Assembly of the forespore membrane (FSM) initiates at the spindle pole body (SPB), and the leading edge of the FSM is a critical factor in the proper shaping of the FSM. We report a novel SPB component Spo7. Our study suggests that Spo7 coordinates formation of the leading edge and initiation of FSM assembly, thereby accomplishing accurate FSM formation.
Abstract
Sporulation in fission yeast represents a unique mode of cell division in which a new cell is formed within the cytoplasm of a mother cell. This event is accompanied by formation of the forespore membrane (FSM), which becomes the plasma membrane of spores. At prophase II, the spindle pole body (SPB) forms an outer plaque, from which formation of the FSM is initiated. Several components of the SPB play an indispensable role in SPB modification, and therefore in sporulation. In this paper, we report the identification of a novel SPB component, Spo7, which has a pleckstrin homology (PH) domain. We found that Spo7 was essential for initiation of FSM assembly, but not for SPB modification. Spo7 directly bound to Meu14, a component of the leading edge of the FSM, and was essential for proper localization of Meu14. The PH domain of Spo7 had affinity for phosphatidylinositol 3-phosphate (PI3P). spo7 mutants lacking the PH domain showed aberrant spore morphology, similar to that of meu14 and phosphatidylinositol 3-kinase (pik3) mutants. Our study suggests that Spo7 coordinates formation of the leading edge and initiation of FSM assembly, thereby accomplishing accurate formation of the FSM.
INTRODUCTION
Sporulation in the fission yeast Schizosaccharomyces pombe is equivalent to gametogenesis in higher eukaryotes, in that this morphogenetic process accompanies meiotic nuclear division and a cell specialization process culminating in formation of ascospores (Shimoda and Nakamura, 2003; Shimoda, 2004). Ascospores are characterized by their dormancy, a high degree of resistance to environmental stress, and increased genetic diversity. S. pombe cells initiate a sporulation program when challenged by nutrient starvation, particularly starvation for nitrogen (Yamamoto et al., 1997). The assembly of a double-unit membrane, called the forespore membrane (FSM), starts during meiosis II. The FSM expands through fusion of membranous vesicles encapsulating a haploid nucleus generated by two rounds of meiotic nuclear division. Spore wall material is then deposited between the inner and outer layers of the FSM (Yoo et al., 1973; Hirata and Tanaka, 1982). The inner layer of the FSM becomes the plasma membrane of newborn spores. The outer layer eventually degrades during sporulation.
One of the most interesting aspects of FSM formation is its initiation because, unlike other biological membranes, the FSM is synthesized de novo within the cytoplasm of the diploid mother cell (Yoo et al., 1973; Hirata and Tanaka, 1982; Tanaka and Hirata, 1982; Nakamura et al., 2001). During meiosis II, FSM formation takes place at the spindle pole body (SPB), which is equivalent to the centrosome of animal cells. The SPB is a proteinaceous structure composed of multiple layers. The S. pombe SPB is located in the cytoplasm very close to the nuclear envelope during interphase, but becomes embedded in the nuclear envelope when cells enter meiosis (Ding et al., 1997). At prophase II, multiple outer plaques are newly formed at the cytoplasmic side of the SPB, as observed by electron microscopy (Hirata and Tanaka, 1982). These morphological alterations of the SPB are referred to as “SPB modification.” SPB modification is also detected by anti-Sad1 antibody as a structural alteration of the SPB from a compact dot to a crescent form at a similar stage of meiosis (Hagan and Yanagida, 1995). Although the relationship between the morphological changes observed by electron microscopy and those seen by fluorescence microscopy remains unclear, this SPB modification is presumed to be indispensable for spore formation.
Isolation and characterization of sporulation-related genes (spo+) have begun to unveil the molecular mechanism of the initiation of FSM formation (Bresch et al., 1968; Kishida and Shimoda, 1986; Shimoda and Nakamura, 2003; Shimoda, 2004). So far, four meiotic SPB components required for FSM formation have been identified (Takeda and Yamamoto, 1987; Ikemoto et al., 2000; Nakase et al., 2008; Itadani et al., 2010). During vegetative growth, the coiled-coil protein Spo15 is a component of the single-plaque SPB (Ikemoto et al., 2000). The calmodulin orthologue Cam1 is essential for localization of Spo15 to the SPB (Itadani et al., 2010). When meiosis I is initiated, two sporulation-specific proteins, Spo2 and Spo13, are newly produced and recruited to the cytoplasmic side of the SPB dependent on the presence of Spo15. When cells enter meiosis II, meiotic outer plaques, probably composed of Spo2 and Spo13, form at the SPB (SPB modification). Localization of Spo13 to the SPB is dependent on Spo2 (Nakase et al., 2008). The FSM, which is continuously associated with the meiotic outer plaque, expands by fusion with membrane vesicles, and eventually forms a nucleated compartment called the “prespore,” a precursor form of the spore. These data show that Spo13 seems to associate closely with the nascent FSM. However, Spo13 is already present at the SPB at meiosis I, considerably earlier than SPB modification. Moreover, Spo13 does not have domains associated with lipid-binding ability (Nakase et al., 2008). Therefore we presume that as yet unknown SPB components that anchor the FSM are involved in meiosis-specific functions.
Morphogenesis of the FSM is also important, as each FSM must accurately encapsulate the dividing nuclei during sporulation. Several proteins involved in FSM morphogenesis have been identified. Pik3/Vps34 is a phosphatidylinositol 3-kinase (PI3K) that catalyzes the production of phosphatidylinositol 3-phosphate (PI3P). Assembly of the FSM still occurs in pik3Δ cells, but the cells exhibit a pleiotropic phenotype in FSM formation, such as aberrant starting positions for expansion, disoriented and insufficient expansion, and failure of closure (Takegawa et al., 1995; Onishi et al., 2003b). Although possible targets of PI3P have been identified (Onishi et al., 2003a, 2007; Koga et al., 2004), how this kinase regulates the initiation of FSM formation (i.e., determination of the starting position for expansion) is still unclear. Septins, a conserved family of GTP-binding proteins, are also involved in spore morphogenesis. In metazoan cells, septins are involved in cytokinesis but are also implicated in a variety of other cellular processes, such as vesicular transport, organization of the actin and microtubule cytoskeletons, and oncogenesis (Spiliotis and Nelson, 2006; Hall et al., 2008). There are seven septins in S. pombe, three of which (spn5+, spn6+, and spn7+) are induced during sporulation. These septins colocalize interdependently to a ring-shaped structure along each FSM, and their mutation results in disoriented FSM expansion. Therefore these septins appear to form a scaffold that helps to guide oriented expansion of the FSM (Onishi et al., 2010). The leading edge structure of the FSM also plays an important role in encapsulating the dividing nucleus. It is thought that the leading edge is coated with leading edge proteins (LEPs) in both Saccharomyces cerevisiae and S. pombe. In S. pombe, meu14+ encodes a LEP. In most meu14Δ cells, the FSM forms in inappropriate places, thus failing to encapsulate the nucleus properly, resulting in ascospores that are abnormal in number and shape. Therefore Meu14 is presumed to guide formation of the FSM (Okuzaki et al., 2003).
For proper FSM formation, morphogenesis of the FSM must be strictly regulated in the early stages of FSM formation. Although a number of proteins responsible for either initiation of FSM assembly or FSM shaping have been identified, the question of how these two events are coordinated is not well understood. In this study, we report a novel meiotic SPB component, Spo7, which contains a pleckstrin homology (PH) domain. Our study suggests that Spo7 coordinates formation of the LEP coat and initiation of FSM assembly, thereby accomplishing accurate FSM formation.
RESULTS
spo7+ encodes a protein that has a PH domain
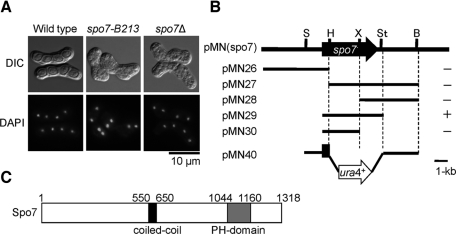
As reported previously, the spo7-B213 mutant exhibits defects in ascospore formation (Bresch et al., 1968; Figure 1A). To identify the spo7+ gene product and its biological function, we isolated the spo7+ gene by functional complementation (Materials and Methods). A plasmid (pMN(spo7)) was isolated that was able to rescue the sporulation deficiency of spo7-B213. Subcloning and partial sequencing revealed that the spo7-complementing ability was due to a single open reading frame (ORF; SPAC6G9.04) of 3.9 kb (Figure 1B). This gene is identical to the previously identified gene mug79+, which was originally identified as a meiosis-up-regulated gene, but its function has not been well analyzed (Martin-Castellanos et al., 2005). We refer to this gene as spo7+ hereafter. The spo7+ gene encodes a 150.9-kDa protein consisting of 1318 amino acids. The predicted Spo7 protein has a coiled-coil domain in its central region and a PH domain in its C-terminal region (Figure 1C). The PH domain is found in proteins related to signal transduction, cytoskeleton, membrane trafficking, and lipid modification, and some of these proteins specifically bind to phospholipids (Yu et al., 2004).
FIGURE 1:
Structure of the spo7+ gene and predicted protein. (A) Differential interference contrast and DAPI-stained images of spo7 mutants. MKW5 (wild type), MN4 (spo7-B213), and MN8 (spo7Δ) strains were incubated at 28°C on SSA for 2 d. Chromosomal DNA was stained with DAPI. (B) Restriction map and subcloning of spo7+. Arrow indicates the region and direction of the spo7+ ORF, which encodes a protein of 1318 amino acids. All of the subclones were derived from pMN(spo7). Complementation of spo7-B213 by each subclone: +, complementation; −, no complementation. Restriction enzyme sites: B, BglII; H, HindIII; S, SpeI; St, StuI. X, XhoI. (C) Schematic diagram of Spo7. The coiled-coil region was predicted by using the program COILS with a 21-residue window setting (Lupas et al., 1991). The putative coiled-coil region (p > 0.8) is shown as a black box; the PH domain is indicated by a gray box.
To investigate the biological function of spo7+, we created a null mutant by conventional gene disruption, using ura4+ as a marker (Figure 1B). The spo7 deletion mutant (spo7Δ) was viable, but displayed a sporulation deficiency similar to the original spo7-B213 mutant (Figure 1A). Because most of the meiosis-defective mutants isolated to date are unable to sporulate (Bresch et al., 1968; Kishida and Shimoda, 1986), we could not rule out the possibility that the spo7Δ mutant had a defect in meiosis. Therefore, we analyzed the meiotic nuclear divisions in spo7Δ. To synchronize meiosis effectively, we used the pat1-114 temperature-sensitive strain, which enters meiosis in a highly synchronous manner when it is shifted to its restrictive temperature, 34°C (Iino et al., 1995). The first and second meiotic divisions of the pat1 spo7Δ cells were found to proceed with kinetics similar to that observed in pat1 cells, with the final yield of tetranucleate cells reaching 90% (Supplemental Figure S1). These results suggest that the spo7Δ mutant is able to complete meiosis but is defective in ascospore formation.
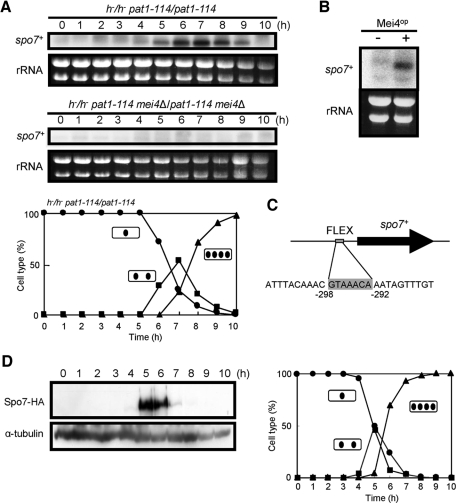
As mentioned above, spo7+/mug79+ was originally identified as a gene that is up-regulated in meiosis (Martin-Castellanos et al., 2005). We therefore examined the expression of the spo7+ gene in greater detail. Northern blot analysis revealed that spo7 mRNA was barely detectable in vegetative cells, but accumulated sharply after shifting to nitrogen-free medium (unpublished data. The exact timing of transcriptional induction during sporulation was further explored using the pat1-114 strain to induce synchronous meiosis. Transcription of spo7+ was induced at 5 h after the temperature shift and peaked at 6–7 h, when cells were in meiosis I (Figure 2A). Because the mei4+ gene encodes a forkhead transcription factor that regulates many genes required for meiosis and sporulation (Horie et al., 1998; Abe and Shimoda, 2000; Mata et al., 2007), we determined whether Mei4 governs spo7+ transcription by examining the induction of spo7+ in the mei4Δ mutant. As shown in Figure 2A, accumulation of spo7+ mRNA was completely abolished in the mei4Δ mutant. Furthermore, ectopic overexpression of mei4+ induced spo7+ mRNA in vegetative cells (Figure 2B). We identified a FLEX-like cis element (GTAAACA), which is used by Mei4 to recognize its target (Horie et al., 1998; Abe and Shimoda, 2000), in the 5′ upstream region of the spo7+ gene (Figure 2C). Taking these results together, we conclude that transcription of spo7+ during meiosis is strictly regulated by Mei4.
FIGURE 2:
Expression of the spo7+ gene. (A) Northern analysis of spo7+ transcripts in pat1-driven meiosis. Synchronous meiosis was initiated in diploid strains homozygous for pat1-114 (JZ670) and pat1-114 mei4 (AB4). At hourly intervals, total RNA was prepared (Jensen et al., 1983) and blotted with a radiolabeled spo7+ DNA fragment. Meiotic nuclear division of pat1-114 (JZ670) was monitored by counting the number of nuclei per cell. Circles, mononucleate cells; squares, binucleate cells; triangles, tetranucleate cells. (B) Effect of ectopic expression of mei4+ on spo7+ transcription. Wild-type cells (TN4) carrying either pREP1 or pREP1(mei4+) were incubated in MM+N at 30°C for 12 h. The approximate quantity of RNA was checked by staining gels with ethidium bromide. (C) Position of the FLEX consensus sequence in the spo7+ promoter region. (D) Changes in Spo7 abundance during meiosis. Cells expressing Spo7-HA (MN48) were allowed to proceed through synchronous meiosis. Aliquots were removed at hourly intervals, and the protein extract was subjected to Western blot analysis with the rat anti-HA antibody 3F10 and the anti–α-tubulin antibody TAT-1 as a loading control. Meiotic nuclear division was monitored by counting the number of nuclei per cell. Circles, mononucleate cells; squares, binucleate cells; triangles, tetranucleate cells.
The abundance of Spo7 during meiosis was monitored using a chromosomally integrated gene expressing Spo7-hemagglutinin (HA). A single copy of the spo7-HA fusion gene completely complemented the sporulation defect of the spo7Δ mutant, showing that Spo7-HA is fully functional. An MN48 strain carrying the pat1-114 mutation and Spo7-HA was cultured at 34°C to induce synchronous meiosis. Spo7-HA was not detected in vegetative cells (at 0 h), but appeared after the temperature shift (at 5 h) as a 160-kDa band on SDS–PAGE (Figure 2D). This apparent molecular mass was consistent with that deduced from the sequence data. Spo7-HA became more abundant coincident with the appearance of tetranucleate cells (5–6 h). Once meiosis was completed, Spo7-HA was no longer detected. We conclude that Spo7 is transiently produced in meiotic cells.
Spo7 is a novel meiotic component of the SPB
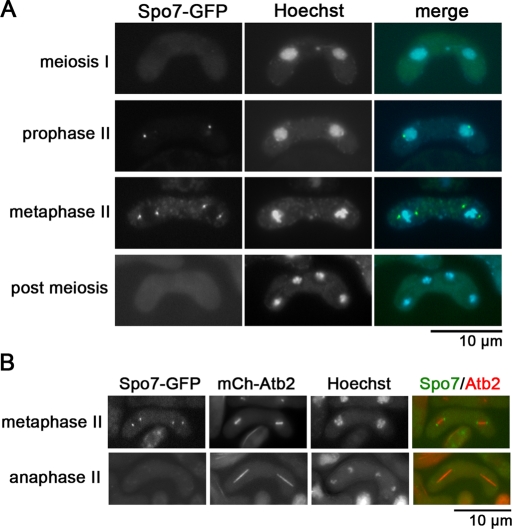
Subcellular localization of Spo7 was observed by fluorescence microscopy. Consistent with the Western blot data, an Spo7–green fluorescent protein (GFP) signal was not observed in vegetative cells or in early meiosis, but appeared in meiosis II as one or two dots in the periphery of nuclei (Figure 3A). These dots were detected at both ends of the spindle microtubules, which were visualized by mCherry-labeled Atb2, at metaphase II (Figure 3B), indicating that Spo7 localizes to the SPB. The Spo7 signal disappeared before spindle breakdown (anaphase II, Figure 3B).
FIGURE 3:
Localization of Spo7 during meiosis and sporulation. (A) Localization of Spo7. Homothallic haploid wild-type cells expressing Spo7-GFP (MN10) were sporulated on SSA to induce meiosis. Chromosomal DNA was stained with Hoechst 33342 and analyzed by fluorescence microscopy. Spo7-GFP (green) and Hoechst 33342 (blue) are overlaid in the merged images. (B) Dual observation of Spo7 and microtubules. The homothallic haploid strain TN450 expressing Spo7-GFP and mCherry-Atb2 was sporulated on SSA for 2 d and analyzed by fluorescence microscopy. Spo7-GFP (green) and mCherry-Atb2 (red) are overlaid in the merged images.
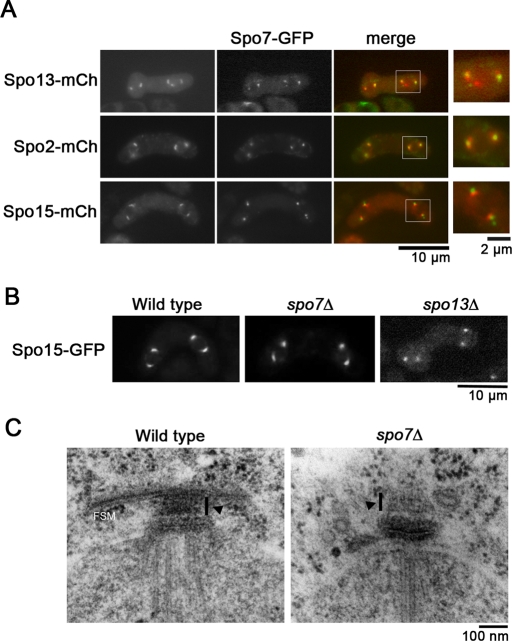
Initiation of FSM formation is regulated by meiotic SPB components, including Spo2, Spo13, and Spo15. To examine the relationship between Spo7 and these other meiotic SPB components, we simultaneously expressed Spo7-GFP and Spo2-mCherry, Spo13-mCherry or Spo15-mCherry in a cell. As shown in Figure 4A, Spo7-GFP was in close contact with each of the three proteins at the SPB. Interestingly, the Spo7-GFP signal was positioned outside the Spo15-mCherry signal at the SPB, but overlapped with Spo2-mCherry and Spo13-mCherry. In addition, the Spo7-GFP signal essentially showed a dot-like pattern but was occasionally accompanied by a faint crescent signal around the dots. However, this pattern was clearly different from those of Spo13-mCherry, Spo2-mCherry, and Spo15-mCherry, which showed a homogeneous crescent signal (Figure 4A). Recruitment of Spo2, Spo13, and Spo15 to the SPB is strictly controlled: localization of Spo13 to the SPB depends on Spo15 and Spo2, whereas that of Spo2 depends only on Spo15 (Nakase et al., 2008). We next examined whether Spo7 localizes to the SPB in mutants of the other meiotic SPB components. As shown in Figure S2A, Spo7 localized to the SPB in each of the spo2, spo13, and spo15 mutant cells. Reciprocally, Spo13, Spo2, and Spo15 each localized to the SPB in spo7 mutants (Figure S2B). Therefore recruitment of Spo7 to the SPB is independent of other meiotic SPB components. Recently we have shown that the calmodulin orthologue Cam1 regulates initiation of FSM formation by recruiting meiotic SPB components to the SPB. In cam1 mutant cells, Spo13, Spo2, and Spo15 all fail to localize to the SPB (Itadani et al., 2010). However, localization of Spo7 at the SPB was not affected by the cam1 mutation (unpublished data).
FIGURE 4:
Morphological changes in the SPB in spo7Δ. (A) Dual observation of Spo7 and other meiotic SPB components. Homothallic haploid strains expressing Spo7-GFP and Spo13-mCherry (TN451), Spo2-mCherry (MN220), or Spo15-mCherry (MN187) were sporulated on SSA for 2 d and analyzed by fluorescence microscopy. Spo7-GFP (green) and Spo13-mCherry, Spo2-mCherry, or Spo15-mCherry (all shown red) are overlaid in the merged images. High-magnification images of the region indicated by the white square are shown on the right. (B) Morphological changes in the spo7Δ SPB during meiosis. Wild-type (MN109), spo7Δ (MN101), and spo13Δ (YN90) were sporulated on SSA. The Spo15-GFP signal in each strain was analyzed by fluorescence microscopy. (C) Fine structures of the SPB in wild-type and spo7Δ strains. Wild-type (MKW5) and spo7Δ (MN8) cells sporulated on SSA at 28°C for 1 d were observed by electron microscopy. Arrowheads indicate the meiotic outer plaque of the SPB.
We examined the physical interaction of Spo7 with other meiotic SPB components by yeast two-hybrid analysis. The colony formation assay indicated a positive interaction between Spo7 and Spo13. However, no interaction of Spo7 with Spo2 or Spo15 was observed (Figure S2C). The interaction between Spo7 and Spo13 was also detected in S. pombe cells (unpublished data). An interaction was also observed between Spo7 and Spo7 (Figure S2C).
We next examined the expression profile of the meiotic SPB components in detail. During meiosis I, an Spo13-GFP signal was observed at the SPB, whereas Spo7-cyan fluorescent protein (CFP) was not detected (Figure S3A), indicating that Spo13 is recruited to the SPB prior to Spo7. Consistently, Western blot analysis showed that both Spo7-HA and Spo13-GFP were expressed exclusively during meiosis II, but Spo13-GFP was expressed slightly earlier than Spo7-HA (Figure S3B).
Spo7 is essential for initiation of FSM formation
Assembly of the FSM initiates in the vicinity of the SPB during meiosis II (Hirata and Tanaka, 1982; Nakamura et al., 2008). Because Spo7 was found to be associated with the SPB prior to spore formation, we presumed that modification of the SPB would be impaired by the spo7Δ mutation. Fluorescence microscopic analysis using Spo15-GFP showed that the modified crescent-shaped SPBs were not present in spo13Δ cells during the second meiotic division (Figure 4B). Interestingly, unlike spo13Δ, most Spo15-GFP signals showed a crescent shape in spo7Δ cells (Figure 4B). The fine structures of the meiotic SPB in spo7Δ cells were compared with those of the SPB in wild-type strains at the same stage. Our previous study showed that spo15Δ cells form no apparent outer plaques (Nakase et al., 2008). Interestingly, as shown in Figure 4C, spo7Δ cells seemed to form outer plaques, although the density of these outer plaques seemed to be slightly reduced. Taken together, these data indicate that SPB modification occurs in spo7Δ cells.
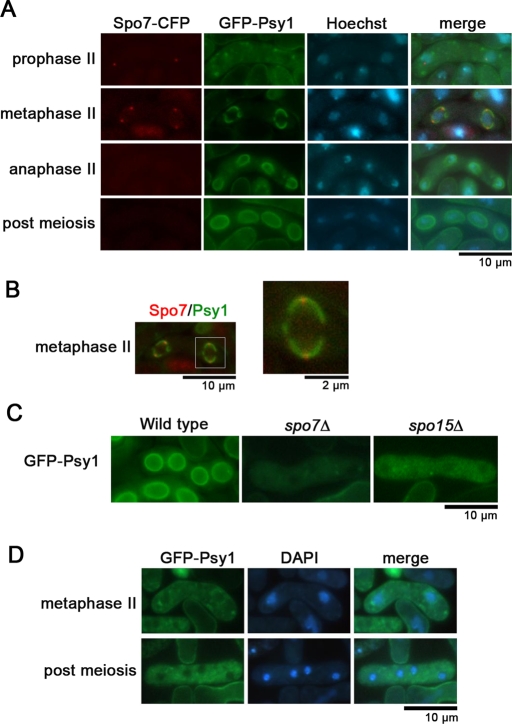
Assembly of the FSM initiates from the outer plaque of the SPB (Hirata and Tanaka, 1982). To observe Spo7 and the FSM simultaneously, the FSM was visualized by using GFP-tagged Psy1, an FSM-resident protein (Nakamura et al., 2001). The nascent FSM was observed to overlap with Spo7-CFP (Figure 5, A and B), suggesting a close relationship between Spo7 and the FSM. We next examined FSM formation in spo7Δ cells. In wild-type cells, the FSM assembled normally as a sac (Figure 5C); by contrast, membrane formation was completely inhibited in spo7Δ cells (Figure 5D). This phenotype was similar to that seen in mutants of the other meiotic SPB components such as spo15 (Figure 5C; Nakase et al., 2008). Furthermore, electron microscopy showed that no FSM was observed near the SPB in spo7Δ cells, although membrane vesicles were observed (Figure 4B). Taken together, these data suggest that Spo7 is indispensable for initiating FSM formation and is associated closely with the nascent FSM.
FIGURE 5:
FSM formation in spo7Δ. (A) Dual observation of Spo7-CFP and the FSM. Wild-type cells (MN56) expressing Spo7-CFP and GFP-Psy1 were sporulated on SSA and were stained with Hoechst 33342. (B) Merged image of Spo7-CFP and the FSM visualized by GFP-Psy1 of (A) (metaphase II). A high-magnification image of the region indicated by the white square is shown on the right. (C) FSM formation in spo7Δ. Wild-type (YN68), spo7Δ (MN103), and spo15Δ (YN67) strains expressing GFP-Psy1 were sporulated on SSA at 28°C for 1 d and were observed by fluorescence microscopy. (D) Initiation of the FSM is defective in spo7Δ. spo7Δ cells (MN103) expressing GFP-Psy1 were sporulated on SSA at 28°C for 1 d and observed by fluorescence microscopy.
Spo7 is essential for recruitment of Meu14 to the SPB
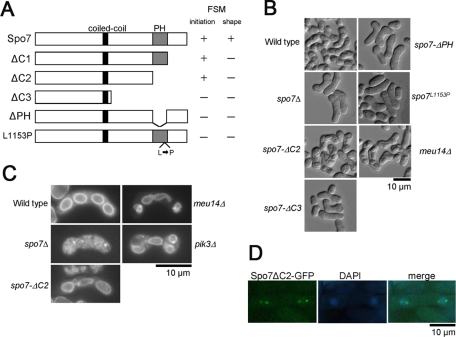
To determine the in vivo function of the PH domain of Spo7, we constructed a series of deletion mutants, as well as a point mutant of spo7 (Figure 6A). These mutant spo7 genes were introduced into the leu1locus in spo7Δ cells. The spo7-ΔC1 mutant, lacking the C-terminal 158 amino acids (Figure 6A), could form spores, but the shapes of the spores were markedly abnormal (unpublished data). Similar results were obtained for the spo7-ΔC2 mutant, in which both the PH domain and the C-terminal region were deleted (Figure 6, B and C). The spo7-ΔC3 mutation, lacking the C-terminal 451 amino acids (Figure 6A), completely inhibited FSM formation, similar to the spo7Δ mutation (Figure 6, B and C). These data indicate that the C-terminal region of Spo7 is not essential for initiation of FSM assembly but is required for proper shaping of the FSM. The spo7-ΔC2 mutant protein localized to the SPB (Figure 6D), indicating that the defect in the spore morphology of spo7-ΔC2 is not due to mislocalization of the protein to the SPB. Interestingly, similar to spo7Δ, the spo7-ΔPH mutant, lacking only the PH domain (Figure 6A), did not form spores. In other words, the spo7-ΔPH mutation exhibited a more severe effect than the spo7-ΔC2 mutation.
FIGURE 6:
Function of the PH domain of Spo7. (A) Schematic illustration of the spo7 deletion mutants. (B) Sporulation in the spo7 mutants. Wild-type (YN68), spo7Δ (MN103), spo7-ΔC2 (MN271), spo7-ΔC3 (MN265), spo7-ΔPH (MN263), spo7L1153P (MN140), and meu14Δ (MN133) cells were sporulated on SSA at 28°C for 2 d. (C) FSM formation in the spo7 mutants. Wild-type (YN68), spo7Δ (MN103), spo7-ΔC2 (MN271), spo7-ΔPH (MN263), spo7-L1153P (MN140), and meu14Δ (MN133) cells expressing GFP-Psy1 were sporulated on SSA at 28°C for 2 d and observed by fluorescence microscopy. (D) Localization of Spo7ΔC2. Wild-type cells expressing Spo7ΔC2-GFP (MN75) were sporulated on SSA at 28°C for 1 d.
To assess the role of the PH domain, we constructed an additional mutant, spo7L1153P, in which a conserved leucine residue (1153) in the PH domain was replaced with proline (Figure 6A). spo7L1153P exhibited a phenotype similar to spo7-ΔPH (Figure 6B). These data indicate that the PH domain of Spo7 is not required for initiation of FSM assembly but is necessary for proper shaping of the FSM. In addition, these data also suggest that the C-terminal region is important for the function of the PH domain. Therefore we subsequently used the spo7-ΔC2 strain to examine the function of the PH domain.
As mentioned above, two-hybrid analysis showed the dimerization of Spo7. To determine whether dimerization of Spo7 is required for sporulation, we examined the interaction of various spo7-deletion mutant proteins with full-length Spo7 using yeast two-hybrid analysis. Whereas Spo7-ΔC2 interacted with Spo7, Spo7-ΔC3 and Spo7-ΔPH did not (Figure S4). Given that spo7-ΔC2, but not spo7-ΔC3 or spo7-ΔPH, initiates FSM formation, these data suggest the possibility that Spo7 dimerization is important for FSM initiation.
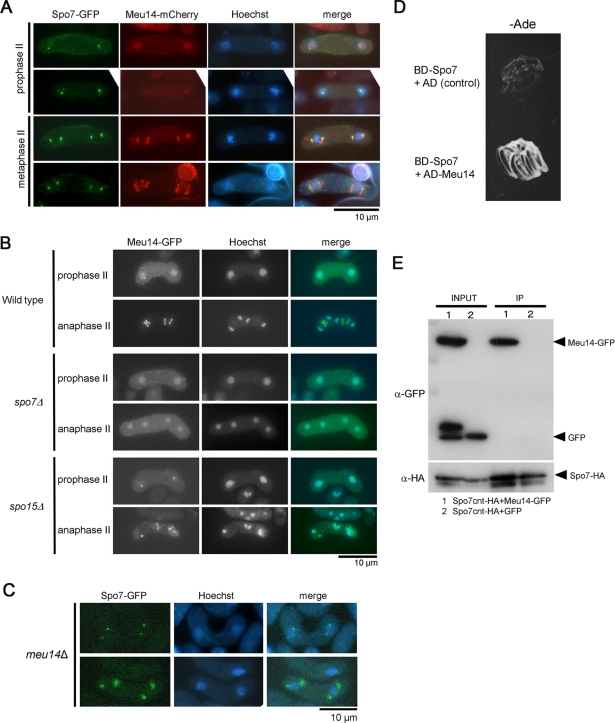
The phenotype of the spo7-ΔC2 mutant seemed reminiscent of the prespores with abnormal spore morphology observed in meu14 mutants. The meu14+ gene was originally identified as the meu gene, the expression of which is up-regulated during meiosis (Watanabe et al., 2001; Okuzaki et al., 2003). Meu14 localizes to the SPB during early meiosis II, and subsequently forms a ring structure at the leading edge of the FSM. The meu14 null mutant forms spores with an abnormal shape (Figure 6B; Okuzaki et al., 2003). Thus Meu14 plays an important role in spore morphogenesis. Therefore, we examined the relationship between Spo7 and Meu14. First, localization of these proteins was observed. At an early stage of prophase II, Spo7-GFP fluorescence was observed at the SPB, whereas Meu14-mCherry was dispersed in the nucleus (Figure 7A). Subsequently, Meu14-mCherry accumulated at the SPB and was overlapped almost completely by Spo7-GFP, indicating that Spo7 is recruited to the SPB prior to Meu14. At metaphase II, Meu14-mCherry formed ring-like structures, whereas Spo7-GFP persisted at the SPB (Figure 7A).
FIGURE 7:
Relationship between Spo7 and Meu14. (A) Dual observation of Spo7 and Meu14. Wild-type cells (MN225) expressing integrated Spo7-GFP and Meu14-mCherry were sporulated on SSA at 28°C for 1 d and analyzed by fluorescence microscopy. (B) Localization of Meu14 in spo7Δ and spo15Δ. Wild-type (YN314), spo7Δ (MN156), and spo15Δ (MN219) mutants expressing Meu14-GFP were sporulated on SSA at 28°C for 1 d. Meu14-GFP (green) and Hoechst 33342 (blue) are overlaid in the merged images. (C) Localization of Spo7 in meu14Δ. meu14Δ cells expressing Spo7-GFP (MN77) were sporulated on SSA at 28°C for 1 d and analyzed by fluorescence microscopy. (D) Yeast two-hybrid analysis between Spo7 and Meu14. spo7+ was cloned into pGBT9 (Clontech), which contains the DNA-binding domain of the GAL4 gene (BD). meu14+ was cloned into pGAD424 (Clontech), which contains the activation domain of the GAL4 gene (AD). Plasmids carrying these fusion constructs were introduced into the S. cerevisiae tester strain (AH109). The assay was done by monitoring growth of the host cells on adenine-depleted medium. “AD (Control)” indicates pGAD424. (E) Physical interaction between Spo7 and Meu14. GFP or Meu14-GFP was coexpressed with Spo7cnt-HA in wild-type cells (TN29). Whole-cell lysates (input) were subjected to immunoprecipitation with the rat anti-HA antibody 3F10 (IP). Precipitates were analyzed by Western blotting using anti-HA (12CA5) and anti-GFP antibodies.
We next determined the localization of Meu14 in spo7Δ cells. Interestingly, in spo7Δ cells, Meu14-GFP fluorescence was observed neither at the SPB nor at the leading edge of the FSM, but in the nucleus (Figure 7B). To examine whether the defect in Meu14 localization to the leading edge is a consequence of the defect in FSM formation, we observed the behavior of Meu14-GFP in spo15Δ cells, in which FSM formation is completely defective. As shown in Figure 7B, dots and ring-like structures of Meu14-GFP were observed in spo15 cells, although the size of rings was markedly reduced. Thus, Spo7 is essential for recruitment of Meu14 to the SPB. By contrast, localization of Spo7-GFP was not affected by the meu14Δ mutation (Figure 7C).
Next the physical interaction between Spo7 and Meu14 was examined by yeast two-hybrid analysis. The colony formation assay showed that Spo7 positively interacted with Meu14 (Figure 7D). These results led us to analyze potential in vivo interactions in S. pombe between Spo7 and Meu14. These polypeptides could not be detected by Western blotting in lysates prepared from sporulating cells. Therefore cell-free extracts were prepared from vegetative cells. Furthermore, because full-length Spo7 was not soluble, a Spo7 cnt polypeptide containing a partial region of Spo7 (aa 363–1043) was coexpressed with Meu14-GFP for these experiments. Spo7cnt-HA was immunoprecipitated with anti-HA antibody, and copurification of Spo7cnt-HA was verified by Western blotting using anti-GFP antibody. As shown in Figure 7E, Meu14-GFP, but not control GFP, was copurified with Spo7cnt-HA, indicating that Spo7 interacts directly with Meu14.
The PH domain of Spo7 has affinity for PI3P
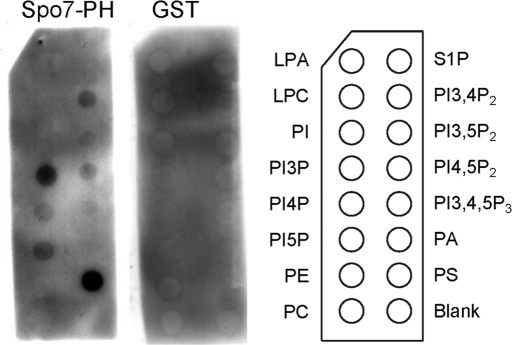
To assess the lipid-binding activity of the Spo7 PH domain, we used a protein–lipid overlay assay (Dower and Mattheakis, 2002). This assay has been used extensively to characterize the lipid-binding specificity of proteins. The PH domain of Spo7 fused with glutathione S-transferase (GST) was used to probe a panel of lipids spotted onto nitrocellulose membranes (PIP Strip; Echelon Biosciences, Salt Lake City, UT). As shown in Figure 8, Spo7 specifically bound to PI3P and phosphatidylserine (PS), whereas GST alone showed no detectable binding.
FIGURE 8:
Protein–lipid overlay assays using PIP Strips for detecting binding between the indicated lipids and the PH domain of Spo7. Membranes containing the indicated phospholipids (100 pmol per spot) were incubated with the purified GST-Spo7-PH fusion protein or control GST at 200 ng/ml. The membranes were washed, and GST fusion proteins were detected by anti-GST antibody. LPA, lysophosphatidic acid; S1P, sphingosine-1-phosphate; LPC, lysophosphatidylcholine; PI; PA, phosphatidic acid; PE, phosphatidylethanolamine; PS; PC, phosphatidylcholine.
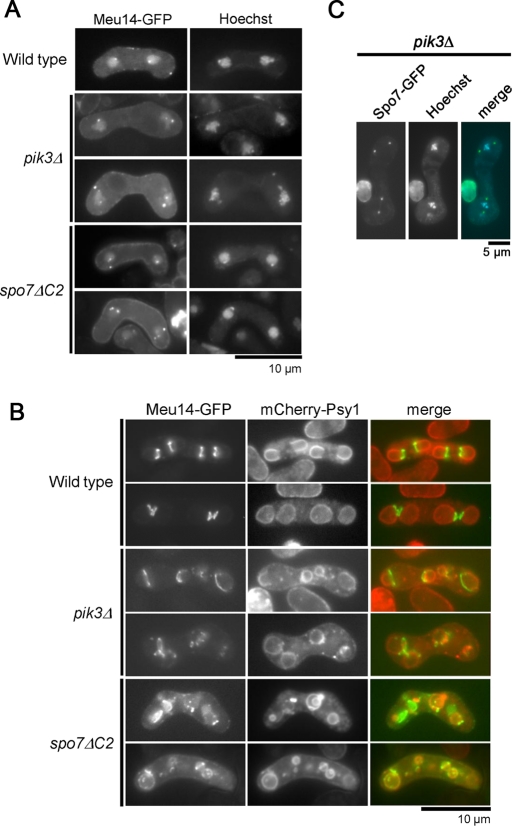
We next examined the role of PI3P in FSM formation. In S. pombe, PI3P is produced from phosphatidylinositol (PI) by Pik3, the sole PI3 kinase. PI3P is not detectable in pik3Δ cells (Takegawa et al., 1995). The pik3Δ mutant shows a pleiotropic phenotype, including a conjugation defect and abnormal vacuole morphology (Takegawa et al., 1995). In addition, pik3Δ cells show a defect in sporulation (Onishi et al., 2003a): growth of the FSM in pik3Δ cells is incorrectly orientated, failing to engulf the nucleus or forming extremely small spores. Therefore, we next observed the behavior of Meu14 in pik3Δ mutants. In wild-type cells, Meu14-GFP was observed as one or two dot(s) at the periphery of each of the two nuclei at early meiosis II (Figure 9A). Although Meu14-GFP dots were also observed in pik3Δ cells, the number of dots in each nucleus was often more than two; in addition, the size of the Meu14-GFP dots in each nucleus differed considerably (Figure 9A). During FSM expansion in wild-type cells, four Meu14-GFP rings localized at the leading edge of the FSM sac and expanded in a synchronous manner (Figure 9B). In pik3Δ cells, by contrast, abnormal Meu14 rings caused by loss of synchrony were observed. Furthermore, the Meu14 rings were often fragmented (Figure 9B). Thus these data indicate that PI3P is important for proper assembly of the Meu14 ring. Spo7-GFP localized properly to the SPB in pik3Δ cells (Figure 9C), suggesting that the abnormal localization of Meu14 is not due to the mislocalization of Spo7 to the SPB.
FIGURE 9:
Abnormal FSM formation in spo7ΔC2 and pik3Δ. (A) Localization of Meu14 in early meiosis II. Wild-type (TN443), spo7ΔC2 (TN444), and pik3Δ (TN445) cells expressing Meu14-GFP were sporulated on SSA at 28°C for 1 d, stained with Hoechst 33342, and analyzed by fluorescence microscopy. (B) FSM formation in spo7ΔC2 and pik3Δ. Wild-type (TN443), spo7ΔC2 (TN444), and pik3Δ (TN445) cells expressing Meu14-GFP and mCherry-Psy1 were sporulated on SSA at 28°C for 1 d and analyzed by fluorescence microscopy. Meu14-GFP (green) and GFP-Psy1 (red) are overlaid in the merged images. (C) Localization of Spo7-GFP in pik3Δ. pik3Δ (TN449) cells expressing Spo7-GFP were sporulated on SSA at 28°C for 1 d, stained with Hoechst 33342, and analyzed by fluorescence microscopy. Spo7-GFP (green) and Hoechst 33342 (blue) are overlaid in the merged images.
We also examined the behavior of Meu14 in the spo7-ΔC2 mutant lacking the PH domain. Unlike in spo7Δ cells, Meu14 rings were observed in spo7-ΔC2 cells. However, formation of the Meu14 dots and rings was apparently abnormal in spo7-ΔC2 cells, similar to pik3Δ cells (Figure 9, A and B). Taken together, these data suggest that the PH domain of Spo7 is important for proper assembly of Meu14 protein, which is critical to shaping of the FSM.
DISCUSSION
The SPB serves as a platform for FSM assembly in addition to its function as a microtubule-organizing center. The present study raises the possibility that Spo7 is the most membrane-proximal protein among the known meiotic SPB components for the following reasons. First, Spo7 was exclusively produced at meiosis II, a stage when SPB modification and initiation of FSM formation occur, whereas other meiotic SPB components, including Cam1, Spo15, Spo2, and Spo13 are present at the SPB from meiosis I onwards. Second, Spo7 was observed by fluorescence microscopy to localize at the outermost region of the cytoplasmic side of the SPB and to overlap with the FSM. Third, the spo7Δ mutation completely inhibited FSM formation, but not SPB modification. In contrast, neither SPB modification nor FSM formation occurs in mutants of other meiotic SPB components (Ikemoto et al., 2000; Nakase et al., 2008; Itadani et al., 2010). At present, however, the mechanism by which Spo7 initiates assembly of the FSM remains unclear. Recently Yang and Neiman (2010) reported that Spo13 acts as a guanine nucleotide exchange factor (GEF) for the small Rab GTPase. Although the in vivo target of Spo13 is unknown, Spo13 binds preferentially to the nucleotide-free form of Ypt2, an orthologue of S. cerevisiae Sec4, which regulates vesicle trafficking from the late Golgi to the plasma membrane (Yang and Neiman, 2010). These results demonstrate that stimulation of Rab GTPase activity is essential for initiation of FSM formation. Because Spo7 interacted with Spo13, Spo7 might be involved in the initiation of FSM assembly by regulating the activity of Rab GTPase. Further studies will be required to identify molecules that physically interact with the Spo7 protein during initiation of FSM formation.
Unexpectedly, the PH domain of Spo7 was not essential for initiation of FSM assembly. The spo7-ΔC2 mutant, which lacks the C-terminal region including the PH domain, showed a phenotype similar to that of meu14Δ and pik3Δ mutants with regard to defects in spore morphology. Meu14 first collects at the SPB, and subsequently forms a ring structure at the leading edge of the FSM sac (Okuzaki et al., 2003). Therefore the accumulation of Meu14 at the SPB might be a prerequisite for localization to the leading edge (Okuzaki et al., 2003). Spo7 physically interacted with Meu14 and was essential for localization of Meu14 to the SPB. However, Meu14 rings, albeit small, were formed in spo15Δ cells, indicating that assembly of leading edge proteins does not require FSM formation. We suggest that Spo7 has at least two independent roles in sporulation: one is the initiation of FSM formation, and the other is the recruitment of Meu14 to the SPB. Because Spo7 can localize to the SPB in spo15Δ cells, we presume that the Meu14 ring can be assembled even in the absence of the FSM. Thus, Spo7 might coordinate the formation of the Meu14 ring and the initiation of FSM assembly, possibly ensuring the precise spatial and temporal control of spore shape.
Our protein–lipid overlay assay showed that the PH domain of Spo7 bound to PI3P. It is known that pik3Δ cells exhibit defects in various steps of FSM formation, such as aberrant starting positions for expansion, disoriented FSM expansion, inefficient FSM expansion, and failure of closure of the leading edge (Onishi et al., 2003a). Some of the possible targets of PI3P during sporulation have been identified, including two sorting nexins, Vps5 and Vps17, and the FYVE domain–containing protein Sst4/Vps27. Vps5 and Vps17 have a phox homology (PX) domain, and analysis of homologues of these proteins in S. cerevisiae suggests that Vps5 and Vps17 in S. pombe act as PI3P-dependent mediators of retrograde trafficking from the endosome to the Golgi apparatus, which in turn acts as an efficient anterograde membrane flux to the FSM (Horazdovsky et al., 1997; Pfeffer, 2001; Onishi et al., 2003a; Koga et al., 2004). In contrast, Sst4 functions at later stages of FSM formation, specifically when the FSM closes (Onishi et al., 2007). However, the target of PI3P that regulates spore morphology at the initial step of FSM formation is still unknown. Interestingly, previous studies by electron microscopy have shown that the tips of the FSM are swollen and fuzzy in pik3Δ cells (Onishi et al., 2003a). These abnormal tips are also observed in meu14Δ cells (Okuzaki et al., 2003). Indeed, our results indicated that the Meu14 ring is often abnormally assembled in pik3Δ cells. Therefore, the most plausible possibility is that PI3P is involved in leading edge ring formation via the PH domain of Spo7. At present, however, we have no evidence that PI3P directly regulates Spo7 in vivo. PH domains have been suggested to bind many proteins, but to date their only clearly demonstrated physiological function is to bind membrane phospholipids (Yu et al., 2004). These facts suggest another possibility: the C-terminal or the PH domain of Spo7 may directly interact with Meu14. However, an Spo7 mutant lacking PH domain still interacted with Meu14 (Figure 7E). We presume that PI3P might influence the interaction between Spo7 and Meu14, thereby facilitating accurate formation of the Meu14 ring and initiation of FSM assembly from the SPB.
The protein–lipid overlay assay also showed that the PH domain interacts with PS, a quantitatively minor phospholipid in eukaryotic cells. S. pombe has a sole PS synthase, Pps1. In medium lacking ethanolamine, pps1Δ mutants exhibit striking cell morphology, with defects in cytokinesis, actin cytoskeleton cell wall remodeling, and integrity (Matsuo et al., 2007). The phenotype of pps1Δ in sporulation has not been investigated. At present, we cannot rule out the possibility that Spo7 is involved in sporulation via binding to PS; however, we presume that PI3P is more important for sporulation, because the phenotype of spo7-ΔC2 cells was similar to that of pik3Δ cells.
It is known that PI3P also plays an important role in autophagy, a major eukaryotic process by which bulk cytoplasmic components are degraded in the vacuole/lysosome. In response to starvation, membrane structures known as autophagosomes nonselectively envelop a fraction of the cytoplasm, including its resident organelles, and target the fraction to the vacuole/lysosome, where the contents are degraded to recycle the components. Like sporulation, formation of the autophagosome is accompanied by de novo assembly of a double-unit membrane (Baba et al., 1994). PI3P is also required for initiation of autophagosomal membrane formation. Thus, regulation of the initiation of membrane assembly by PI3P might be a general mechanism in eukaryotes.
In S. cerevisiae, initiation of the prospore membrane (PSM), which corresponds to the FSM, is very similar to that in S. pombe: PSM formation begins with a modification of the SPB, and the membrane vesicles gather at the modified SPB and fuse to expand the membrane (Byers, 1981; Neiman, 2005). Four meiotic SPB components, Mpc54p, Mpc70p/Spo21p, Spo74p, and Ady4p, are known to localize at the outer plaque of the SPB and are essential for initiation of PSM assembly (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003). Despite their similar roles in sporulation, however, these proteins share no similarity with S. pombe components of the meiotic SPB and do not have domains suggesting lipid-binding ability. A physical interaction between meiotic SPB components and LEPs is also observed in S. cerevisiae. Furthermore, deletion of SSP1, an LEP, causes defects in spore morphogenesis: PSMs are still formed, but they are grossly abnormal. The membranes occasionally grow in the wrong direction, resulting in a failure to capture nuclei (Moreno-Borchart et al., 2001). These phenotypes are very similar to those of meu14Δ cells. Interestingly, however, S. cerevisiae LEPs first appear in the cytoplasm and then gather at the SPB, whereas Meu14 appears in the nucleus. In the present study, Spo7 was essential for recruitment of Meu14 to the SPB, and Meu14 persisted in the nucleus in spo7 mutants, even when meiosis II progressed. Similar to other meiotic SPB components, a protein homologous to Spo7 does not occur in S. cerevisiae. Therefore Spo7 might regulate transport of the LEP Meu14 from the nucleus to the SPB, a process specifically seen in S. pombe.
On the basis of the results from the present study, together with previous data, we propose a model in which Spo7 regulates FSM formation. During meiosis I, Spo2 and Spo13 are expressed and localized to the SPB. When meiosis II starts, Spo7 localizes to the SPB at the outermost side and regulates proper assembly of the Meu14 ring. PI3P might regulate Spo7 function. Coincidently, Spo7 also regulates initiation of membrane vesicle fusion at the SPB, possibly by interacting with Spo13. The leading edge may control morphogenesis of the FSM, and is thus required for proper sporulation.
In summary, our results indicate that Spo7 provides a link between initiation of FSM formation and spore morphogenesis. Future studies of the function of Spo7 and this model system will help elucidate the more general questions of how membrane formation initiates and how the FSM obtains its shape in vivo.
MATERIALS AND METHODS
Yeast strains, media, and plasmids
The S. pombe strains and plasmids used in this study are listed in Tables 1 and 2, respectively. A sporulation-deficient mutant of S. pombe, spo7-B332, was originally isolated by Bresch et al. (1968), and was provided by R. Egel (University of Copenhagen, Copenhagen, Denmark). The media used in this study have been previously described (Egel and Egel-Mitani, 1974; Gutz et al., 1974; Moreno et al., 1991. S. pombe cells were grown at 30°C and sporulated at 28°C, unless stated otherwise. Synchronous meiosis was induced by a temperature shift using strains carrying the pat1-114 allele as described (Iino et al., 1995). S. cerevisiae strain AH109 (Clontech) was used for two-hybrid analysis.
TABLE 1:
Strains used in this study. (Continued)
| Strain (accession no.)a | Genotypeb | Source |
|---|---|---|
| S. pombe | ||
| B213 | h90 spo7-B213 ade6-M210 | Bresch et al., 1968 |
| MKW5 (FY7456) | h90 | Nakamura-Kubo et al., 2003 |
| MN4 (FY21065) | h90 spo7-B213 | This study |
| MN8 (FY21067) | h90 spo7::ura4+ ura4-D18 | This study |
| MN10 (FY21068) | h90 spo7-GFP<<LEU2 leu1-32 | This study |
| MN24 (FY21069) | h90 spo2::ura4+ spo7-GFP<<LEU2 leu1-32 ura4-D18 | This study |
| MN26 (FY21070) | h90 spo13::ura4+ spo7-GFP<<LEU2 leu1-32 ura4-D18 | This study |
| MN28 (FY21071) | h90 spo15::ura4+ spo7-GFP<<LEU2 leu1-32 ura4-D18 | This study |
| MN48 (FY21072) | h−/h− pat1-114/pat1-114 spo7-HA<<LEU2/spo7-HA<<LEU2 ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | This study |
| MN49 (FY21073) | h−/h− pat1-114/pat1-114 spo7::ura4+/spo7::ura4+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | This study |
| MN56 (FY21074) | h90 spo7-CFP<<ura4+ leu1<<GFP-psy1 leu1-32 ura4-D18 | This study |
| MN69 (FY21077) | h90 spo7-CFP<<ura4+ leu1-32 ura4-D18 pAL(spo13-GFP) | This study |
| MN75 (FY21079) | h90 spo7ΔC2-GFP<<LEU2 leu1-32 | This study |
| MN77 (FY21351) | h90 meu14::ura4+ spo7-GFP<<LEU2 leu1-32 ura4-D18 | This study |
| MN78 (FY21352) | h−/h− pat1-114/pat1-114 spo7-HA<<LEU2/spo7-HA<<LEU2 spo13+/spo13-GFP<<<ura4+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | This study |
| MN99 (FY21080) | h90 spo7::ura4+ leu1+<<spo2-GFP leu1-32 ura4-D18 | This study |
| MN100 (FY21081) | h90 spo7::ura4+ leu1+<<spo13-GFP leu1-32 ura4-D18 | This study |
| MN101 (FY21082) | h90 spo7::ura4+ spo15-GFP<<LEU2 leu1-32 ura4-D18 | This study |
| MN103 (FY21083) | h90 spo7::ura4+ leu1+<<GFP-psy1 leu1-32 ura4-D18 | This study |
| MN109 (FY21084) | h90 spo15-GFP<<LEU2 leu1-32 | This study |
| MN133 (FY21086) | h90 meu14::ura4+ leu1+<<GFP-psy1 leu1-32 ura4-D18 | This study |
| MN140 (FY21087) | h90 spo7::ura4+ leu1+<<spo7-L1153P leu1-32 ura4-D18 | This study |
| MN156 (FY21088) | h90 spo7::ura4+ leu1+<<meu14-GFP ade6-M216 leu1-32 ura4-D18 | This study |
| MN187 (FY21353) | h90 spo7-GFP<<ura4+ spo15-mCherry<<LEU2 ade6-M216 leu1-32 ura4-D18 | This study |
| MN219 (FY21091) | h90 spo15::ura4+ leu1+<< meu14-GFP leu1-32 ura4-D18 | This study |
| MN220 (FY21354) | h90 spo7-GFP<<LEU2 spo2-mCherry<<ura4+ ade6-M216 leu1-32 ura4-D18 | This study |
| MN221 (FY21092) | h90 pik3::ura4+ leu1+<<meu14-GFP leu1-32 ura4-D18 | This study |
| MN225 (FY21089) | h90 spo7-GFP<<ura4+ leu1+<<meu14-mCherry ade6-M216 leu1-32 ura4-D18 | This study |
| MN263 (FY21095) | h90 spo7::ura4+ leu1+<<spo7ΔPH ade6+<<GFP-psy1 ade6-M216 ura4-D18 leu1-32 | This study |
| MN265 (FY21096) | h90 spo7::ura4+ leu1+<<spo7ΔC3 ade6+<<GFP-psy1 ade6-M216 ura4-D18 leu1-32 | This study |
| MN271 (FY21097) | h90 spo7::ura4+ leu1+<<spo7ΔC2 ade6+<<GFP-psy1 ade6-M216 ura4-D18 leu1-32 | This study |
| JZ670 (FY7051) | h−/h− pat1-114/pat1-114 ade6-M210/ade6-M216 leu1-32/leu1-32 | Yamamoto |
| AB4 (FY7476) | h−/h− pat1-114/pat1-114 mei4::ura4+/mei4::ura4+ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | Abe and Shimoda, 2000 |
| TN4 (FY7251) | h− leu1-32 | Nakamura et al., 2001 |
| TN29 (FY7816) | h90 leu1-32 ura4-D18 | Ikemoto et al., 2000 |
| TN443 (FY21098) | h90 meu14-GFP his5<<mCherry-psy1::Knr leu1-32 | This study |
| TN444 (FY21099) | h90 spo7::ura4+ ade6+<<meu14-GFP his5+<<mCherry-psy1::Knr leu1+<<spo7ΔC2 ade6-M216 leu1-32 ura4-D18 | This study |
| TN445 (FY21100) | h90 pik3::ura4+ leu1+<< meu14-GFP his5+<<mCherry-psy1::Knr leu1-32 ura4-D18 | This study |
| TN449 (FY21355) | h90 pik3::ura4+ spo7-GFP<<LEU2 leu1-32 ura4-D18 | This study |
| TN450 (FY21356) | h90 spo7-GFP<<LEU2 ade6+<<mCherry-atb2 ade6-M216 leu1-32 ura4-D18 | This study |
| TN451 (FY21357) | h90 spo7-GFP<<LEU2 spo13-mCherry<<ura4+ ade6-M216 leu1-32 ura4-D18 | This study |
| YN67 (FY12205) | h90 spo15::ura4+ leu1+<<GFP-psy1 leu1-32 ura4-D18 | Nakase et al., 2008 |
| YN68 (FY12710) | h90 leu1+<<GFP-psy1 leu1-32 | Nakase et al., 2008 |
| YN90 (FY12318) | h90 spo13::ura4+ spo15-GFP<<LEU2 leu1-32 ura4-D18 | Nakase et al., 2008 |
| YN314 (FY12492) | h90 meu14-GFP leu1-32 | Nakase et al., 2008 |
| S. cerevisiae | ||
| AH109 | Mata, ura3-52, his3-200, ade2-101, trp1-901, leu2-3112, gal4Δ, met−, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS -GAL2TATA -ADE2, ura3::MEL1UAS -MEL1TATA -lacZ | Clontech (Mountain View, CA) |
a Accession numbers are from the Yeast Genetic Resource Center of Japan supported by the National BioResource Project (YGRC/NBRP; http:/yeast.lab.nig.ac.jp/nig). The S. pombe strains constructed in this study have been deposited with the YGRC/NBRP under the accession numbers shown here.
b x << y means that gene y is integrated at gene x.
TABLE 2:
Plasmids.
| Plasmid | Characteristics | Source |
|---|---|---|
| S. pombe | ||
| pAL-KS | ars1, LEU2-based multicopy shuttle vector | Tanaka et al., 2000 |
| pAL(spo13-GFP) | pAL-KS, spo13-GFP | Nakase et al., 2008 |
| pIL | LEU2-based integration vector | Ikemoto et al., 2000 |
| pIL(spo7-HA) | pIL, spo7-HA | This study |
| pIL(spo7-GFP) | pIL, spo7-GFP | This study |
| pIL(spo7ΔC2-GFP) | pIL, spo7ΔC2-GFP | This study |
| pIL(spo7-CFP) | pIL, spo7-CFP | This study |
| pIL(spo15-GFP) | pIL, spo15-GFP | Nakase et al., 2008 |
| pREP1 | ars, LEU2-based expression vector carrying nmt1+ promoter | Maundrell, 1993 |
| pREP1(mei4) | pKEP1, mei4+- coding region | Abe and Shimoda, 2000 |
| pTN197 | ars, LEU2-based expression vector carrying nmt41+ promoter, GFP | Nakamura et al., 2001 |
| pMN231 | pTN197, meu14+ | This study |
| pREP42 | ars, ura4+-based expression vector carrying nmt41+ promoter | Maundrell, 1993 |
| pREP42(spo7cnt-HA) | pREP42, spo7cnt, HA | This study |
| pBR(leu1) | leu1+ in pBR322 | Nakamura-Kubo et al., 2003 |
| pBR(leu1)(spo7) | pBR(leu1), spo7+ | This study |
| pBR(leu1)(spo7ΔC1) | pBR(leu1), spo7ΔC1 | This study |
| pBR(leu1)(spo7ΔC2) | pBR(leu1), spo7ΔC2 | This study |
| pBR(leu1)(spo7ΔC3) | pBR(leu1), spo7ΔC3 | This study |
| pBR(leu1)(spo7ΔPH) | pBR(leu1), spo7ΔPH | This study |
| pBR(leu1)(spo7L1153P) | pBR(leu1), spo7L1153P | This study |
| pBR(leu1)(GFP-psy1) | pBR(leu1), GFP-psy1 | Nakamura-Kubo et al., 2003 |
| pBR(leu1)(mCherry-psy1) | pBR(leu1), mCherry-psy1 | This study |
| pBR(leu1)(spo2-GFP) | pBR(leu1), spo2-GFP | Nakase et al., 2008 |
| pBR(leu1)(spo13-GFP) | pBR(leu1), spo13-GFP | Nakase et al., 2008 |
| pBR(leu1)(mCherry-psy1) | pBR(leu1), mCherry-psy1 | This study |
| pBR(leu1)(meu14-GFP) | pBR(leu1), meu14-GFP | Ito |
| pBR(leu1)(meu14-mCherry) | pBR(leu1), meu14-mCherry | This study |
| pIA | ade6+ in pBluescript II KS(−) | Tamai |
| pIA(mCherry-atb2) | pIA, nmt1 promoter, mCherry-atb2 | This study |
| pIH | his5+ in pBluescript II KS(−) | This study |
| pIH(mCherry-psy1)Knr | pIH, mCherry-psy1, Knr | This study |
| pIU(spo13-GFP) | ura4+ in pBluescript II KS(−), spo13-GFP | This study |
| pIU(spo13-mCherry) | ura4+ in pBluescript II KS(−), spo13-mCherry | This study |
| pIU(spo2- mCherry) | pBR(leu1), spo7+ | This study |
| S. cerevisiae | ||
| pGAD424 | 2 μ origin, LEU2-based vector carrying an activation domain of Gal4 and ADH1 promoter | Clontech (Mountain View, CA) |
| pGAD424(meu14) | pGAD424, meu14+ | This study |
| pGAD424(spo7) | pGAD424, spo7+ | This study |
| ppGAD424(spo2) | pGAD424, spo2+ | This study |
| pGAD424(spo13) | pGAD424, spo13+ | This study |
| pGAD424(spo15) | pGAD424, spo15+ | This study |
| pGBT9 | 2 μ origin, TRP1-based vector carrying a DNA-binding domain of Gal4 and ADH1 promoter | Clontech (Mountain View, CA) |
| pGBT9(spo7) | pGBT9, spo7+ | This study |
| pGBT9(spo7ΔC2) | pGBT9, spo7ΔC2 | This study |
| pGBT9(spo7ΔC3) | pGBT9, spo7ΔC3 | This study |
| pGBT9(spo7ΔPH) | pGBT9, spo7ΔPH | This study |
Cloning of spo7+
The spo7-B332 mutant was transformed with the S. pombe genomic library, pTN-L1 (Nakamura et al., 2001), containing partial Sau3AI fragments constructed in a multi-copy plasmid, pAL-KS (Tanaka et al., 2000). About 105 Leu+ transformants were incubated on sporulation medium (SSA). The plates were exposed to iodine vapor (Gutz et al., 1974), and those colonies that turned brown were selected as candidates for sporulation-proficient transformants. The plasmid DNA pMN(spo7) was responsible for the recovery of sporulation ability in spo7 mutants.
Disruption of the spo7+ gene
The plasmid used for gene disruption was constructed as follows. A 4.0-kb HindIII–StuI fragment in the spo7+ ORF was replaced by ura4+ (1.7 kb; Figure 1). A 6.0-kb SpeI–BglII fragment containing the disrupted spo7::ura4+ gene was transformed into the strain TN29. Disruption was confirmed by genomic Southern hybridization (unpublished data).
Western blotting
MN48 was cultured in liquid sporulation medium (MM−N) at 25°C for 18 h, and the temperature was shifted to 34°C to induce meiosis. At intervals, portions of the culture were collected and crude cell extracts were prepared as described by Masai et al. (1995). Polypeptides were resolved by SDS–PAGE on a 10% gel and then transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Billerica, MA). Filters were probed with rat anti-HA antibody (3F10, Roche Diagnostics, Indianapolis, IN) at a 1:1000 dilution. Blots were also probed with the anti-α-tubulin antibody TAT-1 (Woods et al., 1989) to ensure that approximately equal amounts of protein were loaded. Immunoreactive bands were revealed by chemiluminescence (NEN Life Sciences, Boston, MA) with horseradish peroxidase–conjugated goat anti–rat (Biosource International, Camarillo, CA) or anti–mouse (Promega, Madison, WI) immunoglobulin G (IgG).
In vivo protein interaction assay
To assay binding between Spo7 and Meu14, we cotransformed the wild-type strain TN29 with plasmids pREP42(spo7cnt-HA) and pMN231 (pREP41(meu14-GFP)). pTN197 (pREP41(GFP)) was used as a control strain instead of pMN231. Transformants were precultured on SD medium. The cells were then transferred to liquid minimal medium (MM) and grown to midlog phase. The cells were harvested, resuspended in extraction buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA, 2 mM ethylene glycol tetraacetic acid [EGTA], 200 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and then ruptured with glass beads. The lysate was centrifuged at 13,000 × g for 20 min to prepare a soluble fraction. The cell-free homogenates were incubated with rat anti-HA (3F10) antibody at 4°C for 1 h, and then mixed with protein G Sepharose (GE Healthcare, Waukesha, WI). The target proteins were detected by Western blot analysis using mouse anti-HA (12CA5, Roche) and mouse anti-GFP (Roche) antibodies.
Fluorescence microscopy
For fluorescence microscopy, cells were suspended in liquid synthetic medium (SSL) without fixation. For DNA staining, cells were washed with water and stained with Hoechst 33342 at 5 μg/ml for 10 min. After centrifugation, the cells were suspended in SSL medium and observed. Photomicrographs were obtained by using a BX51 fluorescence microscope (Olympus, San Jose, CA) equipped with a Roper Cool SNAP charge-coupled device camera (see Figures 1A, 4B, 5, 6D, 7, A and C, S2, A and B, and S3A) or Hamamatsu ORCA-R2 (see Figure 3, 4A, 6C, 7B, and 9). The filter sets U-MWU (Olympus), U-MWIB (Olympus), CFP-2432B (Semrock, Rochester, NY), and U-MWIG2 (Olympus) were used for Hoechst 33342, GFP, CFP, and mCherry, respectively. Image acquisition and processing was carried out by using Aquacosmos (Hamamatsu) and Photoshop (Adobe, San Jose, CA) software.
Protein–lipid overlay assay
pGEX constructs encoding the PH domain of Spo7 or GST vector alone were transformed into Escherichia coli BL21, and a 200-ml culture was grown at 30°C in Luria broth containing 100 μg/ml ampicillin until the absorbance at 600 nm was 0.4–0.5. Isopropyl-β-d-thiogalactoside (100 μM) was added, and the cells were cultured for an additional 4 h at 30°C. The cells were collected and washed once with phosphate-buffered saline at pH 7.4. The cells were then resuspended in 25 ml of ice-cold Buffer A (10 mM Tris-HCl, pH 7.4, containing 1 M NaCl, 1% Triton X-100, 1 mM dithiothreitol [DTT], 0.5 mM PMSF) and ruptured by sonication. The lysates were centrifuged at 20,000 × g for 30 min at 4°C. The supernatant was then filtered through a 0.45 μm filter and incubated for 40 min on a rotating platform with 200 μl of glutathione–Sepharose pre-equilibrated in Buffer A (containing 0.5 M NaCl, 20% glycerol). The suspension was centrifuged for 1 min a 3000 × g, and the beads were washed three times with 15 ml of Buffer A containing 0.5 M NaCl, 20% glycerol and then a further six times with 15 ml of Buffer B (20 mM Tris-HCl, pH 7.5, containing 20% glycerol, 100 mM KCl, 0.1% Triton X-100, 1 mM DTT, 0.5 mM PMSF). The protein was eluted from the resin at 4°C by incubation with 0.5 ml of Buffer B containing 20 mM glutathione. The eluate was divided into aliquots and stored at −80°C.
The protein–lipid overlay assay was performed using the GST fusion proteins (GST and GST-Spo7-PH) mentioned above. The membrane was blocked in 3% fatty acid-free bovine serum albumin in TBST (50 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, and 0.1% Tween 20) for 1 h and was then incubated overnight at 4°C with gentle stirring in the same solution containing either 200 ng/ml of GST fusion protein. The membranes were washed six times over 30 min in TBST, and then incubated for 1 h with horseradish peroxidase–conjugated goat with anti–mouse IgG (Sigma). Finally, the membranes were washed 12 times over 1 h in TBST, and GST fusion protein that was bound to the membrane by virtue of its interaction with phospholipid was detected by enhanced chemiluminescence.
Electron Microscopy
Samples for electron microscopy were prepared as described (Ye et al., 2007), and sections were viewed on an electron microscope (H-7600, Hitachi, Tokyo, Japan) at 100 kV.
Supplementary Material
Acknowledgments
We thank Masayuki Yamamoto (University of Tokyo, Tokyo, Japan), Kayoko Tanaka (University of Leicester, Leicester, UK), Hiroshi Nojima (Osaka University, Suita, Japan), Kaoru Takegawa (Kyushu University, Fukuoka, Japan), Richard Egel (University of Copenhagen, Copenhagen, Denmark), and Roger Tsien (University of California, San Diego, CA) for strains and plasmids and Keith Gull (University of Oxford, Oxford, UK) for antibody. This study was supported in part by a Grant-in-Aid for Scientific Research (C) and a Grant-in-Aid for Scientific Research on Priority Area “Cell Proliferation Control” from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Asahi Glass Foundation (to T.N.). M.N.-K. is a recipient of the Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science.
Abbreviations used:
- CFP
cyan fluorescent protein
- DTT
dithiothreitol
- EGTA
ethylene glycol tetraacetic acid
- FSM
forespore membrane
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- HA
hemagglutinin
- LEP
leading edge protein
- MM
minimal medium
- ORF
open reading frame
- PH
pleckstrin homology
- PI
phosphatidylinositol
- PI3P
phosphatidylinositol 3-phosphate
- PMSF
phenylmethylsulfonyl fluoride
- PS
phosphatidylserine
- PSM
prospore membrane
- PX
phox homology
- SPB
spindle pole body
- SSA
sporulation medium
- SSL
liquid synthetic medium.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0125) on July 20, 2011.
REFERENCES
- Abe H, Shimoda C. Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics. 2000;154:1497–1508. doi: 10.1093/genetics/154.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajgier BK, Malzone M, Nickas M, Neiman AM. SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol Biol Cell. 2001;12:1611–1621. doi: 10.1091/mbc.12.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresch C, Muller G, Egel R. Genes involved in meiosis and sporulation of a yeast. Mol Gen Genet. 1968;102:301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. New York: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower WJ, Mattheakis LC. In vitro selection as a powerful tool for the applied evolution of proteins and peptides. Curr Opin Chem Biol. 2002;6:390–398. doi: 10.1016/s1367-5931(02)00332-0. [DOI] [PubMed] [Google Scholar]
- Egel R, Egel-Mitani M. Premeiotic DNA synthesis in fission yeast. Exp Cell Res. 1974;88:127–134. doi: 10.1016/0014-4827(74)90626-0. [DOI] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King RC, editor. Handbook of Genetics 1. New York: Plenum; 1974. pp. 395–446. [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall PA, Russell SEH, Pringle JR. The Septins. London: Wiley-Blackwell; 2008. [Google Scholar]
- Hirata A, Tanaka K. Nuclear behavior during conjugation and meiosis in the fission yeast Schizosaccharomyces pombe. J Gen Appl Microbiol. 1982;28:263–274. [Google Scholar]
- Horazdovsky BF, Davies BA, Seaman MN, McLaughlin SA, Yoon S, Emr SD. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, Abe H, Yamamoto M, Shimoda C. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol Cell Biol. 1998;18:2118–2129. doi: 10.1128/mcb.18.4.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y, Hiramine Y, Yamamoto M. The role of cdc2 and other genes in meiosis in Schizosaccharomyces pombe. Genetics. 1995;140:1235–1245. doi: 10.1093/genetics/140.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto S, Nakamura T, Kubo M, Shimoda C. S. pombe sporulation-specific coiled-coil protein Spo15p is localized to the spindle pole body and essential for its modification. J Cell Sci. 2000;113:545–554. doi: 10.1242/jcs.113.3.545. [DOI] [PubMed] [Google Scholar]
- Itadani A, Nakamura T, Hirata A, Shimoda C. Schizosaccharomyces pombe calmodulin, Cam1, plays a crucial role in sporulation by recruiting and stabilizing the spindle pole body components responsible for assembly of the forespore membrane. Eukaryot Cell. 2010;9:1925–1935. doi: 10.1128/EC.00022-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R, Sprague GF Jr, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci USA. 1983;80:3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida M, Shimoda C. Genetic mapping of eleven spo genes essential for ascospore formation in the fission yeast Schizosaccharomyces pombe. Curr Genet. 1986;10:443–447. doi: 10.1007/BF00419871. [DOI] [PubMed] [Google Scholar]
- Knop M, Strasser K. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 2000;19:3657–3667. doi: 10.1093/emboj/19.14.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga T, Onishi M, Nakamura Y, Hirata A, Nakamura T, Shimoda C, Iwaki T, Takegawa K, Fukui Y. Sorting nexin homologues are targets of phosphatidylinositol 3-phosphate in sporulation of Schizosaccharomyces pombe. Genes Cells. 2004;9:561–574. doi: 10.1111/j.1356-9597.2004.00744.x. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, et al. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol. 2005;15:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Miyake T, Arai K. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Wilbrey A, Bahler J. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 2007;8:R217. doi: 10.1186/gb-2007-8-10-r217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Fisher E, Patton-Vogt J, Marcus S. Functional characterization of the fission yeast phosphatidylserine synthase gene, pps1, reveals novel cellular functions for phosphatidylserine. Eukaryot Cell. 2007;6:2092–2101. doi: 10.1128/EC.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:793–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Moreno-Borchart AC, Strasser K, Finkbeiner MG, Shevchenko A, Knop M. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 2001;20:6946–6957. doi: 10.1093/emboj/20.24.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Asakawa H, Nakase Y, Kashiwazaki J, Hiraoka Y, Shimoda C. Live observation of forespore membrane formation in fission yeast. Mol Biol Cell. 2008;19:3544–3553. doi: 10.1091/mbc.E08-04-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Nakamura-Kubo M, Hirata A, Shimoda C. The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+ encoding syntaxin-like protein. Mol Biol Cell. 2001;12:3955–3972. doi: 10.1091/mbc.12.12.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-Kubo M, Nakamura T, Hirata A, Shimoda C. The fission yeast spo14+ gene encoding a functional homologue of budding yeast Sec12 is required for the development of forespore membranes. Mol Biol Cell. 2003;14:1109–1124. doi: 10.1091/mbc.E02-08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y, Nakamura-Kubo M, Ye Y, Hirata A, Shimoda C, Nakamura T. Meiotic spindle pole bodies acquire the ability to assemble the spore plasma membrane by sequential recruitment of sporulation-specific components in fission yeast. Mol Biol Cell. 2008;19:2476–2487. doi: 10.1091/mbc.E08-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Ascospore formation in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas ME, Schwartz C, Neiman AM. Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot Cell. 2003;2:431–445. doi: 10.1128/EC.2.3.431-445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuzaki D, Satake W, Hirata A, Nojima H. Fission yeast meu14+ is required for proper nuclear division and accurate forespore membrane formation during meiosis II. J Cell Sci. 2003;116:2721–2735. doi: 10.1242/jcs.00496. [DOI] [PubMed] [Google Scholar]
- Onishi M, Iida M, Koga T, Yamada S, Hirata A, Iwaki T, Takegawa K, Fukui Y, Tachikawa H. Schizosaccharomyces pombe Sst4p, a conserved Vps27/Hrs homolog, functions downstream of phosphatidylinositol 3-kinase Pik3p to mediate proper spore formation. Eukaryot Cell. 2007;6:2343–2353. doi: 10.1128/EC.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, et al. Role of septins in the orientation of forespore membrane extension during sporulation in fission yeast. Mol Cell Biol. 2010;30:2057–2074. doi: 10.1128/MCB.01529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Koga T, Morita R, Nakamura Y, Nakamura T, Shimoda C, Takegawa K, Hirata A, Fukui Y. Role of phosphatidylinositol 3-phosphate in formation of forespore membrane in Schizosaccharomyces pombe. Yeast. 2003a;20:193–206. doi: 10.1002/yea.953. [DOI] [PubMed] [Google Scholar]
- Onishi M, Nakamura Y, Koga T, Hirata A, Fukui Y. Importance of phosphatidylinositol 3-phosphate in sporulation of Schizosaccharomyces pombe. Biosci Biotechnol Biochem. 2003b;67:1191–1193. doi: 10.1271/bbb.67.1191. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Membrane transport: retromer to the rescue. Curr Biol. 2001;11:R109–R111. doi: 10.1016/s0960-9822(01)00042-2. [DOI] [PubMed] [Google Scholar]
- Shimoda C. Forespore membrane assembly in yeast: coordinating SPBs and membrane trafficking. J Cell Sci. 2004;117:389–396. doi: 10.1242/jcs.00980. [DOI] [PubMed] [Google Scholar]
- Shimoda C, Nakamura T. Control of late meiosis and ascospore formation. In: Egel R, editor. The Molecular Biology of Schizosaccharomyces pombe, New York: Springer; 2003. pp. 311–327. [Google Scholar]
- Spiliotis ET, Nelson WJ. Here come the septins: novel polymers that coordinate intracellular functions and organization. J Cell Sci. 2006;119:4–10. doi: 10.1242/jcs.02746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Yamamoto M. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1987;84:3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takegawa K, DeWald DB, Emr SD. Schizosaccharomyces pombe Vps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J Cell Sci. 1995;108:3745–3756. doi: 10.1242/jcs.108.12.3745. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hirata A. Ascospore development in the fission yeasts Schizosaccharomyces pombe and S. japonicus. J Cell Sci. 1982;56:263–279. doi: 10.1242/jcs.56.1.263. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Miyashita K, Saito TT, Yoneki T, Kakihara Y, Nabeshima K, Kishi YA, Shimoda C, Nojima H. Comprehensive isolation of meiosis-specific genes identifies novel proteins and unusual non-coding transcripts in Schizosaccharomyces pombe. Nucleic Acids Res. 2001;29:2327–2337. doi: 10.1093/nar/29.11.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Imai Y, Watanabe Y. Mating and sporulation in Schizosaccharomyces pombe. In: Pringle JR, Broach JB, Jones EW, editors. Molecular and Cellular Biology of the Yeast Saccharomyces, New York: Cold Spring Harbor Laboratory,; 1997. pp. 1037–1106. [Google Scholar]
- Yang HJ, Neiman AM. A guanine nucleotide exchange factor is a component of the meiotic spindle pole body in Schizosaccharomyces pombe. Mol Biol Cell. 2010;21:1272–1281. doi: 10.1091/mbc.E09-10-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Fujii M, Hirata A, Kawamukai M, Shimoda C, Nakamura T. Geranylgeranyl diphosphate synthase in fission yeast is a heteromer of farnesyl diphosphate synthase (FPS), Fps1, and an FPS-like protein, Spo9, essential for sporulation. Mol Biol Cell. 2007;18:3568–3581. doi: 10.1091/mbc.E07-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo BY, Calleja GB, Johnson BF. Ultrastructural changes of the fission yeast (Schizosaccharomyces pombe) during ascospore formation. Arch Mikrobiol. 1973;91:1–10. doi: 10.1007/BF00409533. [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.