Abstract
Considerable energetic investment is devoted to altering large stretches of chromatin adjacent to DNA double strand breaks (DSBs). Immediately ensuing DSB formation, a myriad of histone modifications are elicited to create a platform for inducible and modular assembly of DNA repair protein complexes in the vicinity of the DNA lesion. This complex signaling network is critical to repair DNA damage and communicate with cellular processes that occur in cis and in trans to the genomic lesion. Failure to properly execute DNA damage inducible chromatin changes is associated with developmental abnormalities, immunodeficiency, and malignancy in humans and in genetically engineered mouse models. This review will discuss current knowledge of DNA damage responsive histone changes that occur in mammalian cells, highlighting their involvement in the maintenance of genome integrity.
Keywords: DNA Damage Response, Histones, Chromatin, Genome Stability Characters
1. Introduction
The DNA damage response (DDR) represents a specific type of cellular stress response that is initiated by lesions emanating from genomic DNA. While there are many types of DNA damage, each with their own cognate recognition and repair, DNA double strand breaks (DSBs) pose a particular threat to genome integrity as evidenced by the number of human pathophysiologic conditions that occur due to DSB repair deficiency[1,2].
The mammalian genome is comprised of approximately 2 meters of DNA, which is tightly packaged into a nucleoprotein structure known as chromatin. The basic subunit of chromatin is the nucleosome, consisting of approximately 146 base pairs of DNA wound around an octameric histone core, comprised of a dimer of heterotetrameric histones H2A, H2B, H3, and H4. A linker histone H1 may also be present in the internucleosome linker DNA region and can have a profound influence on nucleosome interactions and higher order chromatin structure. Chromatin structure is altered by covalent modification of N- or C-terminal histone tails, which extend from the nucleosome unit. These modifications are synthesized by numerous different histone modifying enzymes in response to a variety of external and internal stimulatory cues[3]. Histone modifications are then recognized by a growing number of chromatin associated proteins that contain evolutionarily conserved binding domains that display specificity for modified histone tail residues. Conversely, enzymes that selectively remove the modification can reverse histone tail modifications. Collectively, these proteins can be thought of as writers, readers, and erasers of the DNA damage responsive “histone code”[4].
DSB recognition and repair are initiated within the first minute of damage induction, revealing temporal and spatial assembly of DNA repair protein complexes at the genomic lesion [5,6]. Fluorescence-based imaging of local damage responses demonstrates the presence of repair factors in intranuclear foci (denoted as ionizing radiation induced foci or IRIF)[7]. IRIF are easily visible on light microscopy due to their presence in supra-stoichiometric ratios to DSB number. High local concentrations of repair protein complexes are postulated to efficiently signal repair and checkpoint responses, as well as maintain sister chromatid cohesion for homology directed DNA repair reactions.
Although not fully appreciated at the time, IRIF provided the initial clues that local chromatin modifications were required for DSB targeting of repair proteins. The seminal discovery by Bonner and colleagues that histone H2A variant, H2AX is rapidly phosphorylated at the γ position in its C-terminal tail (γH2AX) along chromatin tracks flanking DSBs [8,9], paved the way to study IRIF assembly. The Phosphatidylinositol-3 kinase-related kinases (PIKK) ATM (ataxia telangiectasia mutated) and DNA-PK phosphorylate H2AX within minutes of DSB induction at its C-terminus on Ser139[10,11]. Phospho H2AX (γ–H2AX) formation was originally proposed to extend for a megabase from the site of the break in mammalian cells[9]. Higher resolution Chromatin-IP studies in mammalian cells are consistent with these original predictions, albeit with a nonuniform response over the entire length of modified chromatin [12-15]. Indeed, γH2AX proved to be an essential chromatin modification necessary for IRIF formation, as genetically engineered H2AX null mouse cells have dramatically impaired damage inducible focus formation for many DNA repair factors [16,17].
Cellular responses to DSBs involve considerable energetic investment, much of it dedicated to modifying chromatin in cis to the inciting DNA lesion. Here, it is described that extensive modification of chromatin in cis to DSBs not only enables efficient DNA repair, but also allows communication between DSBs and transcriptional responses that resides in cis and in trans within the genome[18]. Such communication may be necessary to maintain genome integrity and to specify cellular states that ensue in response to DNA damage.
2. Histone modifications and the DNA damage Response
2.1 Writers, Readers, and Erasers of the DSB associated chromatin response
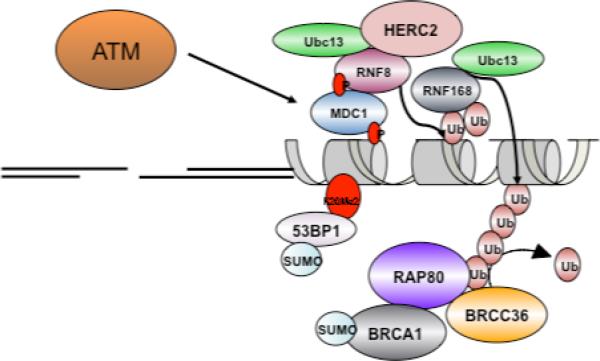
Histone tails on chromatin adjacent to DSBs are covalently altered by numerous post-translational modifications including phosphorylation, ubiquitylation, SUMOylation, methylation, poly(ADP)ribosylation (PAR), and acetylation (Figure 1)[19]. Specific reader proteins exist for each of these marks, utilizing a variety of binding domains to recognize their cognate histone tail modification and access DSB associated chromatin[20-22]. Two prominent examples of DNA repair reader proteins are the breast cancer early onset protein 1 (BRCA1), and the p53 binding protein 1 (53BP1). Each form IRIF in a γH2AX dependent manner and also possess prominent γH2AX independent roles in the DDR [16]. Although sharing γH2AX IRIF dependency, BRCA1 and 53BP1 display different IRIF kinetics and utilize distinct recognition elements for IRIF formation[23-26]. 53BP1 IRIF occur within minutes and utilize a Tudor domain to recognize methylated histone H4K20 residues, while BRCA1 relies on tandem BRCT domains that bind to several different proteins that are serine phosphorylated at a consensus SPXF sequence.
Figure 1. Assembly of DNA Repair Foci at DSBs.
ATM phosphorylates γH2AX, thereby initiating a series of chromatin modifications that are recognized by DNA repair protein complexes. Each DNA repair complex proteins contains binding domain with specificity for a particular modification. This enables stepwise assembly of DNA repair protein complexes over large stretches of chromatin flanking DSBs.
Despite these differences, BRCA1 and 53BP1 share a reliance on γH2AX dependent recruitment of E3 ubiquitin ligases that synthesize lysine63-linked ubiquitin (K63-Ub) chains at DSBs[27-32]. These K63-Ub chains are synthesized at DSBs by E3 ligases RNF8 and RNF168 in association with a K63Ub specific E2 conjugating enzyme Ubc13. The signaling cascade to ubiquitin is initiated by H2AX phosphorylation, which is then bound by the MDC1 protein. MDC1 appears to be the major reader of γH2AX, using its BRCT domains to specifically bind γH2AX and transduce nearly all of its signals[33-37]. MDC1 is extensively phosphorylated at a PIKK consensus SQ/TQ cluster enabling interaction with RNF8, a RING domain E3 ligase that uses its Forkhead-Associated domain (FHA) to bind phosphorylated MDC1[27,30,32]. DSB associated RNF8 binds to the K63-Ub specific E2 conjugating enzyme Ubc13 to begin an ubiquitin ligase cascade. RNF8-Ubc13 interaction is positively regulated by the giant E3 ligase HERC2[38], and negatively regulated by the DUB OTUB1, which binds to Ubc13, thereby limiting access of RING domain E3 ligases[39]. Ligase active RNF8-Ubc13 synthesizes ubiquitin docking sites for RNF168, another E3 ubiquitin ligase that contains ubiquitin binding domains. DSB localized RNF168 also synthesizes K63-Ub chains in conjunction with Ubc13. RNF168 is biallelically mutated in a rare human disorder known as RIDDLE Syndrome (Radiosensitivity Immune Deficiency Developmental Delay)[40]. Cells from these patients lack K63-Ub, BRCA1, and 53BP1 at IRIF[31,40]. Histones H2AX and H2A, known substrates of both ligases, show diminished ubiquitination in RNF8 and RNF168 deficient cells. Interestingly SUMO E3 ligases PIAS1 and PIAS4 are also required for efficient DSB ubiquitylation by RNF8, RNF168, and BRCA1. SUMOylation leads to enhancement of their respective E3 ubiquitin ligase activities, and this is thought to be necessary for efficient DSB associated ubiquitylation[41,42]. The culmination of these reactions creates K63-Ub chains for recognition by ubiquitin binding domain proteins.
BRCA1 interacts with RAP80 (Receptor Associated Protein 80), a protein containing tandem ubiquitin interaction motifs (UIM), in a 6-member protein complex[43-49]. The RAP80 UIM domains specifically recognize K63-ubiquitinated chromatin adjacent to DSBs[43]. The BRCA1-RAP80 complex also contains a K63-Ub specific deubiquitinating enzyme (DUB), BRCC36, which opposes K63-Ub synthesis by RNF8 and RNF168 at DSBs[43,50,51], providing a built-in mechanism to either terminate or edit DSB ubiquitin responses. RAP80 or BRCC36 deficient cells show elevated K63-Ub at DSBs, including higher levels of K63-ubiquitinated histone H2A[50,52,53]. RAP80 is also K63-ubiquitinated in an Ubc13 dependent manner and is a substrate of BRCC36 DUB activity[52] consistent with the notion that DSB ubiquitination is heterogeneous and dictated by a dynamic equilibrium between opposing E3 ubiquitin ligase and DUB activities[21,28,50,54]. RAP80 is among the first of several proteins that utilize ubiquitin binding to recognize DSBs[55], although not the first protein reported to recognize ubiquitin at DNA damage sites. Ubiquitin binding domains are utilized by the Y-family of DNA translesion polymerases to recognize ubiquitinated PCNA at stalled replication forks[56-58], suggesting ubiquitin as a common recognition element for several types of DNA repair.
It is less clear how ubiquitin chains facilitate 53BP1 DSB accumulation. While it was previously thought that H4K20Me2 did not become enriched at DSBs, recent findings suggest otherwise. The histone methyltransferase MMSET is recruited to DSBs in a γH2AX/MDC1 dependent manner to synthesize H4K20Me2[59]. This activity was necessary for 53BP1 DSB recruitment and DNA repair. How RNF8 and RNF168 dependent DSB associated ubiquitin comes into play during this process is still unknown, since RNF8 was not required for MMSET DSB localization or DSB H4K20Me2 enrichment. A plausible, albeit unproven, explanation is that RNF8/RNF168 dependent ubiquitination would relax chromatin constraints to make H4K20Me2 marks more accessible for binding by the 53BP1 Tudor domain.
The functional consequence of ubiquitin and methylated histone recognition by repair proteins at DSBs is still a matter of ongoing investigation. RAP80 or 53BP1 deficiency creates a mild hypersensitivity to IR and other agents that create DSBs. 53BP1 null mice generally exhibit mild DNA repair phenotypes, but display a severe impairment in immunoglobulin rearrangements during class switch recombination, suggesting a failure to join DSB ends that contain large stretches of intervening sequence[16,60,61]. A presumably related finding is that 53BP1 deficiency prevented long range end joining interactions between deprotected telomeres[62]. Both of these findings implicate 53BP1 in allowing movement of DSB termini in nuclear space for end joining reactions.
Interestingly, 53BP1 deficiency rescued BRCA1 mutant cells from genomic instability concomitant with a restoration of DSB end resection, homology directed repair, and resistance to PARP inhibitors[63-65]. To date, 53BP1 is the only known DNA repair gene to exhibit this type of genetic interaction with BRCA1 mutation. These important findings suggest that chromatin bound 53BP1 plays a contributory role to genomic instability and breast and ovarian cancer susceptibility in the context of BRCA1 mutations. RAP80 deficiency also leads to excessive end resection[66,67], although RAP80 ablation exacerbated DNA repair in BRCA1 null cells[47,66,68], indicating clear distinctions between RAP80 and 53BP1. In addition, H2AX knockout cells display excessive end processing[69,70], suggesting that readers of γH2AX dependent ubiquitin modifications help to limit DSB end resection.
Poly(ADP)ribosylation of histone tails is an additional histone modification that has garnered much attention due to the finding that PARP inhibitors show synthetic lethal interactions in BRCA1 and BRCA2 mutated cells[71,72]. This synthetic lethality has borne promising results in the treatment of BRCA mutated cancers in human clinical trials[73]. PARP1 is rapidly and transiently recruited to DSBs to PARylate histones and other substrates. A number of PAR binding domains exist to allow protein recruitment to DNA damage sites. One example is the nucleosome remodeling complex NuRD that requires PARP activity for DSB recruitment[74]. Another chromatin remodeling protein, ALC1 (Amplified in Liver Cancer 1) uses a macro domain to access DNA repair sites and initiate nucleosome sliding[75]. These activities were required for resistance to DNA damaging agents. Finally XRCC1 uses its BRCT domain to access PARylated histones at DSBs for non-homologous end joining type DNA repair[76].
3. How to deal with pre-existing chromatin states
In simple terms, chromatin is defined as either heterochromatic or euchromatic based on simple staining procedures that can be visualized by light or electron microscopy. Chromatin IP coupled to next generation sequencing (ChIP-Seq) approaches has revealed high-resolution portraits of genome wide chromatin modifications, known as the epigenome[77]. Transcriptionally silent heterochromatin and euchromatin possess characteristically different histone modifications. While the total number of chromatin modifications present in the epigenome is well beyond the scope of this review, repressive histone marks commonly demonstrate histone H3 methyl lysine 9 and lysine 27 modifications, Histone H2A ubiquitination at lysine 119 and minimal histone acetylation. Moreover, heterochromatin commonly displays extensive cytosine methylation at CpG dinucleotide clusters within promoter regions. Each of these modifications contributes to the transcriptionally silent status of heterochromatin. Alternatively, active chromatin frequently displays histone H3 and H4 acetylation, lacks methylation on H3 residues 9 and 27 as well as CpG island methylation, and shows H2B ubiquitination. RNA PolII is also associated with characteristic histone modifications. Histone H3 is trimethylated at lysine 4 surrounding transcription start sites, and actively elongating RNA PolII leads to chromatin decondensation and the deposition of histone H3K36 methylation throughout the gene body.
3.1 Dealing with chromatin during repair Part I: Repair in heterochromatin
Nuclease accessibility experiments reveal that heterochromatin is densely packed, while euchromatin can be readily digested to mono and dinucleosomes because its relaxed structure permits nuclease accessibility to internucleosomal DNA. Such observations lead to a prediction that heterochromatin and euchromatin would display different requirements for DNA repair. In line with this assertion is the observation that in both yeast and human cells, heterochromatin displays resistance to γH2AX formation[78]. Moreover, ATM deficiency resulted in persistent DNA damage in regions bordering large heterochromatic domains in mouse and human cells, suggesting that repair in heterochromatin instills a requirement for ATM dependent chromatin relaxation[79].
Recent insights shed light on how the damage response deals with pre-existing chromatin states. DSB responses within heterochromatin are facilitated by TIP60 chromodomain binding to trimethylated H3K9 residues[80]. This activates TIP60 histone acetyltransferase (HAT) activity to acetylate ATM and histone H4 lysine residues to direct an ATM dependent DDR to the vicinity of DSBs within heterochromatin. TIP60 dependent ATM activation therefore requires pre-existing H3K9 methylation and a DNA damage induced dissociation from methylated H3K9 residues by heterochromatin protein HP1β. This HP1β departure from heterochromatin is transient and requires Casein Kinase 2 dependent phosphorylation[81]. HP1β later returns to chromatin and has been reported to accumulate at DSBs[82]. More recent reports document that HP1α also accumulates rapidly and transiently at DSBs that occur in either heterochromatin or euchromatin, requiring the chromatin assembly cofactor p150CAF-1. HP1α deficiency led to reduced homologous recombination mechanisms of DSB repair and hypersensitivity to DNA damaging agents[83]. Activated ATM phosphorylates the heterochromatin associated protein KAP-1 (Krüppel-associated box (KRAB) domain-associated protein 1), which is involved in global relaxation of heterochromatin structures[84]. Either ATM or KAP1 deficiency prevented DNA repair in heterochromatin as did failure to activate TIP60 upon global loss of H3K9 trimethylation.
A modified picture has emerged with respect to DSB repair in heterochromatin in Drosophila[85]. Repair foci formed at similar rates in heterochromatin and in euchromatin in Drosophila cells, however, were more rapidly resolved in heterochromatin. Following end resection, heterochromatic DSBs exhibited extrusion into euchromatin domains for repair by homologous recombination (HR). Heterochromatin DSB repair depended on HR type repair mechanisms and on the SUMO ligase associated SMC5/6 complex. The authors provide evidence that heterochromatin decondensation permits migration of a heterochromatic DSBs to less dense chromatin regions prior to Rad51 nucleofilament formation for HR type repair. It was hypothesized that Rad51 dependent HR repair of highly repetitive DNA found in heterochromatin would occur in a distinct chromatin environment to prevent error prone homeologous repair between repetitive elements and loss of genetic information. It should be noted that Jeggo, Lobrich, and colleagues had previously implicated end resection and HR as a requisite feature of DSB repair within heterochromatin in G2 phase cells, suggesting that heterochromatin may dictate similar repair requirements in mammalian cells [86].
The findings in Drosophila are potentially paradigm changing on several fronts[85]. Namely, they challenge existing models that heterochromatin is refractory to γH2AX formation. Secondly, they imply that not all DSBs in metazoan species display positional stability. Finally, they suggest that DSBs in heterochromatin actually require a specialized form of HR. There are still many unanswered questions regarding these exciting findings. Are responses to heterochromatic DSBs conserved between insect cells and mammalian cells? As the authors point out, a detailed time course of repair foci kinetics with respect to heterochromatin landmarks would be necessary to discern if this is a conserved response. An additional question is whether the foci localization data accurately represent DNA repair? While the data are entirely consistent with this idea, the study did not directly examine repair in heterochromatin, instead using foci localization as a surrogate for repair. Diminished TUNEL positivity over time further supports the assertion that foci analysis is a fair representation of DNA damage[85].
Despite these reservations and the recent nature of this report, striking similarities exist in terms of genetic requirements for recombination-based mechanisms of telomere maintenance known as ALT (Alternative Lengthening of Telomeres). The SMC5/6 proteins form an evolutionarily conserved complex with the MMS21 SUMO E3 ligase. These proteins are required for SUMOylation of telomere binding proteins TRF1 and TRF2 in ALT dependent human cells[87]. This activity is required for movement of telomeres undergoing ALT to PML bodies for recombination. SMC5/6 was also required for Rad51 foci formation at the periphery of heterochromatin in Drosophila cells[85]. While it is not clear these processes are equivalent, the resemblance between DSBs in DAPI dense centromeric DNA and recombinogenic telomeres is uncanny. Both consist of repetitive, heterochromatic DNA that undergoes movement dependent HR type strand invasion reactions. These similarities are suggestive that at least some degree of evolutionary conservation exists between Drosophila heterochromatic DSB repair and repair of DNA damage in certain forms of heterochromatin in mammalian cells.
3.2. Dealing with chromatin during repair Part II: Repair in Euchromatin
An obvious difference between heterochromatin and euchromatin is the presence of active transcription. This difference has a profound impact on genome integrity and there is now evidence that it can explain some of the tissue specificity of genomic alterations found in malignancy. Transcriptional stimuli can promote specific types of genomic instability by enhancing proximity of genetic loci on different chromosomes. Prostate cancer displays characteristic translocations between the TMPRSS2 gene and several different ETS family genes[88]. Both are targets of androgen receptor suggesting a link between transcription and translocation propensity. Indeed, stimulation with dihydrotestosterone (DHT) induces proximity between these loci specifically in prostate epithelium, but not in androgen independent cells[89,90]. DHT alone enhances a detectable level of fusion product between loci as a result of a nonreciprocal translocation. These phenomena were dramatically enhanced when ionizing radiation was added during DHT stimulation.
The basis for transcription associated translocations seems to be multifactorial. Nuclear hormone induced transcription enhances proximity of noncontiguous genomic loci into regions described as transcription factories. RNA Pol II unwinding of DNA for transcription then generates DNA supercoiling related torsional stress, which is relieved by Topoisomerase mediated DNA nicking and DSB formation[91]. Topoisomerase induced DSBs result in γH2AX and DNA repair protein accumulation occurs in the vicinity of nuclear hormone receptor induced transcription start sites. These co-occurrences create a perfect storm of events necessary for non-reciprocal chromosomal translocations. Translocation rates positively correlated with the strength of transcriptional stimulus and required NHEJ type DNA repair [89]. Conversely either ATM deficiency or knockdown of HR related repair proteins dramatically increased translocation frequency. These findings indicate that genomic aberrations in cancer may not occur randomly, instead reflecting the transcriptional programs present in cells of origin for each cancer type.
A second feature of DNA repair specific to euchromatin comes from responses to DNA-RNA hybrids known as R-loops[92]. These hybrid structures are formed at elevated levels when processing of the nascent transcript is impaired. R-loop formation promotes transcription associated mutation and hyperrecombination, both causes of genomic instability. The underlying mechanisms, while still being resolved, are suggestive of higher rates of DSB formation. A siRNA screen for genes that prevent spontaneous γH2AX formation revealed a prominent involvement of splicing factors and regulators of nascent mRNA [93]. Evidence in support of an R loop intermediate came from the observation that γH2AX could largely be prevented by RNase H expression, an enzyme that specifically cleaves RNA-DNA hybrids. The pathway leading from R-loops to DSB formation will likely provide many new insights into transcriptional causes of genomic instability.
4. Chromatin responses and DSBs
4.1 DSB communication to transcription in cis
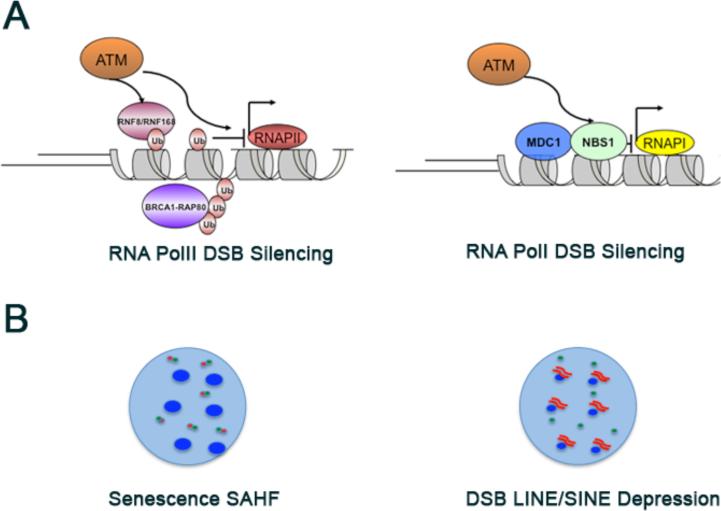
DSBs modify chromatin for up to a megabase adjacent to the lesion, posing the question of what happens to transcription in these flanking regions. The first attempts to address this question suggested that DSB responses extensively silence transcription within γH2AX chromatin domains (Figure 2). Nuclear run-on experiments showed an exclusion of nascent RNA surrounding γH2AX foci [94]. Similar findings were obtained upon monitoring rDNA transcription following IR. Treatment of mouse embryonic fibroblasts with IR led to a transient inhibition of RNA PolI dependent rDNA synthesis[95]. While it is unclear exactly where in the nucleolus the breaks occurred, a series of experiments conclusively showed that DNA damage caused a local shutdown of transcription. Laser induced DSBs targeted to one nucleolar region diminished transcription only within that region and did not affect transcription in trans at other nucleoli. DSB induced transcriptional silencing relied on an ATM, NBS1, and MDC1 dependent pathway, but surprisingly was H2AX independent. Recent reports indicate that RNA PolII dependent transcription is also inhibited by the presence of DSBs on adjacent chromatin. Using a novel reporter system that permits visualization of DSB responses and nascent transcription from a defined location in real time in single cells, it was demonstrated that persistent DSBs silenced transcription in cis for at least 4 kb away form the site of DNA damage [18]. RNA PolII accumulated at the transcription start site, but failed to efficiently enter an elongating state. DSB silencing was dependent on ATM kinase activity. ATM dependent DSB silencing was partially mediated by RNF8 and RNF168 dependent histone H2A ubiquitination, and failure to deubiquitylate H2A-Ub resulted in persistent silencing at the conclusion of repair.
Figure 2. DSBs communicate with transcription.
(A) ATM silences transcription in cis to DSBs[18,92-93]. ATM kinase activity initiates E3 ligase dependent histone H2A ubiquitination, which is responsible for silencing RNA PolII dependent along chromatin in cis to DSBs. ATM, NBS1, and MDC1 are required to silence RNA PolI transcription in the nucleoulus. (B) DSBs communicate to transcription in trans[102-107]. DSBs result in senescence associated heterochromatin formation and derepression of transcription in heterochromatin. (Left) Dysfunctional telomeres (red dots) are recognized as DSBs as evidenced by telomere associated γH2AX foci (adjacent green dots). Persistent telomere damage leads to senescence associated heterochromatin formation (large blue dots). (Right) DSB formation (green dots) results in depression of transcription including LINE and SINE elements (red lines) from heterochromatin (blue dots).
An important question is whether DSB silencing is reversible. The answer seems to be largely yes. RNA PolI dependent rRNA synthesis was restored in 2 hours after IR[95]. RNA PolII dependent nascent transcription also recovered commensurate with the timing of DSB repair[18]. However, there is credible evidence that DSB silencing can persist at a subset of DSBs. The first of which reported that 50% of HR repaired DSBs exhibit DNA methyltransferase 1 (DNMT1) dependent CpG island methylation and transcriptional gene silencing that persisted for many cell divisions in culture[96]. A second report documented histone deacetylase SIRT1 dependent epigenetic silencing occurred at nearly 1% of nuclease-induced DSBs[97]. Considering that tumors exhibit elevated γH2AX and DSB formation at early stages of development[98,99], even a small subset of persistent DSB silencing may lead to heritable chromatin changes and account for some of the epigenetic changes present in malignancy.
Given the span of γH2AX dependent DSB responses from a single DSB, a question of interest is how far DSB silencing extends. Would transcription be suppressed along the entire length of γH2AX chromatin domain in cis to a DSB? The answer to this question is still unclear. Nuclear run on experiments show the entire γH2AX focus to be transcriptionally silent[94]. However, this approach is limited by the resolution of light microscopy and does not examine gene expression from a defined locus. Defined DSBs at endogenous genomic locations has been reported using tamoxifen controlled ER fusion to restriction endonucleases[12,100]. One such system used the AsiSI restriction endonuclease to assess RNAPII occupancy within γH2AX domains[100]. The authors report that γH2AX was excluded from sites containing RNAPII near the transcriptional start site, and was re-established to full levels at 1-2 kb in both 5’ and 3’ directions. Steady state transcription was not affected at 4 hours after nuclease induction unless the DSB occurred very close to the gene body, leading the authors to conclude that DSB silencing does not extend far along γH2AX domains.
The findings with AsiSI seem to contradict nuclear run on experiments that DSBs silence transcription throughout the length of a DSB chromatin domain. However, a closer examination of the data suggests this may not be the case. RNAPI driven transcription was completely restored within 1-2 hours following 5 Gy IR at rDNA repeats in the nucleolus[95]. Complete restoration of transcription was also observed 1-2 hours following termination of nuclease action at a RNAPII driven reporter gene[18]. In both situations, DSB silencing would not have been detected at the 4 hour time point examined for AsiSI DSBs. Moreover, DSB silencing events do not prevent RNAPII association with transcriptional start sites[18,95], and it appears that many transcriptionally silent genes accumulate RNAPII in a promoter proximal state[101]. Collectively, these results suggest that stable silencing is readily detectable at persistent DSBs, but may be too transient for DSBs that are undergoing rapid repair.
A remarkable example of this phenomenon may be sex chromosome silencing in male germ cells. Meiotic sex chromosomes are epigenetically silenced in a process that is required for spermatogenesis known as meiotic sex chromosome inactivation (MSCI). During spermatogenesis, male X and Y chromosomes are unsynapsed and accumulate DDR factors in a manner that recapitulates assembly of chromatin responses surrounding DSBs in somatic cells. γH2AX, MDC1, BRCA1, ubiquitin, and SUMO decorate the entire length of the XY body[102,103]. MDC1 null spermatocytes display greatly diminished H2A-ubiquitin, SUMOylation, and γH2AX signals on X and Y meiotic chromosomes[104]. Interestingly, this results in a failure of MSCI and a 2-4 fold derepression of X and Y chromosome genes in mouse testis compared to MDC1 heterozygous counterparts. γH2AX null mice also fail to execute MSCI, indicating DSB silencing mechanisms require the γH2AX-MDC1 partnership at RNAPII driven genes in this context[103]. In addition, MDC1 was required for γH2AX amplification at sites of replication stress in somatic cells and these damage sites were associated with transcriptional silencing[104]. What still remains to be resolved are whether silencing occurs over large γH2AX chromatin domains at all DSBs, or rather a subset of them in somatic cells. Clearly, more investigation is warranted.
4.2 DSB Communication to transcription in trans
The combination of DNA sequence and chromatin structure controls gene expression and cell fate determination. This is beautifully illustrated by inducible pluripotency cell methodologies that allow conversion of terminally differentiated cells into pluripotent embryonic stem cells[105]. There is now compelling evidence that DSB responses can elicit cell fate changes as well by initiating chromatin alterations at sites distal to DNA damage (Figure 2). Replicative senescence occurs in response to telomere attrition due to the end replication problem. Senescence is characterized by an irreversible cell cycle arrest, distinct cell morphology, and gene expression changes.
Telomere damage induced senescence initiates a p53 and Rb dependent program that generates numerous dense heterochromatic regions (Senescence Associated heterochromatin Formation (SAHF)) characterized by the presence of histone H3K9 trimethylation, HP1 proteins, and the heterochromatin associated H2A variant macro histone H2A[106]. Unlike the DSB silencing described above, SAHF are not reversible and do not occur in cis to the genomic lesion, i.e. they are not adjacent to uncapped telomeres. This observation suggests DSB signals emanating from telomere damage can elicit epigenetic changes in trans to alter cellular states.
Global chromatin alterations also occur in response to IR induced DSBs due to a relocalization of the NAD dependent SIR class of histone deacetylases from heterochromatin to the site of DNA damage. Yeast Sir2 relocalizes from telomeres to nuclease induced DSBs, leading to transcriptional derepression of genes in hetrochromatin[107,108]. Similar findings were reported in mammalian cells, resulting in loss of SIRT1 dependent silencing of heterochromatin [109]. SIRT1 departure from repetitive, heterochromatic elements paralleled gene expression within these regions following DNA damage. The authors speculate that derepression in trans, i.e. gene expression changes at repetitive genomic regions distal to the site of DSBs would be responsible for age-related pathologies. These ideas were supported by observations that derepression of SIRT1 targets occurred in aged mouse brains, and that depression of Sir2 targets led to aging in yeast.
DSB specific forms of DNA damage leads to a global derepression of transposable elements and active transposition of long and short interspersed nuclear elements, LINES and SINES[110]. Similar results are seen during other stress responses. Heat shock causes a strong derepression of repetitive DNA and the formation of centromeric nuclear stress bodies, concomitant with expression of the centromeric α-satellite repeats[111]. Interestingly, cancer genomes show derepression of the same elements, with 100-1000 fold induction of α-satellite RNAs and LINE elements and evidence of active transposition[112,113]. Perhaps persistent DSB responses in tumors play a causal role in expression of noncoding RNA from repetitive elements. Given the specificity of these gene expression changes to tumors, how they influence the overall fitness of cancer cells will be a matter of importance in terms of thinking of novel therapeutic strategies. Nonetheless, RNA expressed from repetitive elements may serve as a novel early biomarker for a broad range of malignancies.
5. Summary and Perspective
It is now clear that extensive histone tail modifications occurring in cis and in trans to DSBs enable high fidelity DNA repair and communication to other cellular processes. The chromatin response therefore not only represents a means to control the temporal and spatial assembly of DNA repair proteins in the vicinity of a DSB, but also a mechanism to control local and global transcriptional responses. The challenge moving forward is to integrate these numerous DSB related chromatin changes into the context of testable models, so that we can begin to understand their full impact genome integrity and cellular state changes in response to genotoxic stress.
Acknowledgements
RAG gratefully acknowledges funding from the NCI (1R01CA138835-01, K08 awards 1K08CA106597-01 and 3K08CA106597-04S1), an American Cancer Society Research Scholar Grant, and funds from the Abramson Family Cancer Research Institute, and from the Penn Genomes Frontiers Institute at the University of Pennsylvania.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 2.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 3.Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 5.Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat Cell Biol. 2003;5:572–7. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- 6.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–35. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 8.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 9.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–16. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–11. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 11.Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 12.Berkovich E, Monnat RJ, Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–90. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 13.Savic V, et al. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier A, et al. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. Embo J. 2007;26:2707–18. doi: 10.1038/sj.emboj.7601719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massip L, Caron P, Iacovoni JS, Trouche D, Legube G. Deciphering the chromatin landscape induced around DNA double strand breaks. Cell Cycle. 9:2963–72. doi: 10.4161/cc.9.15.12412. [DOI] [PubMed] [Google Scholar]
- 16.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassing CH, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 141:970–81. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–8. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 20.Greenberg RA. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 2008;117:305–17. doi: 10.1007/s00412-008-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–26. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huyen Y, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–11. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 24.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 25.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–9. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 26.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–14. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 Ubiquitylates Histones at DNA Double-Strand Breaks and Promotes Assembly of Repair Proteins. Cell. 2007 doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 28.Doil C, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 29.Zhao GY, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell. 2007;25:663–75. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 30.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 Transduces the DNA-Damage Signal via Histone Ubiquitylation and Checkpoint Protein Assembly. Cell. 2007 doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–34. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 32.Kolas NK, et al. Orchestration of the DNA-Damage Response by the RNF8 Ubiquitin Ligase. Science. 2007 doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–6. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 34.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–26. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 35.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 36.Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–6. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 37.Lou Z, Minter-Dykhouse K, Wu X, Chen J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature. 2003;421:957–61. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- 38.Bekker-Jensen S, et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol. 12(sup):80–6. 1–12. doi: 10.1038/ncb2008. [DOI] [PubMed] [Google Scholar]
- 39.Nakada S, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 466:941–6. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 40.Stewart GS, et al. RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci U S A. 2007;104:16910–5. doi: 10.1073/pnas.0708408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–90. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 42.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–9. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 45.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–56. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–54. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–39. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–28. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci U S A. 2009;106:3166–71. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–99. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 52.Patterson-Fortin J, Shao G, Bretscher H, Messick TE, Greenberg RA. Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J Biol Chem. 285:30971–81. doi: 10.1074/jbc.M110.135319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng L, Wang J, Chen J. The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. J Biol Chem. 285:30982–8. doi: 10.1074/jbc.M110.135392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicassio F, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–7. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 55.Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parker JL, Bielen AB, Dikic I, Ulrich HD. Contributions of ubiquitin- and PCNA-binding domains to the activity of Polymerase eta in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:881–9. doi: 10.1093/nar/gkl1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo C, Tang TS, Bienko M, Dikic I, Friedberg EC. Requirements for the interaction of mouse Polkappa with ubiquitin and its biological significance. J Biol Chem. 2008;283:4658–64. doi: 10.1074/jbc.M709275200. [DOI] [PubMed] [Google Scholar]
- 58.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–4. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 59.Pei H, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 470:124–8. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–33. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–63. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–8. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao L, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–41. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 17:688–95. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coleman KA, Greenberg RA. J Biol Chem. The BRCA1-RAP80 Complex Regulates DNA Repair Mechanism Utilization by Restricting End Resection. 286:13669–80. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25:685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iijima J, Zeng Z, Takeda S, Taniguchi Y. RAP80 acts independently of BRCA1 in repair of topoisomerase II poison-induced DNA damage. Cancer Res. 70:8467–74. doi: 10.1158/0008-5472.CAN-10-0267. [DOI] [PubMed] [Google Scholar]
- 69.Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R. Control of Sister Chromatid Recombination by Histone H2AX. Mol Cell. 2004;16:1017–25. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Helmink BA, et al. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 472:247. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 72.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 73.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 74.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. Embo J. 29:3130–9. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahel D, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–3. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–71. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 78.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–18. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–77. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 80.Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–82. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 82.Luijsterburg MS, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–86. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1{alpha} recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–6. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 85.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 144:732–44. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beucher A, et al. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. Embo J. 2009;28:3413–27. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Potts PR, Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol. 2007;14:581–90. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- 88.Tomlins SA, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 89.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–83. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 92.Aguilera A. The connection between transcription and genomic instability. Embo J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paulsen RD, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–39. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Solovjeva LV, Svetlova MP, Chagin VO, Tomilin NV. Inhibition of transcription at radiation-induced nuclear foci of phosphorylated histone H2AX in mammalian cells. Chromosome Res. 2007;15:787–97. doi: 10.1007/s10577-007-1162-x. [DOI] [PubMed] [Google Scholar]
- 95.Kruhlak M, et al. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–4. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- 96.Cuozzo C, et al. DNA damage, homology-directed repair, and DNA methylation. PLoS Genet. 2007;3:e110. doi: 10.1371/journal.pgen.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 99.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 100.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. Embo J. 29:1446–57. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 141:432–45. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–7. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- 103.Turner JM, et al. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–42. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 104.Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 25:959–71. doi: 10.1101/gad.2030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 106.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 107.Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–20. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 108.McAinsh AD, Scott-Drew S, Murray JA, Jackson SP. DNA damage triggers disruption of telomeric silencing and Mec1p- dependent relocation of Sir3p. Curr Biol. 1999;9:963–6. doi: 10.1016/s0960-9822(99)80424-2. [DOI] [PubMed] [Google Scholar]
- 109.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–18. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- 111.Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2:a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ting DT, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 331:593–6. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]