Abstract
Our previous studies revealed that in fibrinogen the αC-domains are not reactive with their ligands, suggesting that their binding sites are cryptic and become exposed upon its conversion into fibrin, in which these domains form αC polymers. Based on this finding, we hypothesized that polymerization of the αC-domains in fibrin results in the exposure of their binding sites and that these domains adopt the physiologically active conformation only in αC-domain polymers. To test this hypothesis, we prepared a recombinant αC region (residues Aα221-610) including the αC-domain (Aα392-610), demonstrated that it forms soluble oligomers in a concentration-dependent and reversible manner, and stabilized such oligomers by covalent cross-linking with factor XIIIa. Cross-linked Aα221-610 oligomers were stable in solution and appeared as ordered linear, branching filaments when analyzed by electron microscopy. Spectral studies revealed that the αC-domains in such oligomers were folded into compact structures of high thermal stability with a significant amount of β-sheets. These findings indicate that cross-linked Aα221-610 oligomers are highly ordered and mimic the structure of fibrin αC polymers. The oligomers also exhibited functional properties of polymeric fibrin since, in contrast to the monomeric αC-domain, they bound tPA and plasminogen and stimulated activation of the latter by the former. Altogether, the results obtained with cross-linked Aα221-610 oligomers clarify the structure of the αC-domains in fibrin αC polymers and confirm our hypothesis that their binding sites are exposed upon polymerization. Such oligomers represent a stable, soluble model of fibrin αC polymers that can be used for further structure/function studies of fibrin αC-domains.
Conversion of fibrinogen into fibrin results in its spontaneous polymerization and formation of a fibrin clot that prevents the loss of blood by sealing the damaged vasculature and subsequently contributes to wound healing by promoting inflammation and angiogenesis. Polymerized fibrin also significantly enhances activation of plasminogen into the active fibrinolytic enzyme plasmin, thereby triggering fibrinolysis and subsequently contributing to propagation of fibrinolysis by keeping plasmin on the fibrin surface. This multitude of fibrinogen functions is connected with the presence in the fibrinogen molecule of multiple regions/domains that allow its specific interactions with various proteins and cell types and thereby its involvement in the above mentioned processes. Among these domains, a pair of the αC-domains, by interacting with each other and with a number of physiologically important proteins and cellular receptors, promotes fibrin polymerization, fibrinolysis, atherogenesis, and angiogenesis (1–5).
The fibrinogen molecule consists of five major structural regions, central region E, two terminal D regions, and two αC regions, each of which is folded into a number of compact domains (6). Crystallographic studies of fibrinogen and its fragments established the three-dimensional structure of more than two-thirds of the molecule including the D and E regions (7–11); however, they failed to resolve the structure of the αC regions. Each of these regions includes the Aα chain amino acid residues 221-610 that form the compact αC-domain (residues 392-610) and the flexible αC-connector (residues 221-391), which tethers the former to the bulk of the molecule (2). Recent studies with the recombinant αC-domain fragments established that the αC-domain consists of two sub-domains, N-terminal and C-terminal, and the former is formed by a β-sheet consisting of two β-hairpins (12–14).
Numerous studies indicate that in fibrinogen two αC-domains form a dimer through the interaction with each other and with the central region of the molecule (1, 15–19). The arrangement of the αC-domains in polymeric fibrin seems to be different. Analysis of cross-linked fibrin revealed that factor XIIIa covalently cross-links fibrin’s α chains to produce α chain polymers (20, 21). Such cross-linking occurs between the αC-domains and αC-connectors of different fibrin molecules (22), suggesting that the αC-domains are closely spaced in fibrin and may interact with each other. In agreement, we have recently found that the isolated αC-domain self-associates at increasing concentrations to form soluble oligomers that may mimic the structure and interactions of the αC-domains in fibrin (14). Thus, although the structure of the isolated αC-domain has been established (13, 14), that of the αC-domains in fibrin remains to be clarified.
Our previous studies revealed that the isolated αC-domain interacts with various plasma proteins including fibronectin (23), apolipoprotein(a) (4), plasminogen, tissue type plasminogen activator (tPA)1, and plasmin inhibitor α2-antiplasmin (2, 24). At the same time, in our experiments, none of these proteins interacted with either fibrinogen or the αC-domain kept in solution; their interactions were observed only when the αC-domain or fibrinogen were adsorbed/immobilized onto a surface or when fibrinogen was converted into fibrin (2, 4, 23, 24). To account for these observations, we hypothesized that the binding sites of the αC-domains are cryptic in fibrinogen and exposed in fibrin. This implies that the αC-domains undergo conformational changes upon conversion of fibrinogen into fibrin and only in fibrin, in which the αC-domains form polymers, do they adopt the physiologically active conformation. The major goals of the present study were to clarify the structure of the αC-domains in fibrin αC-polymers and to prove this hypothesis by testing their interaction with some ligands.
EXPERIMENTAL PROCEDURES
Proteins, Enzymes, Antibodies, Recombinant Fragments
Plasminogen-depleted human fibrinogen, human α-thrombin, human Glu-plasminogen and human FXIII were from Enzyme Research Laboratories (South Bend, IN). Bovine serum albumin was purchased from Pierce. Recombinant single-chain tPA was a Genentech product. The monoclonal antibody TF 359/1-1 directed against αC-region was a gift from B. Kudryk (New York Blood Center, New York). The alkaline phosphatase conjugated ExtrAvidin, aprotinin, and carboxypeptidase B were from Sigma. The recombinant Aα221-610 and Aα392-610 fragments corresponding to the human fibrinogen αC region and αC-domain, respectively, were expressed in Escherichia coli and subsequently purified and refolded by the procedures described previously (25).
Size-Exclusion Chromatography
Analytical size-exclusion chromatography was used to analyze the aggregation state of the prepared recombinant Aα221-610 fragment. The experiments were performed with a fast protein liquid chromatography system (FPLC, Pharmacia) on a Superdex 200 column at a flow rate of 0.5 mL/min and 4 °C. Typically, 100 μl of protein was loaded onto the column equilibrated with 20 mM Tris buffer, pH 7.4, containing 0.15 M NaCl (TBS) or 20 mM Tris buffer, pH 7.4, containing 2 M NaCl followed by elution with the same buffer. Protein elution was monitored by measuring absorbance at 280 nm.
Transmission Electron Microscopy Study
Two preparations of the first and second fractions of cross-linked Aα221-610 oligomers and two preparations of Aα221-610 monomers were visualized by transmission electron microscopy. Samples at 0.1 mg/mL in 20 mM ammonium formate buffer, pH 7.4, were diluted to ~20 μg/mL in a buffer containing 50 mM ammonium formate at pH 7.4, and 30% (v/v) glycerol. Samples were then sprayed onto freshly cleaved mica and rotary shadowed with tungsten in a vacuum evaporator (Denton Vacuum Co., Cherry Hill, NJ) as previously described (18). These samples were examined in a FEI/Philips 400 electron microscope (FEI Co., Hillsboro, OR) operating at 60 kV and a magnification of 60,000×. The grids were thoroughly examined to ensure that the structures observed were representative.
Circular Dichroism Study
Circular dichroism (CD) measurements were taken with a Jasco-810 spectropolarimeter. CD spectra of the Aα221-610 monomer and cross-linked Aα221-610 oligomers, both at 1 mg/mL, or Aα392-610 oligomers at 0.5 mg/mL were recorded using a 0.01 cm path length quartz cuvette. The experiments were performed in TBS at 4 °C. Analysis of the CD spectra was performed using the secondary structure prediction program supplied with the spectropolarimeter, which is based on the previously published method (26). All CD data were expressed as the mean residue ellipticity, [θ], in units of degrees square centimeter per decimole.
Fluorescence Study
Fluorescence measurements of thermally-induced unfolding of the Aα221-610 oligomers and cross-linked Aα221-610 oligomers in TBS or in 20 mM Tris buffer, pH 7.4, containing 2 M NaCl, were performed in an SLM 8000-C fluorometer by monitoring the ratio of the intensity at 370 nm to that at 330 nm with excitation at 280 nm. The temperature was controlled with a circulating water bath programmed to raise the temperature at ~1 °C/min. Fragment concentrations determined spectrophotometrically were ~0.05 mg/mL.
Solid Phase Binding Assay
Solid phase binding was performed in plastic microtiter plates using an enzyme-linked immunosorbent assay (ELISA) as described in (4) with some modifications. Microtiter Immulon 2HB plate wells (Thermo) were coated overnight with 100 μL/well of plasminogen or tPA, both at 10 μg/mL in TBS containing 1 mM Ca2+ (TBS-Ca), followed by washing with the same buffer. The wells were then blocked with 2% bovine serum albumin in TBS-Ca containing 0.01% Tween-20. Followed by washing with TBS-Ca and 0.01% Tween-20, the Aα221-610 monomer, Aα221-610 cross-linked oligomers, or fibrinogen, all at 1 μM, was added to the wells and incubated for 1 h at 37 °C. The monoclonal antibody TF 359/1-1 against αC-region was labeled with biotin using EZ-Link Sulfo-NHS-Biotinylation Kit (Pierce) and bound protein was detected by reaction with the alkaline phosphatase-conjugated avidin. A PPNP Microwell alkaline phosphatase Substrate (Kirkegaard & Perry Laboratories Inc.) was added to the wells and the amount of bound ligand was measured spectrophotometrically at 405 nm.
Surface Plasmon Resonance
The interactions of the Aα221-610 monomer, cross-linked Aα221-610 oligomers, and fibrinogen with plasminogen and tPA were studied by surface plasmon resonance (SPR) using the BIAcore 3000 biosensor (Biacore AB, Uppsala, Sweden), which measures association and dissociation of proteins in real time. Plasminogen or tPA at 5 μg/mL was immobilized to the CM5 sensor chip using the amine coupling kit (BIAcore AB, Uppsala, Sweden) according to the manufacturer’s instructions. In another experiment, the Aα221-610 monomer at 5 μg/mL or cross-linked oligomers at 5 μg/mL was immobilized to the CM5 sensor chip using the amine coupling kit (BIAcore AB, Uppsala, Sweden) according to the manufacturer instructions. Binding experiments were performed in 20 mM HEPES buffer, pH 7.4, containing 150 mM NaCl, 1 mM Ca2+, and 0.01% Tween-20 (HBS-Ca) at 20 μL/min flow rate. The association between the immobilized proteins and added ligands was monitored as the change in the SPR response; the dissociation was measured upon replacement of the ligand solution for HBS-Ca (binding buffer). To regenerate the surface, complete dissociation of the complex was achieved by adding 0.1 M ε-aminocapronic acid in the binding buffer for 30 sec followed by re-equilibration with the same buffer. Experimental data were analyzed using BIAevaluation 4.1 software supplied with the instrument. The dissociation equilibrium constant, Kd, was calculated as Kd = kdiss/kass, where kass and kdiss represent kinetic constants that were estimated by global analysis of the association/dissociation data using the 1:1 Langmuirian interaction model (kinetic analysis). To confirm the kinetic analysis, Kd was also estimated by analysis of the association data using the steady-state affinity model provided by the same software (equilibrium analysis).
Chromogenic Substrate Assay
The stimulating effect of cross-linked Aα221-610 oligomers and other Aα221-610 containing species on the tPA-catalysed conversion of plasminogen into plasmin was evaluated by determination of the amidolytic activity of the newly formed plasmin with chromogenic substrate S-2251 (H-D-valyl-L-leucyl-L-lysine-p-nitroanilide) (Chromogenix) as described in (27). The assay system contained 0.2 μM Glu-plasminogen, 0.14 nM tPA, 0.3 mM S-2251, and 0.5 μM Aα221-610 containing species or 1 μM fibrinogen in TBS with 0.05% Tween 80. The assay was performed in the wells of a microtiter plate at 37 °C. The amidolytic activity was determined by measuring the absorbance at 405 nm using VERSAmax 96-well plate reader (Molecular Devices).
RESULTS
Preparation of a Soluble Model of Fibrin αC-Domain Polymers
Our recent study revealed that the recombinant human and bovine αC-domain fragments self-associate in solution and at increasing concentrations form soluble oligomers that may mimic the structure and properties of fibrin αC polymers (14). Such αC-domain oligomers can be separated from the monomeric fraction by size-exclusion chromatography; however, they are unstable due to dissociation upon lowering their concentration. It is known that in fibrin αC polymers are stabilized by factor XIIIa, which covalently cross-links αC-domains to αC-connectors through their reactive Lys and Gln residues (22). Thus, we hypothesized that if the recombinant αC region (Aα221-610) including the αC-domain (Aα392-610) and αC-connector (Aα221-391) would also form reversible oligomers, such oligomers could be stabilized by factor XIIIa.
To test this hypothesis, we first prepared the recombinant Aα221-610 fragment corresponding to the human αC region and studied its aggregation state in different conditions. Namely, when Aα221-610 was incubated in TBS for two days at two concentrations, 1 mg/mL and 3 mg/mL, and then analyzed by size-exclusion chromatography on Superdex 200, the analysis revealed that this fragment was preferentially monomeric at 1 mg/mL (only ~5% oligomers were detected), while at 3 mg/mL about 22% Aα221-610 formed oligomers (not shown). This confirmed that that Aα221-610 forms oligomers in a concentration-dependent manner. To test if the observed oligomerization is reversible, the sample containing 22% oligomers was diluted with TBS to 1 mg/mL and immediately analyzed by size-exclusion chromatography. The experiment revealed that the amount of oligomers in the diluted sample was reduced to 18% (not shown). Subsequent overnight incubation of this sample resulted in further reduction of oligomeric fraction to 11%. In another experiment performed in the presence of 2 M NaCl, which was previously shown to enhance oligomerization of the recombinant αC-domain and its truncated fragments (14), the amount of oligomers in Aα221-610 incubated at 3 mg/mL overnight was found to be about 60%. When this sample was diluted to 1 mg/mL, dialyzed versus TBS overnight, and then analyzed by size-exclusion chromatography, the amount of oligomers decreased to 15%. Altogether, these experiments confirmed that Aα221-610 form oligomers in a concentration-dependent and reversible manner. They also revealed that dissociation of such oligomers is a relatively slow process.
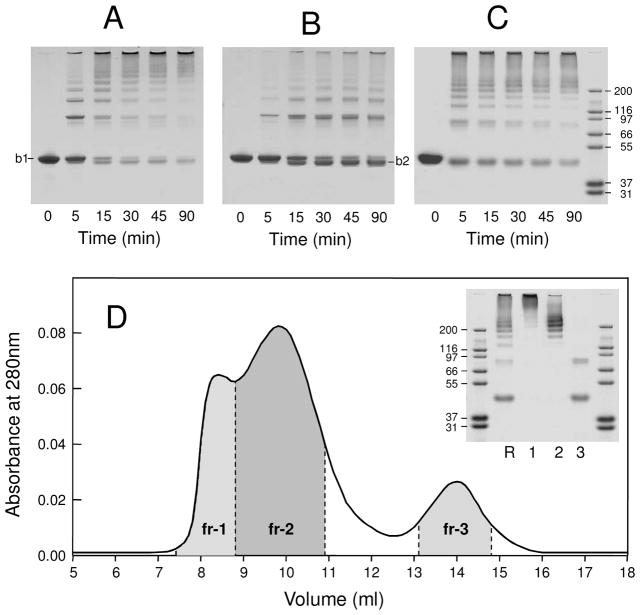
Next, the oligomeric and monomeric fractions of Aα221-610, which was incubated at 3 mg/mL in TBS, were separated on a Superdex 200 column, concentrated to 0.9 mg/mL, incubated with factor XIIIa for 90 min, and time-course of their cross-linking was analyzed by SDS-PAGE (Figure 1, panels A and B). The analysis revealed that most of Aα221-610 oligomers (band 1 denoted “b1” in panel A) were rapidly cross-linked intermolecularly resulting in dimers, trimers, tetramers, and larger multimers. These species were also observed upon cross-linking of the Aα221-610 monomer (panel B); however, its cross-linking was slower. In addition, a large portion of the monomer was converted into a species with higher mobility (band 2 denoted “b2” in panel B), which was previously described as the intramolecularly cross-linked monomeric Aα221-610 fragment (22), and the amount of the multimers was much lower.
Figure 1.
Preparation of cross-linked Aα221-610 oligomers. Panel A and B represent time-course of cross-linking of Aα221-610 oligomers and Aα221-610 monomer, respectively, by activated factor XIII in 20 mM Tris, pH 7.4, with 0.15 M NaCl (TBS) and 5 mM CaCl2 at room temperature; b1 and b2 denote respectively non-cross-linked and internally cross-linked Aα221-610 (see text). Panel C represents time-course of cross-linking of Aα221-610 oligomers by activated factor XIII in 20 mM Tris, pH 7.4, with 2 M NaCl and 5 mM CaCl2, at room temperature; the right outer lane contains protein markers of the indicated molecular masses. All lanes on each gel in panels A, B and C had the same amount of total protein. Panel D shows fractionation of cross-linked Aα221-610 oligomers and SDS-PAGE analysis of individual fractions. The cross-linking was performed in the same conditions as in panel C, the cross-linking reaction was stopped after 30 min by addition of EDTA to final concentration of 10 mM, and the reaction mixture was immediately applied on Superdex 200 column equilibrated with 20 mM Tris, pH 7.4, with 2 M NaCl. The inset shows SDS-PAGE analysis of the reaction mixture applied to the column (lane R), and its individual fractions, fr-1, fr-2 and fr-3 (lanes 1, 2 and 3, respectively); the outer lanes contain protein markers of the indicated molecular masses.
Although SDS-PAGE analysis confirmed that Aα221-610 oligomers were cross-linked more efficiently than the monomer, the final yield of the cross-linked multimers was quite low, since the starting material incubated in TBS contained only 22% oligomers. To increase the yield, we increased the concentration of NaCl in TBS from 0.15 to 2 M. At this NaCl concentration, the amount of oligomers formed by Aα221-610 at 3 mg/mL was about 60%, as mentioned above. These oligomers were separated from Aα221-610 monomer by size-exclusion chromatography on Superdex 200, concentrated to 0.5 mg/mL, and cross-linked with factor XIIIa. The separation and cross-linking were performed in the presence of 2 M NaCl to maximally reduce dissociation of the oligomers. SDS-PAGE analysis of the cross-linking process in these conditions (Figure 1C) revealed a cross-linking pattern similar to that observed in TBS (Figure 1A). These conditions were selected for large-scale preparation of cross-linked Aα221-610 oligomers. The cross-linking reaction was stopped after 30 min by the addition of 10 mM EDTA and the reaction mixture was immediately applied to a Superdex 200 column equilibrated with TBS and eluted with the same buffer. The elution profile exhibited two poorly resolved peaks of different intensity and a well resolved third peak (Figure 1D). Fractions corresponding to these peaks were collected and analyzed by SDS-PAGE. The analysis revealed mostly high molecular mass multimers in the first fraction, a mixture of such multimers and intermediate multimers in the second fraction, and a mixture of the dimer and monomer in the third fraction (Figure 1D, inset). The first and second fractions were used for further structure/function analysis.
Electron Microscopy of the Cross-Linked Aα221-610 Oligomer Fractions
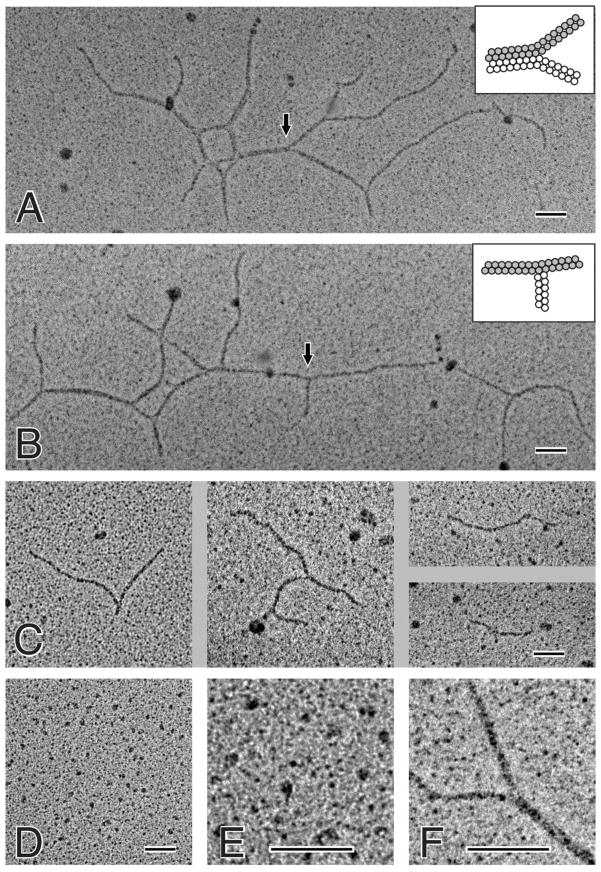
Preparations of the first and second fractions of cross-linked Aα221-610 oligomers, as well as the monomeric fraction of non-crosslinked Aα221-610, were visualized by transmission electron microscopy after preparation by rotary shadowing with tungsten in a vacuum evaporator (Figure 2). The monomer preparations used as controls contained small globular structures with a diameter of 4.5 ± 0.7 nm (n = 1000) (panels D and E) that were similar to those observed earlier with the proteolytically prepared and recombinant αC-fragments (18, 28). In addition, there were larger structures about twice that size. Occasionally, a thin appendage could be seen extending from the globular structures (panel E). In rotary shadowed preparations of the first fraction of the cross-linked oligomers, long thin polymers were observed, as well as some large, complex structures with multiple branch points (panels A and B). It should be noted that no such structures were ever seen in the monomer preparations, in spite of extensive searching. The basic building block of these polymers was a thin filament that was 7.8 ± 0.9 nm (n = 96) in width, i.e., slightly less than twice the diameter of individual monomers. Often individual monomers can be distinguished in these filaments. The globular regions of the monomers sometimes appear to be arranged side-by-side but slightly offset from each other longitudinally. In rotary shadowed preparations of the second fraction of the cross-linked oligomers, similar long thin polymers were observed; however, they were shorter and not as complex in structure as with the first fraction (panel C). In addition, even shorter polymers with no branching were observed in preparations of both fractions (not shown); these polymers most probably represent the aforementioned intermediate multimers identified by SDS-PAGE. To quantify the frequency of different sizes of polymers, they were grouped into structures <350 nm, 350–600 nm, and > 600 nm: first fraction: 21.7% <350 nm, 17.4% 350–600 nm, and 60.9% > 600 nm; second fraction: 83.7% <350 nm, 9.2% 350–600 nm, and 7.1% > 600 nm.
Figure 2.
Electron microscopy of rotary shadowed samples of cross-linked Aα221-610 oligomers and Aα221-610 monomers. Panels A and B show polymeric structures observed for the fraction 1 preparation of cross-linked Aα221-610 oligomers. Two types of branch points were observed, one with all three strands having the same width and the other with one strand being about twice as thick as the other two. The inset in panel A shows a schematic diagram of the second type of branch point (indicated by the arrow), where the strand on the left has twice the width of the two strands on the right. Higher magnification view of the second type of branch point, showing details of the lateral aggregation of two filaments is presented in panel F; the strand at the lower right side of the image is about twice as thick as the other two strands. The inset in panel B shows a schematic diagram of the first type of branch point (indicated by the arrow), where all three strands have the same width. Panel C shows four pictures of polymeric structures observed for the fraction 2 preparation of cross-linked Aα221-610 fragment. These polymers were smaller and less complex than those formed from fraction 1. Panel D shows nonomeric and dimeric structures observed for Aα221-610 monomer; higher magnification of a small portion of panel D, showing more details of the monomeric and dimeric structures, is presented in panel E. All magnification bars indicate 100 nm.
Thorough examination of the branched polymers revealed two types of branch points. The first was made up of three filaments all with the same width (shown by arrow in panel B). The second type of branch points was formed by pair-wise lateral aggregation of two of the thin filaments (shown by arrow in panel A). In other words, each branch point was made up of two 7.8 nm filaments and one filament that was 15.5 ± 1.0 nm (n = 36), or approximately twice the diameter of each individual filament (enlarged image of the second type of branch points is shown in panel F). A histogram of filament diameters showed two clearly separated peaks at 7.8 nm and 15.5 nm, with no peaks at the size of an Aα221-610 monomer or trimer (data not shown). Thus, individual filaments can aggregate laterally in this manner and diverge more than once, producing very complex networks from such basic interactions (panels A and B).
Spectral Study of the Structure and Stability of Cross-Linked Aα221-610 Oligomers
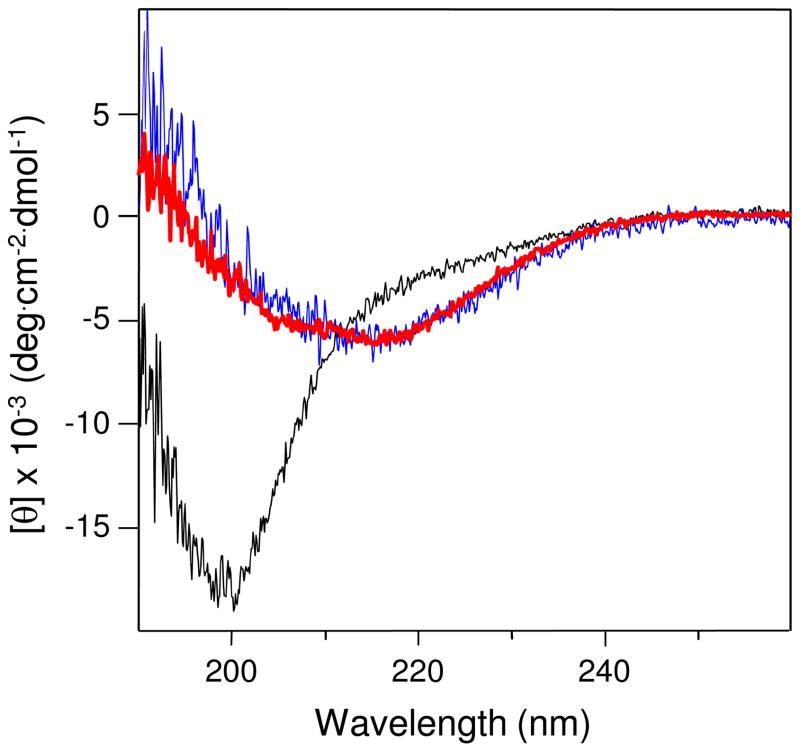
To characterize the structure of cross-linked Aα221-610 oligomers, we used circular dichroism (CD). Visual examination of the CD spectrum of such oligomers revealed a well pronounced negative band at about 217 nm (Figure 3, red curve), suggesting the presence of significant amount of β-structure. Indeed, analysis of this spectrum using the secondary structure prediction program revealed 43% β-sheets, 12% β-turns, 10% α-helices and only 35% random conformation. In contrast, the CD spectrum of the non-cross-linked Aα221-610 monomer, which was purified from the mixture of Aα221-610 monomer/oligomers using a Superdex 200 column, exhibited a weaker band at 217 nm and a dominant negative band at about 200 nm (Figure 3, black curve), suggesting the presence of a substantial amount of random structures. Analysis of this spectrum using the same program revealed 53% random conformation. Thus, as in the case with the isolated αC-domain (14), oligomerization of Aα221-610 resulted in formation of additional regular structures. It should be noted that the CD spectrum of cross-linked Aα221-610 oligomers is comparable with that of the preferentially (about 90%) oligomeric Aα392-610 fragment (Figure 3, blue curve), which was purified from the mixture of Aα392-610 monomers/oligomers using the Superdex 75 column. This is in spite of the fact that the Aα221-610 fragment, in addition to the αC-domains (Aα392-610), contains the αC-connector (Aα221-391), which is considered to be unordered (1, 25).
Figure 3.
CD spectra of cross-linked Aα221-610 oligomers (red), monomeric Aα221-610 fragment (black), and Aα392-610 oligomers (blue), all in 20 mM Tris, pH 7.4, containing 0.15 M NaCl; fragment concentrations were 0.5–1.0 mg/mL. All spectra were obtained in 0.01 cm cuvette at 4 °C and are representative of at least two experiments.
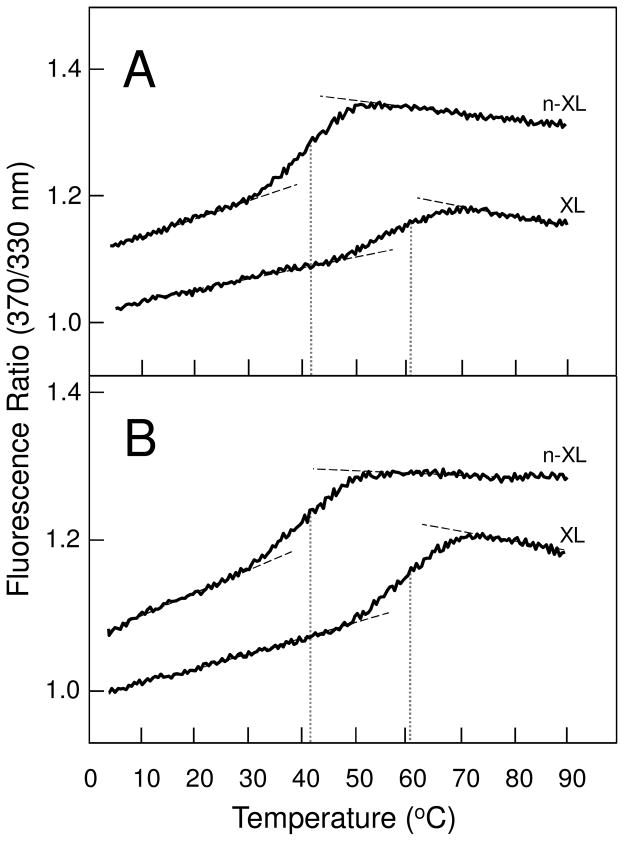
To test the folding status and stability of cross-linked Aα221-610 oligomers we used fluorescence spectroscopy. When heated in the fluorometer while the ratio of fluorescence intensity at 370 nm to that at 330 nm was monitored as a measure of the spectral shift that accompanies unfolding, the cross-linked oligomers in TBS exhibited a well pronounced unfolding transition with a midpoint (Tm) at 60.7 ± 0.2 °C (n = 2) (Figure 4A). In contrast, unfolding of the non-cross-linked oligomeric fraction of Aα221-610, which was prepared by size-exclusion chromatography of this fragment incubated overnight at 3 mg/mL in TBS, was observed at much lower temperature, Tm = 42.5 ± 0.4 °C (n = 2). These results indicate that both non-cross-linked and cross-linked Aα221-610 oligomers contained stable, compact, cooperative structures; however, the thermal stability of non-cross-linked Aα221-610 oligomers was much lower than that of their cross-linked counterparts.
Figure 4.
Fluorescence-detected thermal unfolding of cross-linked (XL) and non-cross-linked (n-XL) Aα221-610 oligomers in 20 mM Tris, pH 7.4, containing 0.15 M NaCl (panel A) and in 20 mM Tris, pH 7.4, containing 2 M NaCl (panel B). The unfolding curves have been arbitrary shifted along the vertical axis to improve visibility; the dashed straight lines represent linear extrapolations of the values of the fluorescence ratio at 370/330nm before and after transitions to highlight their sigmoidal character; the dotted vertical lines show midpoint temperature (Tm) of the unfolding transitions. The curves are representative of at least two independent experiments.
Since oligomerization of Aα221-610 is a reversible process and dilution of the oligomeric fraction to a concentration required for fluorescence study (~0.05 mg/mL) could result in oligomer dissociation, we tested the distribution of oligomers and monomers in the diluted fraction by size-exclusion chromatography. The experiment, in which oligomeric fraction of Aα221-610 was collected from Superdex 200 column, diluted to 0.05 mg/mL, incubated for 60 min at room temperature, and re-applied to the same column, revealed substantial amount of the monomer (about 60%). Thus, one cannot exclude that, like in case with Aα392-610 oligomers whose thermal stability was reduced upon dissociation (14), the observed lower thermal stability of non-crosslinked Aα221-610 oligomers could be connected with their dissociation upon dilution. To test this possibility, we compared thermal stability of cross-linked and non-crosslinked Aα221-610 oligomers in 2 M NaCl, in which we expected more non-cross-linked oligomers to be preserved from dissociation. Indeed, size-exclusion chromatography of such oligomers diluted to 0.05 mg/mL and incubated in 2 M NaCl for 60 min revealed only about 10% monomers in the mixture. In these conditions, the non-cross-linked and cross-linked oligomers unfolded practically in the same temperature ranges as in TBS with very similar Tms, 42.2 ± 0.6 °C (n = 2) and 60.7 ± 0.7 °C (n = 3) °C, respectively. These experiments suggest that a connection between the lower thermal stability of non-cross-linked Aα221-610 oligomers and their dissociation is highly unlikely.
Altogether, the spectral studies described above indicate that cross-linked Aα221-610 oligomers contain compact, cooperative structures with high content of β-sheets whose thermal stability is significantly higher than that in the non-cross-linked oligomers. They also suggest that the increased thermal stability of cross-linked Aα221-610 oligomers is connected with their covalent cross-linking by factor XIIIa.
Interaction of the αC monomer and Cross-linked αC-oligomers with Plasminogen and tPA
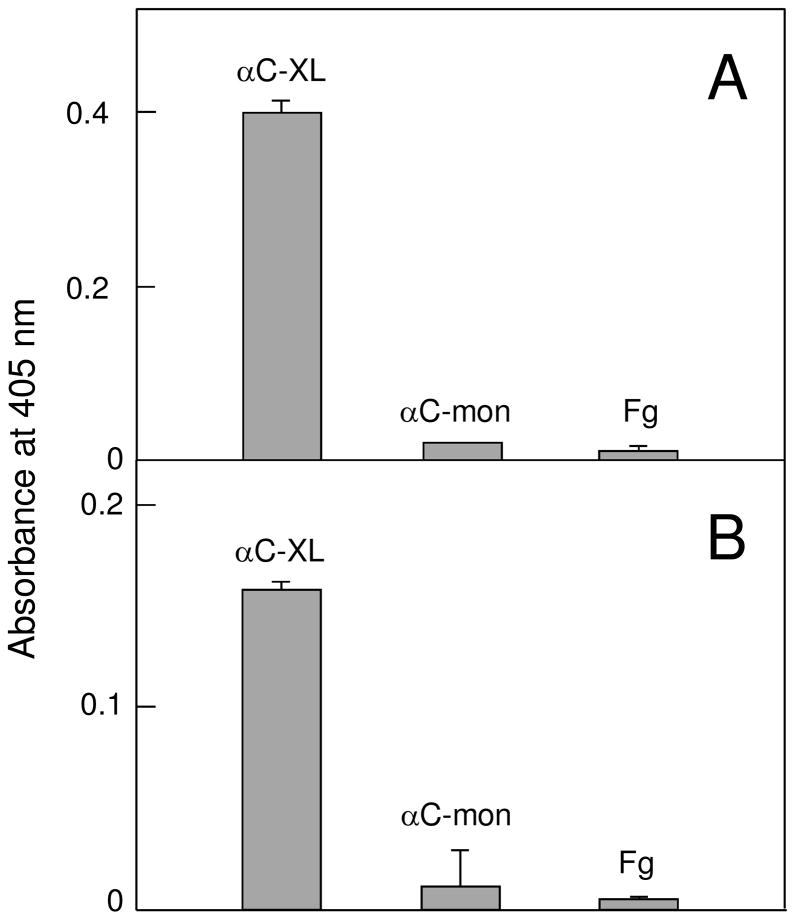
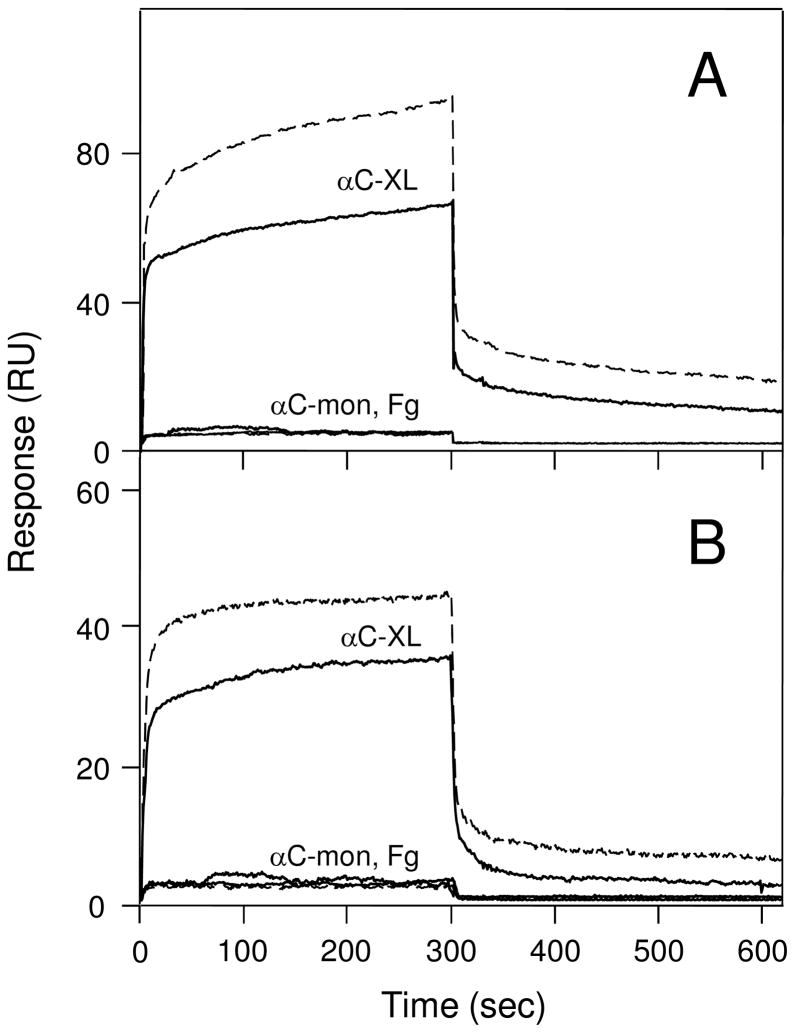
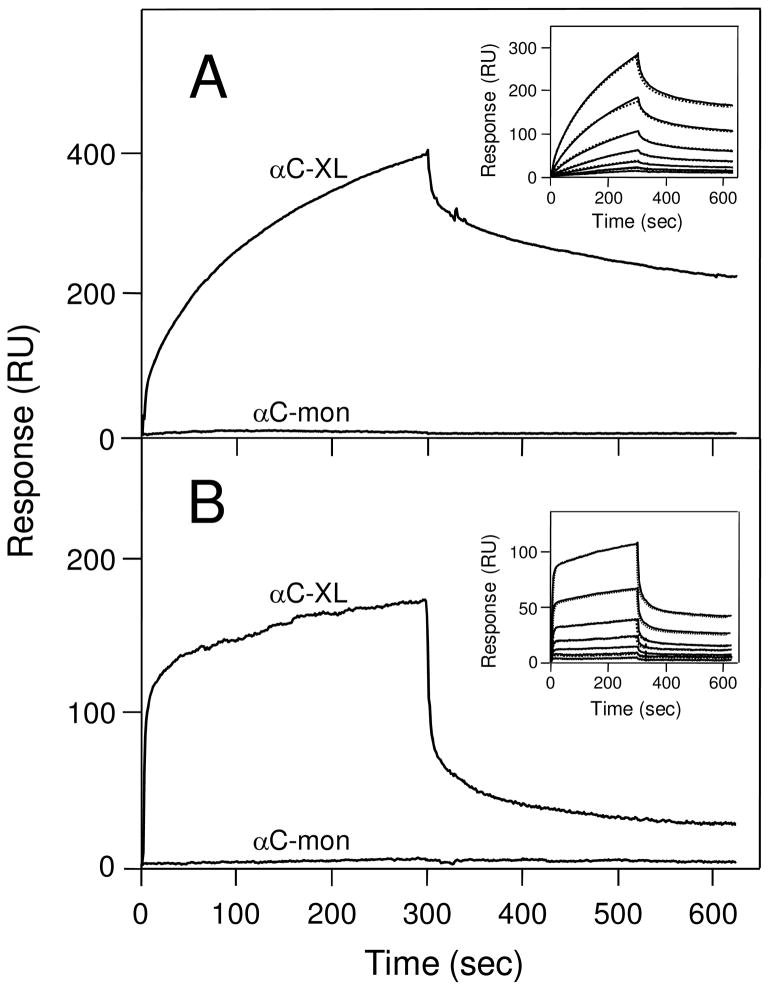
To test our hypothesis that αC-domain cryptic binding sites are exposed in cross-linked Aα221-610 oligomers, we studied the interaction of these oligomers with two αC-domain ligands, plasminogen and tPA, by ELISA and surface plasmon resonance (SPR). Since it is known that adsorption of fibrinogen or its fragments to a surface alters their conformation and binding properties, the cross-linked oligomers in ELISA experiments were kept in solution to prevent such changes. When microtiter plate wells were coated with plasminogen or tPA and cross-linked Aα221-610 oligomers at 2 μM (calculated per monomer) were added, they exhibited a prominent binding to both ligands while no binding was observed with Aα221-610 monomer added at the same concentration (Figure 5). Fibrinogen at 2 μM used as a control also failed to bind, as expected. These results were confirmed by SPR experiments in which plasminogen or tPA were immobilized to the surface of a sensor chip, Aα221-610 monomer, cross-linked Aα221-610 oligomers, or fibrinogen, all at 1 μM, were injected, and their association/dissociation was measured in real time. Among these species, only the oligomers exhibited prominent binding while Aα221-610 monomer and fibrinogen failed to bind (Figure 6). The binding was dose-dependent since the SPR signal increased when the oligomers were injected at 2 μM (Fig. 6, broken curves). We also used SPR to test the interaction of tPA and plasminogen with immobilized cross-linked Aα221-610 oligomers and Aα221-610 monomer. It should be noted that in these experiments immobilization was performed by chemical cross-linking of Aα221-610 species to a dextran-coated surface of a sensor chip. Since such immobilization, in contrast to that by adsorption to a surface, does not usually perturb protein conformation, we expected the immobilized cross-linked oligomers or Aα221-610 monomer to have the same conformation and binding properties as those in solution. Indeed, in agreement with the results described above, when tPA or plasminogen, both at 2 μM, were added to the immobilized species, they exhibited a prominent binding only to the cross-linked oligomers while the binding to the monomer was negligible (Figure 7). To determine the equilibrium dissociation constant (Kd) for these bindings, tPA or plasminogen were injected at increasing concentrations (Figure 7, insets) and the binding data were analyzed as described in Experimental Procedures. The analysis revealed Kd values of 440 ± 15 and 458 ± 9 nM for the binding of tPA and plasminogen, respectively. Altogether, these experiments clearly indicate that the tPA- and plasminogen-binding sites of the αC-domain are cryptic in fibrinogen and Aα221-610 monomer and exposed in cross-linked Aα221-610 oligomers.
Figure 5.
ELISA-detected binding of cross-linked Aα221-610 oligomers, Aα221-610 monomers and fibrinogen to immobilized plasminogen or tPA. 2 μM of cross-linked Aα221-610 oligomers (αC-XL), Aα221-610 monomer (αC-mon), or fibrinogen (Fg) was added to surface-adsorbed plasminogen (panel A) or tPA (panel B). Bound fragments were detected spectrophotometrically at 405 nm using biotinylated avidin conjugated to alkaline phosphatase, as described in Experimental Procedures. All results are means ± the standard deviation of two independent experiments, each performed in duplicate.
Figure 6.
Surface plasmon resonance-detected binding of cross-linked Aα221-610 oligomers and Aα221-610 monomer to immobilized plasminogen or tPA. Cross-linked Aα221-610 oligomers (αC-XL), Aα221-610 monomer (αC-mon), or fibrinogen (Fg), all at 1 μM, were added to immobilized plasminogen (panel A) or tPA (panel B) and their association/dissociation was monitored in real time. Broken curves in both panels show binding of cross-linked Aα221-610 oligomers (αC-XL) or Aα221-610 monomer (αC-mon) both added at 2 μM. Note that the curves for Aα221-610 monomer at both concentrations and for fibrinogen essentially coincide.
Figure 7.
Analysis of the interaction of plasminogen and tPA with immobilized Aα221-610 monomer and cross-linked Aα221-610 oligomers by surface plasmon resonance. Plasminogen (panel A) or tPA (panel B), each at 2 μM, was added to immobilized cross-linked Aα221-610 oligomers (αC-XL) or Aα221-610 monomer (αC-mon) and their association/dissociation was monitored in real time. The insets in panels A and B show concentration-dependent binding of plasminogen and tPA, respectively, each added at 16, 32, 63, 125, 250, 500 and 1000 nM, to immobilized cross-linked Aα221-610 oligomers; the dotted curves in both insets that practically coincide with the solid curves represent the best fit of the data using the kinetic analysis of the association/dissociation data (see Experimental Procedures).
Stimulating effect of the monomeric and oligomeric αC-domains on activation of plasminogen by tPA
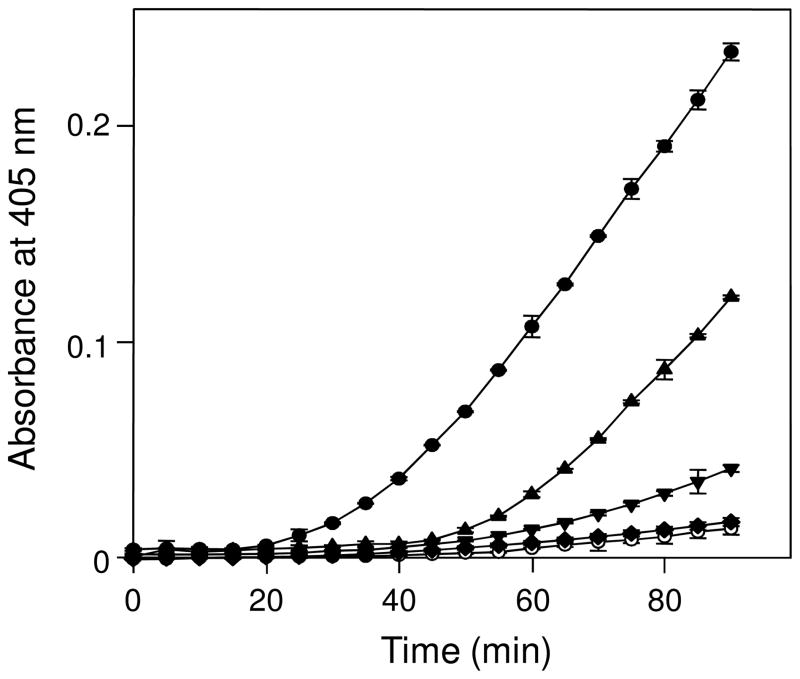
Having established that tPA- and plasminogen-binding sites are cryptic in the monomeric Aα221-610 fragment and exposed in its cross-linked oligomers, we tested the stimulating effect of these species on plasminogen activation using a chromogenic substrate assay. In this assay, plasminogen was activated by tPA and newly generated plasmin was detected by measuring its proteolytic activity towards the specific chromogenic substrate S-2251. The experiments revealed a very dramatic difference in the activation of plasminogen in the presence of different stimulators (Figure 8). The cross-linked oligomers exhibited a prominent stimulating effect while that of the monomer, as well as fibrinogen that was used as a control, was very low. The cross-linked Aα221-610 monomer, which was prepared by treatment of the monomeric Aα221-610 fragment with factor XIIIa as described earlier (2), stimulated plasminogen activation; however, its effect was lower than that of cross-linked Aα221-610 oligomers. The observed superior stimulating effect of the cross-linked oligomers indicates that their αC-domains adopt a physiologically active conformation only upon oligomerization. This further confirms the hypothesis that polymerization of the αC-domains results in the exposure of their cryptic binding sites.
Figure 8.
Stimulating effect of various αC-fragments on activation of plasminogen by tPA. Stimulating effect of cross-linked Aα221-610 oligomers (circles), cross-linked Aα221-610 monomer (triangles up), Aα221-610 monomer (triangles down), and fibrinogen (diamonds) was measured by hydrolysis of chromogenic substrate S-2251 with newly formed plasmin as described in Experimental Procedures. The activation of plasminogen in the absence of stimulators is shown by empty circles. Each point represents the mean ± S.D. of 3 independent experiments.
DISCUSSION
According to the current view, upon conversion of fibrinogen into fibrin, its αC-domains switch from intra- to intermolecular interaction to form αC-domain polymers (1). Numerous data suggest that such a switch is connected with conformational changes that result in physiologically active conformations of the αC-domains in fibrin αC-polymers (2, 4, 23, 24, 29). While the structure of the individual fibrinogen αC-domain has been established (13, 14), little is known about their structure and function in polymeric fibrin. The major problems in studying of fibrin αC-domain polymers are the complexity of fibrin structure and the presence of multiple domains, some of which duplicate the activity of the αC-domain. In addition, αC-domain polymers cannot be prepared, like some other fibrin(ogen) domains or regions, by limited proteolysis of fibrin, due to their rapid degradation into smaller fragments. In the present study, we overcame these problems by preparing a soluble model of αC-domain polymers using the recombinant Aα221-610 fragment corresponding to the fibrinogen αC region, which includes the αC-domain (Aα392-610). The study revealed that in this model, Aα221-610 fragment forms ordered linear oligomers that are stable in solution and contain compact structure whose thermal stability is similar to that of the αC-domains in fibrin(ogen). Furthermore, in contrast to the monomeric αC-domain, such oligomers exhibit prominent binding to plasminogen and tPA supporting our hypothesis that the αC-domain binding sites become exposed upon formation of αC polymers in fibrin.
Our previous electron microscopy experiments revealed that an Aα223-539 fragment containing the αC-connector and truncated αC-domain, which was prepared by limited proteolysis of bovine fibrinogen, may form ordered oligomers (18) that could mimic the arrangement of the αC-domains in fibrin. However, preparation of that fragment was complicated by a very low yield due to high susceptibility of the αC-domains to proteolysis and the C-terminal portion of the αC-domain in the resulting fragment was missing (18). To overcome those problems, we prepared the recombinant bovine Aα224-568 fragment containing the full-length αC-domain and αC-connector, as well as its human analog, the Aα221-610 fragment (25). While such fragments exhibited some ordered oligomers observed by electron microscopy, those oligomers were unstable and, therefore, not detected in solution where the fragments were preferentially monomeric (28). Subsequent treatment of these fragments with factor XIIIa resulted in stable, soluble oligomers; however, electron microscopy revealed that they were neither linear nor ordered (28, 30), suggesting that their structure does not mimic that of αC polymers in fibrin. Thus, although our previous attempts to prepare soluble model of fibrin αC polymers failed, they provided valuable information that prompted us to further searched for a more adequate soluble model of fibrin αC polymers, which would be ordered and stable in solution.
Our search for such a model was facilitated by our recent finding that the recombinant αC-domain, as well as its N-terminal sub-domain, forms ordered oligomers in a concentration-dependent and reversible manner (13, 14). Another finding that the αC-domain interacts with the αC-connector (19) suggested that such interaction may further promote formation of oligomers by the full-length αC region and provide a proper alignment of these portions in such oligomers for their efficient cross-linking with factor XIIIa. Therefore, in the present study, we prepared the recombinant Aα221-610 fragment corresponding to this region, confirmed that it forms oligomers, purified such oligomers, and then stabilized them by covalent cross-linking with factor XIIIa to prevent their dissociation in solution.
Several lines of evidence indicate that the prepared cross-linked Aα221-610 oligomers were highly ordered. First, oligomerization of Aα221-610 was concentration-dependent and reversible, indicating highly specific interactions between the αC-domains in the oligomers. Second, the fact that their cross-linking with factor XIIIa was much more rapid than that of the Aα221-610 monomer further confirms the ordered arrangement of the αC-connectors and αC-domains in the oligomers before the cross-linking occurs. Third, electron microscopy revealed that the oligomers appeared as well organized, almost linear arrays with a width of two monomeric molecules. Finally, spectral studies confirmed that the αC-domains in such oligomers were folded into compact cooperative units having a high content of regular structures and their thermal stability was comparable with that of the αC-domains in fibrin (31, 32). Thus, such highly ordered and reversible oligomerization of isolated Aα221-610 suggests that in fibrin the corresponding αC regions form αC polymers in a similar manner. This implies that cross-linked Aα221-610 oligomers mimic fibrin αC polymers.
Electron microscopy images of cross-linked Aα221-610 oligomers revealed linear arrays, or long, thin filaments. The diameter of the filaments making up these polymers was slightly less than twice the diameter of the monomers, and the globular regions of each pair of side-by-side monomers were slightly offset longitudinally. In addition, the uncross-linked monomer preparations also contained dimeric structures. These results suggest that the filaments are possibly made up of monomers interacting end-to-end and side-by-side requiring at least three interacting sites for each monomer (front, back, and side), and common for long biological filaments (33). Electron micrographs of the cross-linked oligomers demonstrate that besides individual filaments, they also contain branched polymers. Analysis of the width of the branched network revealed that some of the polymers had the same thickness before branching as the individual filaments (two monomers thick) while others were twice as thick. This suggests at least two mechanisms of branching. One mechanism may involve each of the two monomers at the end of a single filament initiating a new filament (see inset in Figure 2B). These monomers could be cross-linked to each other by factor XIIIa through unoccupied reactive Gln and Lys residues. The other type of branching points would occur through lateral association of two individual filaments (see inset in Figure 2A). This mechanism may require specific lateral interactions between individual αC filaments. The existence of any such specific interactions to form such a uniform network of filaments in vitro is evidence that they are intrinsic to the binding and cross-linking sites of the αC region itself. Furthermore, the existence of such regular interactions suggests that they could also occur between αC polymers in fibrin, in which they are located close to each other, although this remains to be demonstrated.
Our CD study indicates that individual αC-domains in cross-linked Aα221-610 oligomers are also highly ordered. While Aα221-610 monomer exhibited a substantial amount of unordered structure (53%), the content of such structures in the oligomers did not exceed 35%. The increase in regular structure content upon oligomerization is, most probably, connected with folding of the C-terminal sub-domains, which was shown earlier to adopt a folded conformation (preferentially consisting of β-sheet structure) in oligomers (14). However, this sub-domain and the N-terminal sub-domain together represent only about a half of the Aα221-610 fragment (αC region) while another half belongs to the αC-connector, which is considered to be flexible and unordered (1, 25). These imply that the αC-connector or its portion(s) may also be ordered in cross-linked Aα221-610 oligomers. This is in agreement with the results of the fluorescence study that were obtained by monitoring the fluorescence of Trp residues upon unfolding of such oligomers. Although our previous experiments with the Aα221-610 fragment performed by CD confirmed the presence of a compact cooperative structure in its αC-domain, in the present study we did not observe any sigmoidal transition when heat-induced unfolding of Aα221-610 monomer was monitored by fluorescence (not shown). This is because the αC-domain of the Aα221-610 fragment does not contain any Trp residues; all of them are located in the unordered αC-connector (34) and, therefore, completely exposed to the solvent. In contrast, Trp residues were responsive to unfolding in the oligomers (Figure 4), most probably because they, or at least some of them, were in an ordered environment. Thus, the results of the present study suggest that the αC-connector or portion(s) of it adopt a regular conformation upon formation of αC polymers. This is in agreement with the previous finding that conversion of fibrinogen into fibrin is accompanied by a significant increase of the β-sheet structure, which was speculated could occur due to interactions between the C-terminal parts of the α chains in polymeric fibrin (35, 36).
Having developed a model mimicking fibrin αC polymers, we used this model to test our hypothesis that polymerization of the αC-domains in fibrin results in the exposure of their multiple binding sites. Since it was established that the αC-domains interact with plasminogen and tPA (2), these two ligands were used as molecular probes for testing such exposure. Our ELISA and SPR experiments revealed that these ligands did not interact with the monomeric Aα221-610 fragments kept in solution. In SPR, no interaction was also observed with monomeric Aα221-610 immobilized on the surface of a sensor chip. This is in contrast to our previous SPR study that revealed a significant binding of tPA and plasminogen to immobilized Aα221-610 (2). One of the possible reasons for such a discrepancy could be that in the previous study the aggregation state of the Aα221-610 fragment was not tested and we cannot exclude the possibility that the immobilized fragment was in an oligomeric state while in this study immobilized Aα221-610 was monomeric. Whatever the reason for the discrepancy is, the present study clearly indicates that the monomeric Aα221-610 fragment including the αC-domain does not interact with plasminogen or tPA. In contrast, these ligands exhibited a prominent binding to cross-linked Aα221-610 oligomers, supporting the above hypothesis. Moreover, these oligomers exhibited a prominent stimulating effect on activation of plasminogen by tPA while that of Aα221-610 monomer was very low, further supporting the above hypothesis. Altogether, these results indicate that the αC-domains adopt a physiologically active conformation only upon their polymerization in fibrin.
In summary, in the present study, we prepared soluble Aα221-610 oligomers containing fibrin(ogen) αC-domains and stabilized their structure by covalent cross-linking with factor XIIIa. Physico-chemical and biochemical studies of these oligomers revealed that their oligomerization occurs through highly specific interactions between monomeric units and results in formation of compact, ordered, linear polymers that most probably reflect the structure αC polymers in fibrin. They also confirmed our hypothesis that the αC-domains adopt a physiologically active conformation in such polymers. Thus, cross-linked Aα221-610 oligomers represent a simple model that mimics structural and functional properties of the αC-domain in fibrin αC polymers. This model can be used for further studying the structure and function of fibrin αC-domains.
Footnotes
This work was supported by National Institutes of Health Grant HL056051 to L.M. and grants HL030954 and HL090774 to J.W.W.
Abbreviations and textual footnotes: tPA, tissue-type plasminogen activator; ELISA, enzyme-link immunosorbent assay; SPR, surface plasmon resonance; CD, Circular Dichroism; TBS, tris buffer saline (20 mM tris, pH 7.4, containing 150 mM NaCl).
References
- 1.Weisel JW, Medved L. The structure and function of the αC domains of fibrinogen. Ann NY Acad Sci. 2001;936:312–327. doi: 10.1111/j.1749-6632.2001.tb03517.x. [DOI] [PubMed] [Google Scholar]
- 2.Tsurupa G, Medved L. Identification and characterization of novel tPA- and plasminogen- binding sites within fibrin(ogen) αC-domains. Biochemistry. 2001;40:801–808. doi: 10.1021/bi001789t. [DOI] [PubMed] [Google Scholar]
- 3.Medved L, Nieuwenhuizen W. Molecular mechanisms of initiation of fibrinolysis by fibrin. Thromb Haemost. 2003;89:409–419. [PubMed] [Google Scholar]
- 4.Tsurupa G, Ho-Tin-Noe B, Angles-Cano E, Medved L. Identification and characterization of novel lysine-independent apolipoprotein(a)-binding sites in fibrin(ogen) αC-domains. J Biol Chem. 2003;278:37154–37159. doi: 10.1074/jbc.M305154200. [DOI] [PubMed] [Google Scholar]
- 5.Kaijzel EL, Koolwijk P, Van Erck MGM, Van Hinsbergh VWM, De Maat MPM. Molecular weight fibrinogen variants determine angiogenesis rate in a fibrin matrix in vitro and in vivo. J Thromb Haemost. 2006;4:1975–1981. doi: 10.1111/j.1538-7836.2006.02081.x. [DOI] [PubMed] [Google Scholar]
- 6.Medved L, Weisel JW on behalf of Fibrinogen and Factor XIII Subcommittee of Scientific Standardization Committee of International Society on Thrombosis and Haemostasis. Recommendations for nomenclature on fibrinogen and fibrin. J Thromb Haemost. 2009;7:355–359. doi: 10.1111/j.1538-7836.2008.03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spraggon G, Everse SJ, Doolittle RF. Crystal structures of fragment D from human fibrinogen and its crosslinked counterpart from fibrin. Nature. 1997;389:455–462. doi: 10.1038/38947. [DOI] [PubMed] [Google Scholar]
- 8.Madrazo J, Brown JH, Litvinovich S, Dominguez R, Yakovlev S, Medved L, Cohen C. Crystal structure of the central region of bovine fibrinogen (E5 fragment) at 1.4-Å resolution. Proc Natl Acad Sci USA. 2001;98:11967–11972. doi: 10.1073/pnas.211439798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Kollman JM, Pandi L, Doolittle RF. Crystal structure of native chicken fibrinogen at 2.7 Å resolution. Biochemistry. 2001;40:12515–12523. doi: 10.1021/bi011394p. [DOI] [PubMed] [Google Scholar]
- 10.Pechik I, Madrazo J, Mosesson MW, Hernandez I, Gilliland GL, Medved L. Crystal structure of the complex between thrombin and the central “E” region of fibrin. Proc Natl Acad Sci USA. 2004;101:2718–2723. doi: 10.1073/pnas.0303440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kollman JM, Pandi L, Sawaya MR, Riley M, Doolittle RF. Crystal structure of human fibrinogen. Biochemistry. 2009;48:3877–3886. doi: 10.1021/bi802205g. [DOI] [PubMed] [Google Scholar]
- 12.Burton RA, Tsurupa G, Medved L, Tjandra N. Identification of an ordered compact structure within the recombinant bovine fibrinogen αC-domain fragment by NMR. Biochemistry. 2006;45:2257–2266. doi: 10.1021/bi052380c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton R, Tsurupa G, Hantgan RR, Tjandra N, Medved L. NMR solution structure, stability, and interaction of the recombinant bovine fibrinogen αC-domain fragment. Biochemistry. 2007;46:8550–8560. doi: 10.1021/bi700606v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsurupa G, Hantgan RR, Burton RA, Pechik I, Tjandra N, Medved L. Structure, stability, and interaction of the fibrin(ogen) αC-domains. Biochemistry. 2009;48:12191–12201. doi: 10.1021/bi901640e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medved LV, Gorkun OV, Privalov PL. Structural organization of C-terminal parts of fibrinogen Aα-chains. FEBS Lett. 1983;160:291–295. doi: 10.1016/0014-5793(83)80985-5. [DOI] [PubMed] [Google Scholar]
- 16.Erickson HP, Fowler WE. Electron microscopy of fibrinogen, its plasmic fragments and small polymers. Ann NY Acad Sci. 1983;408:146–163. doi: 10.1111/j.1749-6632.1983.tb23242.x. [DOI] [PubMed] [Google Scholar]
- 17.Weisel JW, Stauffacher CV, Bullitt E, Cohen C. A model for fibrinogen: domains and sequence. Science. 1985;230:1388–1391. doi: 10.1126/science.4071058. [DOI] [PubMed] [Google Scholar]
- 18.Veklich YI, Gorkun OV, Medved LV, Nieuwenhuizen W, Weisel JW. Carboxyl-terminal portions of the α chains of fibrinogen and fibrin. Localization by electron microscopy and the effects of isolated αC fragments on polymerization. J Biol Chem. 1993;268:13577–13585. [PubMed] [Google Scholar]
- 19.Litvinov RI, Yakovlev SV, Tsurupa G, Gorkun OV, Medved L, Weisel J. Direct evidence for specific interactions of the fibrinogen αC-domains with the central E region and with each other. Biochemistry. 2007;46:9133–9142. doi: 10.1021/bi700944j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKee PA, Mattock P, Hill RL. Subunit structure of human fibrinogen, soluble fibrin, and cross-linked insoluble fibrin. Proc Natl Acad Sci USA. 1970;66:738–744. doi: 10.1073/pnas.66.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz ML, Pizzo SV, Sullivan JB, Hill RL, McKee PA. A comparative study of crosslinked and noncrosslinked fibrin from the major classes of vertebrates. Thromb Diath Haemorrh. 1973;29:313–338. [PubMed] [Google Scholar]
- 22.Matsuka YV, Medved LV, Migliorini MM, Ingham KC. Factor XIIIa-catalyzed cross-linking of recombinant αC fragments of human fibrinogen. Biochemistry. 1996;35:5810–5816. doi: 10.1021/bi952294k. [DOI] [PubMed] [Google Scholar]
- 23.Makogonenko E, Tsurupa G, Ingham K, Medved L. Interaction of fibrin(ogen) with fibronectin: further characterization and localization of the fibronectin-binding site. Biochemistry. 2002;41:7907–7913. doi: 10.1021/bi025770x. [DOI] [PubMed] [Google Scholar]
- 24.Tsurupa G, Yakovlev S, McKee P, Medved L. Noncovalent interaction of α2-antiplasmin with fibrin(ogen): localization of α2-antiplasmin-binding sites. Biochemistry. 2010;49:7643–7651. doi: 10.1021/bi1010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsurupa G, Tsonev L, Medved L. Structural organization of the fibrin(ogen) αC-domain. Biochemistry. 2002;41:6449–6459. doi: 10.1021/bi025584r. [DOI] [PubMed] [Google Scholar]
- 26.Yang JT, Wu CS, Martinez HM. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- 27.Verheijen JH, Mullaart E, Chang GT, Kluft C, Wijngaards G. A simple, sensitive spectrophotometric assay for extrinsic (tissue-type) plasminogen activator applicable to measurements in plasma. Thromb Haemost. 1982;48:266–269. [PubMed] [Google Scholar]
- 28.Tsurupa G, Veklich Y, Hantgan R, Belkin AM, Weisel JW, Medved L. Do the isolated fibrinogen αC-domains form ordered oligomers? Biophys Chem. 2004;112:257–266. doi: 10.1016/j.bpc.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 29.Medved L, Tsurupa G, Yakovlev S. Confomational changes upon convertion of fibrinogen into fibrin. The mechanisms of exposure of cryptic sites. Ann NY Acad Sci. 2001;936:185–204. doi: 10.1111/j.1749-6632.2001.tb03505.x. [DOI] [PubMed] [Google Scholar]
- 30.Belkin AM, Tsurupa G, Zemskov E, Veklich Y, Weisel JW, Medved L. Transglutaminase-mediated oligomerization of the fibrin(ogen) αC domains promotes integrin-dependent cell adhesion and signaling. Blood. 2005;105:3561–3568. doi: 10.1182/blood-2004-10-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mihalyi E, Donovan JW. Clotting of fibrinogen. 2 Calorimetry of the reversal of the effect of calcium on clotting with thrombin and with ancrod. Biochemistry. 1985;24:3443–3448. doi: 10.1021/bi00335a008. [DOI] [PubMed] [Google Scholar]
- 32.Procyk R, Medved L, Engelke KJ, Kudryk B, Blomback B. Nonclottable fibrin obtained from partially reduced fibrinogen: characterization and tissue plasminogen activator stimulation. Biochemistry. 1992;31:2273–2278. doi: 10.1021/bi00123a009. [DOI] [PubMed] [Google Scholar]
- 33.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates; Sunderland, MA: 2001. Polymerization of cytoskeletal filaments; pp. 151–163. [Google Scholar]
- 34.Doolittle RF, Watt KWK, Cottrell BA, Strong DD, Riley M. The amino acid sequence of the α-chain of human fibrinogen. Nature. 1979;280:464–468. doi: 10.1038/280464a0. [DOI] [PubMed] [Google Scholar]
- 35.Marx J, Hudry-Clergeon G, Capet-Antonini F, Bernard L. Laser Raman spectroscopy study of bovine fibrinogen and fibrin. Biochim Biophys Acta. 1979;578:107–115. doi: 10.1016/0005-2795(79)90118-1. [DOI] [PubMed] [Google Scholar]
- 36.Hudry-Clergeon G, Freyssinet JM. Orientation of fibrin in strong magnetic fields. Ann NY Acad Sci. 1983;408:380–387. doi: 10.1111/j.1749-6632.1983.tb23258.x. [DOI] [PubMed] [Google Scholar]