Abstract
Ligand-dependent activation of gene transcription by nuclear receptors is dependent on the recruitment of coactivators, including a family of related NCoA/SRC factors, via a region containing three helical domains sharing an LXXLL core consensus sequence, referred to as LXDs. In this manuscript, we report receptor-specific differential utilization of LXXLL-containing motifs of the NCoA-1/SRC-1 coactivator. Whereas a single LXD is sufficient for activation by the estrogen receptor, different combinations of two, appropriately spaced, LXDs are required for actions of the thyroid hormone, retinoic acid, peroxisome proliferator-activated, or progesterone receptors. The specificity of LXD usage in the cell appears to be dictated, at least in part, by specific amino acids carboxy-terminal to the core LXXLL motif that may make differential contacts with helices 1 and 3 (or 3′) in receptor ligand-binding domains. Intriguingly, distinct carboxy-terminal amino acids are required for PPARγ activation in response to different ligands. Related LXXLL-containing motifs in NCoA-1/SRC-1 are also required for a functional interaction with CBP, potentially interacting with a hydrophobic binding pocket. Together, these data suggest that the LXXLL-containing motifs have evolved to serve overlapping roles that are likely to permit both receptor-specific and ligand-specific assembly of a coactivator complex, and that these recognition motifs underlie the recruitment of coactivator complexes required for nuclear receptor function.
Keywords: Nuclear receptors, NCoA/SRC factors, LXXLL motifs, CBP/p300
Nuclear receptors are ligand-activated transcription factors that are critical for development and homeostasis, regulating gene expression, in most cases, via interaction with cis-acting DNA elements (Mangelsdorf et al. 1995; Katzenellenbogen 1996; Torchia et al. 1998). Members of the nuclear receptor family share a conserved structural and functional organization with the carboxyl terminus mediating ligand-binding. Upon ligand binding, these transcription factors undergo distinct conformational changes, depending on the type of ligand (Fritsch et al. 1992; Beekman et al. 1993). Crystal structures of the carboxy-terminal ligand-binding domain of several receptors (Bourguet et al. 1995; Renaud et al. 1995; Wagner et al. 1995; Brzozowski et al. 1997; Nolte et al. 1998) reveal that the ligand is almost entirely buried within the conserved core of α helices 3, 7, and 10. A conserved carboxy-terminal helix, referred to as the AF2 domain, required for ligand-dependent gene activation (Danielian et al. 1992; Barettino et al. 1994; Durand et al. 1994; Tone et al. 1994), becomes folded against the ligand-binding domain of agonist-bound retinoic acid receptor (RAR), thyroid hormone receptor (TR), estrogen receptor (ER), and PPARγ receptor structures. This change in conformation is thought to allow a productive interaction with transcriptional coactivators and thus results in an activated transcription factor (Fritsch et al. 1992; Beekman et al. 1993). For many nuclear receptors there is a ligand-dependent exchange of a corepressor complex, containing histone deacetylase activity, for a coactivator complex possessing histone acetylase activity (Horlein et al. 1995; Ogryzko et al. 1996; Alland et al. 1997; Heinzel et al. 1997; Nagy et al. 1997; Spencer et al. 1997; Wong et al. 1997).
The precise repertoire of coactivators and coactivator complexes required for physiological nuclear receptor function remains unresolved. Nuclear receptors can interact with, and variably require, CBP/p300 for their transcriptional activity, both in vitro and in vivo (Chakravarti et al. 1996; Hanstein et al. 1996; Kamei et al. 1996; Yao et al. 1996, 1998; Kraus and Kadonaga 1998). On the basis of direct biochemical interactions (Halamachi et al. 1994; Cavailles et al. 1994; Kurokawa et al. 1995), a series of p160 factors requiring both ligand and the AF2 domain for association were identified and, at least in some cell types, are required for receptor activation (Halamachi et al. 1994; Oñate et al. 1995; Torchia et al. 1997). These p160 factors include NCoA-1SRC-1 (Oñate et al. 1995; Kamei et al. 1996), the highly related factor TIF2/GRIP-1/NCoA-2 (Hong et al. 1996; Voegel et al. 1996; Torchia et al. 1997), and a third family member p/CIP/AIB1/ACTR (Anzick et al. 1997; Chen et al. 1997; Li et al. 1997; Torchia et al. 1997). These factors, which together constitute the biochemically identified p160 factors (Kamei et al. 1996), are components of a larger complex, apparently assembled upon binding of ligand that is responsible, at least in part, for critical aspects of receptor function (Korzus et al. 1998). There are a large number of additional factors that exert ligand-dependent receptor interactions and serve as potential coactivators (e.g., Lee et al. 1995); a role for CBP/p300 has been clearly established. Further, on the basis of coimmunoprecipitation and chromatography approaches with the thyroid hormone or vitamin D receptors, a large multicomponent complex was identified, the members of which appear distinct from the CBP/p160/p/CAF proteins (Fondell et al. 1996; Ogryzko et al. 1998; Rachez et al. 1998).
Intriguingly, the p160 factors such as NCoA-1/SRC-1, and many of the other factors capable of interacting with liganded nuclear receptors, share a common motif containing a core consensus sequence LXXLL (L, leucine; X, any amino acid; LeDouarin et al. 1996; Heery et al. 1997; Torchia et al. 1997). These motifs are sufficient for ligand-dependent interaction with nuclear receptors, and were predicted to assume helical conformation (Heery et al. 1997; Torchia et al. 1997). NCoA-1/SRC-1, TIF-2/GRIP-1/NCoA-2, and p/CIP/AIB1/ACTR all contain three LXXLL motifs in a conserved central sequence, which has been defined to serve as the nuclear receptor interaction domain. In addition, NCoA-1/SRC-1 has two LXXLL motifs within a more carboxy-terminal domain capable of interacting with CBP/p300, and a single splicing variant has an additional carboxy-terminal LXXLL-containing motif (Oñate et al. 1996; Torchia et al. 1997).
The co-crystal structure of a region of the NCoA-1/SRC-1 nuclear receptor interaction domain (amino acids 623–760) containing two LXXLL motifs with liganded PPARγ revealed a ternary complex in which one LXXLL binds to one PPARγ ligand-binding domain (LBD), while the second LXXLL binds to the other PPARγ LBD of the dimer, with the connecting, unstructured NCoA-1/SRC-1 sequence spanning the receptor homodimer (Nolte et al. 1998). The crystallographic evidence suggests a model in which the conserved glutamic acid of the AF2 helix (E471) hydrogen bonds to the backbone amides of the first leucine of the motifs and the amino-terminal adjacent residue, while a critical lysine in helix 3 of the receptor (K301) hydrogen bonds to the backbone carbonyls of leucines at position 4 and 5 of the LXXLL motif (Nolte et al. 1998), with the hydrophobic face of LXXLL packed into a hydrophobic pocket formed from the helix packed interfaces of receptor helices 3, 4, and 5 and the AF2 helix. Thus, E471 and K301 in the AF2 and H3 helices, respectively, define a charge clamp that allows the orientation and placement of the LXXLL motif into the coactivator-binding site. The nonconserved amino acids of the core motif (LXXLL) are solvent exposed but would not be expected to exhibit specificity. These observations also imply that other LXXLL-containing factors would exhibit similar binding to the ligand-dependent coactivator-binding site, and raise intriguing questions with regard to LXXLL motif specificity and the role of the multiple LXXLL-containing motifs within the p160 factors.
Here, we report that the three LXXLL-containing helical motifs, referred to as LXDs, within the nuclear receptor-interaction domain of NCoA-1/SRC-1 are differentially required, with the second LXD required and sufficient for action of the ER, while TR and RAR require both the second and third LXXLL helical domains and correct spacing between them. In contrast, PPARγ receptor and progesterone receptor (PR) require both LXD1 and LXD2, again with appropriate spacing. The critical amino acid residues required for interactions with ER, RAR, and TR prove in vivo to reside carboxy-terminal to the LXXLL core motif and have been defined to the level of a single amino acid, revealing receptor specificity of required amino acid interactions. Specificity is, surprisingly, further regulated by ligand itself, as different carboxy-terminal residues of LXD2 were required with distinct PPARγ ligands. Finally, LXXLL-containing helices in the CBP/p300 interaction domain of NCoA-1/SRC-1 have proved to also be required for interactions with CBP/p300, and structure/function analysis is consistent with the hypothesis that these interactions involve a hydrophobic pocket defined by multiple helical motifs in the CBP/p300 interaction domain, which is required to mediate the apparent, obligatory requirement for the CBP/p300 cofactor in nuclear receptor function.
Results
Receptor specificity of the LXXLL helical motifs
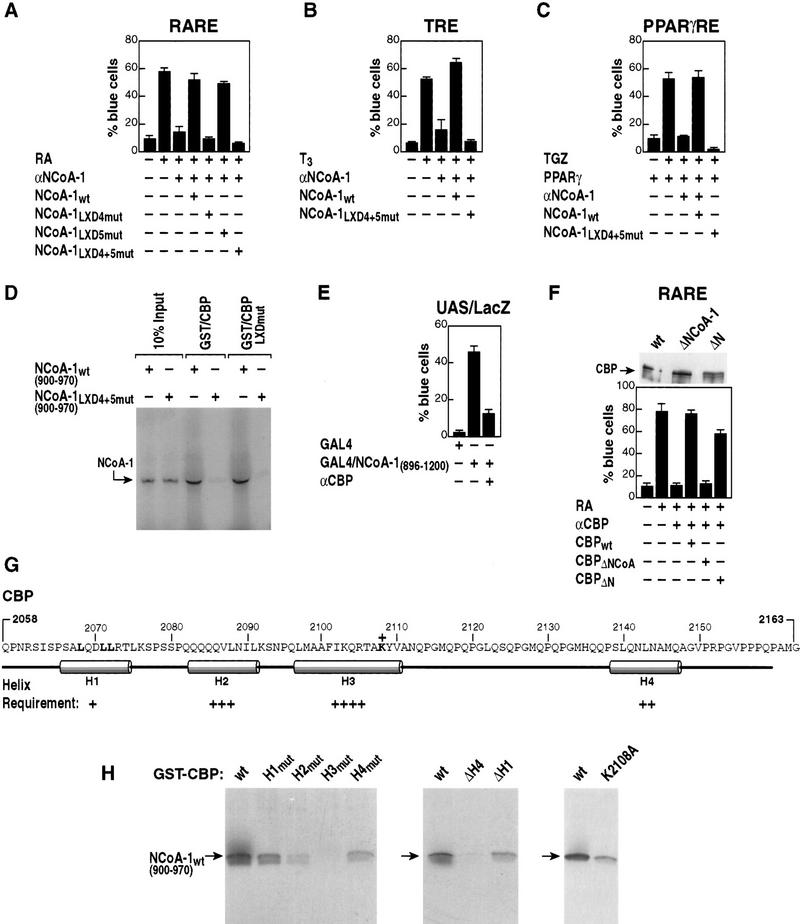
The three members of the p160 coactivator family contain several regions of high homology, and these include the distinct nuclear receptor- and CBP-interaction domains; the sequences of the 21 amino acids that encompass each component LXD are more related between family members than they are between motifs within a given p160 protein (Fig. 1A). Because the region encompassing LXD1, LXD2, and LXD3 does not interact with CBP/p300, while conversely the region encompassing LXD4 and LXD5 does not effectively interact with liganded nuclear receptors (Torchia et al. 1997), we explored the possibility that there might be a preferential recognition code of LXXLL motifs required for actions of specific nuclear receptors. This hypothesis was investigated initially at a functional level, determining the ability of NCoA-1/SRC-1 mutated in specific LXDs to rescue receptor function following nuclear microinjection of specific anti-NCoA-1/SRC-1 IgG. In these experiments, we evaluated the function of each LXD; we altered the fourth and fifth (leucine) residue of LXXLL (+1 to +5) to alanines, or altered residues 2, 3, 4, and 5 to alanine, obtaining identical results in all experiments. The mutation of L4 and L5 was chosen on the basis of the critical roles of these leucine residues in binding to the liganded nuclear receptor (Heery et al. 1997; Torchia et al. 1997) and the co-crystal structure of PPARγ with a portion of the NCoA-1/SRC-1 interaction domain, confirming the specificity of the requirement for leucine at the +5 position (Nolte et al. 1998).
Figure 1.
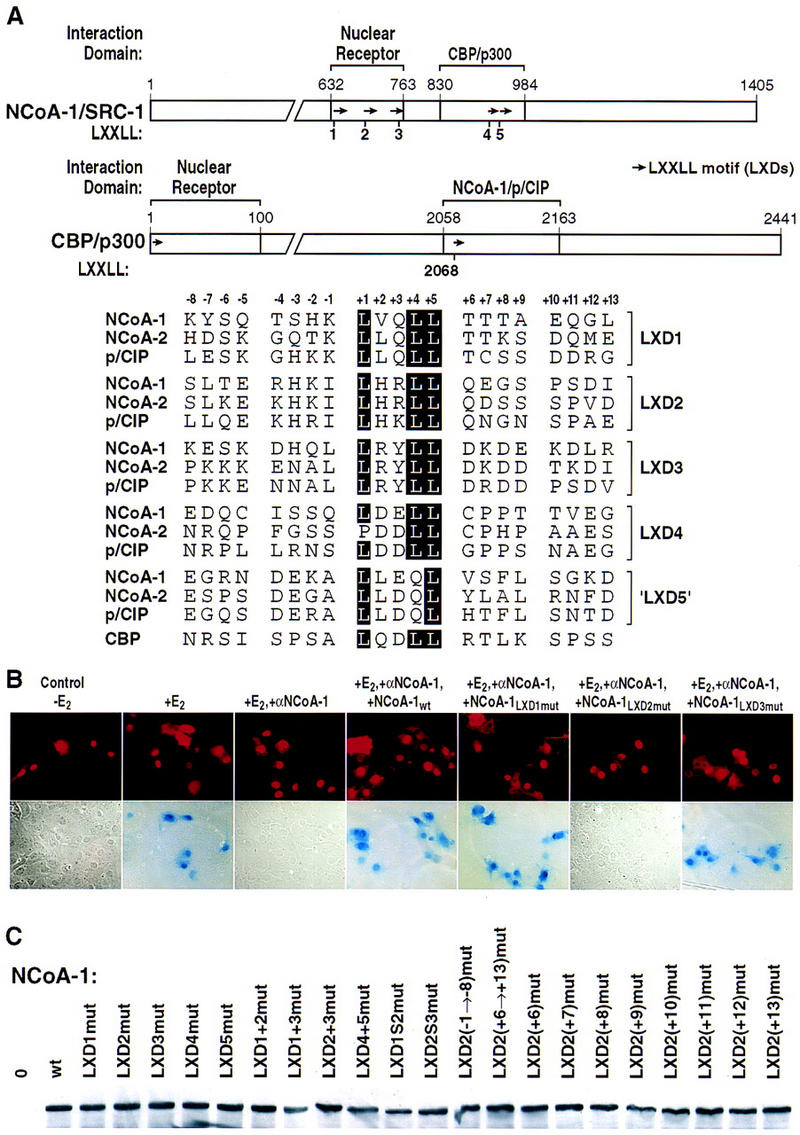
LXXLL helical motif requirements for actions of ER. (A) Map of NCoA-1/SRC-1 with LXXLL helical motif (LXDs) indicated, and of CBP, with a portion of a LXXLL motif in the NCoA-1/p/CIP interaction domain. (Arrows) LXXLL motifs; (bottom) sequences of these motifs are compared. (B) Effects of mutation of LXD1, LXD2, or LXD3 (LXXLL → LAAAA) on ability to rescue 17β estradiol (E2)-dependent ER activation abolished by nuclear microinjection of anti-NCoA-1/SRC-1 affinity-purified IgG. (C) Western blot analysis of expression of NCoA-1/SRC-1, wild type (wt), or the indicated mutations, in extracts of cells transfected with specific IgG. No significant differences in protein levels were observed for the mutations studied.
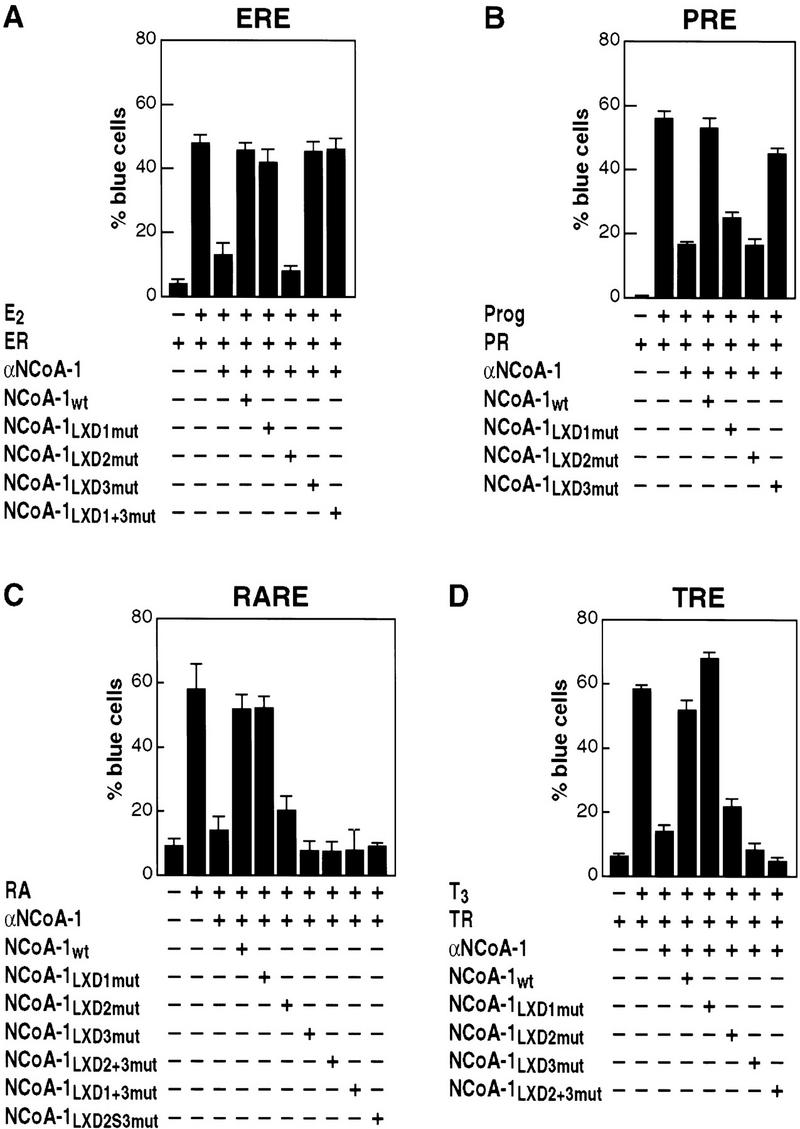
We first performed rescue experiments following injection of anti-SRC-1/NCoA1 IgG on 17β-estradiol-stimulated estrogen receptor function, using wild-type NCoA-1/SRC-1 or NCoA-1/SRC-1 derivatives encoding the indicated, specific mutations in LXD1, LXD2, or LXD3. These experiments indicated that only LXD2, but neither LXD1 nor LXD3, were required for function of NCoA-1/SRC-1 on this receptor (Figs. 1B and 2A). Even with mutation of both LXD1 and LXD3, the ER activation function of NCoA-1/SRC-1 remained intact (Figs. 1B and 2A). The mutant forms of NCoA-1/SRC-1 were expressed at apparently equivalent levels as determined by Western blot analysis of transfected cells, as were all other mutated proteins used in these studies (Fig 1C). More definitively, the function of each mutation was established by its ability to rescue other nuclear receptors (see below). Similar analysis with the PR revealed a requirement for both LXD1 and LXD2 (Fig. 2B), indicating distinct patterns of LXD usage for different nuclear receptors.
Figure 2.
Differential role of LXD motifs in actions of ER (A), PR (B), RAR (C), or TR (D) receptors. In each case, ligands were added at 10−6 m, and reporters were under control of the appropriate response element (Torchia et al. 1997). (ERE) Estrogen-responds element; (PRE) progesterone-response element; (RARE) retinoic acid-response element; (TRE) thyroid hormone-response element. Rat-1 cells were microinjected with anti-NCoA-1/SRC-1 IgG and the CMV-expression vectors encoding the indicated proteins (Torchia et al. 1997). In addition to the mutations in leucine residues +4 and +5 of each motif, a deletion of 30 amino acids between LXD2 and LXD3 (LXD2S3mut) were created, leaving the 10 amino acids immediately flanking the LXD intact. Ligands were retinoic acid (RA), triiodothyronine (T3), 17β-estradiol (E2), or progesterone (Prog). Where indicated, receptors were also expressed; similar results were obtained in at least three independent experiments with >300 cells microinjected for each data point.
For RAR and TR function, two helical motifs were again required, but now LXD2 and LXD3 proved to be the motifs required, while mutation of LXD1 did not affect NCoA-1/SRC-1 function (Fig. 2C,D). On the basis of the conserved spacing of 50 amino acids between LXD2 and LXD3, we deleted 30 amino acids, leaving 10 amino acids carboxy- and amino-terminal to LXD2 and LXD3, respectively. Rescue experiments with the spacing variant of NCoA-1/SRC-1 (LXD2S3) revealed that, while equivalently expressed and fully functional on PPARγ receptor (see Fig. 3A), this mutant was unable to serve in activation function for either the TR or RAR (Fig. 2C and data not shown). Therefore, we conclude that, in contrast to the requirements for a single LXD by ER, both RAR and TR, as well as PR, require the actions of two, critically spaced LXDs, rather than a single LXD. The critical LXXLL-containing helices, however, are distinct between the PR or PPARγ receptor and the RAR or TR.
Figure 3.
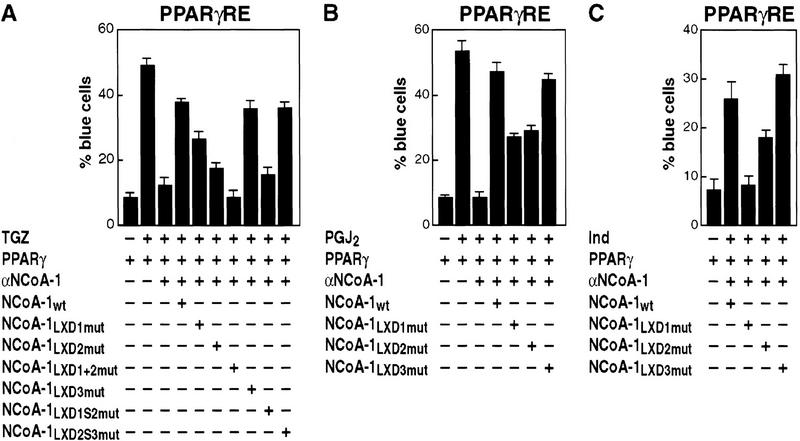
Requirements of LXD domains of NCoA-1/SRC-1 for activation of PPARγ on a response element (PPARγRE)-dependent promoter (AOX/LacZ; Korzus et al. 1998), repeated with different ligands. Nuclear microinjection of at least 300 Rat-1 cells was performed for each data point with αNCoA-1/SRC-1 IgG, and CMV-expression plasmids encoding wild-type NCoA-1/SRC-1 (NCoA1wt) or NCoA-1/SRC-1 with point mutations in LXXLL motifs 1, 2 or 3 (LXD1mut, LXD2mut, and LXD3mut). Ligands used were TGZ (10−6 m), 15-deoxyΔ12,14- prostaglandin J2 (PGJ2, 10−6 m), or indomethacin (Ind, 10−3 m). Results were repeated in three separate experiments; mean ± s.e.m.
A similar microinjection analysis for activity of the PPARγ receptor (Fig. 3A) revealed that mutation of either LXD1 or LXD2, but not of LXD3, resulted in a partial loss of transactivation in response to addition of a thiazolidenedione (troglitazone; TGZ). Mutation of both LXD1 and LXD2 caused complete loss of function (Fig. 3A), while even with simultaneous mutation of LXD1 and LXD3 or LXD2 and LXD3, NCoA-1/SRC-1 remained partially competent to rescue TGZ-stimulated PPARγ function (data not shown). Reduction of the conserved spacing (from 52 to 20 amino acids) between LXD1 and LXD2 abolished the ability of NCoA-1/SRC-1 to serve as a PPARγ coactivator, while altered spacing between LXD2 and LXD3 had no effect, confirming both the effective expression and function of this variant NCoA-1/SRC-1 protein (Fig. 3A).
On the basis of the X-ray crystal structures of liganded receptor carboxy-terminal domains (Bourguet et al. 1995; Renaud et al. 1995; Wagner et al. 1995; Nolte et al. 1998), which document distinct, ligand-dependent placement of the AF2 helix, we considered the possibility that, as a consequence, different ligands might alter the LXD requirements for a particular receptor. This possibility was tested in the case of PPARγ receptor by evaluating several different classes of ligands for PPARγ, each of which would be predicted to bind quite differently in the large ligand-binding pocket and might be expected to alter surface contacts by coactivators (Nolte et al. 1998). Mild distinctions were observed in the quantitative requirements for LXD1 and LXD2; with prostaglandin J2 metabolites (PGJ2) there were equivalent, partial requirements for both LXD1 and LXD2 (PGJ2; Fig. 3B), and with indomethacin, there was a greater quantitative importance of LXD1, but still some effect of LXD2 (Fig. 3C).
Determinants of LXD specificity
While the co-crystal structure of PPARγ with a portion of the SRC-1-interaction domain (amino acids 623–710) provided insight into the critical role of the leucine residues and hydrophobic helix, the molecular basis of specificity of helical choice has remained unclear. The co-crystal of PPARγ LBD with the SRC-1 receptor interaction domain revealed no detectable ordered structure beyond eight amino acids amino- or carboxy-terminal to the conserved LXXLL core (Nolte et al. 1998). Therefore, we investigated the role of these amino- and carboxy-terminal flanking residues as a potential basis for specificity.
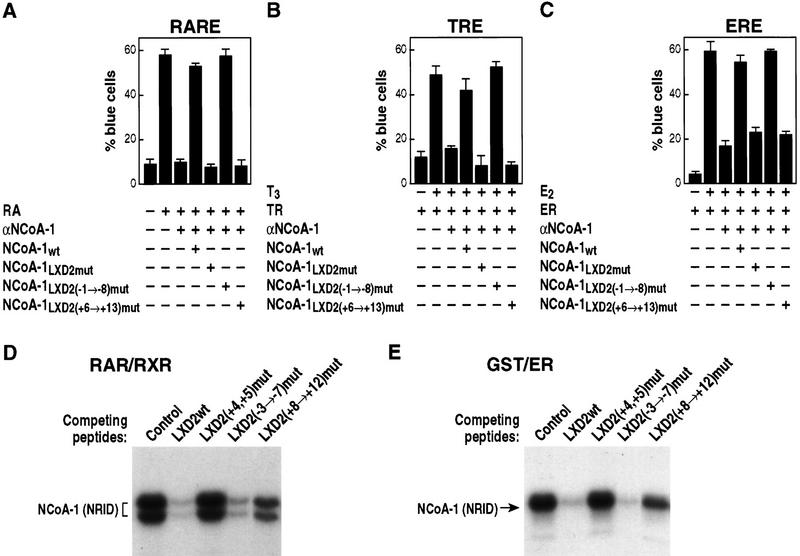
In initial experiments, the sequences encompassing the entire eight amino-terminal or carboxy-terminal flanking amino acids of LXD2 were mutated to alanine codons, to preserve helical conformation of the NCoA-1/SRC-1 LXD. Subsequently, mutation of residues spanning −1 to −3 (see Fig. 1A) was also introduced, and the efficacy of these mutant NCoA-1/SRC-1 molecules to rescue retinoic acid-, thyroid hormone-, and estrogen-dependent transcription was assessed. Whereas there was no effect of mutation of the flanking amino-terminal residues, mutation of the eight carboxy-terminal residues abolished function on all receptors tested (Fig. 4A–C). A similar result was obtained with biochemical experiments that evaluated the ability of LXD peptides (21-mers flanking LXXLL sequences −7 to +8) to compete for the binding of the nuclear receptor interaction domain (NRID) of NCoA-1/SRC-1 on DNA bound RAR–retinoic X receptor (RXR) heterodimers. Altering residues +8 through +12 to alanines markedly diminished the ability to compete for binding (Fig 4D). As expected, mutation of L+4 and L+5 abolished function entirely (Fig. 4D). However, a cluster of alanine substitutions from −3 to −7 in the context of the 21-mer peptide also led to loss of competition, which may reflect interference with the formation of the amino-terminal cap signal and, hence, decreased LXD2 helical stability. A similar study with GST–ER carboxyl terminus also suggested the importance of the +8 to +12 residues in LXD2 to compete for binding (Fig. 5E).
Figure 4.
Carboxy-terminal flanking regions dictate specificity of LXXLL domain function. Nuclear microinjection studies in Rat-1 cells were performed with NCoA-1/SRC-1 proteins in which LXD2-flanking residues (−1 → −8) or (+6 → 13) were mutated to alanine and were evaluated RAR, TR, and ER, as shown in A, B, and C, respectively. Results of the average ± s.e.m. of two sets of nuclear microinjected cells; three independent experiments gave similar results. (D) Avidin–biotin DNA complex assay with thrombin-cleaved, bacterially expressed RARβ and RXRα proteins were bound to biotinylated direct repeat core sequence spaced by 5 bp (DR+5) oligonucleotide, and the NRID of NCoA-1/SRC-1 (amino acids 700–763) was prepared as a 32P-labeled bacterial protein. Competition was assessed with wild-type LXD2 (21-mer) synthetic peptides (LXD2wt), or peptides containing alanine substitutions in the indicated amino acids used in excess (1 μm) to compete for binding. Binding of the NCoA-1 NRID indicates the efficiency of the peptide competition; with more binding indicating loss of function, as observed with mutation of L4 and L5. (E) Similar analysis performed with a bacterially expressed GST–ER carboxy-terminal protein, to evaluate the effects of residue substitution. Mutation of +8 through +12 caused considerable loss of function (i.e., less competition).
Figure 5.
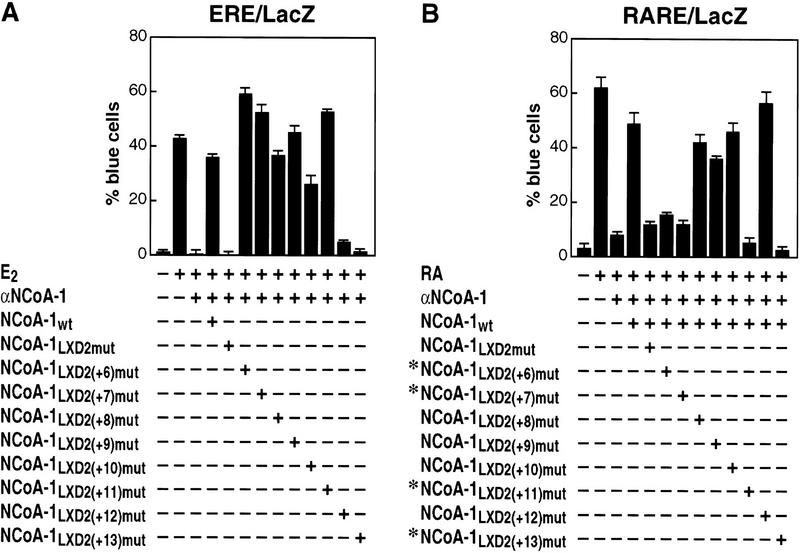
Identification of critical carboxy-terminal residues in the NCoA-1/SRC-1 LXD2 motif for function in transcriptional activation by ER from an expression plasmid (A), or endogenous RAR (B). In each case, the ability of NCoA-1/SRC-1 wild-type, LXD2 mutant (LAAAA), or NCoA-1 proteins with single amino acid alanine substitutions [LXD2(+6)mut through LXD2(+13)mut] were evaluated by use of the single cell microinjection assay. The critical amino acids differed in the case of ER and RAR; mean ± s.e.m.
Therefore, we evaluated the effect of mutation of each individual amino acid from +6 to +13, initially on the ER, which relied entirely on LXD2 for function. This analysis revealed that, for the ER, there was only a minimal effect of mutation at positions +6 through +11; however, mutation at either +12 or +13 (D, I) abolished the ability of the LXD2 to mediate SRC-1 function on liganded ER entirely (Fig. 5A). Therefore, a similar analysis was performed with RAR to evaluate whether critical residues in LXD2 would prove to be invariant. Surprisingly, when this analysis was performed for RAR–RXR heterodimers on a DR +5 DNA element, distinct residues (+6, +7, and +11), as well as +13, were now required for effective function (Fig. 5B). Thus, the carboxy-terminal amino acids immediately flanking the LXXLL core motif have proved to be responsible for mediating receptor-specific interactions, with preferential interactions required for biological function of the coactivator in the intact cell.
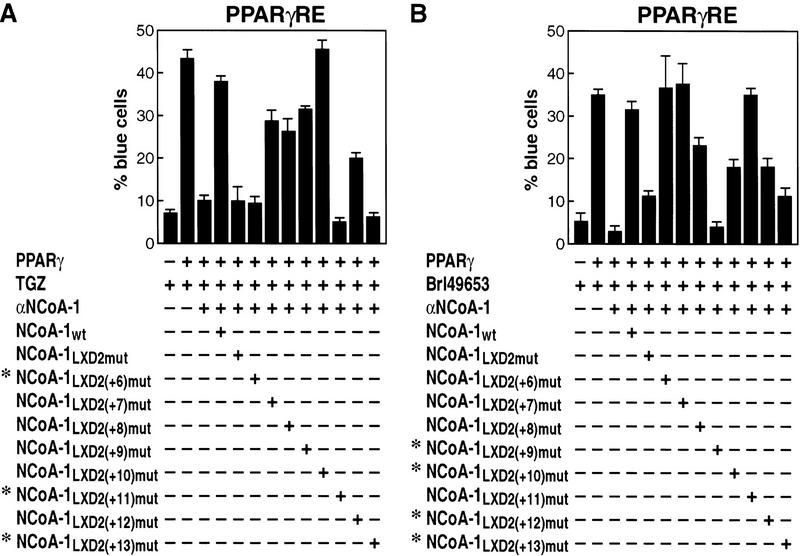
Therefore, we evaluated the residues critical for function of PPARγ, because this receptor can bind multiple structurally distinct ligands, and because of the availability of the co-crystal structure of BRL49653-liganded PPARγ LBD with an 88 amino acid fragment of SRC-1 encompassing LXD1 and LXD2 (amino acids 623–710). With TGZ as ligand, the critical residues were found to be +6, +11, and +13 (Fig. 6A). When a similar analysis was performed with BRL49653 as ligand, to permit direct comparison with co-crystal structure, now residues +9, +10, +12, and +13 proved to be the critical determinant (Fig. 6B). These studies further suggest ligand-specific alterations of receptor structure actually imposes a requirement for different LXD residues to achieve high-affinity interactions.
Figure 6.
Mapping of critical carboxy-terminal residues in the NCoA-1/SRC-1 LXD2 required for function in transcriptional activation by PPARγ with either TGZ (10−6 m) (A), or BRL49653 (10−6 m) (B) as ligand. The entire series of alanine substitutions was evaluated; (*) critical amino acid residues identified.
LXXLL-containing motifs are required for CBP/p300 recruitment to the coactivator complex
Next, it was of particular interest to determine whether LXD motifs in the CBP/p300-interaction domain (Fig. 1A) were important for function and interactions with CBP/p300. As shown in Figure 7A–C, mutation of LXD4 and LXD5 abolished functional activation of the TR, RAR, and PPARγ receptor. Therefore, the effect of these motifs on the strong interactions of NCoA-1/SRC-1 with CBP/p300 was assessed biochemically. As shown in Figure 7D, the interaction with CBP was abolished with mutation of both LXD4 and LXD5.
Figure 7.
Role of LXDs in the CBP/p300-interaction domain of NCoA-1/SRC-1 in interaction and receptor activation function. (A) The LXD motifs (4, 5; Fig. 1A) were mutated to place alanines in positions 2, 3, 4, 5 alone, or together, and evaluated for function on RAR-dependent gene activation. Mutations of LXD4 or LXD4 and LXD5 abolished the ability of SRC-1/NCoA-1 to function in retinoic acid-dependent activation events in Rat-1 single cell nuclear microinjection studies. (B,C) The requirement for the LXD4 and LXD5 in the CBP/p300-interaction domain of NCoA-1/SRC-1 is demonstrated for coactivation of the TR and PPARγ. (D) Role of LXDs in the interactions between 35S-labeled NCoA-1/SRC-1 CBP-interaction domain and CBP (wild type) or CBP in which the CBP LXD is mutated (LXXLL → LAAAA; CBP LXDmut). The mutation of LXD4 and LXD5 virtually abolished interactions by GST pull-downs, but mutation of the CBP LXXLL motif in the interaction domain did not affect interactions. (E) Gal4–NCoA-1 (896–1200) fusion protein activates transcription from the UAS p36 promoter (Torchia et al. 1997) and was blocked by addition of anti-CBP IgG (Kamei et al. 1996). (F) Effect of deletion of the NCoA-1 interaction domain of CBP on function of CBP in RAR activation. Anti-CBP IgG (Kamei et al. 1996) was used to block RAR activation, and CMV-expression vectors encoding wild-type CBP, CBPΔN (Δ 2–468), or CBP ΔNCoA-1 (Δ 2098–2163) were evaluated for their ability to rescue. The three CBP protein variants were expressed at comparable levels in transcripted cells as detected by anti-Flag IgG (top). (G) Predicted structure of the CBP region interacting with SRC-1/NCoA-1, with helix 3 as a predicted hydrophobic helix. (H) Requirements of each helix were tested by mutation to break the helix (helix 1, QDLL → PDPG; helix 2, QQQV → PQPG; helix 3, FIKQ → PIPG; helix 4, NLNA → PLPG), or by helix removal (ΔH4, amino acids 2058–2133; ΔH1, amino acids 2078–2163). Regions were tested in vitro for their ability to interact with amino acids 900–970 of NCoA-1/SRC-1, 35S-labeled by in vitro transcription and translation.
In concert with these findings, we and others have noted that the major activation domain of NCoA-1/SRC-1 is the sequence encompassing the CBP/300 interaction domain. The role of CBP/p300 in this activation event was further evaluated by use of a GAL4 DNA-binding domain–NCoA-1/SRC-1 fusion protein containing the CBP/p300-interaction domain. The activation properties of this domain were largely abolished by anti-CBP IgG (Fig. 7E). We evaluated whether, conversely, the interaction domain of CBP was required for activation by nuclear receptors; deletion of this interacting region in CBP (CBP ΔNCoA) was sufficient to abolish the ability of CBP/p300 to mediate receptor activation function without affecting expression. In contrast, an amino-terminal region deletion in CBP (CBP ΔN) did not affect its activation function (Fig. 7F). Together, these data suggest that the LXDs of NCoA-1/SRC-1 are themselves required for activation function and that their functional importance is indeed likely to be linked, at least in part, to their requirement for interaction with CBP/p300. Conversely, mutation of the LXXLL-containing motif in the CBP/p300 interaction domain to LAAAA had no effect on interactions (Fig. 7D). The predicted structure of CBP suggests that hydrophobic helices could form a binding pocket, analogous to that of the nuclear receptor-binding pocket, by which the LXXLL motifs of NCoA-1/SRC-1 bind to CBP/p300 (Fig. 7G). To begin to explore this possibility, we mutated each of the four predicted helices by various algorithms introducing a PXPG motif into each helix, as well as creating a deletion of helix 1 (ΔH1) or helix 4 (ΔH4). As shown in Figure 7H, helix 3 proved to be absolutely required for interaction; helix 2 was less quantitatively important, but still significant; and lesser roles were exerted by helices 4 and 1. Even a single point mutation of helix 3 (K2109A) significantly impaired the interaction of CBP with NCoA-1/SRC-1, consistent with the critical importance of this hydrophobic helix.
Discussion
LXDs as a code for receptor, coactivator assembly
The finding that nuclear receptors require coactivators to mediate their regulation of gene transcription and that many potential coactivators share a conserved LXXLL motif in the nuclear receptor-interaction domain has suggested intriguing potential regulatory strategies with respect to control of nuclear receptor function. Members of the p160/NCoA/SRC family of coactivators, which can associate in the cell and in vitro with liganded nuclear receptors appear, at least for some promoters, to be required for ligand-dependent transcription (Halamachi et al. 1994; Oñate et al. 1995; Kamei 1996; Torchia et al. 1997). The p160 proteins interact with nuclear receptors via an internal region containing three LXDs with core LXXLL motifs that are modestly conserved amongst family members, with respect to the amino- and carboxy-terminal residues that immediately flank the core LXXLL residues (Heery et al. 1997; Torchia et al. 1997) and which are found in other nuclear receptor-associated factors (LeDouarin et al. 1996). Structural algorithms predicting these motifs would be helical were confirmed by co-crystallization of a sequence encompassing two motifs of NCoA-1/SRC-1 with liganded PPARγ (Nolte et al. 1998). This structure revealed that the LXXLL helix was oriented by a conserved lysine residue in helix 3 and a conserved glutamic acid residue in the AF2 helix of the PPARγ ligand-binding domain, permitting the leucine residues of the LXXLL to pack into a hydrophobic pocket formed by helices 3, 4, and 5, and AF2. This mechanism of interaction, however, fails to explain differences in the specific LXDs required for transcriptional actions by different nuclear receptors. In this report, we have provided several independent types of evidence that there is a receptor-specific code of interaction that reflects usage with distinct NCoA-1/SRC-1 LXXLL-containing motifs (LXDs). These observations are in concert with the finding that the distinct LXXLL-containing motifs in the carboxyl terminus of NCoA-1/SRC-1 account for recruitment of CBP/p300 and are also required components of the coactivator machinery (Chakravarti et al. 1996; Hanstein et al. 1996; Kamei et al. 1996; Yao et al. 1998). The finding that LXDs serve as protein–protein interaction interfaces that assemble the coactivator receptor complex is consistent with reports over the past several years of the critical importance of a series of small regulatory motifs, such as SH2, SH3, PTB, and PBZ domains that have been identified in a series of platform and adaptor proteins that serve to assemble and coordinate complexes regulating signaling events at the plasma membrane, in protein trafficking, and in nuclear transport (Koch et al. 1991; van der Geer and Pawson 1995; Pawson and Scott 1997).
Furthermore, we have documented that amino acids flanking the LXXLL-containing helices subserve specific functions, and this specificity provides several insights into the serial events by which nuclear receptor–coactivator interactions operate to activate gene expression. The presence of multiple LXXLL motifs in the p160 factors, and in several other potential coactivators, has suggested the possibility that there may be an underlying code of differential utilization and an advantage to the presence of more than one motif. There has proved to be at least two layers of specificity to the LXD preference code. The first involves a differential requirement for the number of LXXLL-containing helices utilized by the different nuclear receptors. Thus, in the case of the ER, the second LXD is required and sufficient. The requirement by ER for only a single LXD, and the inability of other, otherwise effective motifs to functionally substitute, implies that a single motif is capable of high-affinity interactions with the estrogen receptor in the intact cell, and suggests that each liganded estrogen receptor of the DNA-bound homodimer is likely to associate with one NCoA-1/SRC-1/p/CIP family member. In contrast, receptors binding as heterodimers with RXR and PR homodimers, require two LXDs. This requirement is consistent with the co-crystal structure of the PPARγ LBD with a region of the NCoA-1/SRC-1-interaction domain containing two LXXLL motifs, in which the two LBDs of the dimer were contacted by LXXLL helices from a single NCoA-1/SRC-1 molecule (Nolte et al. 1998) and the observation that two LXDs are required for cooperative binding to RAR–RXR and PPARγ–RXR heterodimers bound to DNA (Westin et al. 1998). In addition, the requirement for activation of two LXDs for RAR or TR heterodimers with RXR, might reflect the observation that the RXR AF2 domain can interact directly with the LXXLL-binding pocket and competes with NCoA-1/SRC-1 (Westin et al. 1998). It is tempting to speculate that, although a single LXD can displace the RXR AF2 from RAR, the presence of a second LXXLL motif, looping over the receptor heterodimer on DNA and binding to the activation surface of unliganded RXR, might hold the AF2 of RXR in an active configuration.
The second layer of the specificity code concerns the requirements of specific residues adjacent to the LXXLL core motif for function of a particular nuclear receptor. The present findings indicate that receptor-specific differences are dictated by flanking carboxy-terminal residues (+6 to +13), with different residues modulating specific interactions with the ligand-binding domains of different receptors. Thus, the presence of multiple LXXLL-containing motifs has provided a mechanism for receptor-specific interactions. For example, in the case of ER, residues +12 and +13 of LXD2 provide specificity; in contrast, residue +6 proved critical for RAR function. However, the most striking specificity is the difference in residues in LXD2 required by PPARγ when TGZ (amino acids +6, +11, and +13) or BRL49653 (amino acids +9, +10, +12, and +13) are used as ligands. The carboxy-terminal amino acids of LXD2 interact primarily with four separate helices in the PPARγ LBD. As shown in Fig. 8, the +12 and +13 side chains are solvent exposed and the +4 and +6 side chains primarily form interactions with amino acids at the carboxyl terminus of helix H3 in PPARγ. The +7 and +8 side chains point out toward solvent, and the +9 and +10 amino acids form interactions with the small helix, H3′, between H3 and H4. The +11 side chain is again solvent exposed and the +12 and +13 amino acids form weak interactions at the amino-terminus of H1 in PPARγ. Taken together, the amino acids carboxy-terminal to the LXXLL motif could provide specificity at positions +4, +5, +6, +9, +10, +12, and +13, although a small conformational change in this region of SRC-1 could result in different amino acid specificity for other nuclear receptors or for PPARγ with different ligands. Our data (Fig. 6B) indicate a functional importance of positions +9, +10, +12, +13 of LXD2 in vivo for BRL49653-liganded PPARγ, in accordance with these structural predictions. Conformational alterations in this region of SRC-1 are likely to occur when it contacts other nuclear receptors, and these variations are suggested to underlie the differential usage of specific LXD2 residues for different nuclear receptors. We suggest that there may be sufficient flexibility in LXD2 residue placement between NCoA-1/SRC-1 family members that despite the switch in conserved residues between +12 D, +13 I (NCoA-1/SRC-1) to +12 A, +13 E (p/CIP), they may subserve similar specificity functions. Finally, the regions of H1 and H3 in PPARγ with which specific contacts are made with LXD2 are poorly conserved between different nuclear receptors, consistent with the observation that there are receptor-specific contacts; these data imply further that there will be receptor specificity of recognition of LXXLL motifs in other putative coactivators.
Figure 8.
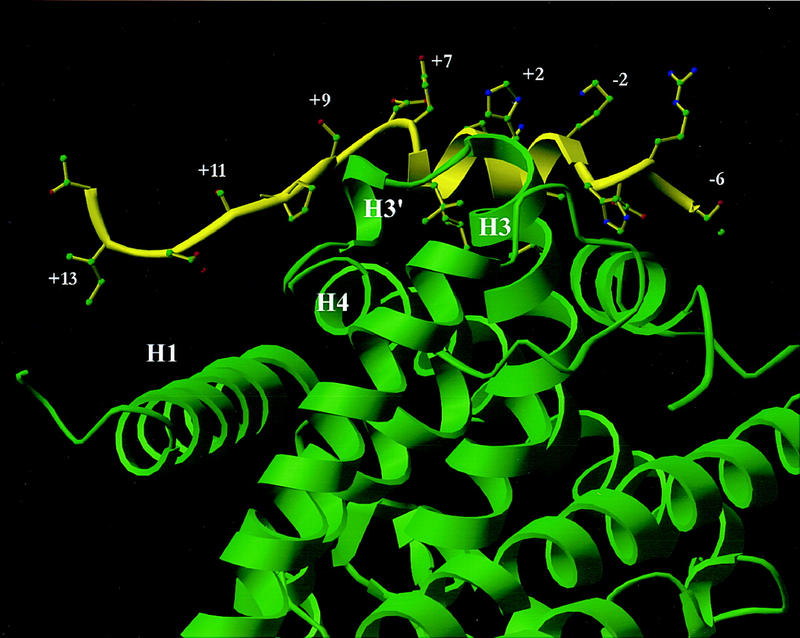
Model of SRC-1 LXD2 (−6) through (+14) bound to BRL49653-liganded PPARγ LBD based on the co-crystal structure with SRC-1 amino acids 623–710 (Nolte et al. 1998). A ribbon drawing of the LXD2 motif of human SRC-1 is shown in yellow with the human PPARγ LBD shown in green. When the electron density maps of the co-crystal structure of the SRC-1 heterodimer with liganded PPARγ (Nolte et al. 1998) are examined and modeled, the +12, +13 amino acids form weak interactions at the amino terminus of helix 1; the +6 side chains contact the carboxyl terminus of helix 3; amino acids +9 and +10 form interactions with the small helix (H3′) between helices 3 and 4.
LXXLL motifs recruit CBP/p300 as well as nuclear receptors
In parallel, distinct LXXLL motifs in NCoA-1/SRC-1 are themselves required for the interactions with CBP/p300, and in this report, we have provided several independent types of evidence that it is CBP/p300 itself that is specifically required for at least a portion of the activation functions imparted by NCoA-1/SRC-1. It is predicted that the region carboxy-terminal to the LXXLL core sequence will also exert important roles in determining whether it will interact with nuclear receptors or with CBP/p300. The precise structure of the interacting region of CBP is unknown, but our data suggest that the interaction is likely to involve a hydrophobic pocket formed by several helices that might, in a fashion analogous to the coactivator-binding domain pocket of the nuclear receptor carboxyl terminus that selectively binds LXXLL motifs (Fig. 7G). Interactions between CREB and another specific region of CBP (KIX domain) are dependent on phosphorylation and induced conformational structuring of this CBP interaction domain (Radhakrishnan et al. 1997); analogous induced-fit events may apply to NCoA-1/SRC-1-CBP/p300 interactions.
Thus, it is suggested that the duplication/mutation of primordial LXXLL motifs over evolutionary time has permitted the appearance of coactivator proteins with enhanced repertoires of both nuclear receptors with which they can interact, as well as the proper interfaces to recruit additional cofactors to a DNA-bound transcription factor. On the basis of altered specificity in response to ligands, it is tempting to speculate that other cofactors containing LXXLL motifs will be differentially recruited on the basis of cell type, levels of coactivators, and ligand. Thus, structural features, duplication, and spacing between LXXLL motifs have evolved to provide specificity in the assembly of critical components of a coactivator complex required for function of a large family of regulated transcription factors.
Materials and methods
DNA-dependent protein–protein interaction and GST pull-down assays
GST–ERα(302–595), GST–RXRα(45–462), and GST–RARα(20–462) proteins were prepared as crude bacterial lysates, immobilized on a glutathione affinity matrix, and washed twice in buffer H [20 mm HEPES (pH 8.0) 50 mm KCl, 20% glycerol, 0.1% NP-40] and once in CHAPS buffer [8 mm Tris-phosphate (pH 7.4), 0.12 m KCl, 8% glycerol, 4 mm DTT, 0.5% CHAPS]. Where indicated, thrombin-cleaved products were prepared. Then, the receptors were incubated in CHAPS buffer with the appropriate ligand for 30 min of competition experiments, incubated in NET-N buffer [50 mm Tris (pH 8.0), 5 mm EDTA, 150 mm NaCl, 0.1% NP-40] in the absence or presence of a NCoA-1/SRC-1 fragment. Peptides were synthesized to contain a tyrosine residue at the carboxyl terminus and, where indicated, labeled with 125I by use of Iodo-gen (Westin et al. 1998). Of each peptide, 3 × 105 cpm was incubated with the liganded receptors. The sequences of the wild-type LXD2 peptide was LTERHKILHRLLQEGSPSDIY; variants involved substitutions with alanine (A) residues and were as follows: LXD2(+4,+5)mut, LTERHKILHRAAQEGSPSDIY; LXD2(−3 → −7), AAAAAKILHRLLQEGSPSDIY; LXD2(+8 → +12), LTERHKILHRLLQEAAAAAIY. For DNA-dependent ABCD assays, oligonucleotides were synthesized to contain biotin residues at the 5′ end. The sequence of the sense strand of the retinoic acid response element was: 5′biotin-AAGGGGATCCGGGTAGGGTTCACCGAAGTTCACGAGATCT-3′. Purified, double-stranded oligonucleotides (1 μg) were incubated with 100 ng each of purified RARα or PPARγ and RXR produced as GST-fusion proteins in Escherichia coli and cleaved from the GST portion with thrombin. DNA binding was performed in CHAPS buffer. Receptor–DNA complexes were captured by use of 25 μl of a slurry of streptavidin–agarose and washed two times in buffer H and once in CHAPS buffer. Receptors were then incubated in the binding buffer with the indicated ligands or solvent alone for 30 min at room temperature. Following ligand binding, the receptor–DNA complexes were incubated with either 1 × 105 cpm 35S-labeled NCoA-1/SRC-1 proteins generated by in vitro transcription and translation, or with 1 × 105 cpm 32P-labeled NCoA-1/SRC-1 protein labeled by protein kinase A, for 1 hr at 4°C in NET buffer. Competing peptides were present at 1 μm. Complexes were washed five times in buffer H, resolved by SDS-polyacrylamide gels and either dried and autoradiographed, or transferred to nitrocellulose membranes for Western blotting. The generated NCoA-1/SRC-1 fragments were amino acids 626–783 containing LXD1, LXD2, and LXD3, or amino acids 626–716 containing LXD1 and LXD2.
GST–CBP fusion proteins were prepared as crude bacterial lysates and immobilized on glutathione–agarose affinity beads. The GST-fusion protein bound to glutathione–agarose was washed and incubated with in vitro-transcribed and translated NCoa-1/SRC-1 as indicated for 2 hr at 4°C. Following washing, proteins were eluted from beads and analyzed by SDS–PAGE.
Single-cell microinjection assay and immunoblots
Microinjection assays of coactivator function were performed essentially as described previously (Torchia et al. 1997). Briefly, insulin-responsive Rat-1 fibroblasts were seeded on acid-washed cover slips and rendered quiescent by incubation in serum-free medium for 24–36 hr. Plasmids were injected into the nuclei of cells at 100 μg/ml. Preimmune IgG or anti-NCoA-1/SRC-1 IgG was coinjected and the injected cells unambiguously identified by immunofluorescence microscopy. Cells were stimulated with ligand 6 hr after injection to allow expression of NCoA-1/SRC-1 from coinjected plasmids. After overnight incubation, cells were fixed and stained to detect injected IgG and β-galactosidase expression. At least 200 injected cells were quantitated for each condition; experiments were repeated at least three times. A large series of mutant NCoA–1/SRC-1 expression plasmids were generated, including mutations of L4 and L5 of LXD1, LXD2, LXD3, LXD4, and LXD5, deletions of 32 and 30 amino acids, respectively, from LXD1S2 and LXD2S3 leaving 10 flanking residues adjacent to each LXXLL motif. All mutants were constructed by a standard protocol from the Quick Change Mutagenesis Kit (Stratagene), and were confirmed by complete DNA sequencing. Western blots were performed as described previously (Xu et al. 1998); anti-HA antibodies (Babco) were utilized for SRC-1 detection, as all NCoA-1/SRC-1 proteins used contain an HA epitope tag.
Structural model
The electron density of the PPARγ ligand-binding domain–SRC–TGZ ternary complex (Nolte et al. 1998) was used to identify and build the structure of helical domain 2 of SRC-1 around the PPARγ ligand-binding domain. The SRC-1 model was built with the O software package (Jones et al. 1989), and refined with X-PLOR (Brunger 1992).
Acknowledgments
We thank Mathias Treier, Degeng Wang, and Riki Kurokawa for discussions and reagents, Peggy Meyer for her expertise in the preparation of illustrations, and Marie Fisher for assistance with the manuscript preparation. E.M.M. is supported by a National Research Service Award (National Institute of Diabetes and Digestive and Kidney Diseases); D.W.R by an American Diabetes Association Career Development Award; S.W. by the Swedish Cancer Society and U.S. Army Medical Research Program Award (E.K.). C.K.G. is an Established Investigator of the American Heart Association and M.G.R. is an Investigator in the Howard Hughes Medical Institute. These studies were supported by grants from CAP CURE and the National Institutes of Health to C.K.G. and a U.S. Army Medical Research Program Award to M.G.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mrosenfeld@ucsd.edu; FAX (619) 534-8180.
References
- Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for NCoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Barettino D, Vivanco Ruiz MM, Stunnenberg HG. Characterization of the ligand-dependent transactivation by steroid hormone receptors. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman JM, Allan GF, Tsai SY, O’Malley BW. Transcriptional activation by the estrogen receptor requires a conformational change in the ligand binding domain. Mol Endocrinol. 1993;7:1266–1274. doi: 10.1210/mend.7.10.8264659. [DOI] [PubMed] [Google Scholar]
- Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson J-A, Carlquist M. Molecular basis of agonism and antagonism of oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- Brunger A. X-PLOR Version 3.0: A system for crystallography and NMR. New Haven, CT: Yale University Press; 1992. [Google Scholar]
- Cavailles V, Dauvois S, Danielain PS, Parker MG. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chkravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with p/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–776. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Danielian PS, White R, Lees JA, Parker MG. Identification of a conserved region required for hormone-dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function-2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: Presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch M, Leary CM, Furlow JD, Ahrens H, Schuh TJ, Mueller GC, Gorski J. A ligand-induced conformational change in the estrogen receptor is localized in the steroid binding domain. Biochemistry. 1992;31:5303–5311. doi: 10.1021/bi00138a009. [DOI] [PubMed] [Google Scholar]
- Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: Possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional coactivators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang W M, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing NCoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Jones TA, Bergdoll M, Kjeldgaard M, Ealick S. In: Crystallographic computing and modeling methods in molecular design. Bugg C, editor. New York, NY: Springer; 1989. [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;7:1266–1274. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen BS. Estrogen receptors: Bioactivities and interactions with cell signaling pathways. Biol Reprod. 1996;54:287–293. doi: 10.1095/biolreprod54.2.287. [DOI] [PubMed] [Google Scholar]
- Koch A, Anderson D, Moran MF, Ellis C, Pawson T. SH2 and SH3 domains: Elements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT. p300 and estrogen receptors cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes & Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa R, Soderstrom M, Horlein A, Halamachi S, Brown M, Rosenfeld MG, Glass CK. Polarity-specific activities of retinoic acid receptors determined by a corepressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- LeDouarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Choi HS, Gyuris J, Brent R, Moore DD. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Nolte RT, Wisely B, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and coactivator assembly of the peroxisome proliferator-activated receptor γ. Nature. 1998;395:137–144. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the pCAF histone deacetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Oñate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2081. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Rachez C, Suldan Z, Ward J, Chang CPB, Burakov D, Erdjument-Bromage H, Tempst P, Freedman LP. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes & Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson H J, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91:741–753. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-γ ligand binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Tone Y, Collinwood TN, Adams M, Chattergee VK. Functional analysis of a transactivation domain in the thyroid hormone receptor. J Biol Chem. 1994;269:31157–31161. [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional coactivator p/CIP binds CBP and mediates nuclear receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Torchia J, Glass CK, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- Tzukerman MT, Esty A, Santiso-Mere D, Danielian P, Parker MG, Stein RB, Pike JW, McDonnell DP. Human estrogen receptor transactivation capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- van der Geer, P. and T. Pawson. 1995. The PTB domain: A new protein module implicated in signal transduction (phosphotyrosine-binding). Trends Biochem. Sci. 20. [DOI] [PubMed]
- Voegel JJ, Heine MJS, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Wagner RL, Apriletti J W, McGrath ME, West BL, Baxter JD, Fletterick RJ. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- Westin S, Kurokawa R, Nolte RT, Wisely B, McInerney EM, Rose DW, Milburn MV, Rosenfeld MG, Glass CK. Allosteric interactions governing the assembly, structure and function of nuclear receptor heterodimer-coactivator complexes. Nature. 1998;395:199–203. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- Wong J, Shi YB, Wolffe AP. Determinants of chromatin disruption and transcriptional regulation instigated by thyroid hormone receptor: Hormone-regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J. 1997;11:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Lavinsky RM, Dasen JS, Flynn SE, McInerney EM, Mullen T-M, Heinzel T, Szeto D, Korzus E, Kurokawa R, Aggarwal AK, Rose DW, Glass CK, Rosenfeld MG. Distinct histone acetyltransferases and CBP domains are required for activation of Pit-1 by different signalling pathways. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- Yao T-P, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou N-D, Ch’ng L-E, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–472. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]