The level of c-Myb is a determining factor in the response of leukemia cells to GSK3β kinase inhibiton, which is of particular interest for the therapy of leukemia and cancers that have c-Myb amplifications.
Abstract
Glycogen synthase kinase 3β (GSK3β) regulates diverse physiological processes, including metabolism, development, oncogenesis, and neuroprotection. GSK3β kinase activity has been reported to be critical for various types of cancer cells, but the mechanism has remained elusive. In this study we examine the mechanism by which GSK3β regulates the survival of leukemia cells. We demonstrate that upon GSK3β kinase inhibition different types of leukemia cells show severe proliferation defects as a result of apoptosis. The transcription factor c-Myb is found to be the main target of GSK3β inhibition in cell survival. GSK3β inactivation reduces the expression of c-Myb by promoting its ubiquitination-mediated degradation, thereby inhibiting the expression of c-Myb–dependent antiapoptotic genes Bcl2 and survivin. Coimmunoprecipitation, reporter assays, chromatin immunoprecipitation, and knockdown studies show that c-Myb needs to interact and cooperate with transcription factor LEF-1 in the activation of Bcl2 and survivin and that both transcription factors are required for cell survival. These data reveal an as-yet-unknown mechanism by which GSK3β controls cell survival.
INTRODUCTION
GSK3β, a multifunctional serine/threonine (Ser/Thr) kinase, was originally discovered as a key enzyme in the control of glucose metabolism. In addition, it was identified as a component of the Wnt signaling pathway, forming a complex with APC and Axin, which inactivates the pathway in the absence of Wnt ligands by phosphorylating and degrading β-catenin. In the presence of Wnt ligand this complex is disrupted (Harwood, 2001). GSK3β is also involved in Alzheimer's disease since it associates with presenilin and phosphorylates the microtubule-associated protein tau (Takashima et al., 1998; Baki et al., 2004). Disruption of the murine GSK3β gene results in embryonic lethality due to severe liver degeneration, and the GSK3β-deficient fibroblasts are more sensitive to tumor necrosis factor-α–induced apoptosis as a result of reduced nuclear factor (NF)-κB activity (Hoeflich et al., 2000). In certain pancreatic cancers, prostate cancers, and gliomas the activity of GSK3β is required for cancer cell survival (Ougolkov et al., 2005; Kotliarova et al., 2008; Vene et al., 2008; Korur et al., 2009; Mai et al., 2009; Mamaghani et al., 2009). In addition, in leukemia cells GSK3β can enhance proliferation and/or survival (Holmes et al., 2008; Wang et al., 2008). The mechanism of these GSK3β functions remains elusive and might depend on cell type, context, and nature of the oncogenic mutations.
The transcription factor c-Myb was discovered as a protooncogene and is expressed predominantly in immature hematopoietic cells (Westin et al., 1982; Luscher and Eisenman, 1990; Thompson and Ramsay, 1995). Forced expression of c-Myb inhibits erythroid differentiation, whereas antisense c-Myb oligonucleotides cause growth arrest and inhibition of hematopoietic colony formation (Clarke et al., 1988; Gewirtz and Calabretta, 1988; Shivdasani and Orkin, 1996). c-Myb–deficient mice show normal hematopoiesis in the yolk sac but defective hematopoiesis in the fetal liver (Mucenski et al., 1991). Oncogenically activated versions of c-Myb have been found in two different avian leukemia viruses—avian myeloblastosis virus and E26—which can transform immature hematopoietic cells in vitro and induce acute myeloid or erythroid leukemia in chickens (Beug et al., 1979; Radke et al., 1982; Oh and Reddy, 1999). c-Myb regulates hematopoietic gene expression in combination with other transcription factors, cofactors, or accessory proteins, such as core-binding factor/PEBP2, C/EBPalpha, C/EBPε, PU.1, and cAMP response element–binding (CREB)–binding protein (Hernandez-Munain and Krangel, 1995; Dai et al., 1996; Verbeek et al., 1999). Oncogenic changes in c-Myb have also been found in colorectal and breast cancers (Ramsay and Gonda, 2008). However, the signaling pathway(s) regulating c-Myb activity are largely unclear.
Lymphoid enhancer–binding factor-1 (LEF-1), a member of the high-mobility group box family, was originally identified as a pre–B and T lymphocyte–specific protein that regulates the T cell receptor α enhancer (Giese et al., 1991; Travis et al., 1991). By binding to the minor groove it was found to induce a sharp bend in the DNA, thereby facilitating the assembly of a higher-order multiprotein enhancer complex containing other lymphoid-specific proteins and Ets-1, PEBP2-α, and ATF/CREB (Giese et al., 1992; Love et al., 1995). LEF-1 is also an essential regulator of canonical Wnt/β-catenin signaling and a leukemogenesis factor (Behrens et al., 1996; Hsu et al., 1998; Petropoulos et al., 2008; Gutierrez et al., 2010).
In the present study, we examined the mechanism by which GSK3β maintains leukemia cell growth. In different types of leukemia cells, GSK3β kinase inhibitors caused severe proliferation defects due to apoptosis. We found the survival genes Bcl2 and survivin to be inhibited upon GSK3β inactivation, as a result of decreased binding of c-Myb and LEF-1 to their promoters. GSK3β inactivation increased c-Myb degradation, and overexpression of c-Myb could block the inhibitory effect of GSK3β inactivation on cell proliferation. We also show that c-Myb and LEF-1 interact and cooperate in the activation of Bcl2 and survivin in leukemia cells and that both are required for cell survival. These data reveal an as-yet-unknown function of GSK3β and suggest that the effect of GSK3β on leukemia cell proliferation depends on its regulation of c-Myb.
RESULTS
GSK3β activity is required for the survival of leukemia cells
To examine the role of GSK3β activity in leukemia cells, we treated acute T cell leukemia Jurkat, chronic myelogenous leukemia K562, and myeloma RPMI-8226 cells with the GSK3β kinase inhibitor LiCl or SB216763. Both inhibitors strongly reduced the proliferation of these cells as measured by cell counting and cell viability analysis. In contrast, proliferation of HEK293 cells was not inhibited (Figure 1, A and B). The kinase activity of GSK3β was found to be critical for the proliferation of the leukemia cells since overexpression of the constitutively active GSK3β mutant S9A rescued the growth inhibition by SB216763, whereas wild-type GSK3β and the kinase-dead mutant K85M did not (Figure 1, C and D).
FIGURE 1:
GSK3β inactivation causes growth inhibition of leukemia cells. (A) Jurkat, K562, RPMI 8226, and HEK293 cells were treated with or without LiCl (10 mM) or SB216763 (10 μM). Cell numbers were determined with a CASY model TT cell counter (Schärfe System, Reutlingen, Germany) on days 0 and 3; the fold change is depicted. (B) Cells were treated with or without SB216763 as described, and their viability was measured with the CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS; Promega, Madison, WI) at indicated time points. The relative viability (±SB216763) is depicted. (C) Jurkat cells were infected with lentivirus expressing GSK3β WT, GSK3β S9A, and GSK3β K85M and treated with or without SB216763 (10 μM). Cell numbers were determined as described for A. (D) Immunoblotting shows the expression levels of wild-type and mutant GSK3β in the experiment shown in C and verification of their effects on β-catenin expression. Data are represented as mean and SD of triplicates and are representative of at least two independent experiments.
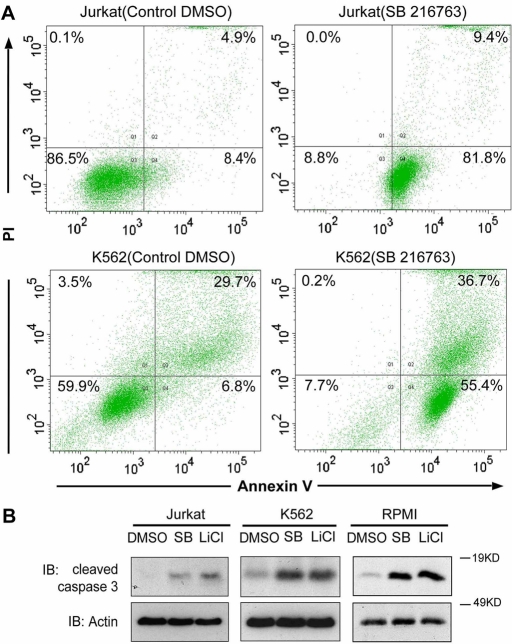
We next tested whether the decreased viability observed after SB216763 or LiCl treatment in the leukemia cells was due to increased apoptosis. Indeed, as shown in Figure 2A, in both Jurkat and K562 cells, SB216763 treatment strongly increased the amount of annexin V–positive cells. Similar results were obtained in Jurkat, K562, and RPMI-8226 when the amount of sub-G1 cells was measured (Supplemental Figure S1 and unpublished data). The levels of active caspase 3 were also increased after SB216763 or LiCl treatment in these three cell lines (Figure 2B). This demonstrates that GSK3β activity protects leukemia cells from apoptosis.
FIGURE 2:
GSK3β inactivation induces apoptosis in leukemia cells. (A) Jurkat and K562 cells were treated with or without SB216763 (10 μM) for 20 h, stained with annexin V and PI, and analyzed by fluorescence-activated cell sorting. (B) Jurkat, K562, and RPMI 8226 cells were treated with SB216763 (SB; 10 μM) or LiCl (10 mM) for 20 h, and cell lysates were analyzed by immunoblotting.
GSK3β inactivation inhibits the activation of the survival genes Bcl2 and survivin by c-Myb and LEF-1
To elucidate the mechanism by which GSK3β regulates proliferation and survival of leukemia cells, we first analyzed the expression of various proliferation- and apoptosis-related genes. Quantitative real-time PCR (qRT-PCR) assays in Jurkat and K562 cells showed that LiCl or SB216763 strongly reduced the mRNA levels of the apoptosis-related genes Bcl2, survivin, c-Myc, and CEBP/α (Figure 3A and unpublished data). In contrast, the levels of Bim, Bax, and Bcl-xL did not show any difference, whereas the cell cycle inhibitors p21 and p27 showed up-regulation and down-regulation, respectively (Supplemental Figure S2). Activation of the Wnt target genes DKK1 and Axin2 by LiCl or SB216763 indicated that Wnt signaling was strongly induced under these conditions (Figure 3A). Western analysis confirmed that the protein levels of Bcl2 and survivin also were inhibited by GSK3β inactivation in Jurkat, K562, and RPMI-8226 cells (Figure 3B).
FIGURE 3:
GSK3β inactivation inhibits the expression of Bcl2 and survivin. (A) Jurkat cells were treated with SB216763 (10 μM) or LiCl (10 mM) for 20 h, and RNA was reverse transcribed and analyzed by qRT-PCR. Values and error bars represent the mean and SD of triplicates and are representative of at least two independent experiments. (B) Jurkat, K562, and RPMI 8226 cells were treated with SB216763 (SB) (10 μM) or LiCl (10 mM) for 20 h, and cell lysates were analyzed by immunoblotting. (C, D) Jurkat cells were treated with or without SB216763 (10 μM) for 8 h. Cells were lysed, and ChIP was performed with anti–LEF-1, anti–c-Myb, or a control antibody (immunoglobulin G [IgG]). The precipitated promoter fragments of Bcl2 (C) and survivin (D) were amplified by qRT-PCR (top). The positions of the identified LEF-1– and c-Myb–binding sites in the promoters are shown in a sketch map (bottom). Values and error bars represent the mean and SD of triplicates and are representative of at least two independent experiments.
Because Bcl2 and survivin are target genes of the transcription factors c-Myb and LEF-1, respectively, whereas c-Myc is regulated by both c-Myb and LEF-1, we next investigated whether GSK3β inhibition affects the binding of these transcription factors to the Bcl2 and survivin promoters. Chromatin immunoprecipitation (ChIP) analysis showed that after SB216763 treatment, both the binding of c-Myb to the Bcl2 promoter and the binding of LEF-1 to the survivin promoter were dramatically decreased (Figure 3, C and D). Moreover, LEF-1 also bound to the Bcl2 promoter, whereas c-Myb also bound to survivin promoter, and these interactions were reduced upon SB216763 treatment as well. Taken together, GSK3β inactivation suppresses the transcriptional activation of apoptosis-related genes Bcl2 and survivin by c-Myb and LEF-1.
GSK3β inactivation reduces c-Myb protein levels and promotes its ubiquitination
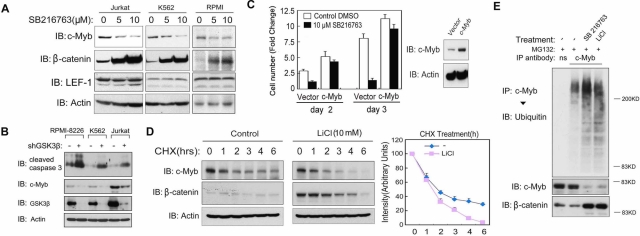
We next tested the effect of GSK3β inactivation on the c-Myb and LEF-1 proteins. Of importance, in all leukemia cells examined, the levels of c-Myb were reduced upon SB216763 or LiCl treatment, whereas the levels of LEF-1 did not show significant differences (Figure 4A and Supplemental Figure S3). This decrease in c-Myb was already prominent after 4 h of GSK3β kinase inhibition and coincided with Wnt signaling activation as measured by β-catenin induction, which indicates that c-Myb might be regulated by GSK3β kinase activity directly (Supplemental Figure S4). Consistent with results published in the literature, we could not detect endogenous c-Myb expression in HEK293 cells (Supplemental Figure S3) (Corradini et al., 2005; Zhao et al., 2009), which might explain why the proliferation of these cells is not inhibited by GSK3β (Figure 1, A and B). Of importance, and consistent with results obtained with pharmacological GSK3β inhibitors, short hairpin RNA (shRNA)–mediated depletion of GSK3β in all three kinds of leukemia cell lines leads to an obvious decrease in c-Myb and up-regulation of cleaved caspase 3 (Figure 4B). c-Myb overexpression could rescue the inhibition of cell proliferation by SB216763 in Jurkat cells, demonstrating that c-Myb is a critical growth-regulatory target of GSK3β kinase activity in these cells (Figure 4C). Moreover, in a small-scale screen in which 52 kinase inhibitors were tested for their ability to inhibit Jurkat cell proliferation, almost all of the inhibitors that suppressed GSK3β kinase activity were found also to decrease the levels of c-Myb and to suppress proliferation (Supplemental Figure S5).
FIGURE 4:
The c-Myb protein level is critical for GSK3β-dependent cell survival. (A) Jurkat, K562, and RPMI 8226 cells were treated with or without 5 or 10 μM SB216763 for 8 h, and cell lysates were analyzed by immunoblotting. (B) RPMI 8226, K562, and Jurkat cells were infected with lentivirus expressing GSK3β shRNA or a nonspecific control shRNA. Four days after infection, cells were harvested for immunoblotting analysis. (C) Jurkat cells were infected with lentiviral-overexpressed c-Myb or a control vector and after 48 h treated with or without SB216763 (10 μM) for another 2 or 3 d. Cell numbers were determined at days 0, 2, and 3 with a CASY Model TT cell counter (left). Data are represented as mean and SD. The overexpression of c-Myb was verified by immunoblotting (right). (D) Jurkat cells were treated with cycloheximide (20 μg/ml) in the absence or presence of LiCl (10 mM) for the indicated time points, and cell lysates were analyzed by immunoblotting. For the quantification on the right, the c-Myb protein level at time 0 was set at 100%. (E) Jurkat cells were treated with or without SB216763 (10 μM) or LiCl (10 mM) for 6 h, after which MG132 (10 μM) was added for another 4 h. c-Myb and nonspecific control immunoprecipitations were analyzed by SDS–PAGE and immunoblotting.
We subsequently investigated how GSK3β inactivation could lead to a rapid decrease (within 4 h) in the amount of c-Myb. qRT-PCR analysis of c-Myb mRNA in SB216763- or LiCl-treated Jurkat and K562 cells revealed only significant down-regulation after 8 h (unpublished data), which is probably an indirect consequence of decreased c-Myb autoinduction (Nicolaides et al., 1991). Inhibition of de novo protein synthesis by cycloheximide (CHX) showed that c-Myb degraded faster in cells treated with SB216763 or LiCl (Figure 4D and unpublished data). In addition, we found that the ubiquitination of c-Myb was increased after GSK3β inactivation (Figure 4E). Proteosome inhibition by MG132 could partially rescue c-Myb degradation (unpublished data). This suggests that GSK3β kinase may protect c-Myb against proteasome-mediated degradation by inhibiting its ubiquitination.
LEF-1 and c-Myb cooperatively regulate transcription of Bcl2 and survivin
Because both the LEF-1 and c-Myb fractions bound to the survivin and Bcl2 promoters were decreased upon GSK3β inactivation (Figure 3, C and D), whereas only the (total) c-Myb levels were reduced, we next examined whether c-Myb and LEF-1 could cooperate in LEF-1– or c-Myb–dependent transcription activation. Indeed, both c-Myb and LEF-1 were able to activate the LEF-1–responsive reporter LEF-Luc and c-Myb–responsive pTRHR-Luc, and the activation was strongly increased when the two proteins were coexpressed (Figure 5, A and B). Moreover, knockdown of c-Myb reduced the activation of LEF-Luc by LEF-1, and knockdown of LEF-1 reduced the activation of pTRHR-Luc by c-Myb (Figure 5, C and D). We next examined whether c-Myb was required for LEF-1 binding to and/or activation of the endogenous survivin and Bcl2 promoters and vice versa. ChIP assays in c-Myb–depleted or LEF-1–depleted cells showed that both c-Myb and LEF-1 were required for optimal binding of each of them to the Bcl2 and survivin upstream regulatory regions in Jurkat cells. c-Myb knockdown in fact completely blocked the binding of LEF-1 to the Bcl2 promoter, whereas LEF-1 knockdown strongly impaired the binding of c-Myb to the survivin promoter (Figure 5E). Coimmunoprecipitation assays showed that endogenous c-Myb and LEF-1 can bind each other, suggesting that they form a complex regulating these survival genes (Figure 5F). Moreover, qRT-PCR analysis in the c-Myb or LEF-1 knockdown cells showed that both c-Myb and LEF-1 are required for the expression of Bcl2 and survivin (Figure 5G), whereas overexpression of c-Myb or LEF-1 increased the mRNA levels of these two survival genes (Supplemental Figure S6). Together, these data show that c-Myb and LEF-1 interact and cooperate in the activation of Bcl2 and survivin in leukemia cells.
FIGURE 5:
LEF-1 and c-Myb cooperatively regulate transcription. (A–D) K562 cells were transiently transfected with the indicated plasmids and analyzed for luciferase assay after 36 h. Data are represented as mean and SD. (E) Jurkat cells were infected with lentivirus expressing c-Myb or LEF-1 shRNA or a nonspecific control (ctrl) shRNA. After 60 h, cells were lysed and ChIP was performed with anti–LEF-1, anti–c-Myb, or a control antibody (IgG). The precipitated promoter fragments were amplified by qRT-PCR (left). Data are represented as mean and SD. The knockdown efficiency of the c-Myb and LEF-1 shRNAs was validated by immunoblotting analysis (right). (F) To detect the interaction between c-Myb and LEF-1 in Jurkat cells, LEF-1 and nonspecific control (ns) immunoprecipitations (IPs) were analyzed by SDS–PAGE and immunoblotting (IB). (G) RNA from the Jurkat cells treated as described for E was analyzed by qRT-PCR analysis. Values and error bars represent the mean and SD of triplicates and are representative of at least two independent experiments.
C-Myb and LEF-1 are both required for leukemia cell survival
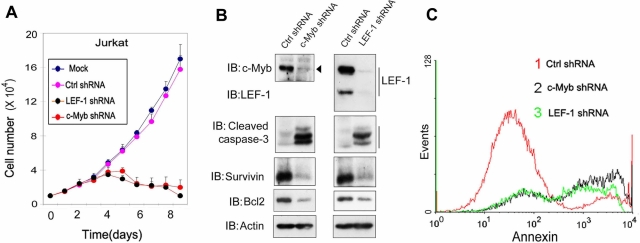
The results described earlier suggested that both c-Myb and LEF-1 are critical in GSK3β-dependent cell survival. We therefore examined the effect of c-Myb and LEF-1 knockdown on cell proliferation and cell death. Knockdown of c-Myb or LEF-1 was found to strongly inhibit the proliferation of Jurkat and K562 cells (Figure 6A and Supplemental Figure S7). Western analysis showed that this inhibition was accompanied by a decrease in the levels of Bcl2 and survivin and a strong increase in active caspase 3 (Figure 6B). The amount of annexin V–positive cells also increased (Figure 6C). These data demonstrate that by cooperatively activating Bcl2 and survivin, both c-Myb and LEF-1 are essential factors responsible for GSK3β activity–mediated cell survival in leukemia cells.
FIGURE 6:
C-Myb and LEF-1 are both required for the survival of leukemia cells. (A) Jurkat cells were infected with lentivirus expressing c-Myb or LEF-1 shRNA, or nonspecific control (ctrl) shRNA or were mock infected. The cell numbers were counted at the indicated days. (B) Lysates of Jurkat cells treated as described for A were isolated 60 h after infection and analyzed by immunoblotting. (C) Jurkat cells treated as described for A were harvested 60 h after infection, stained with annexin V and PI, and analyzed by fluorescence-activated cell sorting.
DISCUSSION
In this study, we analyzed the effect of GSK3β inactivation in several kinds of leukemia cell lines. In all of these cells GSK3β inhibitors induced apoptosis and reduced the expression of the survival factors Bcl2 and survivin. Moreover, GSK3β was found to control Bcl2 and survivin expression by preventing ubiquitin-dependent degradation of c-Myb and thereby to stimulate c-Myb– and LEF-1–dependent transcriptional activation. In contrast to the leukemia cells examined, we found that GSK3β inactivation did not affect the proliferation of HEK293 cells, which do not express c-Myb (Supplemental Figure S3). Previous studies showed that GSK3β can control cell proliferation and/or survival by positively regulating NF-κB function (Hoeflich et al., 2000; Ougolkov et al., 2005) or by destabilizing the cyclin-dependent kinase inhibitor p27Kip1 (Wang et al., 2008). However, we found no evidence that GSK3β altered NF-κB activity and p27Kip1 stability in the cell lines that we investigated (unpublished data). Our results therefore reveal a novel mechanism by which GSK3β stimulates cell survival and proliferation and suggest that the levels of c-Myb can be a determining factor in GSK3β inhibition–induced apoptosis of certain leukemia cell lines.
We show here that in leukemia cells GSK3β inactivation promotes the ubiquitination of c-Myb. Because phosphorylation of c-Myb by NLK can regulate its ubiquitin-dependent degradation (Kanei-Ishii et al., 2004; Ramsay and Gonda, 2008), we tested the effect of GSK3β inactivation on c-Myb phosphorylation status. Of interest, c-Myb threonine phosphorylation was significantly increased after treatment with SB216763 or LiCl, whereas phosphorylation on serine and tyrosine remained unchanged (unpublished data). However, it remains to be established whether GSK3β kinase activity protects c-Myb against proteosome-mediated degradation by inhibiting its threonine phosphorylation, and whether NLK is involved in this. We could not detect a stable interaction between endogenous GSK3β and c-Myb.
Because GSK3β inactivation is a direct consequence of Wnt signaling, we also examined whether other components of the Wnt signaling pathway affect the levels and ubiquitination of c-Myb in leukemia cells. Consistent with a previous report that Wnt1 induces ubiquitination and proteasome-dependent degradation of c-Myb (Kanei-Ishii et al., 2004), Wnt1 and Wnt3a indeed decreased c-Myb protein level in Jurkat cells, but wild-type β-catenin and a constitutively active mutant did not (Supplemental Figure S8A). Ubiquitination of c-Myb was also increased by Wnt1 and decreased by DKK1 but not by β-catenin (Supplemental Figure 8B and unpublished data). This indicates that GSK3β is the key component of the Wnt pathway in the control of c-Myb protein and that the negative regulation by GSK3β occurs independent of canonical Wnt/β-catenin signaling.
By decreasing the level of c-Myb, inhibition of GSK3β affects the function of the transcription factor LEF-1. Here we show that c-Myb and LEF-1 cooperatively bind to and activate the growth-regulatory genes Bcl2, survivin, and c-Myc in leukemia cells and thereby protect these cells against apoptosis. Overexpression of LEF-1 and c-Myb also showed cooperative effects in HEK293 cells (unpublished data). However, in HEK293 cells endogenous c-Myb is undetectable (Supplemental Figure S3) (Corradini et al., 2005; Zhao et al., 2009). Of interest, the other two TCF/LEF family members, TCF1 and TCF4, which play a similar role as LEF-1 in Wnt signaling, did not interact with or regulate the transactivation of c-Myb (unpublished data), indicating that the cooperation between LEF-1 and c-Myb is rather specific in controlling the growth of leukemia cells.
As a component of multiple signaling pathways, GSK3β has been implicated in metabolic defects, developmental problems, cancers, and neuroprotective disorders. Therefore, various types of GSK3β kinase inhibitors are being tested for their specificity and suitability in therapeutic applications. Our results suggest that the level of c-Myb is a determining factor in the response of leukemia cells to GSK3β kinase inhibitors, which is of particular interest for the therapy of leukemia and cancers that have c-Myb amplifications.
MATERIALS AND METHODS
Reagents and plasmids
SB216763 was obtained from Tocris Bioscience (Ellisville, MO). The expressing vectors for LEF-1, GSK3β WT, GSK3β K85M, and LEF-luciferase and TopFlash-luciferase transcriptional reporter constructs were described previously (Zhang et al., 2006; Zhou et al., 2008). GSK3β S9A (Guo et al., 2008) was kindly provided by Xiao-Fan Wang (Duke University, Durham, NC) and HA-c-Myb and pTHRH-Luci (Saether et al., 2007) by Odd Stokke Gabrielsen (University of Oslo, Oslo, Norway). Lentiviral viral vectors expressing shRNA targeting human LEF-1, c-Myb, and GSK3β were obtained from Sigma-Aldrich (St. Louis, MO) Mission shRNA (LEF-1, TRCN0000020163; c-Myb, TRCN0000040062; GSK3β, TRCN0000000822). The lentiviral vectors that direct the ectopic expression of LEF-1 and c-Myb were generated by inserting the LEF-1 and c-Myb cDNA fragments into pLV.CMV.bc.puro by PCR and MluI and XhoI digestion. Lentiviral vectors for ectopic expression of GSK3β WT, GSK3β S9A, and GSK3β K85M mutants were generated by cloning the corresponding cDNAs into pLV.CMV.bc.puro FLAG vector by PCR and SalI and XbaI digestion. All the primers used for making constructs are listed in Supplemental Table S1.
Cell culture
RPMI 8226, Jurkat, and K562 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. HEK293T cells were cultured in DMEM supplemented with 10% fetal calf serum and l-glutamine. All the cells were incubated at 37°C in a humidified atmosphere of 5% CO2.
Reporter assay
Cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and harvested after 36 h to determine luciferase activities using a PerkinElmer (Waltham, MA) luminometer. In all transfections, 20 ng of a Renilla expression vector was cotransfected as an internal transfection control. Each experiment was performed in triplicate, and the data represent the mean ± SD of three independent experiments after normalization to Renilla activity.
Flow cytometry (fluorescence-activated cell sorting)
Aliquots of 106 cells were washed twice with phosphate-buffered saline (PBS) and resuspended in binding buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid/NaOH, pH 7.5, 0.14 M NaCl, 2.5 mM CaCl2). Propidium iodide (PI) (3.3 μg/ml)/annexin V–fluorescein isothiocyanate (0.55 μg/ml) was added, and the samples were incubated for 10 min at room temperature in the dark. Fluorescence was detected using BD LSR II flow cytometry (BD Biosciences, San Diego, CA). The number of cells that were negative for PI and positive for annexin V was scored as apoptotic. The results were analyzed by BD FACSDiva software.
Immunoprecipitation and immunoblotting
As previously described (Zhang et al., 2004, 2006), cells were lysed with 1 ml of lysis buffer (20 mM Tris-HCl, pH 7.4, 2 mM EDTA, 25 mM NaF, 1% Triton X-100) plus protease inhibitors (Sigma-Aldrich) for 10 min at 4°C. After centrifugation at 12,000 × g for 15 min, the lysates were immunoprecipitated with antibodies and protein A–Sepharose (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) for 3 h at 4°C. Thereafter, the precipitates were washed three times with washing buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) and the immune complexes were eluted with sample buffer containing 1% SDS for 5 min at 95°C and analyzed by SDS–PAGE.
Immunoblotting was performed as described (Zhou et al., 2008) using secondary anti–mouse or anti–rabbit antibodies conjugated to horseradish peroxidase (GE Healthcare, Buckinghamshire, United Kingdom) and visualized by chemiluminescence.
Antibodies used for immunoprecipitation and immunoblotting were as follows: LEF-1 (C12A5; Cell Signaling, Beverly, MA), c-Myb (clone 1-1; Upstate, Millipore, Billerica, MA), β-catenin (BD Transduction Laboratories, Lexington, KY), β-actin (A5441; Sigma-Aldrich), Bcl2 (C-2; Santa Cruz Biotechnology, Santa Cruz, CA), survivin (sc-17779; Santa Cruz Biotechnology), cleaved caspase 3 (Asp175; 5A1E; Cell Signaling), and ubiquitin (P4D1; Santa Cruz Biotechnology).
Ubiquitination detection
Cells were washed with PBS and lysed in two pellet volumes of RIPA buffer (20 mM sodium phosphate pH 7.4, 150 mM NaCl, 1% Triton, 0.5% sodium deoxycholate, and 1% SDS) supplemented with protease inhibitors and 10 mM N-ethylmaleimide. Lysates were sonicated, boiled at 95°C for 5 min, diluted to RIPA buffer containing 0.1% SDS, and then centrifuged at 4°C (16 × 103 g for 15 min). The supernatant was incubated with c-Myb antibody and protein A–Sepharose for 3 h at 4°C. After extensive washing, bound proteins were eluted with 2× SDS sample buffer and separated on SDS–PAGE, followed by Western blotting.
qRT-PCR
Total RNA was isolated using the NucleoSpin RNA II Kit (Macherey-Nagel, Düren, Germany). A 1-μg amount of RNA was reverse transcribed using a RevertAid First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany). qRT-PCR was performed by SYBR Green incorporation (Applied Biosystems, Foster City, CA) using a StepOne Plus real-time PCR system (Applied Biosystems). Results were normalized to those obtained with glyceraldehyde-3-phosphate dehydrogenase and presented as fold induction over control cells. All the primers used for qRT-PCR are listed in Supplemental Table 1.
Chromatin immunoprecipitation assay
Cells were chemically cross-linked by addition of 1/10 volume of freshly prepared 11% formaldehyde solution to the culture medium and incubated for 15 min at room temperature. Cells were subsequently rinsed twice with PBS, harvested using a silicon scraper, lysed (1% SDS, 50 mM Tris, pH 8.0, 5 mM EDTA, and proteinase inhibitors), and sonicated to obtain DNA fragments of ∼300–500 base pairs on average. Samples were then centrifuged at 14,000 rpm for 10 min. The supernatant was diluted (20 mM Tris, pH 8.0, 2 mM EDTA, 1% Triton X-100, 150 mM NaCl, proteinase inhibitors) and then incubated overnight at 4°C with 2.5 μg of antibodies (anti–c-Myb and anti–LEF-1). The immunocomplex was collected with 50 μl of protein A beads by 1-h coincubation and then washed sequentially with TSE I (0.1% SDS, 20 mM Tris, pH 8.0, 2 mM EDTA, 1% Triton X-100, 150 mM NaCl, proteinase inhibitors), TSE II (0.1% SDS, 20 mM Tris, pH 8.0, 2 mM EDTA, 1% Triton X-100, 500 mM NaCl, proteinase inhibitors), LiCl buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 0.25 mM LiCl, 0.1% NP-40, 1% deoxycholate sodium), and TE (10 mM Tris, pH 8.0, 1 mM EDTA, pH 8.0). The bound immunocomplex was eluted with 400 μl of fresh elution buffer (25 mM Tris, pH 8.0, 10 mM EDTA, 0.5% SDS) by heating at 65°C with occasional vortex for 15 min, and cross-linking was reversed by overnight incubation at 65°C. Whole-cell-extract DNA (input fraction obtained from the sonication step) was also treated for cross-linking reversal. Immunoprecipitated DNA and whole-cell-extract DNA were then purified by treatment with RNaseA, proteinase K, and multiple phenol:chloroform:isoamyl alcohol extraction. All of the samples were examined by qRT-PCR. All of the primers used for ChIP assay are listed in Supplemental Table 1.
Statistical analysis
Statistical analyses were performed with a two-tailed unpaired t test. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We are grateful to Xiao-Fan Wang and Odd Stokke Gabrielsen for reagents and Martijn Rabelink, Midory Thorikay, Maarten van Dinther, Melissa van Pel, and Frank Staal for experimental assistance and valuable discussions. This work was supported by the Netherlands Organization for Scientific Research (NWO 918.66.066) and Center for Biomedical Genetics.
Abbreviations used:
- Bax
BCL2-associated X protein
- Bcl2
B-cell CLL/lymphoma 2
- Bcl-xL
BCL2-like 1
- Bim
BCL2-like 11 (apoptosis facilitator)
- C/EBP
CCAAT/enhancer binding protein
- DKK1
dickkopf homologue 1
- GSK3β
glycogen synthase kinase 3β
- HA
hemagglutinin
- LEF-1
lymphoid enhancer-binding factor-1
- shRNA
small hairpin RNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0483) on July 27, 2011.
*These authors equally contributed to this work.
The authors declare no conflict of interest.
REFERENCES
- Baki L, Shioi J, Wen P, Shao Z, Schwarzman A, Gama-Sosa M, Neve R, Robakis NK. PS1 activates PI3K thus inhibiting GSK-3 activity and tau overphosphorylation: effects of FAD mutations. EMBO J. 2004;23:2586–2596. doi: 10.1038/sj.emboj.7600251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Beug H, von Kirchbach A, Doderlein G, Conscience JF, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–390. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- Clarke MF, Kukowska-Latallo JF, Westin E, Smith M, Prochownik EV. Constitutive expression of a c-myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol Cell Biol. 1988;8:884–892. doi: 10.1128/mcb.8.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradini F, Cesi V, Bartella V, Pani E, Bussolari R, Candini O, Calabretta B. Enhanced proliferative potential of hematopoietic cells expressing degradation-resistant c-Myb mutants. J Biol Chem. 2005;280:30254–30262. doi: 10.1074/jbc.M504703200. [DOI] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Hou DX, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- Gewirtz AM, Calabretta B. A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science. 1988;242:1303–1306. doi: 10.1126/science.2461588. [DOI] [PubMed] [Google Scholar]
- Giese K, Amsterdam A, Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991;5:2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Guo X, Ramirez A, Waddell DS, Li Z, Liu X, Wang XF. Axin and GSK3- control Smad3 protein stability and modulate TGF-signaling. Genes Dev. 2008;22:106–120. doi: 10.1101/gad.1590908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A Jr, Tschumper RC, Wu X, Shanafelt TD, Eckel-Passow J, Huddleston PM 3rd, Slager SL, Kay NE, Jelinek DF. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B-cell lymphocytosis. Blood. 2010;116:2975–2983. doi: 10.1182/blood-2010-02-269878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood AJ. Regulation of GSK-3: a cellular multiprocessor. Cell. 2001;105:821–824. doi: 10.1016/s0092-8674(01)00412-3. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munain C, Krangel MS. c-Myb and core-binding factor/PEBP2 display functional synergy but bind independently to adjacent sites in the T-cell receptor delta enhancer. Mol Cell Biol. 1995;15:3090–3099. doi: 10.1128/mcb.15.6.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- Holmes T, O'Brien TA, Knight R, Lindeman R, Shen S, Song E, Symonds G, Dolnikov A. Glycogen synthase kinase-3beta inhibition preserves hematopoietic stem cell activity and inhibits leukemic cell growth. Stem Cells. 2008;26:1288–1297. doi: 10.1634/stemcells.2007-0600. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei-Ishii C, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18:816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korur S, Huber RM, Sivasankaran B, Petrich M, Morin P Jr, Hemmings BA, Merlo A, Lino MM. GSK3beta regulates differentiation and growth arrest in glioblastoma. PLoS One. 2009;4:e7443. doi: 10.1371/journal.pone.0007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotliarova S, et al. Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res. 2008;68:6643–6651. doi: 10.1158/0008-5472.CAN-08-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- Luscher B, Eisenman RN. New light on Myc and Myb. Part II. Myb. Genes Dev. 1990;4:2235–2241. doi: 10.1101/gad.4.12b.2235. [DOI] [PubMed] [Google Scholar]
- Mai W, Kawakami K, Shakoori A, Kyo S, Miyashita K, Yokoi K, Jin M, Shimasaki T, Motoo Y, Minamoto T. Deregulated GSK3b sustains gastrointestinal cancer cells survival by modulating human telomerase reverse transcriptase and telomerase. Clin Cancer Res. 2009;15:6810–6819. doi: 10.1158/1078-0432.CCR-09-0973. [DOI] [PubMed] [Google Scholar]
- Mamaghani S, Patel S, Hedley DW. Glycogen synthase kinase-3 inhibition disrupts nuclear factor-kappaB activity in pancreatic cancer, but fails to sensitize to gemcitabine chemotherapy. BMC Cancer. 2009;9:132. doi: 10.1186/1471-2407-9-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ Jr, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Gualdi R, Casadevall C, Manzella L, Calabretta B. Positive autoregulation of c-myb expression via Myb binding sites in the 5′ flanking region of the human c-myb gene. Mol Cell Biol. 1991;11:6166–6176. doi: 10.1128/mcb.11.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- Ougolkov AV, Fernandez-Zapico ME, Savoy DN, Urrutia RA, Billadeau DD. Glycogen synthase kinase-3beta participates in nuclear factor kappaB-mediated gene transcription and cell survival in pancreatic cancer cells. Cancer Res. 2005;65:2076–2081. doi: 10.1158/0008-5472.CAN-04-3642. [DOI] [PubMed] [Google Scholar]
- Petropoulos K, et al. A novel role for Lef-1, a central transcription mediator of Wnt signaling, in leukemogenesis. J Exp Med. 2008;205:515–522. doi: 10.1084/jem.20071875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radke K, Beug H, Kornfeld S, Graf T. Transformation of both erythroid and myeloid cells by E26, an avian leukemia virus that contains the myb gene. Cell. 1982;31:643–653. doi: 10.1016/0092-8674(82)90320-8. [DOI] [PubMed] [Google Scholar]
- Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nature Rev Cancer. 2008;8:523–534. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- Saether T, Berge T, Ledsaak M, Matre V, Alm-Kristiansen AH, Dahle O, Aubry F, Gabrielsen OS. The chromatin remodeling factor Mi-2alpha acts as a novel co-activator for human c-Myb. J Biol Chem. 2007;282:13994–14005. doi: 10.1074/jbc.M700755200. [DOI] [PubMed] [Google Scholar]
- Shivdasani RA, Orkin SH. The transcriptional control of hematopoiesis. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- Takashima A, et al. Presenilin 1 associates with glycogen synthase kinase-3beta and its substrate tau. Proc Natl Acad Sci USA. 1998;95:9637–9641. doi: 10.1073/pnas.95.16.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MA, Ramsay RG. Myb: an old oncoprotein with new roles. Bioessays. 1995;17:341–350. doi: 10.1002/bies.950170410. [DOI] [PubMed] [Google Scholar]
- Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- Vene R, Larghero P, Arena G, Sporn MB, Albini A, Tosetti F. Glycogen synthase kinase 3beta regulates cell death induced by synthetic triterpenoids. Cancer Res. 2008;68:6987–6996. doi: 10.1158/0008-5472.CAN-07-6362. [DOI] [PubMed] [Google Scholar]
- Verbeek W, Gombart AF, Chumakov AM, Muller C, Friedman AD, Koeffler HP. C/EBPepsilon directly interacts with the DNA binding domain of c-myb and cooperatively activates transcription of myeloid promoters. Blood. 1999;93:3327–3337. [PubMed] [Google Scholar]
- Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin EH, Gallo RC, Arya SK, Eva A, Souza LM, Baluda MA, Aaronson SA, Wong-Staal F. Differential expression of the amv gene in human hematopoietic cells. Proc Natl Acad Sci USA. 1982;79:2194–2198. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gao X, Wen J, Ning Y, Chen YG. Dapper 1 antagonizes Wnt signaling by promoting dishevelled degradation. J Biol Chem. 2006;281:8607–8612. doi: 10.1074/jbc.M600274200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhou H, Su Y, Sun Z, Zhang H, Zhang Y, Ning Y, Chen YG, Meng A. Zebrafish Dpr2 inhibits mesoderm induction by promoting degradation of nodal receptors. Science. 2004;306:114–117. doi: 10.1126/science.1100569. [DOI] [PubMed] [Google Scholar]
- Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb proto-oncogene and microRNA-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2009;113:505–516. doi: 10.1182/blood-2008-01-136218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Zhang L, Wang A, Song B, Gong K, Zhang L, Hu M, Zhang X, Zhao N, Gong Y. The association of GSK3 beta with E2F1 facilitates nerve growth factor-induced neural cell differentiation. J Biol Chem. 2008;283:14506–14515. doi: 10.1074/jbc.M706136200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.