantification of immunoblots from five independent experiments with antibodies to HA or FLAG epitopes on the CFP (IRES-mediated initiation, black bars) or YFP (5′ cap-dependent initiation, white bars) marker proteins. Results are represented as means ± SD; p values are derived from Student's t test (null hypothesis).
To further investigate the contribution of Eg5 in the different stages of translation, polysome profiles were obtained to reveal the levels of ribosomal subunits (40S and 60S) and ribosomes (80S complexes and polysomes) within cells after Eg5 inhibition. Changes in the polysome profiles can be used to differentiate between initiation and postinitiation translational defects. Defects in translation initiation result in a reduction of polysomes and a corresponding increase in 80S ribosomes, consisting of vacant 80S ribosomes and/or mRNA-bound 80S ribosomes ( Ashe et al., 2000). In contrast, defects during translational elongation or termination lead to a reduction in the 80S ribosomes with an accumulation of polysomes because slower-moving ribosomes accumulate on the mRNA ( Saini et al., 2009; Shin et al., 2009). CHX is usually added during the purification process for polysome profiling to prevent polysome disassembly by freezing ribosomes on the mRNA ( Saini et al., 2009).
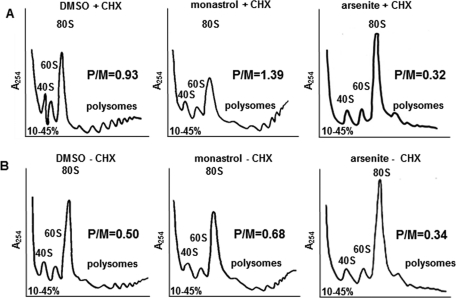
After a 4-h monastrol treatment, RPE1 cells exhibited a typical translation elongation/termination–defect phenotype for the polysome profiles ( Figure 4A, monastrol + CHX). We observed both an accumulation of polysomes and a decrease in 80S ribosomes as compared with the dimethyl sulfoxide (DMSO) control (DMSO + CHX). In each of these profiles, we measured the area under each curve to calculate a polysomes/80S monosomes (P/M) ratio to avoid variance between data sets due to sample size. Translation initiation defects result in decreased P/M ratios as compared with controls, whereas translation elongation/termination defects result in increased P/M ratios ( Shin et al., 2009). When Eg5 was inhibited by monastrol, the P/M ratio increased ∼50% compared with the DMSO control, consistent with a defect in elongation/termination ( Figure 4A). Other Eg5 inhibitors, such as S-trityl-l-cysteine, confirmed this conclusion by producing a similar phenotype of enhanced polysomes, a decrease in the 80S ribosomes, and an ∼43% increase in the P/M ratio (Supplemental Figure S5). Thus these data are most consistent with a defect in translation elongation and/or termination after Eg5 inhibition. These phenotypic changes after monastrol treatment were distinct from those observed when cells became senescent and nondividing. When RPE1 cells were serum starved for 32 h, a decrease in both the 80S ribosomes and the polysomes was observed in the polysome profiles (Supplemental Figure S6).
FIGURE 4:
Polysome profiling after loss of Eg5 activity leads to a decrease in the 80S ribosomes and an accumulation of polysomes, indicative of an elongation and/or termination defect. (A) Polysome profiling in RPE1 cells before (left, DMSO + CHX) or after a 4-h 130 μM monastrol treatment (middle, monastrol + CHX) or in the presence of 0.05 mM arsenite (right, arsenite + CHX). CHX was added 10 min prior to cell trypsinization. DMSO + CHX and monastrol + CHX were completed six times, whereas arsenite + CHX was completed twice, all yielding similar results. (B) Same experimental design as in A, except that CHX was omitted. DMSO − CHX and monastrol – CHX were completed four times, whereas arsenite − CHX was completed twice, all yielding similar results. All six of the profiles shown were completed at the same time. Polysome profiling was completed as described in Figure 2A and Materials and Methods. P/M, polysomes/monosomes ratio calculated as described in Materials and Methods.
As stated earlier, CHX was used while isolating ribosomes to preserve the polysomes and prevent them from ribosome runoff due to continued elongation during purification ( Shin et al., 2009). The absence of CHX can help to identify postinitiation defects, as slowed ribosome transit will reduce ribosome runoff and maintain polysomes, even without the addition of CHX. The experimental conditions from Figure 4A were repeated in the absence of CHX to confirm a postinitiation defect after Eg5 inhibition ( Figure 4B). As one would expect, in the absence of CHX, ribosomes in control cells continued to elongate and ran off the mRNA. This is demonstrated by the decreased polysomes, increased 80S ribosomes, and decreased P/M ratio of the DMSO − CHX profile compared with the DMSO + CHX profile ( Figure 4). If a buildup of polysomes still occurred after Eg5 inhibition, it would be consistent with the interpretation that Eg5 is required for optimal translation elongation/termination. Four hours after monastrol treatment and in the absence of CHX ( Figure 4B, monastrol − CHX), polysomes were found to persist and the P/M ratio increased by ∼36% compared with the DMSO − CHX, as expected if a defect in elongation or termination occurs when Eg5 is inhibited. The persistence of polysomes in the absence of CHX can be explained by slowed ribosome runoff and reduced ribosome transit as a consequence of Eg5 inhibition. Thus the polysome profiles in the absence of CHX further support a role for Eg5 in the postinitiation stages of translation ( Figure 4B). In contrast, polysome profiles of cells treated with the translation initiation inhibitor arsenite revealed decreased polysomes, increased 80S ribosomes, and decreased P/M ratios in the presence or in the absence of CHX, as expected for an initiation inhibitor ( Dang et al., 2006) and the opposite of the Eg5-deficient phenotype ( Figure 4; right, arsenite ± CHX). Therefore the contrast in polysome profiles after monastrol and arsenite treatments suggests that Eg5 is not required for the initiation stage of translation.
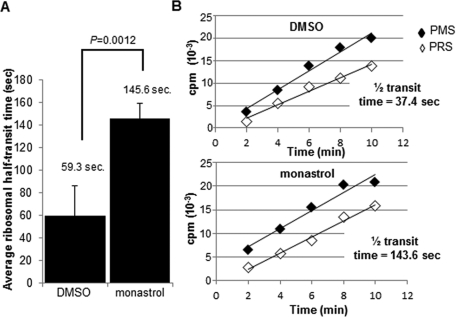
To provide additional experimental evidence of Eg5 functioning in ribosome transit (elongation and/or termination), the ribosome half-transit time was measured, an indication of the time it takes for one ribosome to traverse an average-sized mRNA ( Fan and Penman, 1970). This assay was completed by measuring 35S Met/Cys incorporation into total proteins (completed polypeptides and nascent proteins attached to the ribosomes [PMS]) and into completed polypeptides (proteins released from the ribosomes [PRS]). These data were plotted as a function of time, and the half-transit time was calculated as the displacement in time between the two lines (total proteins and completed proteins). If Eg5 is required for optimal elongation/termination, an increase in the ribosome half-transit time would be expected because ribosomes are slower to move along the mRNA. After a 4-h monastrol treatment, the average ribosome half-transit time of four independent experiments completed in RPE1 cells increased significantly from 59.3 s (transit time, 1 min, 59 s) in control cells to 145.6 s (transit time, 4 min, 51 s) in the monastrol-treated cells, an ∼2.5-fold increase ( Figure 5A). A single representative experiment with the calculated ribosome half-transit times is shown in Figure 5B. This assay demonstrated that the ribosome transit rate decreased when Eg5 was inhibited, consistent with a role for Eg5 in elongation/termination.
FIGURE 5:
Inhibition of Eg5 slows the rate of ribosome transit. Ribosome half-transit assays before (DMSO) or after (4 h, 130 μM monastrol) Eg5 inhibition in RPE1 cells, as described further in Materials and Methods and Results. Briefly, incorporation of 35S Met/Cys into total proteins (postmitochondrial supernatants [PMS]; black diamonds) and completed proteins (postribosomal supernatants [PRS]; white diamonds) were graphed over time. The half-transit time was determined by linear regression analysis from the displacement in time between the two x-intercepts. (A) Bar graph represents the average half-transit time of four independent experiments before (DMSO) or after (monastrol) Eg5 inhibition and demonstrated an ∼2.5-fold increase after Eg5 inhibition. (B) A single representative experiment is shown with the calculated half-transit times for this experiment. The DMSO half-transit time was calculated to be 37.4 s (transit time, 1 min, 15 s), whereas after a 4-h monastrol treatment the half-transit time was 143.6 s (transit time, 4 min, 47 s), resulting in an ∼3.8-fold increase for this representative experiment. Results in A are represented as means ± SD; p values are derived from Student's t test (null hypothesis).
DISCUSSION
This study makes two related conclusions: mammalian cells require the molecular motor Eg5 for normal levels of protein synthesis, and Eg5 has functions outside of mitosis in diverse cell types. We conclude that Eg5 activity is important during the postinitiation phase of polypeptide synthesis, which includes elongation and/or termination. When Eg5 is inhibited, delayed ribosomal half-transit times, increased polysome peaks, and higher P/M ratios were observed. Together with the association of Eg5 with ribosomes and its requirement to link ribosomes to microtubules in vitro, these properties implicate Eg5 as an agent that promotes ribosome elongation and/or termination by linking ribosomes to microtubules during translation.
Overall, five different specific inhibitors of Eg5 caused an ∼40% reduction in nascent polypeptide synthesis, ruling out off-target effects as the explanation for the translation defects. In addition, translation inhibition phenotypes were seen as little as 1 h after inhibition of Eg5, with fewer than 5% of the cells in mitosis and no significant increase in the mitotic frequency over controls (in four of five Eg5 inhibitors), demonstrating that the translational phenotypes were not a result of mitotic arrest. Eg5 expression is decreased after mitosis but is expressed throughout the cell cycle ( Levesque and Compton, 2001; Rapley et al., 2008), consistent with a role for Eg5 in interphase.
Although Eg5 inhibition resulted in an ∼40% decrease in protein synthesis, microtubule disruption resulted in only an ∼20–25% reduction. Because microtubule disruption should inhibit all microtubule motors, one might anticipate that microtubule inhibitors would also result in at least an ∼40% reduction in protein synthesis. However, it is possible that there may be other, unidentified microtubule-associated proteins, including additional motors, that have inhibitory or stimulatory actions on polypeptide synthesis. Loss of microtubules will influence all of these activities, resulting in a collective effect on translation that could result in less than ∼40% decrease in translation. Therefore the microtubule phenotype may not match the phenotype of the loss of a single motor.
Eg5 has been shown to be a homotetrameric bipolar complex in Drosophila oocytes by velocity centrifugation and rotary shadow electron microscopy ( Cole et al., 1994; Kashina et al., 1997), indicating that Eg5 cross-links and slides microtubules during spindle assembly ( Tanenbaum and Medema, 2010; Peterman and Scholey, 2009). A homotetrameric bipolar molecular structure is not the typical kinesin configuration associated with cargo binding, as the cargo usually binds opposite to the motor domain in most kinesins ( Verhey et al., 2011). However, in Drosophila oocytes only 60–70% of the Eg5 molecules were immunolabeled at both ends of the minifilament with antibodies to the motor domain, as would be observed if Eg5 was a bipolar homotetramer ( Kashina et al., 1996). In addition, during mitosis CDK1 phosphorylation might give the Eg5 complex a specialized structure and activity during division ( Cahu et al., 2008). As the oligomeric structure of vertebrate Eg5 has not yet been determined, these observations leave open possible additional configurations for Eg5 during interphase of the mammalian cell cycle.
In this study, we demonstrated that Eg5 is required to link ribosomes to microtubules in in vitro binding assays and that ATPase activity of Eg5 is essential for normal translation. We propose that Eg5 serves as a motile link between the ribosome and microtubules to enhance the efficiency of translation ( Figure 6). We do not know whether Eg5 directly binds the ribosome ( Figure 6A) or associates with ribosomes via undefined linker molecules ( Figure 6B). Eg5 could serve as a traditional elongation or termination factor to promote enzymatic assembly of the polypeptide chain, but there is no evidence to support that conclusion. We suggest that Eg5 may play a nontraditional role in translation elongation and/or termination. Our preferred model is that Eg5 functions as a motor, consistent with its known roles in mitosis, and interacts with ribosomes as a macromolecular cargo to maintain association of ribosomes with the cytoskeleton and to promote ribosome transit. Although the detailed structural aspects of the association of Eg5 with ribosomes remain to be determined, these results identify for the first time a role for the Eg5 molecular motor in polypeptide synthesis.
FIGURE 6:
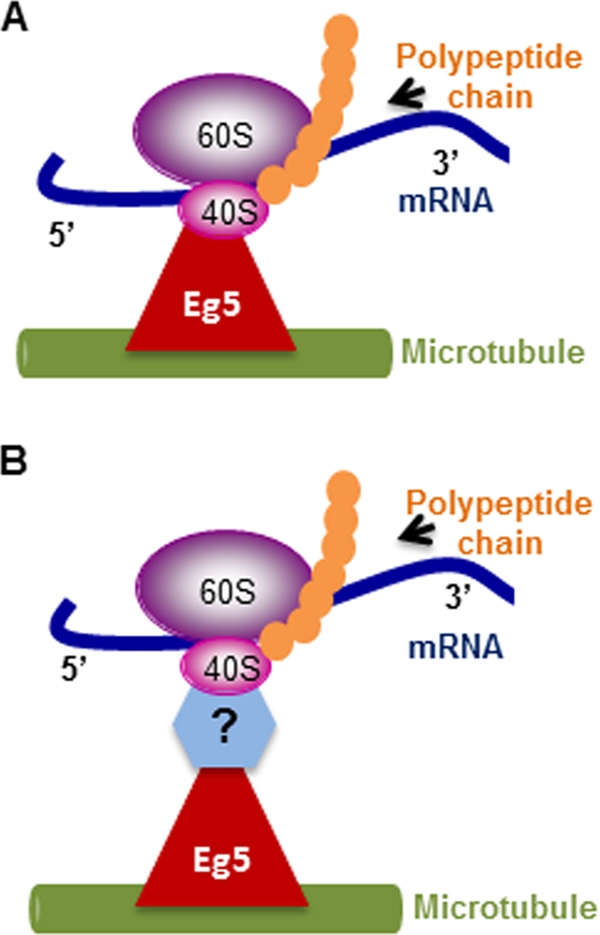
Schematic representation of the proposed function of Eg5. Eg5 is suggested to serve as a mobile molecular link between the ribosomes and microtubules to enhance the efficiency of polypeptide synthesis. (A) Eg5 may directly link the 80S ribosomes to microtubules or (B) Eg5 may be indirectly associated with ribosomes via unidentified linker molecules.
Molecular motors, and in particular Eg5, are potential anticancer drug targets, as motor-driven forces are critical for proper cell division. Small-molecule inhibitors of mitosis offer a novel and effective means of cancer cell proliferation inhibition. Critical to the rationale of choosing mitotic motors as targets is the belief that they function only in mitosis and their inhibition should only minimally interfere with interphase cells ( Sakowicz et al., 2004; Bergnes et al., 2005; Duhl and Renhowe, 2005; Lad et al., 2008; Sarli and Giannis, 2008; Huszar et al., 2009; Nakai et al., 2009; Burris et al., 2011). On a cautionary note, our study and those completed by Baas and colleagues show that Eg5 functions in essential interphase processes. These discoveries for a role of Eg5 in protein synthesis have important implications for phase I and phase II clinical trials targeting Eg5 for cancer treatment ( Bergnes et al., 2005; Lad et al., 2008; Sarli and Giannis, 2008; Zhang and Xu, 2008; Huszar et al., 2009; Burris et al., 2011). These new interphase functions of Eg5 should be considered in the design and application of future anti-Eg5 therapeutics.
MATERIALS AND METHODS
Cell lines
RPE1 (human retinal pigmented epithelial cells stably transfected with hTERT) were cultured in DMEM/F-12 (D6421) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA). HFF-hTERT (human foreskin fibroblasts) and NIH-3T3 (mouse fibroblasts) cells were maintained in DMEM media with 2 mM l-glutamine and 10% FBS. U2OS (human osteosarcoma) cells were maintained in McCoy's media (16600082; Invitrogen, Carlsbad, CA) with 10% FBS. All cultures were grown at 37°C with 5% CO2. All culture media and supplements were purchased from Sigma-Aldrich (St. Louis, MO), and cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA).
Antibodies
The following antibodies were used in this study: Eg5, rabbit anti-Eg5 (AKIN03; Cytoskeleton, Denver, CO); actin, rabbit anti-actin (AAN01; Cytoskeleton); Eg5, mouse anti-Eg5 (627802; BioLegend, San Diego, CA); RPS5, mouse anti-RPS5 (AB58345; Abcam, Cambridge, MA); RPL10A, mouse anti-RPL10A (Ab55544; Abcam); GAPDH, rabbit anti-GAPDH (14C10) (2118; Cell Signaling, Beverly, MA); calnexin, rabbit anti-calnexin (Stressgen, Enzo Life Sciences, San Diego, CA); caspase-3, rabbit anti–caspase 3 (8G10) (9665; Cell Signaling); FLAG, mouse anti-FLAG (4049; Sigma-Aldrich); HA, mouse anti-HA (1 583 816; Roche, Indianapolis, IN); RPL4, rabbit anti-RPL4 (Proteintech Group, Chicago, IL); and phospho-H3, rabbit anti–phospho-H3 (Upstate, Millipore, Billerica, MA).
Small-molecule treatments
The following times and concentrations were used: 4 h, 130 μM monastrol (Tocris Bioscience, Ellisville, MO; batch 3); 2 h, 12 μM nocodazole; 2 h, 0.002 mg/ml Colcemid; 0.1 mg/ml CHX; 1 h, 0.05 mM arsenite; 1 h, 3 μM dimethylenastron (Alexis Biochemicals, San Diego, CA); and 4 h, 1.5 μM S-trityl-l-cysteine (Alexis Biochemicals). Final DMSO concentration for control experiments were as follows: experiments using monastrol and nocodazole, 0.04%; for dimethylenastron, 0.009%; for S-trityl-l-cysteine, 0.0003%. All reagents were purchased from Sigma-Aldrich, unless otherwise noted, and dissolved in DMSO, except for arsenite and Colcemid, which were purchased predissolved. For washout experiments, cells were treated for 4 h with monastrol, followed by three washes in FBS and a 4-h recovery.
Transfections
siRNA.
Cells were reverse transfected for 24 h (unless specified) with 1.5 μg/60 mm tissue culture plate of siRNA using HiPerFect Transfection Reagent (Qiagen, Valencia, CA), following the manufacturer's protocol. Fluorescently labeled scrambled siRNA (siControl) was used (1022563) as the control. All reagents were purchased from Qiagen unless specified otherwise. Eg5 siRNA#1 (SI02653770) and Eg5 siRNA#2 (s7904) (Ambion, Austin, TX) were used.
Plasmids.
Bicistronic plasmid transfection (plasmid 18673; Addgene, Cambridge, MA) into U2OS cells was completed using FuGene6 (1814443; Roche) transfection reagent for 24 h following the manufacturer's protocol after a 12-h knockdown of Eg5. Eg5 was knocked down for a total of 36 h. Antibodies to HA or FLAG were used for immunoblot analysis, and quantitation of immunoblots was completed using Adobe Photoshop (San Jose, CA).
35S Met/Cys incorporation assays.
Cells were grown in 60-mm plates, and the media were replaced with DMEM without methionine and cysteine (D0244; Sigma-Aldrich) plus 5% dialyzed FBS (F0392; Sigma-Aldrich), 2 mM l-glutamine, and the addition of 100 μCi/ml of 35S Met/Cys (NEG072007MC; PerkinElmer, Waltham, MA) and incubated for 30 min at 37°C. To stop reactions, 0.1 mg/ml CHX was added, and cells were trypsinized and washed in PBS prior to cell lysis in RIPA buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% NP-40). Protein samples were split in half; the first half was subjected to a Lowry assay and/or separated on SDS–PAGE for confirmation of equal loading or of knockdown of the specified protein. The second half was subjected to scintillation counting, in which duplicate samples of each lysate were placed on GF/C filters (28497-743; VWR, Radnor, PA), washed three times with 2 ml of 10% TCA and once with 100% ethanol, and dried before analysis.
Fractionation of cell lysates.
For fractionation of cell lysates into cytosolic and ER fractions, a digitonin fractionation protocol was used. Briefly, after the 35S Met/Cys incorporation assay cells were trypsinized and washed in PBS and the cell membrane was broken open by pipetting 25 times with a cut 200-μl pipette tip in digitonin buffer solution (10 mM 1,4-piperazinediethanesulfonic acid [PIPES; pH 6.8], 300 mM sucrose, 3 mM MgCl2, 5 mM EDTA, 0.01% digitonin, 1 mM phenylmethylsulfonyl fluoride [PMSF]). Lysates were incubated for 8 min on ice and centrifuged at 3000 × g for 4 min, and the cytosolic fraction was removed for analysis. The pellet was washed once in PBS, centrifuged, and resuspended in RIPA buffer to retain the membrane fraction for analysis.
Polysome profiling: 10–45% sucrose gradients
Between 20 and 30 million RPE1 cells were incubated with or without 0.1 mg/ml CHX for 10 min prior to trypsinization. (Samples that were treated with CHX are labeled +CHX, whereas samples that were not treated with CHX are labeled −CHX.) Cells were lysed (20 mM Tris-HCl [pH 7.2], 130 mM KCl, 30 mM MgCl2, 2.5 mM DTT, 0.2% NP-40, 0.5% sodium deoxycholate, 0.1 mg/ml cycloheximide, 0.2 mg/ml heparin, 1 mM PMSF), incubated for 15 min on ice, the DNA pellet was removed by centrifugation, and a Lowry assay was completed to ensure equal loading onto the gradient. The lysates were placed on top of a 10–45% (wt/wt) sucrose gradient (10 mM Tris-HCl [pH 7.2], 60 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1 mg/ml heparin), and samples were centrifuged at 27,000 rpm for 2.5 h at 4°C using a Beckman L7 Ultracentrifuge (model L7-65) in a Sorvall AH629 rotor. Gradients were fractionated by upward displacement through an ISCO UA-5 with constant UV monitoring at an absorbance of 254 nm.
In the absence of MgCl2 and in the presence of EDTA, the experiment was completed as described except that MgCl2 was omitted from the lysis buffer and the sucrose gradients and 2 mM of EDTA was added.
Immunoblot analysis of polysome profiling
For immunoblot analysis of 10–45% sucrose gradients, fractions representing each of the ribosomal subunits and/or ribosomes were pooled together. For extraction of proteins, a final concentration of 20 mM Tris, pH 7.5, was added, followed by the addition of 15–30 μl StrataClean resin (Stratagene, Santa Clara, CA). Samples were then rotated at room temperature for 30 min prior to centrifugation; pelleted beads were resuspended in 2× SDS loading dye, and samples were boiled for 10 min to elute proteins before subjection to SDS–PAGE (15% gel).
Polysomes/monosomes ratio calculations
For the calculation of P/M ratio, each polysome profile graph was photocopied and enlarged to 151%. Next, the area under each ribosomal peak (40S, 60S, 80S, and polysomes) was estimated by weighing paper cutouts of the profiles. The baseline was chosen based on the lowest point on each profile. Each peak was cut out (in triplicate) and weighed (in triplicate) on an analytical balance (Adventurer SL AS64; Ohaus, Pine Brook, NJ). Averages of the area under each ribosomal peak were calculated, and the average weight of the polysomes was divided by the average weight of the monosomes (80S ribosomes) per profile to calculate the P/M ratio. P/M ratios represent the exact polysome profile shown.
Serum starvation
RPE1 cells were serum starved for 32 h in DMEM media without FBS. Fresh DMEM was added every 6 h, prior to CHX addition, cell lysis, and polysome profiling.
Immunoprecipitation
Immunoprecipitation (IP) was completed following the manufacturer's protocol, with these exceptions: 10–15 million RPE1 cells were lysed (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES; pH 7.5], 150 mM NaCl, 0.1% Triton X-100, 1 mM EDTA, 2.5 mM ethylene glycol tetraacetic acid [EGTA], 10% glycerol, 1 mM NaF, 0.1 mM Na3VO4, 10 mM β-glycerophosphate, 1 mM PMSF), and the DNA pellet was removed by centrifugation. Each tube received 2 μg of rabbit anti-Eg5 antibody, nonimmune serum, or 4 μl of 30% glycerol (−AB; Eg5 antibody was reconstituted in 30% glycerol) for the immunoprecipitation. For the rpS5 IP, 2.5 μg of antibody was added. Samples were subjected to SDS–PAGE (12 or 15% gel). Each IP represented 50% of the total; input represented 10% for Eg5 immunoprecipitations and 15% for the rpS5 immunoprecipitation. Nonimmune serum was used as a negative control. Protein A beads were used in the reaction (Amersham Biosciences, GE Healthcare, Piscataway, NJ).
Ribosome half-transit time assay
Thirty million cells were trypsinized, washed, and resuspended in 3 ml of BME (B1522; Sigma-Aldrich) plus 10% dialyzed FBS and 2 mM l-glutamine for 20 min prior to the addition of 10 μCi/ml 35S Met/Cys. At each time point, 500 μl of cells were removed, placed in an ice-cold tube with 500 μg/ml CHX, and incubated on ice. Cells were centrifuged at 4°C, washed with ice-cold PBS containing CHX, recentrifuged, lysed in 1 ml of polysome profiling buffer with the following changes (0.02 M Tris [pH 7.2], 0.130 M KCl, 0.03 M MgCl2, 1% NP-40, 0.05% sodium deoxycholate, 0.2 mg/ml heparin, 0.25 mg/ml CHX, 1 mM DTT, 1 mM PMSF, RNasin Inhibitor [Promega]), and incubated on ice for 15 min, and the DNA was removed by centrifugation prior to splitting the lysates: 500 μl of the lysate was saved (PMS fraction) containing total proteins, whereas the other 500 μl was placed on a stepwise 20% sucrose buffer and 60% sucrose cushion. Samples were centrifuged in an S100-AT5 ultracentrifuge rotor at 55,000 rpm for 27 min, after which 500 μl of the sample was removed (PRS) containing completed proteins released from the ribosomes. The PMS and PRS fractions were then TCA precipitated on GF/C filters (as described earlier) and subjected to scintillation counting. Half-transit times were calculated by comparing the incorporation of radioactivity into total proteins and completed proteins graphed over time. This assay was completed by combining various protocols from different publications ( Ruvinsky et al., 2005; Saini et al., 2009).
In vitro microtubule-binding assays
Purified tubulin (isolated from bovine brains; Moyer et al., 1996) was thawed on ice with the addition of an equal volume of 1× PM (10 mM PIPES [pH 7.0], 5 mM magnesium acetate, and 1 mM EGTA [pH 7.0)] buffer and 2 mM GTP, followed by a 30-min centrifugation at 10,000 × g (4°C). The supernatant was removed, grow buffer (1× PM buffer, 0.1 mM taxol [Sigma-Aldrich], 5 mM GTP) was added to it in a 4:1 ratio (tubulin:grow buffer), followed by a 15-min incubation (34°C) with rotation. Fractions representing the 80S ribosome from polysome profiling were pooled together, inverted, and split: half received the binding reaction (5× PM buffer, 100 mM NaCl, 0.04 mM taxol, 1 mM GTP) plus 12% tubulin and was incubated with rotation for 45 min (34°C), whereas the other half received the binding reaction without tubulin, taxol, or GTP and remained at 4°C. After incubation, cells were centrifuged at 10,000 × g for 30 min at 34 or 4°C. The supernatant was removed (containing non–microtubule-bound proteins), pellets were washed twice in PBS and recentrifuged, and the supernatant was discarded. The pellet contained microtubules and microtubule-bound proteins; 20% of the supernatant and 50% of the pellet were loaded on 12% SDS–PAGE gels.
Mitotic index analyses by DAPI
Cells were fixed in 0.2% Triton-X 100 in 4% paraformaldehyde for 15 min prior to the addition of DAPI. A minimum of 300 cells was counted per trial, and the experiment was completed in triplicate. All cells were analyzed on an Olympus (Center Valley, PA) BX60 epifluorescence microscope with a 100× oil immersion objective, unless specified. A Hamamatsu (Hamamatsu, Japan) Argus-20 charge-coupled device (CCD) camera was used to record images.
Mitotic index analyses by phospho-H3 staining
Cells were fixed in 0.2% Triton-X 100 in 4% paraformaldehyde for 30 min prior to a 30-min block in 1.5% BSA in PBST. Phospho-H3 antibody was diluted (1:500) in blocking buffer and incubated on cells for 30 min prior to three washes in PBS, the addition of secondary for 30 min, three additional washes in PBS, and DAPI staining. A minimum of 300 cells was counted per trial, and the experiment was completed in triplicate. All cells were analyzed on an Olympus BX60 epifluorescence microscope with a 100× oil immersion objective, unless specified. A Hamamatsu Argus-20 CCD camera was used to record images.
Cell cycle analysis
RPE1 cells were grown to 70% confluency in 100-mm Petri dishes before monastrol treatment. Cells were incubated for either 4 or 24 h in monastrol prior to trypsinization, two washes in PBS, and fixation in 100% ice-cold ethanol and storage overnight at 4°C. Next, the cells were pelleted by centrifugation, followed by two washes in PBS and incubation with 100 μl of 100 μg/ml RNase A for 8 h. Finally, 500 μl of a 50 μg/ml solution of propidium iodide was added to cells immediately before analysis by flow cytometry.
Apoptosis assay
Caspase-3 antibody was used to determine apoptosis after a 4- to 16-h monastrol treatment; 16 h of 1 μM staurosporine was used as a positive control to demonstrate caspase-3 cleavage.
Metabolic activity assay
A CellTiter 96 AQueous One Solution Cell Proliferation Assay was performed following the manufacturer's protocol (Promega, Madison, WI) after a 24-h knockdown of Eg5 (siEg5#1 or siEg5#2) or after Eg5 inhibition or recovery by monastrol.
Supplementary Material
Acknowledgments
We thank the Biochemistry and Molecular Genetics Program at the University of Pittsburgh School of Medicine and acknowledge support from National Institutes of Health Grants GM28301 to J.L.W. and R01DE016086 to W.S.S. We thank Thomas E. Dever of the National Institutes of Health, Terri Goss Kinzy of the University of Medicine and Dentistry of New Jersey Robert Wood Johnson Medical School, and LingQian Xu of the University of Pittsburgh for careful and critical reading of the manuscript.
Abbreviations used:
- CFP
cyan fluorescent protein
- CHX
cycloheximide
- DMSO
dimethyl sulfoxide
- Eg5
kinesin 5/KIF 11
- hTERT
human telomerase reverse transcriptase
- IP
immunoprecipitation
- Met/Cys
methionine and cysteine
- MT
microtubules
- PMS
postmitochondrial supernatant
- PRS
postribosomal supernatant
- RPE1
retinal pigmented epithelial cells stably transfected with hTERT
- siC
siControl siRNA
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-03-0211) on July 27, 2011.
REFERENCES
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergnes G, Brejc K, Belmont L. Mitotic kinesins: prospects for antimitotic drug discovery. Curr Top Med Chem. 2005;5:127–145. doi: 10.2174/1568026053507697. [DOI] [PubMed] [Google Scholar]
- Bernstam VA, Gray RH, Bernstein IA. Effect of microtubule-disrupting drugs on protein and RNA synthesis in Physarum polycephalum amoebae. Arch Microbiol. 1980;128:34–40. doi: 10.1007/BF00422302. [DOI] [PubMed] [Google Scholar]
- Bisbal M, Wojnacki J, Peretti D, Ropolo A, Sesma J, Jausoro I, Caceres A. KIF4 mediates anterograde translocation and positioning of ribosomal constituents to axons. J Biol Chem. 2009;284:9489–9497. doi: 10.1074/jbc.M808586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford CT, Sullivan DT. Isolation of polysomes from larval and adult Drosophila melanogaster. Anal Biochem. 1981;112:176–181. doi: 10.1016/0003-2697(81)90277-3. [DOI] [PubMed] [Google Scholar]
- Burris HA 3rd, Jones SF, Williams DD, Kathman SJ, Hodge JP, Pandite L, Ho PT, Boerner SA, Lorusso P. A phase I study of ispinesib, a kinesin spindle protein inhibitor, administered weekly for three consecutive weeks of a 28-day cycle in patients with solid tumors. Invest New Drugs. 2011;29:467–472. doi: 10.1007/s10637-009-9374-x. [DOI] [PubMed] [Google Scholar]
- Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C, Doherty-Kirby A, Lajoie G, Surrey T. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS One. 2008;3:e3936. doi: 10.1371/journal.pone.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin GM, Herbst R. Induction of apoptosis by monastrol, an inhibitor of the mitotic kinesin Eg5, is independent of the spindle checkpoint. Mol Cancer Ther. 2006;5:2580–2591. doi: 10.1158/1535-7163.MCT-06-0201. [DOI] [PubMed] [Google Scholar]
- Cole DG, Saxton WM, Sheehan KB, Scholey JM. A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- DeBonis S, Skoufias DA, Lebeau L, Lopez R, Robin G, Margolis RL, Wade RH, Kozielski F. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol Cancer Ther. 2004;3:1079–1090. [PubMed] [Google Scholar]
- Duhl DM, Renhowe PA. Inhibitors of kinesin motor proteins—research and clinical progress. Curr Opin Drug Discov Dev. 2005;8:431–436. [PubMed] [Google Scholar]
- Fan H, Penman S. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J Mol Biol. 1970;50:655–670. doi: 10.1016/0022-2836(70)90091-4. [DOI] [PubMed] [Google Scholar]
- Ferhat L, Cook C, Chauviere M, Harper M, Kress M, Lyons GE, Baas PW. Expression of the mitotic motor protein Eg5 in postmitotic neurons: implications for neuronal development. J Neurosci. 1998;18:7822–7835. doi: 10.1523/JNEUROSCI.18-19-07822.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton AB, Wan KM, Penman S. The spatial distribution of polyribosomes in 3T3 cells and the associated assembly of proteins into the skeletal framework. Cell. 1980;20:849–857. doi: 10.1016/0092-8674(80)90331-1. [DOI] [PubMed] [Google Scholar]
- Gartner M, Sunder-Plassmann N, Seiler J, Utz M, Vernos I, Surrey T, Giannis A. Development and biological evaluation of potent and specific inhibitors of mitotic kinesin Eg5. Chembiochem. 2005;6:1173–1177. doi: 10.1002/cbic.200500005. [DOI] [PubMed] [Google Scholar]
- Hamill D, Davis J, Drawbridge J, Suprenant KA. Polyribosome targeting to microtubules: enrichment of specific mRNAs in a reconstituted microtubule preparation from sea urchin embryos. J Cell Biol. 1994;127:973–984. doi: 10.1083/jcb.127.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Yu J, Guo F, Watkins SC. Polysomes are associated with microtubules in fertilized eggs of Chinese pine (Pinus tabulaeformis) Protoplasma. 2006;227:223–227. doi: 10.1007/s00709-005-0150-2. [DOI] [PubMed] [Google Scholar]
- Haque SA, Hasaka TP, Brooks AD, Lobanov PV, Baas PW. Monastrol, a prototype anti-cancer drug that inhibits a mitotic kinesin, induces rapid bursts of axonal outgrowth from cultured postmitotic neurons. Cell Motil Cytoskeleton. 2004;58:10–16. doi: 10.1002/cm.10176. [DOI] [PubMed] [Google Scholar]
- Hesketh JE, Pryme IF. Evidence that insulin increases the proportion of polysomes that are bound to the cytoskeleton in 3T3 fibroblasts. FEBS Lett. 1988;231:62–66. doi: 10.1016/0014-5793(88)80703-8. [DOI] [PubMed] [Google Scholar]
- Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28:197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- Jansen RP. RNA-cytoskeletal associations. FASEB J. 1999;13:455–466. [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Rogers GC, Scholey JM. The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim Biophys Acta. 1997;1357:257–271. doi: 10.1016/s0167-4889(97)00037-2. [DOI] [PubMed] [Google Scholar]
- Lad L, Luo L, Carson JD, Wood KW, Hartman JJ, Copeland RA, Sakowicz R. Mechanism of inhibition of human KSP by ispinesib. Biochemistry. 2008;47:3576–3585. doi: 10.1021/bi702061g. [DOI] [PubMed] [Google Scholar]
- Lenk R, Ransom L, Kaufmann Y, Penman S. A cytoskeletal structure with associated polyribosomes obtained from HeLa cells. Cell. 1977;10:67–78. doi: 10.1016/0092-8674(77)90141-6. [DOI] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque AA, Compton DA. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol. 2001;154:1135–1146. doi: 10.1083/jcb.200106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci. 2009;122:3973–3982. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor HC, Stebbings H. A massive system of microtubules associated with cytoplasmic movement in telotrophic ovarioles. J Cell Sci. 1970;6:431–449. doi: 10.1242/jcs.6.2.431. [DOI] [PubMed] [Google Scholar]
- Maliga Z, Kapoor TM, Mitchison TJ. Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem Biol. 2002;9:989–996. doi: 10.1016/s1074-5521(02)00212-0. [DOI] [PubMed] [Google Scholar]
- Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Miyamoto DT, Perlman ZE, Burbank KS, Groen AC, Mitchison TJ. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J Cell Biol. 2004;167:813–818. doi: 10.1083/jcb.200407126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon RT, Nicosia RF, Olsen C, Hille MB, Jeffery WR. The cytoskeletal framework of sea urchin eggs and embryos: developmental changes in the association of messenger RNA. Dev Biol. 1983;95:447–458. doi: 10.1016/0012-1606(83)90046-5. [DOI] [PubMed] [Google Scholar]
- Moyer ML, Gilbert SP, Johnson KA. Purification and characterization of two monomeric kinesin constructs. Biochemistry. 1996;35:6321–6329. doi: 10.1021/bi960017n. [DOI] [PubMed] [Google Scholar]
- Myers KA, Baas PW. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J Cell Biol. 2007;178:1081–1091. doi: 10.1083/jcb.200702074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadar VC, Ketschek A, Myers KA, Gallo G, Baas PW. Kinesin-5 is essential for growth-cone turning. Curr Biol. 2008;18:1972–1977. doi: 10.1016/j.cub.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai R, et al. K858, a novel inhibitor of mitotic kinesin Eg5 and antitumor agent, induces cell death in cancer cells. Cancer Res. 2009;69:3901–3909. doi: 10.1158/0008-5472.CAN-08-4373. [DOI] [PubMed] [Google Scholar]
- Nie M, Htun H. Different modes and potencies of translational repression by sequence-specific RNA-protein interaction at the 5′-UTR. Nucleic Acids Res. 2006;34:5528–5540. doi: 10.1093/nar/gkl584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman EJ, Scholey JM. Mitotic microtubule crosslinkers: insights from mechanistic studies. Curr Biol. 2009;19:R1089–R1094. doi: 10.1016/j.cub.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Ramaekers FC, Benedetti EL, Dunia I, Vorstenbosch P, Bloemendal H. Polyribosomes associated with microfilaments in cultured lens cells. Biochim Biophys Acta. 1983;740:441–448. doi: 10.1016/0167-4781(83)90093-3. [DOI] [PubMed] [Google Scholar]
- Rapley J, Nicolas M, Groen A, Regue L, Bertran MT, Caelles C, Avruch J, Roig J. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J Cell Sci. 2008;121:3912–3921. doi: 10.1242/jcs.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakowicz R, et al. Antitumor activity of a kinesin inhibitor. Cancer Res. 2004;64:3276–3280. doi: 10.1158/0008-5472.can-03-3839. [DOI] [PubMed] [Google Scholar]
- Sarli V, Giannis A. Targeting the kinesin spindle protein: basic principles and clinical implications. Clin Cancer Res. 2008;14:7583–7587. doi: 10.1158/1078-0432.CCR-08-0120. [DOI] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125–138. doi: 10.1083/jcb.144.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Acker MG, Maher KN, Lorsch JR, Dever TE. rRNA suppressor of a eukaryotic translation initiation factor 5B/initiation factor 2 mutant reveals a binding site for translational GTPases on the small ribosomal subunit. Mol Cell Biol. 2009;29:808–821. doi: 10.1128/MCB.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu-Hiza M, Perlman ZE, Wittmann T, Karsenti E, Mitchison TJ. Eg5 causes elongation of meiotic spindles when flux-associated microtubule depolymerization is blocked. Curr Biol. 2004;14:1941–1945. doi: 10.1016/j.cub.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol. 2007;27:6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Calliari A, Cardenas M, Koenig E, Sotelo JR. Myosin Va and kinesin II motor proteins are concentrated in ribosomal domains (periaxoplasmic ribosomal plaques) of myelinated axons. J Neurobiol. 2004;60:187–196. doi: 10.1002/neu.20015. [DOI] [PubMed] [Google Scholar]
- Suprenant KA. Microtubules, ribosomes, and RNA: evidence for cytoplasmic localization and translational regulation. Cell Motil Cytoskeleton. 1993;25:1–9. doi: 10.1002/cm.970250102. [DOI] [PubMed] [Google Scholar]
- Suprenant KA, Tempero LB, Hammer LE. Association of ribosomes with in vitro assembled microtubules. Cell Motil Cytoskeleton. 1989;14:401–415. doi: 10.1002/cm.970140310. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ. The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol. 2008;18:1860–1864. doi: 10.1016/j.cub.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Kaul N, Soppina V. Kinesin assembly and movement in cells. Annu Rev Biophys. 2011;40:267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- Wang H, Dictenberg JB, Ku L, Li W, Bassell GJ, Feng Y. Dynamic association of the fragile X mental retardation protein as a messenger ribonucleoprotein between microtubules and polyribosomes. Mol Biol Cell. 2008;19:105–114. doi: 10.1091/mbc.E07-06-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xu W. Progress on kinesin spindle protein inhibitors as anti-cancer agents. Anticancer Agents Med Chem. 2008;8:698–704. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.