Abstract
The estrogen receptor (ER) is an important regulator of growth and differentiation of breast epithelium. Transactivation by ER depends on a leucine-rich motif, which constitutes a ligand-regulated binding site for steroid receptor coactivators (SRCs). Cyclin D1 is frequently amplified in breast cancer and can activate ER through direct binding. We show here that cyclin D1 also interacts in a ligand-independent fashion with coactivators of the SRC-1 family through a motif that resembles the leucine-rich coactivator binding motif of nuclear receptors. By acting as a bridging factor between ER and SRCs, cyclin D1 can recruit SRC-family coactivators to ER in the absence of ligand. A cyclin D1 mutant that binds to ER but fails to recruit coactivators preferentially interferes with ER activation in breast cancer cells that have high levels of cyclin D1. These data support that cyclin D1 contributes significantly to ER activation in breast cancers in which the protein is overexpressed. Our present results reveal a novel route of coactivator recruitment to ER and establish a direct role for cyclin D1 in regulation of transcription.
Keywords: Estrogen receptor, cyclin D1, breast cancer, SRC-1, coactivator
The estrogen receptor (ER) belongs to the steroid/nuclear receptor family of ligand-regulated transcription factors. Members of this superfamily display a modular structure with six distinct functional regions (termed A–F), which includes domains for DNA binding, ligand binding, and transcriptional activation. Like other members of the nuclear hormone receptor superfamily, ER harbors two transcriptional activation functions (AFs) that act synergistically in transactivation (Kumar et al. 1987; Tzukerman et al. 1994). Transcriptional activation is mediated by means of the autonomous activation function (AF-1) in the amino-terminal A/B domain and the ligand-dependent activation function (AF-2) in the carboxy-terminal hormone-binding domain (Evans 1988; Kumar and Chambon 1988; Beato 1989). These two regions flank the DNA-binding domain of the receptor. On ligand binding, ER binds to estrogen-responsive elements, which results in activation of specific ER target genes (Beato 1989).
It is generally thought that nuclear receptors stimulate transcription through direct binding to several of the basal transcription factors, thereby enhancing the formation of a stable transcription pre-initiation complex (Mitchell and Tjian 1989). This notion is supported by in vitro protein-binding studies that demonstrated that several steroid receptors interact directly with components of the basal transcriptional apparatus, including the TATA-box-binding protein TBP (Sadovsky et al. 1995), TFIIB (Ing et al. 1992; Baniahmad et al. 1993), and human TAFII30 (Jacq et al. 1994). Several lines of evidence, however, suggest that efficient transactivation requires additional, positively acting factors termed coactivators (Pugh and Tjian 1990). Several candidate steroid receptor coactivators (SRCs) have been identified. The first coactivator identified based on its ability to interact with the progesterone receptor was SRC-1 (Onate et al. 1995; Yao et al. 1996). This protein is the founding member of a family of related SRCs that include TIF-2/GRIP-1 (Voegel et al. 1996; Hong et al. 1997) and AIB-1/ACTR/RAC-3/p/CIP (Anzick et al. 1997; Chen et al. 1997; Li et al. 1997; Torchia et al. 1997). Several functional domains are highly conserved in all members of this family. For instance, the amino-terminal regions contain basic helix-loop-helix (bHLH) and Per-ARNT-Sim (PAS) domains. Both motifs are thought to be involved in protein–protein interactions and DNA–protein interactions (Yao et al. 1996). Interestingly, the bHLH–PAS domain is dispensable for SRC-1 activity, including receptor interaction and receptor activation (Onate et al. 1995; Yao et al. 1996). In addition, all SRCs contain multiple LxxLL motifs (L is leucine; x is any amino acid) in the central region of the protein. These motifs were shown recently to be involved in nuclear receptor interaction (Le Douarin et al. 1996; Heery et al. 1997; Torchia et al. 1997). Besides ER, SRC-1 also interacts with another coactivator of steroid receptors, CBP/p300, and both types of coactivators act synergistically to enhance ER transactivation (Chakravarti et al. 1996; Hanstein et al. 1996; Kamei et al. 1996; Smith et al. 1996; Yao et al. 1996; Chen et al. 1997). Both coactivators of the SRC-1 family and the p300/CBP family have intrinsic histone acetyl transferase (HAT) activity, which is widely believed to be involved in chromatin remodeling during transcriptional activation (Ogryzko et al. 1996; Jenster et al. 1997; Spencer et al. 1997).
Transactivation by steroid/nuclear receptors involves the well-conserved AF-2 domain located in helix 12 of the carboxyl terminus of the receptors. It has been demonstrated that helix 12 harbors a leucine-rich motif that constitutes a ligand-regulated binding site for coactivators, like SRC-1 (Danielian et al. 1992; Le Douarin et al. 1995; Voegel et al. 1996; vom Baur et al. 1996). Consequently, transactivation by nuclear receptors is dramatically reduced in receptors that contain mutations in helix 12 (Danielian et al. 1992; White et al. 1997). The leucine-rich motif in helix 12 of nuclear receptors is involved in binding to the LxxLL motifs of the steroid receptor coactivators (Le Douarin et al. 1996; Heery et al. 1997).
Cyclin D1 is induced in response to mitogenic stimulation of quiescent cells and acts as an activator of CDK4 and CDK6. These cyclin D1/CDK complexes are key regulators of progression through the G1 phase of the cell cycle and are involved in functional inactivation of the retinoblastoma family proteins (for review, see Beijersbergen and Bernards 1996). cyclin D1 is amplified or overexpressed in a number of human malignancies, the most prominent being breast cancer, in which up to 50% of all cases have elevated levels of cyclin D1 (Schuuring et al. 1992a,b; Buckley et al. 1993; van Diest et al. 1997). The relevance of cyclin D1 overexpression is underscored by the finding that tissue-specific transgenic expression of cyclin D1 in mice results in mammary hyperplasia and adenocarcinoma (Wang et al. 1994).
Furthermore, cyclin D1 knockout mice show a marked defect in breast epithelium development during pregnancy and cyclin D1 reduces mitogen requirement of breast cancer cell lines (Musgrove et al. 1994; Fantl et al. 1995; Sicinski et al. 1995; Zwijsen et al. 1996). cyclin D1 is overexpressed preferentially in ER-positive breast cancers, suggesting that cyclin D1 derives (part of) its oncogenic activity in breast cancer by acting on ER (Gillett et al. 1996; van Diest et al. 1997). We and others have recently made a connection between ER and cyclin D1 by showing that cyclin D1 can interact directly with the ligand-binding domain of ER and can stimulate ER transactivation in a ligand-independent and CDK-independent fashion (Neuman et al. 1997; Zwijsen et al. 1997). In this study, we describe an unexpected relationship between cyclin D1 and SRCs that places cyclin D1 at the center of a complex transcription regulatory network of nuclear hormone receptors and their coactivators.
Results
ER and cyclin D1 share a coactivator binding motif
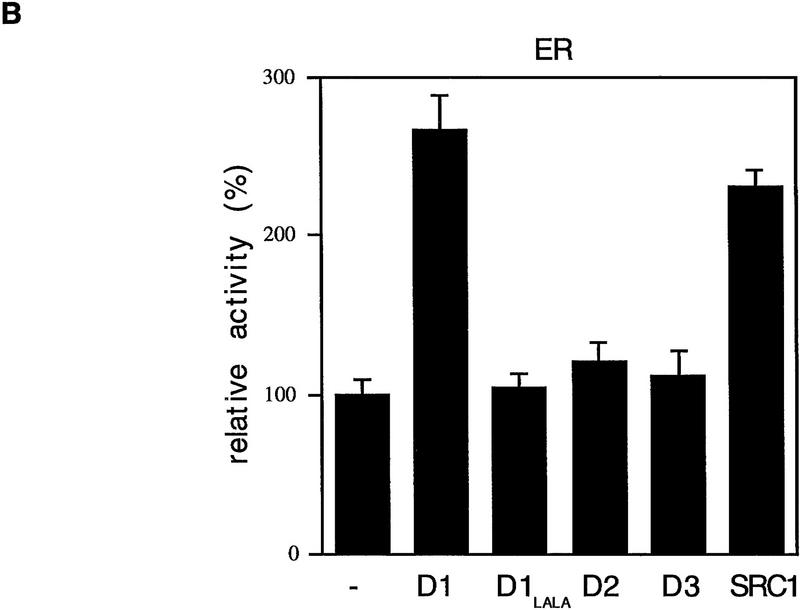
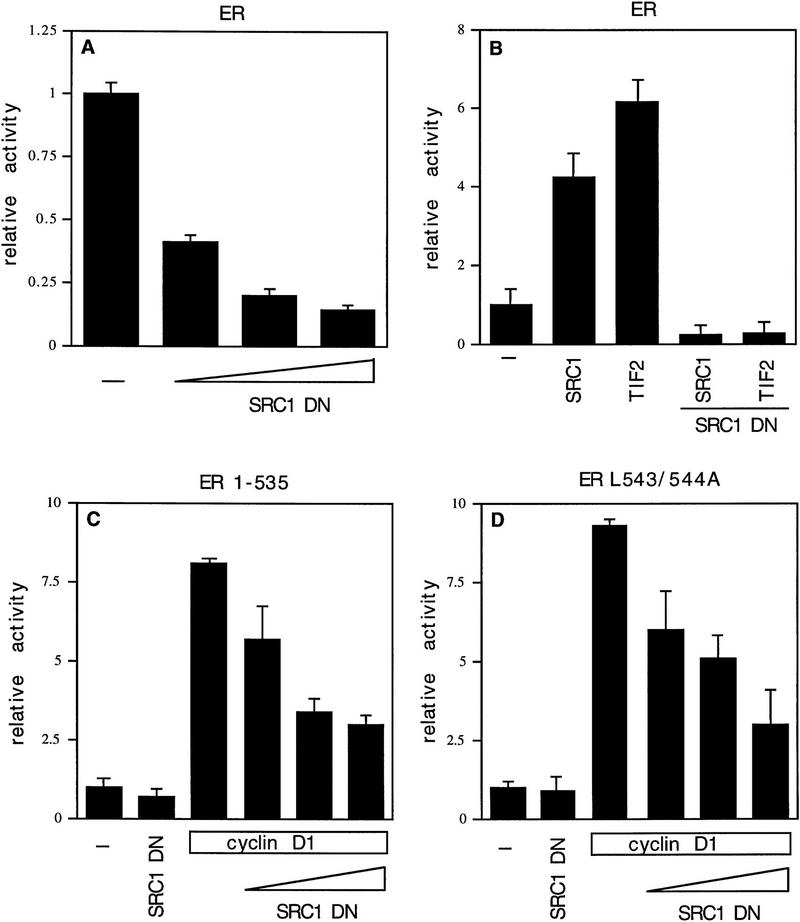
To study how cyclin D1 activates ER, cyclin D1 deletion mutants were tested for their effect on ER transactivation. Cos-7 cells were transfected with cyclin D1 mutants, together with ER and a luciferase reporter gene construct driven by a minimal TATA promoter linked to an estrogen response element (ERE). Figure 1A shows that an amino-terminal deletion mutant of cyclin D1 (D1, amino acids 91–295) was still able to activate ER, whereas two carboxy-terminal deletion mutants of cyclin D1 (D1, amino acids 1–202; D1, amino acids 1–247) lack ER transactivation capacity. A cyclin D1 mutant lacking the extreme carboxyl terminus (D1, amino acids 1–267) partially retained ER activation. Together, these data indicate that the domain required for ER activation is located in the carboxy-terminal 48 amino acids of cyclin D1. This part of the protein is not involved in CDK interaction and is poorly conserved among the different cyclins. Alignment of sequences in this part of cyclin D1 with ER revealed that a motif that resembles the highly conserved leucine-rich coactivator binding motif in AF-2 of ER is present within the domain of cyclin D1 implicated in ER transactivation at the amino acid positions 254–259 (Fig. 1A). This motif is only partially conserved in cyclins D2 and D3, two cyclins that are far less active in ER transactivation (Neuman et al. 1997; Zwijsen et al. 1997). To test the relevance of this leucine-rich domain of cyclin D1 in ER activation, a cyclin D1 mutant was constructed in which leucines 254 and 255 were mutated to alanines (D1 L254/255A). This mutation in cyclin D1 is similar to the mutation in ER (ER L543/544A) that interferes with coactivator binding to AF-2 (Danielian et al. 1992). In contrast to wild-type cyclin D1, the L254/255A mutant cyclin D1 was virtually unable to activate wild type ER even though this mutant was equally well expressed and was fully active in other assays (Fig. 1B and see below). Cyclins D2 and D3, which lack this leucine-rich motif, behaved similar to the D1 L254/255A mutant in ER activation (Fig. 1B). These data suggest that cyclin D1 can activate ER through an AF-2-like motif.
Figure 1.
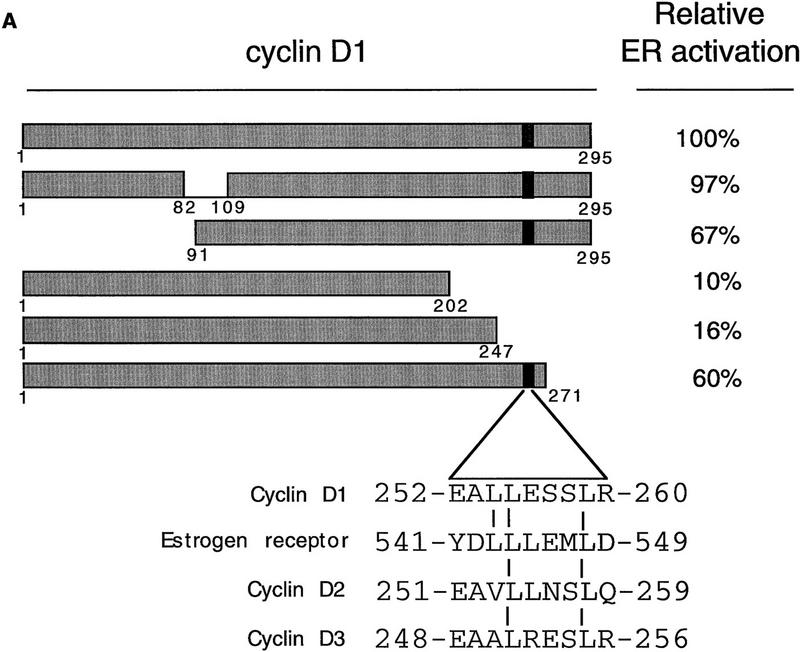
Mapping of the region of cyclin D1 required for ER-mediated transactivation. (A) The effect of cyclin D1 deletion mutants on ER activation in the presence of ligand. The cyclin D1 derivatives used in this study are shown at left; (right) the relative capacity of the mutants to potentiate ERE-dependent transcription. ER-negative Cos-7 cells were transfected with expression vectors for wild-type ER (200 ng), cyclin D1, or cyclin D1 mutants (2.5 μg), an internal control β-galactosidase plasmid (0.5 μg), and an ERE–TATA–luciferase reporter (3 μg). The effect on ER transactivation of wild-type cyclin D1 was set to 100%. These studies were performed in three separate experiments and expressed as mean values with s.d. < 10 % (data not shown). The alignment of leucine-rich motif of ER with D-type cyclins is shown on the bottom; the leucine-rich motif in cyclin D1 is indicated as a solid box. (B) ER activation by D-type cyclins, cyclin D1 L254/255A point mutant (D1–LALA), and SRC-1 in the presence of ligand. Cos-7 cells were transfected with D-type cyclin expression vectors, cyclin D1 leucine-to-alanine point mutant (D1–LALA), or SRC-1 expression vector together with wild-type ER expression vector, an ERE–TATA–luciferase reporter plasmid, and the internal control β-galactosidase construct. Data are expressed as relative luciferase activity compared with basal ERE–luciferase activity of wild-type ER and are normalized for transfection efficiencies.
Activation of AF-2 mutant ERs by cyclin D1
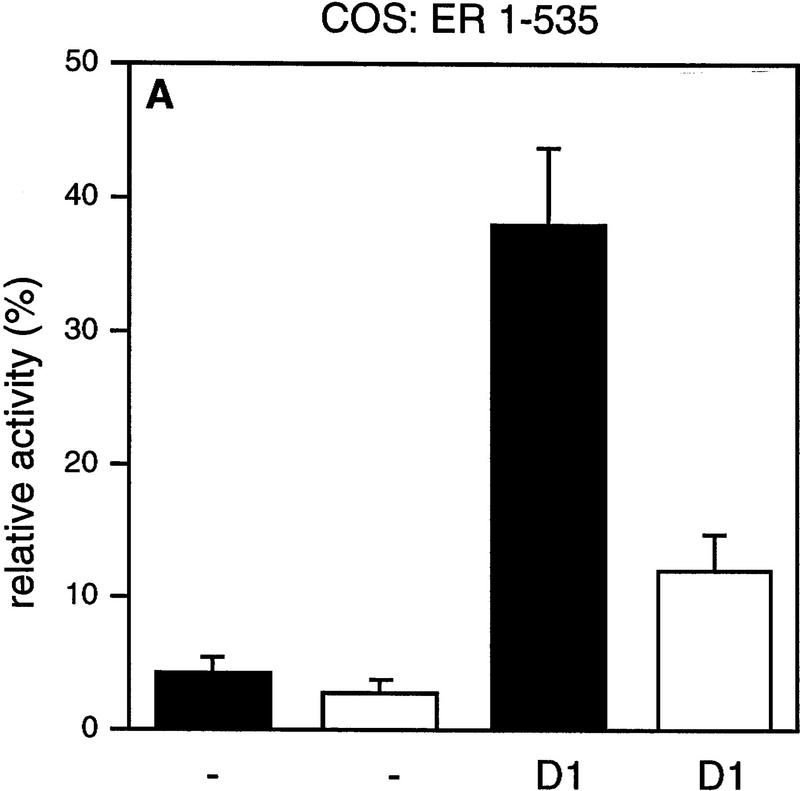
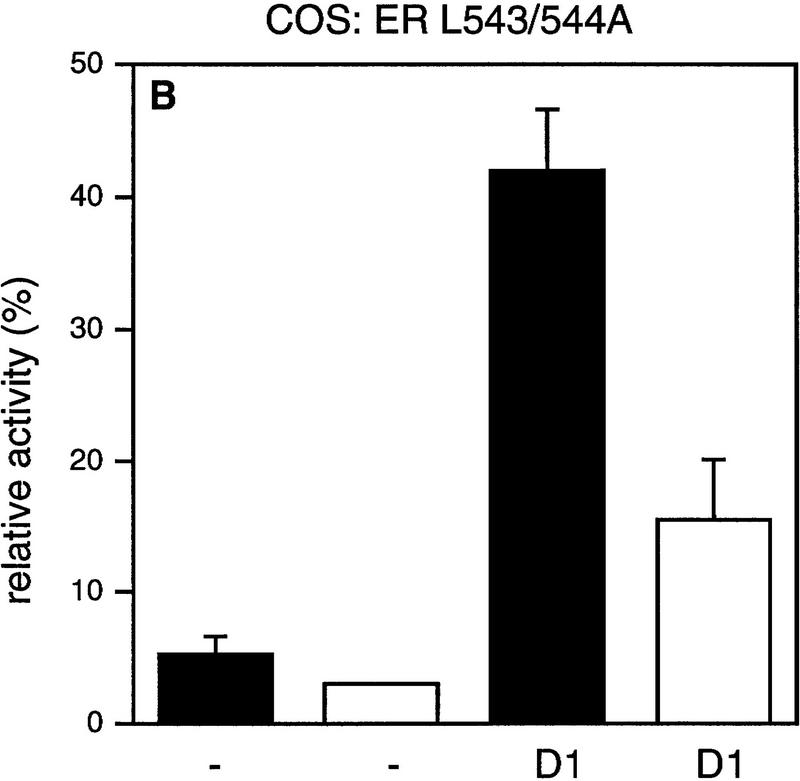
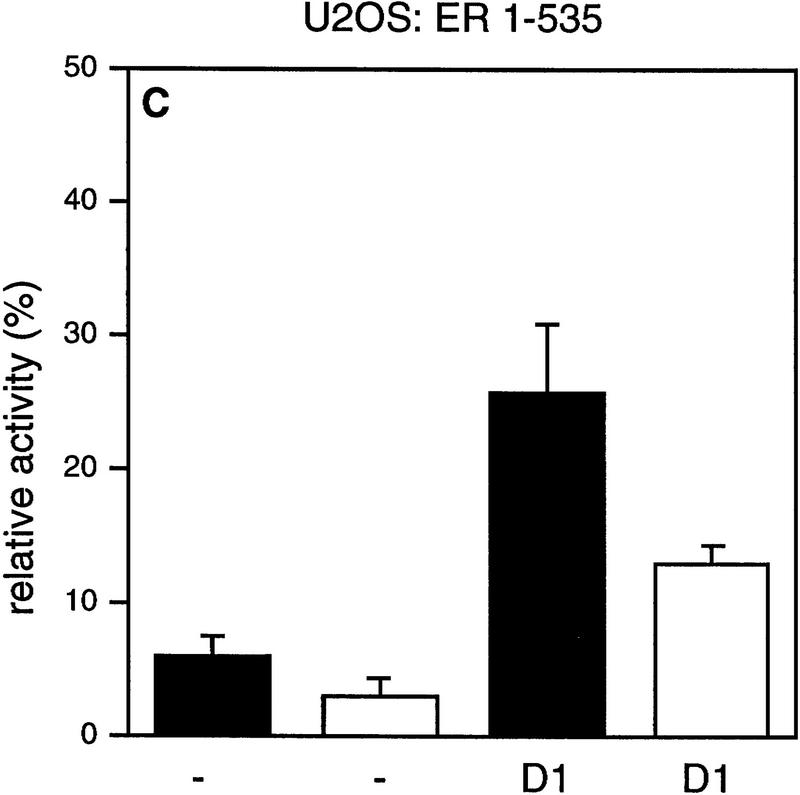
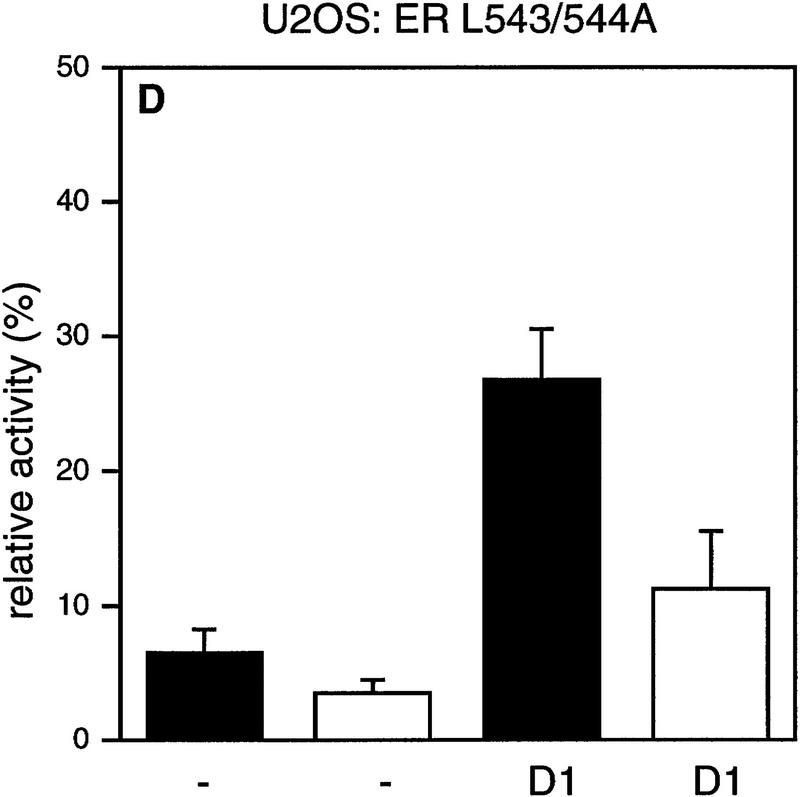
It has been demonstrated that AF-2 mutant ERs are unable to activate transcription, because they cannot recruit SRCs efficiently (Danielian et al. 1992; vom Baur et al. 1996). To determine the role of the AF-2 domain of ER in cyclin D1-mediated transactivation, a deletion mutant and a point mutant in the ligand-regulated carboxy-terminal activation domain (AF-2) of ER were tested in Cos-7 cells. Figure 2 shows that, as reported previously, the activity of these AF-2 mutant ERs reflects background levels (Danielian et al. 1992; vom Baur et al. 1996). Surprisingly, coexpression of cyclin D1 resulted in a significant activation of the ER AF-2 deletion mutant (ER 1-535) to levels that were up to 40% of ligand-activated wild-type receptor (Fig. 2A). Cyclin D1 was also able to induce transcription in the absence of ligand, although this increase was less pronounced. Similarly, ER L543/544A, which harbors a mutation in the leucine-rich coactivator-binding site in AF-2 (LLxxxL to AAxxxL), could be activated by cyclin D1 (Fig. 2B). Comparable results were obtained in ER-negative U2-OS osteosarcoma cells (Fig. 2C,D), indicating that the effect of cyclin D1 is not cell type-specific. These results indicate that cyclin D1 can mediate activation of ER mutants that are unable to interact with SRCs efficiently.
Figure 2.
Effect of cyclin D1 on helix 12 mutants of ER. The effect of cyclin D1 on ER mutants was tested in Cos-7 cells (A,B) and in U2-OS cells (C,D) in the presence of ligand. An ERE–TATA–luciferase reporter construct was used in transient transfections together with cyclin D1 and ER 1–535 (A,C) or ER L543/544A mutants (B,D). β-Galactosidase served as an internal control. The reporter activity was determined both in the presence (solid bars) and in the absence (open bars) of 10 nm 17β-estradiol. The relative luciferase activity was calculated by normalizing to the β-galactosidase activity. The relative reporter activity of wild-type ER in the presence of ligand was used as a reference and set at 100%. In the absence of transfected ER plasmid, cyclin D1 did not induce transcriptional activity of the reporter (data not shown). At least five separate transfection experiments were performed (expressed as average ±s.d.).
To ask whether SRCs are involved in the cyclin D1-mediated activation of the mutant ERs, a dominant-negative mutant of SRC-1 (SRC1–DN, encoding amino acids 1245–1441 of SRC-1) was used. This mutant harbors a LxxLL motif, which mediates binding to the leucine-rich coactivator-binding site in ER (Le Douarin et al. 1995; Heery et al. 1997) but lacks a transactivation domain (Jenster et al. 1997; Spencer et al. 1997). As shown in previous studies (Onate et al. 1995), this construct served as a dominant inhibitor for endogenous SRC-1 function on wild-type ER (Fig. 3A). As expected, SRC1–DN inhibited the ability of SRC-1 and of the closely related coactivator TIF2 on ER transactivation (Fig. 3B), whereas it was inactive on the nonrelated E2F-1 transcription factor (data not shown). Importantly, SRC1–DN markedly repressed the cyclin D1-induced activation of the ER AF-2 mutants (Fig. 3C,D). These data suggest that the cyclin D1-mediated activation of these mutant ERs somehow requires SRC activity.
Figure 3.
Role of coactivators in cyclin D1-induced transactivation. (A) Effect of SRC1–DN on ER transactivation. SRC1–DN encoding amino acids 1245–1441 of SRC-1 (1, 2.5, and 5 μg) was introduced by transient transfection, together with wild-type ER (200 ng), and tested for its ability to modulate ERE-dependent transcription. (B) Effect of SRC1–DN on SRC-1- and TIF-2-mediated ER transactivation. SRC-1 (3 μg) or TIF-2 (3 μg) were transfected with SRC1–DN (3 μg) and tested for ER transactivation. (C) Effect of SRC1–DN on cyclin D1-induced transactivation of ER 1–535 mutant. Cyclin D1 was cotransfected with SRC1–DN (1, 2.5, 5 μg) and tested for its effect on an ER harboring a deletion of coactivator-binding site in helix 12 (ER 1–535). (D) Effect of SRC1–DN on cyclin D1-induced transactivation of ER L543/544A mutant. Cyclin D1 and SRC1–DN (1, 2.5, 5 μg) were transfected and tested for the activity of ER helix 12 point mutant (ER L543/544A). The transient transfections (A–C) were performed in Cos-7 cells, which were maintained in DMEM with 10 nm ligand. The relative activity was calculated by normalizing to the internal control and was divided by luciferase activity of ER (mutant) in the presence of ligand. The transfections for each set of conditions were done in at least four independent experiments and expressed as average ±s.d.
Direct binding of cyclin D1 to SRCs
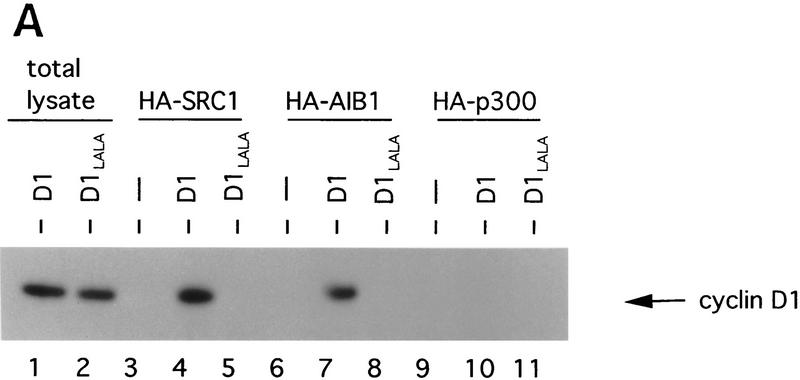
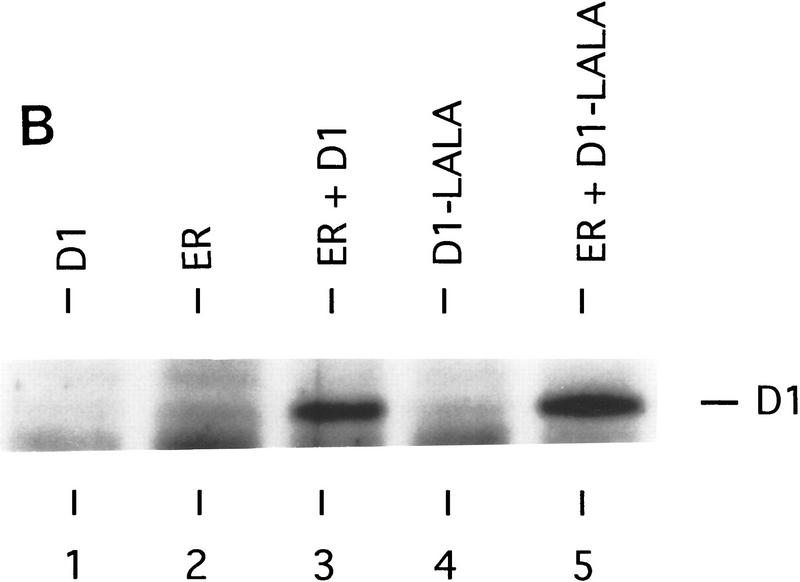
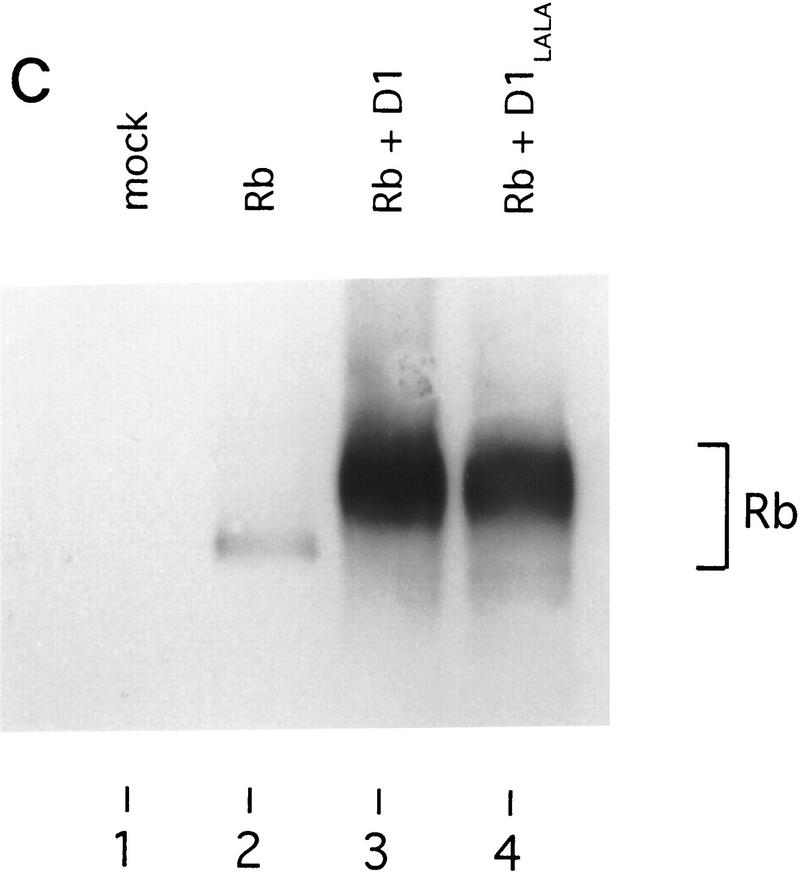
Because the leucine-rich motifs of nuclear receptors have been shown to recruit SRC family coactivators (Danielian et al. 1992; Heery et al. 1997), we tested whether cyclin D1 is also able to interact with SRCs through its leucine-rich motif. Cos-7 cells were cotransfected with cyclin D1 and HA-tagged constructs encoding the nuclear receptor coactivators SRC-1, AIB-1, or p300 (Onate et al. 1995; Chakravarti et al. 1996; Hanstein et al. 1996; Kamei et al. 1996; Anzick et al. 1997). Immunoprecipitation using antibodies directed against HA-tag and subsequent immunoblot analysis revealed that cyclin D1 coimmunoprecipitated with both SRC-1 and AIB-1, but hardly with p300 (Fig. 4A). Significantly, the cyclin D1 mutant L254/255A, which failed to activate ER (Fig. 1B), did not interact with SRC family proteins (Fig. 4A), even though this mutant was expressed equally and was unaffected in its ability to bind ER (Fig. 4B) and to phosphorylate pRb in cells that lack cyclin D1-associated kinase activity (Fig. 4C). These data suggest that the AF-2-like leucine-rich motif of cyclin D1 mediates binding to SRC-family proteins.
Figure 4.
Direct interaction between cyclin D1 and SRCs. (A) In vivo binding of cyclin D1 to SRCs. HA-tagged SRC-1, AIB-1, or p300 cDNA expression vectors were introduced into Cos-7 cells together with control plasmid (−), plasmids directing the synthesis of wild-type cyclin D1, or D1 L254/255A mutant (D1–LALA), as indicated. Lanes 1 and 2 contain total lysate of cells (5% of amount used in IP) transfected with cyclin D1 and cyclin D1–LALA, respectively. 12CA5 HA antibodies were used for immunoprecipitation of whole-cell extracts prepared from these cells and coimmunoprecipitation of cyclin D1 (mutants) was examined by Western blot analysis using anti-cyclin D1 antibody. Note that wild-type cyclin D1, but not the leucine-to-alanine mutant D1–LALA, coimmunoprecipitates with SRC-1 and AIB-1, whereas binding of both wild-type cyclin D1 and D1–LALA mutant to p300 was hardly detectable. (B) Binding of cyclin D1 and cyclin D1 L254/255A mutant to ER. Cos-7 cells were transfected with ER expression vector, cyclin D1 (mutant), and/or control plasmids. Monoclonal ER antibodies were used for immunoprecipitation of ER of whole-cell extracts prepared from these cells and coimmunoprecipitation of cyclin D1 (mutant) was examined by Western blot analysis using monoclonal cyclin D1 antibodies. (C) Activity of cyclin D1 and D1 L254/255A mutant in phosphorylation of pRb in Rb−/− 3T3 cells. Cells were transfected with pRb expression vector, cyclin D1 (mutant), and/or control plasmids and were maintained in low serum conditions. After 40 hr, cells were lysed and proteins were separated by low-percentage polyacrylamide gel electrophoresis. Differentially phosphorylated species of pRb were detected by Western blotting using the polyclonal pRb antibody (C15, Santa Cruz).
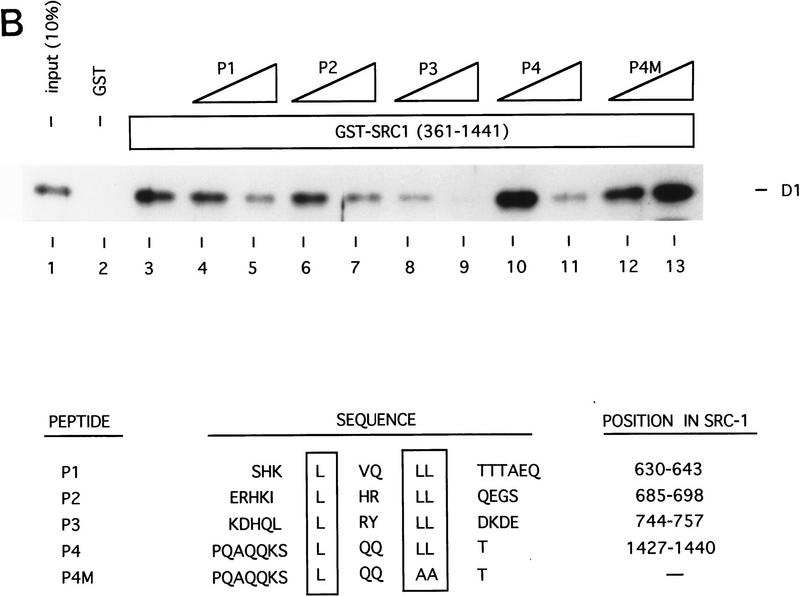
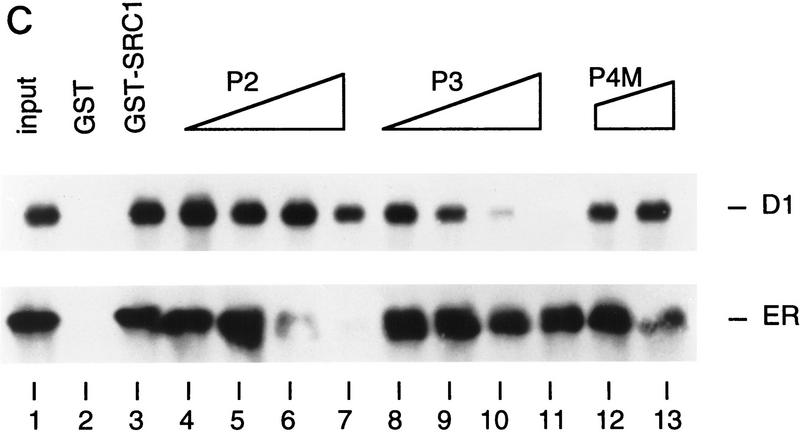
To test whether the interaction between cyclin D1 and SRC-1 is direct, we used bacterially expressed GST–SRC1 and Escherichia coli-produced 6× histidine-tagged cyclin D1 in an in vitro protein-binding assay. As shown in Figure 5A, His-cyclin D1 binds strongly binds to GST–SRC1 protein, but not to GST alone, indicating that this association is specific and direct. To ask which domain or motif of SRC-1 is involved in cyclin D1 binding, we generated a series of GST–SRC1 deletion mutants. Figure 5A shows that amino acids 568–782 of SRC-1 are required for cyclin D1 binding. Interestingly, this region of SRC-1 harbors three LxxLL motifs, which are involved in binding to nuclear receptors (Le Douarin et al. 1995; Heery et al. 1997; Torchia et al. 1997). To ask whether the LxxLL motifs of SRC-1 are involved in cyclin D1 binding, a peptide competition experiment was performed. Figure 5B shows that LxxLL peptides, but not a LxxAA mutant peptide, interfered with binding of cyclin D1 to SRC-1. Of the four SRC-1 LxxLL peptides tested, the P3 peptide, which corresponds to the third of the three centrally located LxxLL motifs, was the best competitor. Consistent with previous studies (Heery et al. 1997), the P2 peptide competed most efficiently the interaction between SRC-1 and ER (Fig. 5C). These data indicate that the various LxxLL motifs of SRC-1 display specificity for protein interactions.Therefore, SRC-1 uses the LxxLL motifs not only to interact with nuclear receptors (through the P2 motif), but also to bind cyclin D1 (through the P3 motif). Because these LxxLL motifs interact with the leucine-rich motifs on nuclear receptors (Heery et al. 1997; Torchia et al. 1997), these data are in agreement with our experiments shown in Figure 4A, which suggested a major role for the leucine-rich AF-2-like motif of cyclin D1 in SRC binding. Because both the in vitro and the in vivo association experiments shown above were performed in the absence of 17β-estradiol, these data also indicate that the cyclin D1/SRC-1 interaction, in contrast to the ER/SRC-1 interaction, is hormone-independent.
Figure 5.
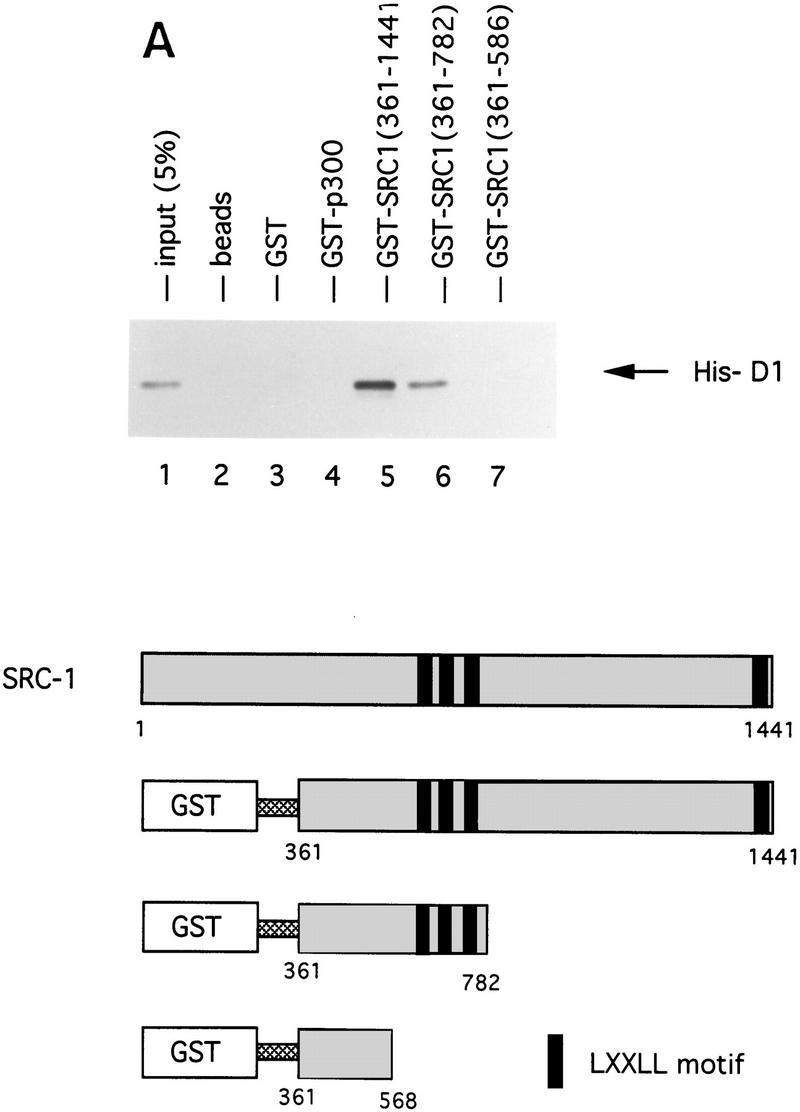
Cyclin D1 interacts directly with SRC-1. (A) In vitro interaction between SRC-1 and cyclin D1. A series of GST fusion proteins containing SRC-1 (GST–RC1 361–441; GST–RC1 361-782; GST-SRC1 361-568) or GST–p300 (1–95) were tested for direct binding to His-tagged cyclin D1 (His–D1); the GST–SRC1 derivatives are shown at bottom. The conserved LxxLL motifs are boxed and the amino acid boundaries are demonstrated. In the in vitro binding assay, a series of GST-containing proteins were incubated with bacterially expressed His-tagged cyclin D1 and immobilized on glutathione–agarose. Cyclin D1 binding was detected by Western blot analysis using anti-cyclin D1 antibody. (B) Competition of cyclin D1–SRC1 binding by SRC1 peptides. Peptides (0.3 and 3 μg) derived from the four LxxLL motifs of SRC1 were used in a GST pull-down assay using GST–SRC1(361–1441) and His-tagged cyclin D1 as described in A. The sequence and the position of the peptides in SRC-1 are shown at bottom. (C). Competition of P2 and P3 in cyclin D1–SRC-1 and ER–SRC-1 interaction. Peptides P2 and P3 (0.1, 0.2, 0.4, or 0.8 μg) and control peptide P4M (0.4 and 0.8 μg) were tested for their ability to compete the binding between GST–SRC1 and cyclin D1 (top) or GST–SRC1 and ER (bottom). Input of cyclin D1 and ER proteins shown represents 20% of the amount of protein used in the binding assay.
Cyclin D1 act as a physical bridge between ER and SRC-1
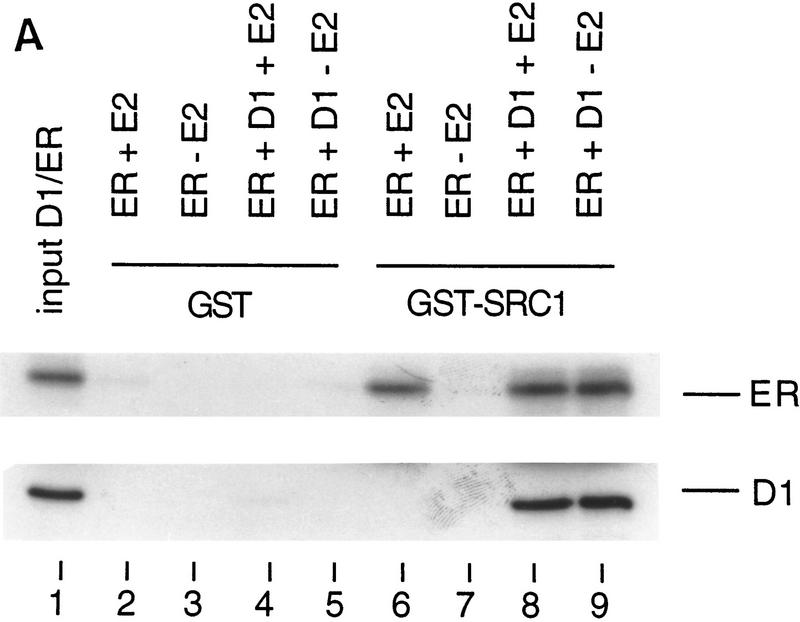
Cyclin D1 activates the ER in a ligand-independent fashion through direct binding to ER (Neuman et al. 1997; Zwijsen et al. 1997). This study demonstrates that cyclin D1, besides ER, also interacts directly with SRC-1 in vivo and in vitro. These data suggest a model in which cyclin D1 can act as a bridging factor between ER and SRC-1, allowing ligand-independent recruitment of coactivators to ER in the presence of cyclin D1. To test this model directly, we used bacterially expressed 6× histidine-tagged cyclin D1, E. coli-produced GST–SRC1, and baculovirus-produced ER in an in vitro GST pull-down assay. Protein binding to GST–SRC1 was identified by Western blotting analysis using monoclonal antibodies directed against cyclin D1 and ER. GST protein served as a control for binding specificity. In agreement with several earlier studies, we found that ER binds to GST–SRC1 in a ligand-dependent manner in vitro (Cavailles et al. 1994) (Fig. 6A). Significantly, cyclin D1 could also recruit ER to GST–SRC1 in the absence of ligand (Fig. 6A). These data indicate that cyclin D1 can cause ER activation by acting as a ligand-independent adapter molecule between ER and its coactivator SRC-1.
Figure 6.
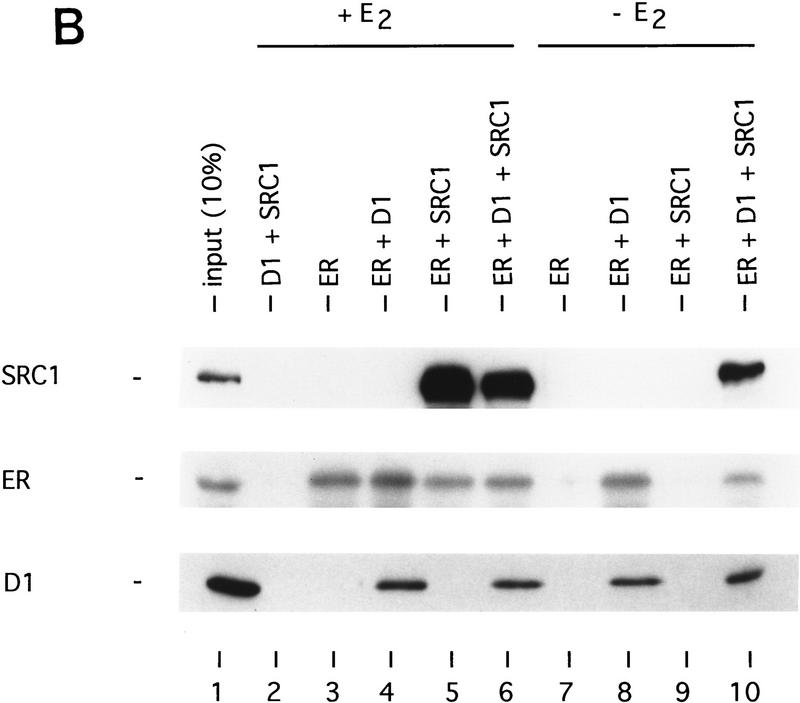
Cyclin D1 mediates ligand-independent recruitment of SRC-1 to ER. (A) Ligand-independent in vitro binding of SRC-1, ER, and cyclin D1. The purified proteins GST–SRC1, His-tagged cyclin D1, and baculovirus-produced ER were tested for in vitro binding in a GST pull-down assay. GST protein served as negative control. Cyclin D1 and ER were incubated with GST–SRC1 in the presence or absence of 1 μm 17β-estradiol and binding was detected by Western blot analysis using anti-cyclin D1 and anti-ER monoclonal antibodies. Lane 1 represents 10% of the input for cyclin D1 and 20% of input of ER proteins. (B) Cyclin D1 and SRC-1 can interact with DNA-bound ER. Oligonucleotide-containing ER binding sequence was biotin 5′-end labeled and bound to paramagnetic particles coated with streptavidin. Purified GST–SRC1, baculovirus-produced ER, and His-tagged cyclin D1 proteins were tested for DNA binding using these ERE-containing beads and analyzed by Western blotting using antibodies directed against GST, ER, and cyclin D1, respectively.
To test whether a ternary complex can also be formed when ER is bound to its cognate DNA-binding site (the ERE), we performed a similar ternary complex assay with purified proteins as described above, with the modification that the ternary complex was pulled down with ERE oligonucleotides coupled to paramagnetic beads. Figure 6B shows that ER binds to ERE in the presence of ligand (lanes 3–6), but also in the absence of ligand when cyclin D1 is present (lanes 8 and 10). Therefore, cyclin D1 allows ligand-independent DNA binding of ER to its ERE. More importantly, this assay clearly shows that a ternary complex consisting of ER, cyclin D1, and ER can be formed on DNA both in the presence and in the absence of 17β-estradiol (Fig. 6B, lanes 6 and 10). These data suggest that cyclin D1 is a bridging factor between ER and SRC-1 also when ER is bound to DNA.
The role of cyclin D1 in ER activation in breast cancer
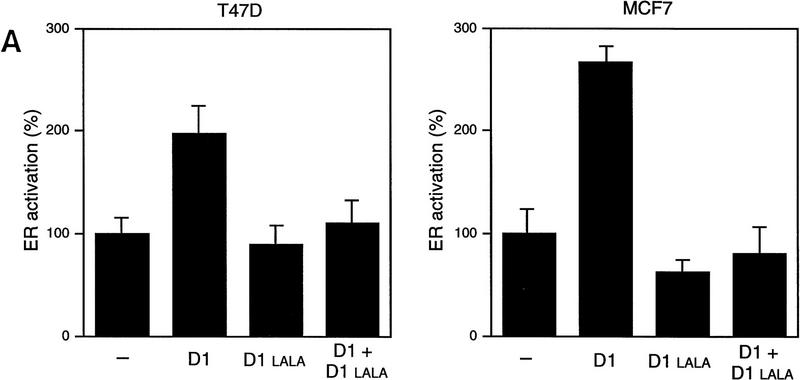
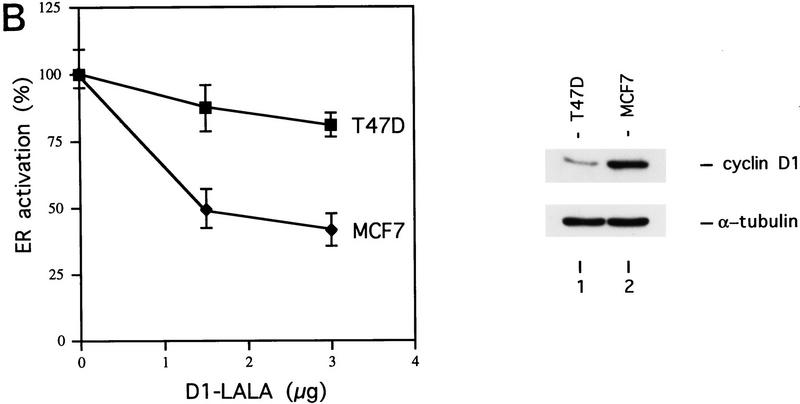
To get more insight in the role of cyclin D1 in ER transactivation in breast cancers with elevated levels of cyclin D1, we tested the effect of the mutant cyclin D1 L254/255A on ER activation in two different breast cancer cell lines. This cyclin D1 mutant does bind to ER (Fig. 4B) but fails both to interact with SRC-1 (Fig. 4A) and consequently fails to activate ER (Fig. 1B). When co-expressed with wild-type cyclin D1, the mutant cyclin D1 L254/255A inhibited the cyclin D1-mediated activation of ER in both T47D and in MCF-7 breast cancer cells (Fig. 7A). Therefore, this mutant acts as a dominant negative in cyclin D1-mediated ER transactivation. We tested the effect of this cyclin D1 mutant on ER activation in two breast cancer cells that contain endogenous wild-type ER but differ in their cyclin D1 protein levels (Fig. 7B). In T47D, which contains relative low levels of cyclin D1, co-expression of this cyclin D1 mutant resulted in a slight inhibition of ER activity. In contrast, in MCF-7, which contains relative high levels of cyclin D1 (Fig. 7B), the cyclin D1 mutant inhibited ≤50% of ER activity. Therefore, this dominant-negative cyclin D1 mutant preferentially interferes with ER activation in breast cancer cells that have high levels of cyclin D1 protein. These data strongly support the notion that elevated cyclin D1 protein levels in breast cancer contribute significantly to ER activation.
Figure 7.
Role of cyclin D1 in ER transactivation in breast cancer cells. (A) Dominant-negative activity of cyclin D1‘LALA’. The ER-containing T47D and MCF-7 breast cancer cells were maintained in 17β-estradiol-enriched medium with 10% fetal calf serum after cotransfection with cyclin D1 (1.5 μg) in the presence and absence of the cyclin D1 L254/255A mutant (cyclin D1–LALA, 1.5 μg), together with an ERE-reporter gene for testing its effect on ER activation. ER transactivation in the absence of coexpression of cyclin D1 was set at 100%. (B). Cyclin D1–LALA inhibits ER activation preferentially in cyclin D1-overexpressing breast cancer cells. T47D and MCF-7 breast cancer cells were maintained in 17β-estradiol-enriched medium with 10% fetal calf serum and transfected with increasing amounts of cyclin D1–LALA expression vector. ER activity was measured by cotransfection with the ERE-reporter plasmid. Transactivation in the absence of co-expression of the cyclin D1–LALA was set at 100%. (Right) The expression levels of endogenous cyclin D1 in both breast tumor cell lines compared with α-tubulin, which served as an internal control.
Discussion
This study indicates that cyclin D1 can act as a bridging factor between ER and SRCs, which allows the formation of a transcriptionally active ternary complex in the absence of ligand (Fig. 8). It is generally thought that coactivator recruitment by nuclear receptors results from a ligand-induced conformational change in the AF-2 domain of the receptor (Renaud et al. 1995; Brzozowski et al. 1997). Our present data for the first time show an alternative route of coactivator recruitment to ER that can take place in the absence of ligand. As such, these data reveal a novel mechanism of ER activation and establish a direct role for cyclin D1 in regulation of transcription.
Figure 8.
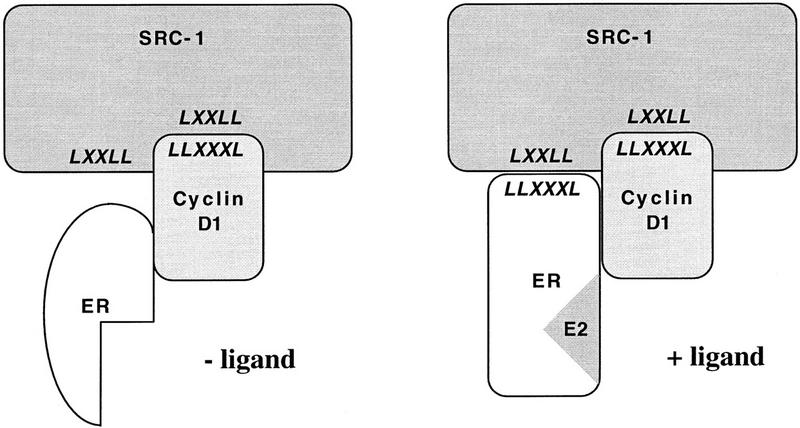
Model for cyclin D1-mediated ER transactivation. In the absence of ligand, ER is unable to interact with SRCs directly as its leucine-rich coactivator interaction motif is sterically unavailable for interaction. Ligand-independent binding of cyclin D1 to ER provides a single leucine-rich interaction motif for SRCs which is present in the carboxyl terminus of cyclin D1. This results in partial activation of ER (left). Subsequent ligand binding of ER induces a conformational change in ER that also exposes the leucine-rich motif in AF-2 of ER for SRC interaction, allowing higher affinity binding of SRCs to the liganded D1/ER complex (right). The observed synergism between estradiol and cyclin D1 in ER activation results from their cooperative recruitment of SRCs to the D1/ER complex. The protein interaction motifs are shown in italics.
Our work led to the identification of a novel functional domain in the carboxyl terminus of cyclin D1 that mediates direct binding to SRCs like SRC-1 and AIB-1. This leucine-rich motif of cyclin D1 is very similar in character to the ligand-regulated SRC-binding motif that is present in helix 12 of ER and in many other nuclear receptors. Several lines of experimental evidence indicate that the leucine-rich motif of cyclin D1 is required for coactivator recruitment to ER and subsequent activation of ER. First, cyclin D1 interacts directly with SRC-1 both in vivo and in vitro and introduction of two point mutations in this motif of cyclin D1 abolishes SRC-1 interaction and prevents ER activation by cyclin D1 (Figs. 1 and 4). Second, cyclins D2 and D3 have only a partial conservation of the leucine-rich motif and are hardly active in ER activation (Neuman et al. 1997; Zwijsen et al. 1997). Third, a dominant-negative mutant of SRC-1 prevents cyclin D1 activation of ER, indicating that SRCs are required for cyclin D1-mediated activation of ER (Fig. 3). Fourth, in in vitro binding studies, cyclin D1 could recruit ER to SRC-1 in the absence of ligand (Fig. 6). Together these data suggest a model in which cyclin D1 can recruit SRCs to ER, which results in a transcriptionally productive interaction between ER and its coactivators.
The functional similarity between the leucine-rich motifs of cyclin D1 and ER is also supported by structural analysis. Even though a crystal structure of cyclin D1 is not available at present, the crystal structures of cyclins A and H have been solved (Jeffrey et al. 1995; Andersen et al. 1997). Alignment of the sequence of cyclin D1 with cyclin A and comparison with the structures of cyclins A and H indicates that the leucine-rich motif in the carboxyl terminus of cyclin D1 aligns at the carboxyl terminus of helix 5′ of cyclins A and H. Importantly, the PHD program indicates that this part of cyclin D1 has a >90% probability to be α-helical and is markedly amphipathic (Rost and Sander 1993). Because the leucine-rich coactivator binding motif of ER is also an amphipathic helix, it is possible that the leucine-rich motifs of cyclin D1 and ER are capable of making similar protein interactions with SRCs.
Consistent with the notion that cyclin D1 and ER have a similar coactivator interaction surface, we found that binding of cyclin D1 to SRC-1 also requires the highly conserved LxxLL motifs in SRC-1 (Fig. 5). These motifs were recently shown to mediate binding to the leucine-rich coactivator binding site in the amphipathic helix 12 of ER (Le Douarin et al. 1995; Heery et al. 1997; Torchia et al. 1997). Significantly, depending on the pattern of splicing, SRC-1 has three or four LxxLL motifs, three of which are in close proximity (Kalkhoven et al. 1998). In principle, this could allow simultaneous interaction of SRC-1 with the leucine-rich motifs of both ER and cyclin D1. Consistent with this, we observed that a peptide that spans the third LxxLL motif of SRC-1 competed most efficiently the binding between cyclin D1 and SRC-1, whereas the second LxxLL motif of SRC-1 is the preferred site of interaction for ER (Fig. 5C; Heery et al. 1997; Kalkhoven et al. 1998). Based on these observations, we propose that in the absence of ligand, expression of cyclin D1 provides a single interaction site for coactivators on the cyclin D1/ER complex as both binding of cyclin D1 to ER and binding of cyclin D1 to SRC-1 is ligand-independent. This provides a rationale for the ligand-independent activation of ER in the presence of high levels of cyclin D1 (Zwijsen et al. 1997). After ligand binding of ER, the leucine-rich domain in AF-2 is exposed, which constitutes a second binding site for SRCs. The presence of two SRC-1 binding sites on the liganded cyclin D1/ER complex provides a rationale for the observed synergism between estradiol and cyclin D1 in ER activation (Fig. 8; Zwijsen et al. 1997).
The model represented in Figure 8 does not take into account that binding of cyclin D1 to ER also allows ligand-independent DNA binding by ER in vitro and in vivo (Fig. 6B; Zwijsen et al. 1997). Therefore, cyclin D1 can not only stimulate coactivator recruitment to ER but also act to enhance DNA binding of ER. Therefore, the synergistic action between cyclin D1 and ligand in ER activation may also be attributable, in part, to synergistic induction of ER DNA binding (Fig. 2; Zwijsen et al. 1997).
This study showed that cyclin D1 can bind to SRC-1 and AIB-1 but not to p300 (Fig. 4A). Therefore, cyclin D1 can discriminate between the different coactivator families. Apparently, a LxxLL motif (present in both SRCs and p300) is required for cyclin D1 binding, but flanking regions contribute to binding specificity (see also Fig. 5B). The finding that cyclin D1 interacts with at least two members of the SRC family, SRC-1 and AIB-1, would allow, in principle, for promiscuous activation of steroid receptors by cyclin D1. Cyclin D1, however, does not activate the progesterone receptor, nor a number of other steroid hormone receptors (Zwijsen et al. 1997; R.M.L. Zwijsen and R. Bernards, unpubl.). It is likely that the ability of cyclin D1 to interact with ER directly contributes to the specificity of nuclear receptor activation by cyclin D1.
To date, interaction between nuclear receptors and steroid receptor coactivators like SRC-1 has not been detected in vivo in the absence of overexpression. This is probably the result of the low affinity of the hydrophobic interactions between the leucine-rich motifs in both proteins and the short half-life of the SRC family proteins. This may also explain why a cyclin D1/SRC-1 complex is not readily detectable in the absence of transient overexpression.
Cyclin D1 is an important regulator of growth and differentiation of breast epithelium (Musgrove et al. 1994; Wang et al. 1994; Fantl et al. 1995; Sicinski et al. 1995; Zwijsen et al. 1996; van Diest et al. 1997). Significantly, both the genes encoding cyclin D1 and the SRC AIB-1 are amplified or overexpressed frequently in breast cancer (Schuuring et al. 1992b; Buckley et al. 1993; Gillett et al. 1994; Anzick et al. 1997; van Diest et al. 1997). Because this study indicates that both cyclin D1 and SRCs are components of a multimeric complex involved in ER-mediated transcription, it is conceivable that overexpression of limiting factors in this complex results in deregulation of ER-mediated growth. In agreement with this, we found that a mutant of cyclin D1 that can bind to ER but fails to recruit coactivators acted as a dominant-negative mutant for ER activation primarily in breast cancer cells with elevated levels of cyclin D1 (Fig. 7). Therefore, cyclin D1 is likely to contribute significantly to ER activation in breast cancers in which the protein is overexpressed. An important question that we wish to address next is how much of the oncogenic activity of cyclin D1 in breast cancer is mediated through the ‘classical’ cdk4 route and how much through ER activation. The availability of specific mutants of cyclin D1 in which these activities can be separated should allow us to assess the contribution of each of these two activities of cyclin D1 to mammary carcinogenesis separately.
Materials and methods
Cell culture and transient transfection assays
Cos-7 cells and U2-OS cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. Twenty-four hours before transfection, cells were maintained in DMEM without phenol red containing 5% charcoal-treated fetal bovine serum (FBS). Cells were transfected with 3 μg of ERE–TATA–luciferase expression vector, 500 ng of β-galactosidase expression vector (internal control), 200 ng of ER expression plasmid and 2.5 μg of cyclin D1, coactivators and/or empty vectors as indicated. After 16 hr, cells were rinsed in PBS and re-fed with fresh medium and ligand (10 nm 17β-estradiol) or vehicle was added. One day later, cells were harvested and assayed for luciferase and β-galactosidase activities. β-Galactosidase activity was used to correct for differences in transfection efficiency.
Immunoprecipitation and Western blotting
Cells were lysed in ELB containing 250 mm NaCl, 0.1% NP-40, 50 mm HEPES at pH 7.0, 5 mm EDTA, and protease inhibitors. The cell lysate was precleared three times with 5 μl of normal mouse serum coupled to protein A–Sepharose beads. For immunoprecipitations, the supernatant was incubated with 100 μl of 12CA5 hybridoma supernatant or 10 μl of monoclonal antibody to the ER (TE111.5D11, Neomarkers), which was coupled to protein A–Sepharose beads at 4°C. After 1 hr, beads were washed in ELB buffer and boiled in Laemmli buffer. Samples were separated on a 10% SDS/polyacrylamide gel and transferred to nitrocellulose. After blocking with PBS containing 5% milk and 0.1% Tween 20, proteins were detected with monoclonal antibodies directed against cyclin D1 (DCS-6, Neomarkers) and peroxidase-conjugated goat anti-mouse IgG. The blots were washed in PBS containing 0.1% Tween 20 and developed by enhanced chemiluminescence (ECL) reactions (Amersham).
DNA-binding assay
For the DNA-binding assay, we used DNA affinity beads coated with streptavidin (Dynal A/S) and (5′-biotin-labeled) DNA oligonucleotides containing a binding sequence for ER as described before (Zwijsen et al. 1997). The complementary DNA strands were annealed in TE buffer containing 100 mm KCl at 75°C for 10 min followed by cooling to room temperature over a period of 2 hr. Dynabeads were mixed with biotinylated oligonucleotides in TE buffer containing 1 m NaCl for 15 min, washed and incubated with cell extract in 8 mm Tris-phosphate at pH 7.4, 0.12 KCl, 8% glycerol, 4 mm DTT, and 0.5% CHAPS for 1 hr at 4°C. Subsequently, beads were washed in 20 mm HEPES at pH 7.7, 50 mm KCl, 20% glycerol, and 0.1% NP-40. The beads were boiled in Laemmli buffer and the proteins were separated on 10% polyacrylamide gels and identified by Western blotting.
GST pull-down and peptide competition assay
GST protein, GST–SRC1 fusion protein, and His-tagged cyclin D1 protein were purified as described previously (Zwijsen et al. 1997). Binding between 500 ng GST–SRC1 and 100 ng His–D1 was performed in binding buffer (50 mm NaCl, 50 mm HEPES–KOH at pH 7.6, 0.1 mm 0.1% (wt/vol) NP-40, 0.1 mm PMSF, 0.5% charcoal-stripped serum) bound to glutathione–Sepharose for 1 hr at 4°C. The beads were washed three times and bound proteins were eluted by boiling for 10 min in sample buffer and separated on 10% SDS–polyacrylamide gels. The binding of His–cyclin D1 to GST–SRC1 was detected by Western blot analysis using monoclonal antibodies directed against cyclin D1 (DCS-6, Neomarkers). For testing a ternary complex, a baculovirus-produced ER (750 ng, Pan Vera) was added to GST–SRC1 (500 ng) and His–D1 (100 ng) in the presence or absence of 1 μm 17β-estradiol using similar conditions as described above. In Western blot analysis, monoclonal antibodies directed against cyclin D1 (DCS-6, Neomarkers) and ER (LH2, Novocastra) were used.
For the peptide inhibition assay, 150 ng GST/GST–SRC1 and 50 ng of His-tagged cyclin D1 were used. Peptides were pre-incubated with target protein for 40 min at room temperature, before addition of the bait. A mixture of GST fusion peptides, His-tagged cyclin D1, and peptides was incubated for an additional 20 min at room temperature. The amounts of peptide added in competition studies were 0.3 and 3 μg (Fig. 5B) or 0.1, 0.2, 0.4, and 0.8 μg (Fig. 5C).
Acknowledgments
We thank B. O’Malley, P. Chambon, N. La Thangue, M. Parker, H. Gronemeyer, and B. Katzenellenbogen for the kind gift of plasmids; L. Vernie for peptide synthesis; and T. Sixma for help in protein structure analyses. This work was supported by a grant from the Dutch Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Bernards@nki.nl; FAX (31) 20-512 1954.
References
- Andersen G, Busso D, Poterszman A, Hwang JR, Wurtz JM, Ripp R, Thierry JC, Egly JM, Moras D. The structure of cyclin H: Common mode of kinase activation and specific features. EMBO J. 1997;16:958–967. doi: 10.1093/emboj/16.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai MJ, O’Malley BW. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- Beijersbergen RL, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochem Biophys Acta Rev Cancer. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson J-A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- Buckley MF, Sweeney KJ, Hamilton JA, Sini RL, Manning DL, Nicholson RI, deFazio A, Watts CK, Musgrove EA, Sutherland RL. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8:2127–2133. [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, Danielian PS, Parker MG. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Danielian PS, White R, Lees JA, Parker MG. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantl V, Stamp G, Andrews A, Rosewell I, Dickson C. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes & Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, Peters G. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994;54:1812–1817. [PubMed] [Google Scholar]
- Gillett C, Smith P, Gregory W, Richards M, Millis R, Peters G. Cyclin D1 and prognosis in human breast cancer. Int J Cancer. 1996;69:92–99. doi: 10.1002/(SICI)1097-0215(19960422)69:2<92::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ing NH, Beekman JM, Tsai SY, Tsai MJ, O’Malley BW. Members of the steroid hormone receptor superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich N. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Jenster G, Spencer TE, Burcin MM, Tsai SY, Tsai MJ, O’Malley BW. Steroid receptor induction of gene transcription: a two-step model. Proc Natl Acad Sci. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhoven E, Valentine JE, Heery DM, Paerker MG. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Glass B, Lin S-C, Heyman RA, Rose DW, Glass CK, Rosenfeld MG. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- Le Douarin B, Zechel C, Garnier J-M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–388. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Musgrove EA, Lee CS, Buckley MF, Sutherland RL. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci. 1994;91:8022–8026. doi: 10.1073/pnas.91.17.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman E, Ladha MH, Lin N, Upton TM, Miller SJ, DiRenzo J, Pestell RG, Hinds PW, Dowdy SF, Brown M, Ewen ME. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: Evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Sadovsky Y, Webb P, Lopez G, Baxter JD, Fitzpatrick PM, Gizang GE, Cavailles V, Parker MG, Kushner PJ. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol Cell Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuuring E, Verhoeven E, Mooi WJ, Michalides RJ. Identification and cloning of two overexpressed genes, U21B31/PRAD1 and EMS1, within the amplified chromosome 11q13 region in human carcinomas. Oncogene. 1992a;7:355–361. [PubMed] [Google Scholar]
- Schuuring E, Verhoeven E, van Tinteren H, Peterse JL, Nunnink B, Thunnissen FB, Devilee P, Cornelisse CJ, van de Vijver M, Mooi WJ, et al. Amplification of genes within the chromosome 11q13 region is indicative of poor prognosis in patients with operable breast cancer. Cancer Res. 1992b;52:5229–5234. [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, Haslam SZ, Bronson RT, Elledge SJ, Weinberg RA. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Smith CL, Onate SA, Tsai MJ, O’Malley BW. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, McKenna NJ, Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Tzukerman MT, Esty A, Santiso MD, Danielian P, Parker MG, Stein RB, Pike JW, McDonnell DP. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol Endocrinol. 1994;8:21–30. doi: 10.1210/mend.8.1.8152428. [DOI] [PubMed] [Google Scholar]
- van Diest PJ, Michalides RJ, Jannink L, van der Valk P, Peterse HL, de Jong J, Meijer CJ, Baak JP. Cyclin D1 expression in invasive breast cancer. Correlations and prognostic value. Am J Pathol. 1997;150:705–711. [PMC free article] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJS, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation of AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- vom Baur E, Zechel C, Heery D, Heine MJ, Garnier JM, Vivat V, Le DB, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- White R, Sjoberg M, Kalkhoven E, Parker MG. Ligand-independent activation of the oestrogen receptor by mutation of a conserved tyrosine. EMBO J. 1997;16:1427–1435. doi: 10.1093/emboj/16.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwijsen RM, Klompmaker R, Wientjens EB, Kristel PM, van der Burg B, Michalides RJ. Cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol Cell Biol. 1996;16:2554–2560. doi: 10.1128/mcb.16.6.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwijsen RML, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJAM. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]