Abstract
Membrane lipid composition can vary dramatically across the three domains of life and even within single organisms. Here we review evidence that the lipid-exposed surfaces of membrane proteins have generally evolved to maintain correct structure and function in the face of major changes in lipid composition. Such tolerance has allowed evolution to extensively remodel membrane lipid compositions during the emergence of new species without having to extensively remodel the associated membrane proteins. The tolerance of membrane proteins also permits single-celled organisms to vary membrane lipid composition in response to their changing environments and enables dynamic and organelle-specific variations in the lipid compositions of eukaryotic cells. Membrane protein structural biology has greatly benefited from this seemingly intrinsic property of membrane proteins: the majority of structures determined to date have been characterized under model membrane conditions that little-resemble native membranes. Nevertheless, with a few notable exceptions most experimentally-determined membrane protein structures appear, to a good approximation, to faithfully report on native structure.
It has long been appreciated that membrane proteins (MPs) are not always fully functional following purification into model membranes such as detergent micelles or lipid-detergent mixed micelles. Reduced functionality can reflect destabilization, misfolding, or perturbation of MP structure in model membranes relative to native bilayer conditions. Loss or perturbation of MP function can also reflect a requirement by certain MPs to form specific complexes with lipids, which may play co-factor roles in promoting function and/or serve to buttress native structure (reviews in (1–10)). Recent biophysical and structural studies of integral MPs have also highlighted the degree to which some model membranes such as micelles or lipid-detergent mixed micelles can fail to fully support native MP stability or structure. For example, homodimerization of single-span MPs such as the receptor tyrosine kinases is sometimes weaker in detergent micelles than in bilayers (review in (11)), reflecting a reduction in the free energy for dimerization in micelles relative to bilayers by as much as 5 kcal/mol (12). Another example is provided by the initial high resolution structure determined for a voltage-gated potassium channel, KvAP, which was crystallized from micelles (13). KvAP was seen to have a distorted disposition of the voltage sensor domain relative to the channel domain, a fact that was appreciated later when a more native-like structure was crystallized from lipid-containing mixed micelles (14).
Data such as that cited above has helped to drive the development of model membranes such as bicelles, lipidic cubic mesophases, and nanodiscs that capture some of the advantages of working with micelles and mixed micelles while at the same time providing a bilayered environment for MPs reconstituted therein (15–18). Moreover, techniques such as electron 2-D crystallography and solid state NMR are increasingly being used to directly probe MP structure in actual bilayered lipid vesicles, sometimes to high resolution (see later section).
In the laudable impetus to carry out quantitative structural and functional measurements under membrane conditions as close to native as possible, it can be tempting to view current and previous studies of MPs conducted in model membranes with a skepticism that extends beyond well-justified caution regarding extrapolating results obtained using model membranes to the situation in native membranes. However, we suggest that there are a number of reasons that an overly judgmental view of the relevance of model-membrane-derived results is not well-justified. One reason is the tremendously useful body of information gleaned from the many dozens of high resolution structures of MPs determined during recent years, the vast majority of which employed detergent micelles as the model membrane medium. Structural advances have been matched by tremendous progress in studies of MP folding/stability and of MP function, work that has also relied heavily on results derived using non-bilayered model membranes. It can also be observed that “native membrane conditions” represent an ideal rather than a fixed reality in light of the facts that (i) a given MP will often traffic through several different organelles, each with a distinct lipid composition, en route to its destination membrane, (ii) membrane lipid composition is dynamic even within a single plasma or organellar membrane, (iii) even within a single membrane, all components are not uniformly and randomly mixed, but some lipids and proteins will transiently form domains, the components of which are likely to also spend significant time in the bulk (unorganized) membrane domain, and (iv) there can be even more dramatic variations in lipid composition from organism to organism (see sections below for more on this topic). Even studies conducted directly on unpurified MPs in natural membranes often rely on conditions in which the protein of study is highly overexpressed relative to normal physiological conditions.
In this review we explore a more fundamental reason why studies of MPs in model membranes have been and are likely to continue to be informative. We propose that many MPs are remarkably tolerant of significant variations in membrane composition, reflecting the outcome of strong evolutionary selective pressure to be so. In the following sections we cite bodies of evidence that this is the case. We emphasize that the purpose of this review is NOT in any way to either discount previous work documenting specificity of MP-lipid interactions or to encourage irresponsible use of model membranes. Rather, the purpose is to highlight evidence that many MPs appear to be quite tolerant of major variations in membrane composition and to reassure those pursuing structural or biochemical studies of MPs in model membranes that good faith efforts to conduct experiments using the best available model membrane medium compatible with a given experimental approach are likely to be rewarded by illuminating data.
E. coli can survive knockout of all major classes of phospholipids
The tools of microbial genetics have been extensively used during the past 30+ years to explore lipid metabolism and homoeostasis in E. coli, with mutations being used to knock out enzyme activities required for biosynthesis of various native lipid species (19–21). A major goal of these studies has been to discover specific roles for various phospholipid species in E. coli physiology and associated cellular biology, biochemistry, and biophysics. For example, elegant work from the Dowhan lab has established the fact that the presence of phosphatidylethanolamine (PE) is required in order for certain transporters such as lactose permease to adopt their correct topology in the plasma membrane and to functional normally (22). A long term focus of the genetic studies has been to unravel specific examples where lipids play specific roles in regulating or sustaining normal structure and function. However, a largely overlooked outcome of these studies is a large body of evidence that E. coli is remarkably tolerant of extensive remodeling of its phospholipid composition.
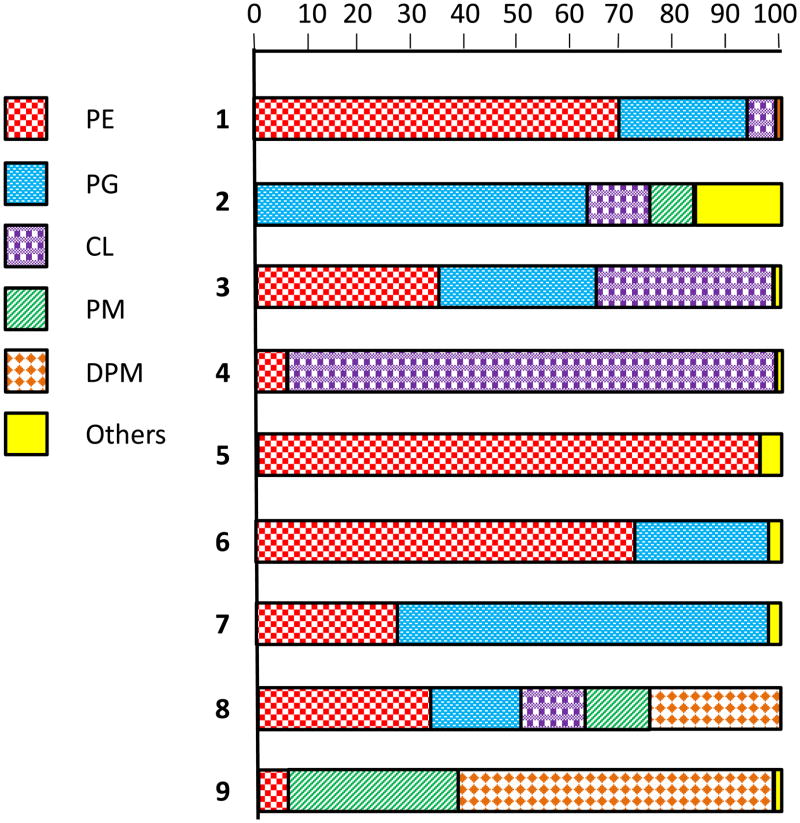
Figure 1 (adapted from (20)) illustrates the plasma membrane (PM) lipid compositions of wild type and phospholipid biosynthesis knock-out mutant strains of E. coli at various stages of growth and/or under unusual culture conditions. Represented in this figure are wild type E. coli (example 1) and strains in which its major phospholipids, phosphatidylethanolamine (zwitterionic), phosphatidylglycerol (PG, anionic), and cardiolipin (CL, anionic) have each been eliminated. Also represented is a strain in which both PG and CL have been removed. Very unusual PM lipid compositions have been observed. One strain has membranes that are almost exclusively zwitterionic (example 5); some have lipids that are almost exclusively anionic (examples 2 and 4); one has membranes composed almost exclusively of cardiolipin (example 4)—a lipid with a non-conventional architecture (see Figure 2); yet another has membranes that are almost exclusively composed of lipids that are present only in very low quantities in wild type cells (example 9). Despite such extreme differences in lipid composition life persists, although the mutant strains often have more stringent growth requirements and are less robust than wild type strains. It can also be pointed out that E. coli and its set of essential MPs are also remarkably tolerant of the introduction of foreign lipids such as phosphatidylcholine (up to at least 70% of total lipid (23)) or mono- or diglucosyldiacylglycerols (at 40% of total lipid, (24–26)). Some foreign lipids can successfully substitute for PE to fulfill its role in facilitating adoption of correct topology of lactose permease (22), among which is glucosyl-diacylglycerol, the headgroup of which resembles PE only in that both have a net charge of zero (26). The results summarized above strongly suggest that a many of the of MPs present in E. coli are tolerant of major changes in membrane lipid composition, remaining correctly threaded into the plasma membrane, reasonably stable, and functional despite the loss of their usual phospholipid neighbors.
Figure 1.
Phospholipid compositions observed for various strains of E. coli, as measured at various growth phases or following culturing in unusual media. Example 1 is wild type strain SD12 during exponential growth. Example 2 contains an interrupted allele of PS synthase in strain AH930 during exponential growth. Example 3 is strain SD10, which contains a temperature sensitive PS synthase, during exponential growth. Example 4 is strain SD10 grown at stationary phase. Example 5 is a double mutant of strain SD312 containing a mutated phosphatidylglycerophosphate synthase and a defective CL synthase during exponential growth. Example 6 is strain CB64-CLI with a knockout of CL synthase during exponential growth. Example 7 is strain SD9 containing a temperature sensitive PS synthase and a defective CL synthase during exponential growth. Example 8 is strain SD10 (see example 3) grown under high D-mannitol conditions during exponential growth. Example 9 is strain SD10 grown under high mannitol conditions during stationary phase. Abbreviations: phosphatidylethanolamine (PE), phosphatidylglycerol (PG), cardiolipin (CL), phosphatidylmannitol (PM), diphosphatidylmannitol (DPM), phosphatidylserine (PS). This figure was adapted from (21).
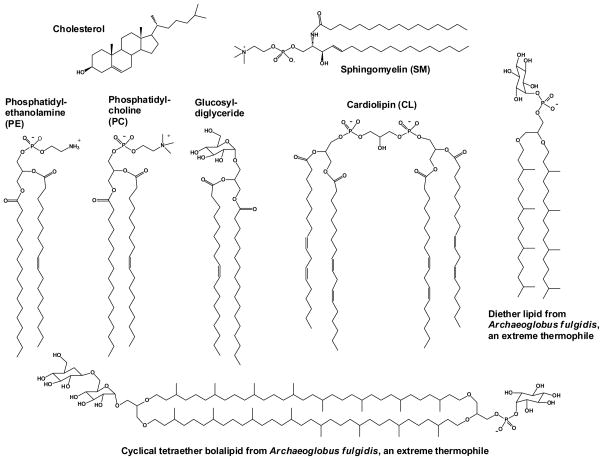
Figure 2.
Structures of representative lipids from different domains of life. Cholesterol and sphingolipids such as sphingomyelin are found primarily in eukaryotes. Phosphatidylcholine is among the most common glycerophospholipids of higher eukaryotes, but is less common in bacteria (and is absent in E. coli). Phosphatidylethanolamine is common both in bacteria and eukaryotes. Glucosyldiglyceride and related neutral glycoglycerolipids are the often the most common lipids of plants. Cardiolipin is common in bacteria and in the mitochondria of eukaryotes. Ether-linked isoprenoid lipids such as those exemplified by the bottom and far right lipids dominate the membraned of thermophilic archaebacteria (50, 62).
Membrane lipid composition is not static but can vary dramatically during the lifetime of a cell
Any given MP inhabits a variety of different lipidic environments during its lifetime. Eukaryotic MPs destined to reside in the PM are first synthesized and inserted into the membrane of the endoplasmic reticulum (ER), transported to the Golgi body, and then finally make their way to the PM. Each of these organelles has a different lipid composition, with levels of cholesterol in the membrane varying from <15% in the ER to 20–50% in the PM of mammalian cells (27, 28). The ratios of different phospholipids often vary greatly between the organelles (27, 29–32). For instance, sphingomyelin makes up a significant percentage of the phospholipids in the mammalian PM while sphingolipid content is very low in the ER (27). The PC:PE:PI:PS ratio in the PM of rat liver is 28:17:6:6, while in the Golgi and the rough ER it is 44:19:7:2 and 54:20:9:3, respectively (31). Similarly, ergosterol:phospholipid ratios are greater at the plasma membrane (1:2) of yeast than in organelles, where they range from 1:50 in the outer mitochondrial membrane and 1:30 in the peroxisomes to 1:3 in lipid particles. Yeast inositol sphingolipids are greatly enriched at the PM (ca. 25%) relative to other organelles (e.g., near 0 in the ER) (27, 33). Even more specific differences between organelles exist. For example, phosphatidylserine and phosphatidylethanolamine in the PM have fatty acid compositions that are 40% and 30% saturated, whereas the corresponding degree of saturation for these lipids at the whole cell level are 29% and 16%, respectively (33). Based on the observations outlined above, it can reasonably be inferred that most MPs remain essentially folded in a variety of lipid environments as they move from the ER to their destination membranes, an inference that does not imply that the functions of these proteins are independent of environment. These environments may vary in their bilayer thickness and bilayer fluidity as a consequence of variations in sphingolipid and cholesterol content as well as acyl chain composition.
In addition to trafficking through the secretory pathway en route to their destination membranes, MPs are subject to internalization by pinocytosis and endocytosis and many are recycled from endosomes back to the plasma membrane (34). Studies of cultured hepatocytes indicate that these cells turnover the equivalent of their plasma membrane surface areas ca. 5 times every hour (35). In plant cells, plasma membrane turnover rates can be as rapid as once every ten minutes (36). MPs internalized and recycled back to the plasma membrane are likely experiencing rapid variations in the local lipid composition and yet retain their essential fold and ultimately remain functional after these transitions.
Cellular lipid composition also changes in response to various developmental and environmental conditions. Alterations of the fluidity of the bilayer are evident from studies of hepatocytes as they progress through the cell cycle, with an increase in membrane fluidity due to decreased cholesterol:phospholipid ratios (dropping from a resting ratio of 3:4 to under 2:4 during early regeneration) accompanying rapid cellular proliferation (37). Neuroblastoma cells, which are often employed as a model for malignant cell differentiation, show significant increases in cholesterol:phospholipid ratios during differentiation—from 1:3 to 1:2 in the whole cell (38). Human epidermal cells also show large changes in lipid composition during differentiation, such as an enrichment of sphingolipids (which comprise from 7% the lipids of the strata basal and strata spinosum to 18% of the lipids of the stratum corneum) and neutral lipids (from 51% in the strata basal/spinosum to 78% in the stratum corneum) coupled with a decrease in the fraction of polar lipids (from 45% in the strata basal/spinosum to 5% in the stratum corneum) (39). E. coli, too, demonstrates lipid remodeling during growth. For example, when grown in minimal medium only 5% of the lipids of E. coli K-12 strain contained cyclopropane-containing fatty acyl lipids in a 4.5 hour culture, while at 17 hours this number rose to 32% (40).
Studies on various plant species show that lipid composition is altered in response to environmental conditions such as mineral exposure, aluminum stress, temperature variation and light exposure (41–44). Similarly, yeast show lipid composition alterations in response to nutrient source and temperature variation (45, 46). These alterations can range from changes in saturation levels, large changes in sterol:phospholipid ratios, and significant differences in the ratios of the most prevalent phospholipids. For instance, in soybean plants, heavily shaded leaves see a decrease in the 18:3 fatty acids coupled with an increase in the 18:1 and 18:2 fatty acids, with the most desaturated 18:3 fatty acids decreasing by over 40% in the most heavily shaded leaves (44). Not surprisingly, heat-stressed plants experience an increase in the saturation levels of the fatty acyl tails of their membrane lipids to maintain appropriate membrane fluidity and stability under hot conditions (43). The peroxisomal membrane lipid composition of the yeast Pichia pastoris is altered by carbon source, with 80% of lipid fatty acyl groups becoming oleoyl when the yeast are cultivated with oleic acid as the major carbon source, relative to only 30% otherwise (45).
The sometimes large variations in lipid and fatty acid content surveyed above are representative of what likely is very common throughout all domains of life. Such variations in lipid composition will, of course, often be accompanied by alterations in MP composition via changes in transcription/translation/degradation as well as a result of altered protein trafficking. Such variations are also likely to alter the functions of many MPs through specific protein-lipid interactions and/or by altering the bulk properties of the surrounding bilayer (fluidity, thickness, etc.). However, it can also be inferred that a great many MPs must be fairly tolerant of swings in membrane composition—remaining membrane-integrated, correctly folded, and functional. Else, life would cease.
While membrane proteins are often highly conserved, lipid compositions across the domains of life can vary spectacularly
Many MPs have been conserved by evolution throughout all domains of life. Based on a limited body of high-resolution structural data (below), it appears that MPs have generally similar 3-D structures throughout all domains. Such high retention of protein structure across eons of evolutionary time is in stark contrast to membrane lipid architecture and composition. While the building block amino and nucleic acids used in proteins, DNA, and RNA are essentially invariant across the furthest extremes of terrestrial and marine life, the building block lipids of membranes can exhibit remarkable diversity from organism to organism (see reviews in (27, 29, 32, 47–51)). The dominant lipids of hyperthermophilic archaebacteria, which include cyclical ether-linked isoprenoid bolalipids (see Figure 2 and (50)), are vastly different from the glycerophospholipid mixtures that dominate E. coli and many other eubacteria, which in turn only partially overlap with the lipid compositions of vertebrates, the latter of which include not only glycerophospholipids but also abundant sphingolipids and cholesterol (see Figure 2). Plant chloroplast lipid compositions, on the other hand, are often rich in neutral glycolipids (52). Diversity is great even among Gram negative bacteria: E. coli is dominated by the diglyceride phospholipids PE, PG, and CL, yet membranes of Trepenoma pallidum contain >50 mol% neutral glycolipid—galactosyl-diglyceride, while >75% of the lipids of Megasphaera elsdenii are plasmalogen-based (48).
Given the widely variant and distinctive membrane compositions that have arisen in different lineages, one might expect that this would place evolutionary pressure on lipid-exposed residues of protein TMDs to diverge in an effort to adapt to relatively rapidly evolving changes in membrane compositions. However, lipid-exposed surfaces of MPs exhibit about the same degree of (relatively low) sequence conservation as water-exposed surfaces of both membrane and water soluble proteins (53, 54). This is despite the fact that the molecular identities of the “solvent” lipid molecules within the membrane vary dramatically, in contrast to the constancy of water.
There are a few examples where the same MP from more than one organism has had its structure determined to high resolution. In some cases, the membranes of these organisms are widely divergent, yet the structures remain similar. Consider thermophilic eubacteria and hyperthermophilic archaebacteria, whose membrane lipid compositions are very different from mesophilic bacteria such as E. coli and eukaryotic membranes. One might expect that there would be substantial differences in MP structure between thermophiles and mesophiles both to confer thermostabilization and also to reflect adaptation to the very significant differences in the structures of the lipids found in thermophiles relative to mesophiles. However, we can cite examples of MPs from thermophiles that are very similar to those found in mesophilic bacteria or higher organisms, at least in terms of static structure.
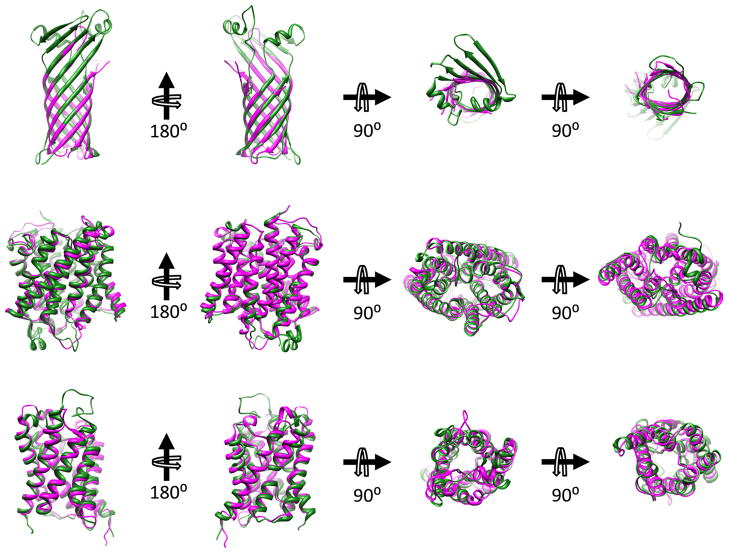
The lipids of the eubacteria genus Thermus have been determined to have high proportions of glycolipids and glycophospholipids in their membranes, with the fatty acyl tails being mainly composed of branched chains (55, 56). A crystal structure of a beta-barrel outer membrane protein TtoA from Thermus thermophilus HB27 (optimal growth temperature 70ºC) shows no obvious structural differences from mesophilic beta-barrel outer membrane proteins(57, 58). An alignment of TtoA’s structure with that of OmpA from the E. coli outer membrane demonstrates high conservation of structure as well as some sequence similarity within the transmembrane regions (Figure 3) (48, 59–61). Indeed, the overall RMSD for the corresponding alpha carbons in the two structures is only 1.1 Å
Figure 3.
Superpositions of structures of thermophilic archaeal membrane proteins on those of mesophilic counterparts reveal high similarity. Views onto the membrane surface are from the extracellular space (second panel from right) and from the cytosol (far right). (Top Panel) Superposition of porins TtoA from Thermus thermophilus (green, PDB ID: 3DZM (58)) and OmpA from E. coli (magenta, PDB ID: 1QJP (59)). (Middle Panel) Superposition of ammonium transporters Amt-1 from the archaeal hyperthermophile A. fulgidus (green, PDB ID: 2B2H (64)) and AmtB from E. coli (magenta, PDB ID: 1U77 (65)). (Bottom Panel) Superposition of aquaporin from A. fulgidus (green, PDB ID: 3NE2) with AqpZ from E. coli (magenta, PDB ID: 1RC2 (66)).
To cite another example, Archaeoglobus fulgidus is an archaebacterial thermophile (optimal growth temperature is 83ºC) that has a very exotic lipid composition, with the two major lipid backbones being ether-linked diglycerides of either conventional (diether) or cyclical tetraether bolalipid architecture ((50, 62) see Figure 2). The chains are composed of saturated isoprenoids. It has been shown that the ratio of the tetraether bolalipid to diether lipids increases with increasing growth temperature (ratio = 1:1 at 83°C), indicating that the bolalipids are involved in imparting extreme thermostability (63). Despite the extreme differences in lipid structure between this organism and E. coli, the ammonium transporter Amt-1 from Archaeoglobus fulgidus is remarkably similar to that of the E. coli transporter AmtB, as illustrated by superposition of the two structures ((64, 65) Figure 3), where the RMSD for the transmembrane domain alpha carbons is 0.9 Å). The crystal structure of a second MP from Archaeoglobus fulgidus, an aquaporin, was recently determined by the Stroud lab (PDB ID: 3NE2). This protein shares ca. 30% sequence identity with the E. coli aquaporin AqpZ, for which a crystal structure is also available (66). A superposition reveals very similar structures (see Figure 3), with an RMSD for the transmembrane domain alpha carbons in the two structures of 0.94 Å.
That lipid structure is so highly variable from organism to organism likely reflects a combination of both evolutionary selective pressure placed upon a successful membrane to adapt to changes in environment (temperature, energy and/or carbon sources, pH, hostile neighbors, etc.) and evolutionary drift. That MPs, in contrast, are often structurally well-conserved between evolutionarily-distant organisms likely reflects the fact that all PMs share some common structural and dynamic properties under organism-specific physiological conditions even though their lipids are highly divergent. At the same time, it appears equally likely that widely-retained MPs have been selectively adapted so as to remain foldable, stable, and functional despite variations in membrane composition. It seems that in the development of new species evolution can substantially alter membrane lipid composition without at the same time having to dramatically alter the organismal complement of membrane proteins.
Aquaporin-0 has a tolerant lipid-exposed surface
Aquaporin-0 (AQP0) is a tetrameric helical MP that serves as a water channel in the fiber cells of the vertebrate ocular lens and also forms intermembrane junctions between layers of flattened cells. T. Walz and co-workers carried out pioneering structural studies that provided atomic level details of the interaction of this protein with its annular layer of interacting lipids. (For other examples of MP structures in which annular lipids have observed see (67) and references therein). 2-D crystals of sheep AQP0 were prepared in lipid bilayers and then structurally characterized using electron crystallography. This led to a 1.9 Å structure of the protein in dimyristoylphosphatidylcholine (DMPC) bilayers (68) and to a 2.5 Å structure in bilayers composed of E. coli phospholipids (69), (see Figure 4). In both structures the annular lipids interacting directly with the exposed surface of the transmembrane domains of the tetramer are observed, revealing specific modes of direct interaction with the protein surface. The annular lipid-to-protein subunit stoichiometry is in each case 7:1. There are a number of interesting observations that can be made about these structures.
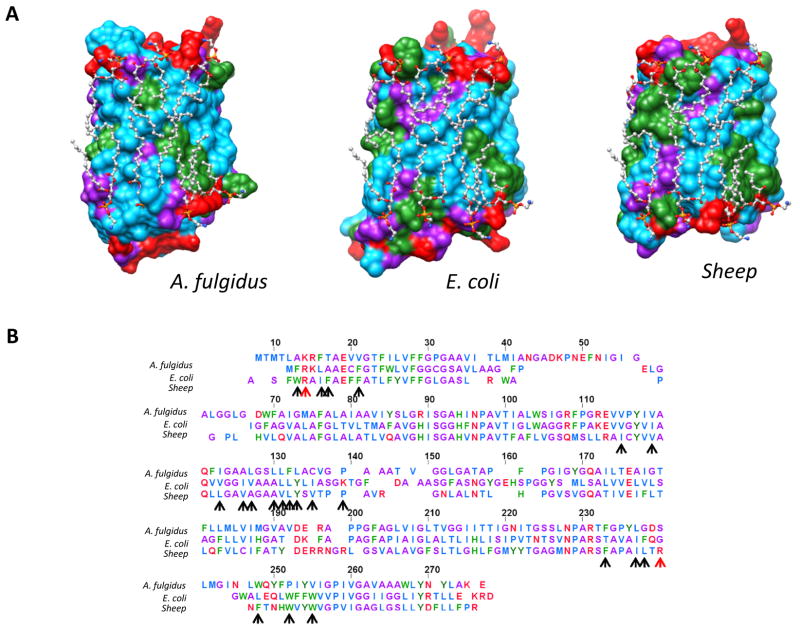
Figure 4.
The lipid-contact faces of aquaporins from three domains of life show some common features. (A) Depiction of the lipid contact faces of aquaporins from an hyperthermophilic archaebacterium, A. fulgidus (PDB ID: 3NE2), E. coli (PDB ID: 1RC2 (66)), and O. airies (sheep) (PDB ID: 3M9I (69)). Residues are colored as follows; red: polar residues; blue: large hydrophobic; green: aromatic/His; purple: small (Gly, Ala, Ser, Cys). In all three cases, the structures have been overlaid with the annular E. coli lipids as found in the structure of sheep aquaporin crystallized in the Walz laboratory (PDB ID: 3M9I). (B) Structure-based sequence alignment of the aquaporins depicted in (A). Arrows denote residues that were in direct contact with the lipid aliphatic chains in the sheep AQP0 structure. Red arrows depict residues in contact with the head groups. Color coding of residues is as given above for (A).
First, even though both structures reveal preferred modes of acyl chain/AQP0 interactions, neither pure DMPC nor E. coli phospholipids reflect compositions that are similar to the lipid composition of native lens fiber membranes. Those membranes are extremely rich in both cholesterol and sphingolipids, to the extent that they can be considered to be homogenously raft-like in composition and biophysical properties (70, 71). It is remarkable that AQP0 exhibits specific and preferred modes of interactions with its surrounding lipids even when those lipids are very different from its neighboring lipids under physiological conditions.
Secondly, while both DMPC and E. coli lipids exhibit preferred modes of interaction with the sheep AQP0 surface, the set of binding modes for the 7 annular DMPC molecules is distinctly different from the 7 E. coli lipids (Figure 4). DMPC differs from the E. coli lipids by having two relatively short and fully saturated acyl chains, whereas E. coli lipids usually have both a saturated chain and an unsaturated chain, both of which are typically longer than the C14 chains of DMPC. In both cases the acyl chains fit into grooves on the surface of the transmembrane domain. It is notable that these grooves are able to accommodate multiple (alternative) modes of acyl chain interactions to suit the properties of its lipid neighbors in terms of chain length and possible sites of cis double bonds—even when these lipid neighbors are non-native.
While E. coli lipids form fluid phase bilayers that are significantly thicker than liquid crystalline phase DMPC, the structure of the protein is seen to be nearly identical regardless of which lipids were used during 2-D crystallization (see Figures 1D and 4 (69)). Indeed, the X-ray crystal structure of bovine AQP0 determined in alkylglycoside micelles is nearly identical to that determined in the electron crystallographic structures (see Figure 4 in reference (72)).
While we do not yet know the degree to which AQP0 can be regarded as a typical MP in terms of its interactions with lipids, the above observations suggest a structural basis for MP tolerance. The hydrophobic grooves on the lipid-exposed surface of AQP0’s transmembrane domain are compatible of participating in multiple alternative modes of intimate interaction with lipid chains, even involving chains that may be rare or completely absent in native membranes. It is as if the surface of AQP0 is similar to a “master key” with one defined set of notches that can nevertheless open many different locks. This surface appears to reflect the Scouting imperative “Always be Prepared” in terms of being able to accommodate a wide range of surrounding lipid compositions.
Despite being from organisms with very different membrane compositions from each other (see Figure 2) and from lens AQP0, the lipid-exposed surfaces of the E. coli and hyperthermophilic A. fulgidus AQPZ proteins exhibit surprisingly similar structural properties to the lipid-contact face of AQP0 (Figure 4). In all three structures, a tract of large non-polar residues crisscrosses the lipid-contact surface found in the AQP0 structure (see Figure 4). In addition, the aquaporins presented here have a similar pattern of aromatic residues on the lipid-contact face. When the E. coli lipids from the O. airies (sheep) AQP0 structure are overlaid across the E. coli and A. fulgidus crystal structures (as shown in Figure 4), it appears that these structures may similarly be able to accommodate these lipids with only minor adjustments in acyl chain and head group positions. In fact, in the E. coli structure, which was crystallized in micelles of n-octylglucoside, the detergent molecules are found on the natively lipid-exposed face, with the hydrophobic tails lying along similar grooves to the E. coli lipid niches in the lens AQP0 structure (PDB ID: 1RC2 (66)). This conservation would imply that the aquaporins did not need to drastically alter their lipid-contact surface properties over the course of evolution, despite the large changes in lipid composition of the evolving membranes. This observation provides further support for the notion that the lipid-exposed surfaces of aquaporins are capable of accommodating multiple modes of lipid-protein interactions.
The AQP0 protein was seen to be resistant to structural changes induced either by differing modes of lipid interactions in membranes of different compositions or to differing preferred bilayer thicknesses for the mixtures of lipids in the membrane surrounding the protein. AQP0’s tolerance is apparently rooted in a healthy intrinsic structural stability.
Final Observations and Conclusions
Numerous additional examples of tolerance are provided by MPs that have been purified and then functionally reconstituted in model membranes that often are very different in composition than their native membranes. To cite an extreme example, E. coli diacylglycerol kinase (DAGK) is a homotrimer with three transmembrane helices per subunit that catalyzes an interfacial reaction between a lipid and a water soluble substrate (73). Over 50% of DAGK’s residues lie within the membrane and its active site is partially membrane-submerged. DAGK is active in a variety of different types of detergent micelles provided that lipid is present, with full activation usually being observed at 10–20 mol% lipid content (74–76). However, the lipid specificity is low—full activation can be accomplished even using lipids not found in E. coli, such as saturated phosphatidylcholine or hexadecylsulfate (74–76). More recently, using a water soluble form of diacylglycerol, it has also been shown that DAGK is fully active in certain lysophospholipid micelles even in the complete absence of any non-substrate lipid (77). Indeed, DAGK has been shown to be fully active in lipid- and detergent-free amphipathic polymers known as “amphipols” (78), in CHAPSO-DMPC bicelles (79) and in lipidic cubic phases composed of monoolein (80). E. coli DAGK has also been shown to retain function following expression in mammalian COS cells (81). DAGK is highly active following reconstitution in lipid vesicles, with maximal activity being observed in vesicles composed of dioleoylphosphatidylcholine (not present in E. coli membranes), and somewhat lower activity being observed in vesicles composed of lipids chosen to mimic the lipid composition of the plasma membrane of E. coli (82, 83). That DAGK is so exceedingly tolerant of extreme variations in its membrane milieu may be closely related to the extremely high thermal stability of this protein in native membranes (cf., (84)). Indeed, the fact that E. coli membranes can be boiled for several minutes without any loss of DAGK activity suggests that DAGK’s thermal stability may have been “overdetermined” by evolution. Perhaps this trait represents a “reserve” of intrinsic stability that is not normally essential to DAGK or its host during the normal life span of an E. coli bacterium, but that could facilitate evolutionary membrane remodeling under conditions of selective pressure. While most membrane proteins are much less stable than DAGK, this enzyme illustrates the extreme degree to which evolution can sometimes confer tolerance to MPs.
In addition to numerous examples of reconstituting MPs into non-native model membranes, there are also examples of functionally expressing a MP from one organism in another organism that has a very different membrane composition. A revealing example is provided by a number of mammalian G protein-coupled receptors that have been expressed into the plasma membrane of E. coli and shown to remain functional, at least to the extent that they retain the ability to specifically bind antagonists with high affinity (85–89). Such retention of function is despite the fact that cholesterol is thought to be important for the stability and function of some of these receptors, but is not present in E. coli (see review in (6)).
Finally, we note that there are a number of MPs that have now been crystallized from both detergent micelles and from bilayered model membrane media (bicelles or lipidic cubic phases). These proteins, which include AQP0 (see above), the rhomboid protease (67), bacteriorhodopsin (17), and VDAC (90) have been seen to adopt similar structures in both detergents and bilayered model membranes, again supporting the notion that MPs are often remarkably tolerant of their membrane or membrane-mimetic environment. There are, of course, relatively rare exceptions such as the KvAP potassium channel, whose flexibly-linked multidomain architecture makes it particularly susceptible to micellar distortion (see Introduction). Also, it is acknowledged that at least some of the membrane proteins (such as AQP0) that have yielded to high resolution structural analysis represent “low hanging fruit” in terms of being relatively stable and rigid membrane proteins. It should also be noted that even for proteins that exhibit the same static structure when determined in both detergent micelles and in lipid-containing model membranes it has been seen that the conformational flexibility of the protein in micelle-derived crystals is higher than in crystals with lipids present (91).
To conclude, we emphasize that it is abundantly clear (1–10) that many membrane proteins require specific lipid cofactors for proper folding, structure, and/or function. Some membrane proteins also have rather specific requirements regarding membrane fluidity, thickness, lateral surface pressure, or other membrane properties. However, the data highlighted in this review suggests that some degree of fundamental tolerance to variations in membrane lipid composition is also a trait that has been conferred in varying degrees by evolution to many—if not all—membrane proteins. In this regard we should note that there is no inherent contradiction between this property and the notion that many membrane proteins also have specific lipid or membrane requirements. For example, a given membrane protein might be generally tolerant of a wide range of membrane compositions and yet still have a specific lipid cofactor requirement in order to function, both of these traits having been evolutionarily-selected. Here, we have suggested a couple of structural and biophysical mechanisms that may have been used by evolution to help confer tolerance to some membrane proteins: protein stabilization and the generation of grooved “master key”-like lipid exposed surfaces. There likely are others. Tolerance is an intrinsic property shared by many membrane proteins that helps to explain the spectacular success of the reductionist approach to membrane biology, which has relied heavily on the assumption that illuminating and biologically-relevant information can be gleaned from carefully-controlled studies of membrane protein structure, folding, and function under model membrane conditions. Such studies should, of course, always employ the most realistic model membranes that are consistent with a given experimental approach and should also include verification of protein functionality, when possible.
Acknowledgments
We thank Catherine Deatherage, Yuanli Song, and the reviewers of an earlier draft of this paper for their useful suggestions.
Abbreviations
- A. fulgidus
Archaeoglobus fulgidus
- CHAPSO
3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate
- CL
cardiolipin
- DAGK
diacylglycerol kinase
- DMPC
dimyristoylphosphatidylcholine
- DPM
diphosphatidylmannitol
- E. coli
Escherichia coli
- ER
endoplasmic reticulum
- MP
membrane protein
- NMR
nuclear magnetic resonance
- O. airies
Ovis airies
- PM
plasma membrane
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PI
phosphatidylinositol
- PS
phosphatidylserine
- RMSD
root mean squared deviation
- TM
transmembrane
- TMD
transmembrane domain
- 2-D
two-dimensional
- 3-D
three-dimensional
Footnotes
This work was supported by US NIH grants RO1 DC007416 and NS058815 and by PO1 GM080513 (to CRS) and by a US NSF Graduate Research Fellowship (to KFM, grant DGE0909667).
References
- 1.Adamian L, Naveed H, Liang J. Lipid-binding surfaces of membrane proteins: evidence from evolutionary and structural analysis. Biochim Biophys Acta. 2011;1808:1092–1102. doi: 10.1016/j.bbamem.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunte C. Specific protein-lipid interactions in membrane proteins. Biochem Soc Trans. 2005;33:938–942. doi: 10.1042/BST20050938. [DOI] [PubMed] [Google Scholar]
- 3.Hunte C, Richers S. Lipids and membrane protein structures. Curr Opin Struct Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Qin L, Sharpe MA, Garavito RM, Ferguson-Miller S. Conserved lipid-binding sites in membrane proteins: a focus on cytochrome c oxidase. Curr Opin Struct Biol. 2007;17:444–450. doi: 10.1016/j.sbi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Opekarova M, Tanner W. Specific lipid requirements of membrane proteins - a putative bottleneck in heterologous expression. Bba-Biomembranes. 2003;1610:11–22. doi: 10.1016/s0005-2736(02)00708-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee AG. Lipid-protein interactions. Biochem Soc Trans. 2011;39:761–766. doi: 10.1042/BST0390761. [DOI] [PubMed] [Google Scholar]
- 8.Ernst AM, Contreras FX, Brugger B, Wieland F. Determinants of specificity at the protein-lipid interface in membranes. Febs Lett. 2010;584:1713–1720. doi: 10.1016/j.febslet.2009.12.060. [DOI] [PubMed] [Google Scholar]
- 9.Nyholm TK, Ozdirekcan S, Killian JA. How protein transmembrane segments sense the lipid environment. Biochemistry-Us. 2007;46:1457–1465. doi: 10.1021/bi061941c. [DOI] [PubMed] [Google Scholar]
- 10.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochim Biophys Acta. 2008;1778:1545–1575. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 11.MacKenzie KR, Fleming KG. Association energetics of membrane spanning alpha-helices. Curr Opin Struct Biol. 2008;18:412–419. doi: 10.1016/j.sbi.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowie JU. Membrane protein folding: how important are hydrogen bonds? Curr Opin Struct Biol. 2011;21:42–49. doi: 10.1016/j.sbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 14.Lee SY, Lee A, Chen J, MacKinnon R. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. P Natl Acad Sci USA. 2005;102:15441–15446. doi: 10.1073/pnas.0507651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 17.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 18.Sanders CR, Prosser RS. Bicelles: a model membrane system for all seasons? Structure. 1998;6:1227–1234. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 19.Bogdanov M, Xie J, Dowhan W. Lipid-protein interactions drive membrane protein topogenesis in accordance with the positive inside rule. J Biol Chem. 2009;284:9637–9641. doi: 10.1074/jbc.R800081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto K. Dispensable nature of phosphatidylglycerol in Escherichia coli: dual roles of anionic phospholipids. Mol Microbiol. 2001;39:1427–1433. doi: 10.1046/j.1365-2958.2001.02320.x. [DOI] [PubMed] [Google Scholar]
- 21.Shibuya I. Metabolic regulations and biological functions of phospholipids in Escherichia coli. Prog Lipid Res. 1992;31:245–299. doi: 10.1016/0163-7827(92)90010-g. [DOI] [PubMed] [Google Scholar]
- 22.Dowhan W, Bogdanov M. Lipid-protein interactions as determinants of membrane protein structure and function. Biochem Soc Trans. 2011;39:767–774. doi: 10.1042/BST0390767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogdanov M, Heacock P, Guan Z, Dowhan W. Plasticity of lipid-protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wikstrom M, Kelly AA, Georgiev A, Eriksson HM, Klement MR, Bogdanov M, Dowhan W, Wieslander A. Lipid-engineered Escherichia coli membranes reveal critical lipid headgroup size for protein function. J Biol Chem. 2009;284:954–965. doi: 10.1074/jbc.M804482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wikstrom M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander A, Dowhan W. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem. 2004;279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 26.Xie J, Bogdanov M, Heacock P, Dowhan W. Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J Biol Chem. 2006;281:19172–19178. doi: 10.1074/jbc.M602565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Bio. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitra K, Ubarretxena-Belandia I, Taguchi T, Warren G, Engelman DM. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. P Natl Acad Sci USA. 2004;101:4083–4088. doi: 10.1073/pnas.0307332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daum G. Lipids of Mitochondria. Biochim Biophys Acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 30.Allan D. Mapping the lipid distribution in the membranes of BHK cells (mini-review) Mol Membr Biol. 1996;13:81–84. doi: 10.3109/09687689609160580. [DOI] [PubMed] [Google Scholar]
- 31.Zambrano F, Fleischer S, Fleischer B. Lipid composition of the Golgi apparatus of rat kidney and liver in comparison with other subcellular organelles. Biochim Biophys Acta. 1975;380:357–369. doi: 10.1016/0005-2760(75)90104-6. [DOI] [PubMed] [Google Scholar]
- 32.Henry SA, editor. Biochemical and Genetic Studies. 11B. Cold Spring Harbor Laboratory Press; 1982. Membrane Lipids of Yeast. [Google Scholar]
- 33.Schneiter R, Brugger B, Sandhoff R, Zellnig G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G, Paltauf F, Wieland FT, Kohlwein SD. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wileman T, Harding C, Stahl P. Receptor-Mediated Endocytosis. Biochem J. 1985;232:1–14. doi: 10.1042/bj2320001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scharschmidt BF, Lake JR, Renner EL, Licko V, Vandyke RW. Fluid Phase Endocytosis by Cultured Rat Hepatocytes and Perfused-Rat-Liver - Implications for Plasma-Membrane Turnover and Vesicular Trafficking of Fluid Phase Markers. P Natl Acad Sci USA. 1986;83:9488–9492. doi: 10.1073/pnas.83.24.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steer MW. Plasma-Membrane Turnover in Plant-Cells. J Exp Bot. 1988;39:987–996. [Google Scholar]
- 37.Cheng S, Levy D. Effects of Cell-Proliferation on the Lipid-Composition and Fluidity of Hepatocyte Plasma-Membranes. Arch Biochem Biophys. 1979;196:424–429. doi: 10.1016/0003-9861(79)90293-5. [DOI] [PubMed] [Google Scholar]
- 38.Gulaya NM, Volkov GL, Klimashevsky VM, Govseeva NN, Melnik AA. Changes in Lipid-Composition of Neuro-Blastoma C1300-N18 Cell during Differentiation. Neuroscience. 1989;30:153–164. doi: 10.1016/0306-4522(89)90361-8. [DOI] [PubMed] [Google Scholar]
- 39.Lampe MA, Williams ML, Elias PM. Human Epidermal Lipids - Characterization and Modulations during Differentiation. J Lipid Res. 1983;24:131–140. [PubMed] [Google Scholar]
- 40.Cronan JE. Phospholipid Alterations during Growth of Escherichia Coli. J Bacteriol. 1968;95:2054. doi: 10.1128/jb.95.6.2054-2061.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuiper PJC, Kahr M, Stuiver CEE, Kylin A. Lipid-Composition of Whole Roots and of Ca2+, Mg2+-Activated Adenosine Triphosphatases from Wheat and Oat as Related to Mineral Nutrition. Physiol Plantarum. 1974;32:33–36. [Google Scholar]
- 42.Zhang GC, Slaski JJ, Archambault DJ, Taylor GJ. Alteration of plasma membrane lipids in aluminum-resistant and aluminum-sensitive wheat genotypes in response to aluminum stress. Physiol Plantarum. 1997;99:302–308. [Google Scholar]
- 43.Larkindale J, Huang BR. Changes of lipid composition and saturation level in leaves and roots for heat-stressed and heat-acclimated creeping bentgrass (Agrostis stolonifera) Environ Exp Bot. 2004;51:57–67. [Google Scholar]
- 44.Burkey KO, Wilson RF, Wells R. Effects of canopy shade on the lipid composition of soybean leaves. Physiol Plantarum. 1997;101:591–598. [Google Scholar]
- 45.Wriessnegger T, Gubitz G, Leitner E, Ingolic E, Cregg J, de la Cruz BJ, Daum G. Lipid composition of peroxisomes from the yeast Pichia pastoris grown on different carbon sources. Bba-Mol Cell Biol L. 2007;1771:455–461. doi: 10.1016/j.bbalip.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Hunter MIS, Thirkell D. Variation in Fatty Acid Composition of Sarcina-Flava Membrane Lipid with Age of Bacterial Culture. J Gen Microbiol. 1971;65:115. doi: 10.1099/00221287-65-1-115. [DOI] [PubMed] [Google Scholar]
- 47.Luzzati V, Gambacorta A, Derosa M, Gulik A. Polar Lipids of Thermophilic Prokaryotic Organisms - Chemical and Physical Structure. Annu Rev Biophys Bio. 1987;16:25–47. doi: 10.1146/annurev.bb.16.060187.000325. [DOI] [PubMed] [Google Scholar]
- 48.Goldfine H. Lipids of Prokaryotes - Structure and Distribution. Curr Top Membr Trans. 1982;17:1–43. [Google Scholar]
- 49.Kaneshiro ES. Lipids of Paramecium. J Lipid Res. 1987;28:1241–1258. [PubMed] [Google Scholar]
- 50.Ulrih NP, Gmajner D, Raspor P. Structural and physicochemical properties of polar lipids from thermophilic archaea. Appl Microbiol Biotechnol. 2009;84:249–260. doi: 10.1007/s00253-009-2102-9. [DOI] [PubMed] [Google Scholar]
- 51.Prasad R. Lipids in the structure and function of yeast membrane. Adv Lipid Res. 1985;21:187–242. doi: 10.1016/b978-0-12-024921-3.50012-5. [DOI] [PubMed] [Google Scholar]
- 52.Benning C. Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu Rev Cell Dev Biol. 2009;25:71–91. doi: 10.1146/annurev.cellbio.042308.113414. [DOI] [PubMed] [Google Scholar]
- 53.Oberai A, Joh NH, Pettit FK, Bowie JU. Structural imperatives impose diverse evolutionary constraints on helical membrane proteins. P Natl Acad Sci USA. 2009;106:17747–17750. doi: 10.1073/pnas.0906390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mokrab Y, Stevens TJ, Mizuguchi K. A structural dissection of amino acid substitutions in helical transmembrane proteins. Proteins. 2010;78:2895–2907. doi: 10.1002/prot.22809. [DOI] [PubMed] [Google Scholar]
- 55.Paskhughes RA, Shaw N. Glycolipids from Some Extreme Thermophilic Bacteria Belonging to the Genus Thermus. J Bacteriol. 1982;149:54–58. doi: 10.1128/jb.149.1.54-58.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heinen W, Klein HP, Volkmann CM. Fatty Acid Composition of Thermus-Aquaticus at Different Growth Temperatures. Arch Mikrobiol. 1970;72:199. doi: 10.1007/BF00409525. [DOI] [PubMed] [Google Scholar]
- 57.Oshima M, Yamakawa T. Chemical-Structure of a Novel Glycolipid from an Extreme Thermophile, Flavobacterium-Thermophilum. Biochemistry-Us. 1974;13:1140–1146. doi: 10.1021/bi00703a014. [DOI] [PubMed] [Google Scholar]
- 58.Brosig A, Nesper J, Boos W, Welte W, Diederichs K. Crystal Structure of a Major Outer Membrane Protein from Thermus thermophilus HB27. J Mol Biol. 2009;385:1445–1455. doi: 10.1016/j.jmb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Pautsch A, Schulz GE. High-resolution structure of the OmpA membrane domain. J Mol Biol. 2000;298:273–282. doi: 10.1006/jmbi.2000.3671. [DOI] [PubMed] [Google Scholar]
- 60.Vandeputte-Rutten L, Bos MP, Tommassen J, Gros P. Crystal structure of Neisserial surface protein A (NspA), a conserved outer membrane protein with vaccine potential. J Biol Chem. 2003;278:24825–24830. doi: 10.1074/jbc.M302803200. [DOI] [PubMed] [Google Scholar]
- 61.Rahman MM, Kolli VSK, Kahler CM, Shih G, Stephens DS, Carlson RW. The membrane phospholipids of Neisseria meningitidis and Neisseria gonorrhoeae as characterized by fast atom bombardment mass spectrometry. Microbiol-Sgm. 2000;146:1901–1911. doi: 10.1099/00221287-146-8-1901. [DOI] [PubMed] [Google Scholar]
- 62.Monbouquette HG, Lai D, Springstead JR. Effect of growth temperature on ether lipid biochemistry in Archaeoglobus fulgidus. Extremophiles. 2008;12:271–278. doi: 10.1007/s00792-007-0126-6. [DOI] [PubMed] [Google Scholar]
- 63.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrade SLA, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. P Natl Acad Sci USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khademi S, O’Connell J, Remis J, Robles-Colmenares Y, Miericke LJW, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.3.5 angstrom. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 66.Savage DF, Egea PF, Robles-Colmenares Y, O’Connell JD, 3rd, Stroud RM. Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z. PLoS Biol. 2003;1:E72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bondar AN, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hite RK, Li Z, Walz T. Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J. 2010;29:1652–1658. doi: 10.1038/emboj.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McIntosh TJ, Tong JH, Briggs MM, Mlaver D, Vidal A. Sorting of Lens Aquaporins and Connexins into Raft and Nonraft Bilayers: Role of Protein Homo-Oligomerization. Biophys J. 2009;97:2493–2502. doi: 10.1016/j.bpj.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borchman D, Yappert MC. Lipids and the ocular lens. J Lipid Res. 2010;51:2473–2488. doi: 10.1194/jlr.R004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harries WE, Akhavan D, Miercke LJ, Khademi S, Stroud RM. The channel architecture of aquaporin 0 at a 2.2-A resolution. Proc Natl Acad Sci U S A. 2004;101:14045–14050. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian C, Sonnichsen FD, Sanders CR. Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badola P, Sanders CR., 2nd Escherichia coli diacylglycerol kinase is an evolutionarily optimized membrane enzyme and catalyzes direct phosphoryl transfer. J Biol Chem. 1997;272:24176–24182. doi: 10.1074/jbc.272.39.24176. [DOI] [PubMed] [Google Scholar]
- 75.Bohnenberger E, Sandermann H., Jr Lipid dependence of diacylglycerol kinase from Escherichia coli. Eur J Biochem. 1983;132:645–650. doi: 10.1111/j.1432-1033.1983.tb07412.x. [DOI] [PubMed] [Google Scholar]
- 76.Walsh JP, Bell RM. sn-1,2-Diacylglycerol kinase of Escherichia coli. Structural and kinetic analysis of the lipid cofactor dependence. J Biol Chem. 1986;261:15062–15069. [PubMed] [Google Scholar]
- 77.Koehler J, Sulistijo ES, Sakakura M, Kim HJ, Ellis CD, Sanders CR. Lysophospholipid micelles sustain the stability and catalytic activity of diacylglycerol kinase in the absence of lipids. Biochemistry. 2010;49:7089–7099. doi: 10.1021/bi100575s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gorzelle BM, Hoffman AK, Keyes MH, Gray DN, Ray DG, Sanders CR. Amphipols can support the activity of a membrane enzyme. J Am Chem Soc. 2002;124:11594–11595. doi: 10.1021/ja027051b. [DOI] [PubMed] [Google Scholar]
- 79.Czerski L, Sanders CR. Functionality of a membrane protein in bicelles. Anal Biochem. 2000;284:327–333. doi: 10.1006/abio.2000.4720. [DOI] [PubMed] [Google Scholar]
- 80.Caffrey M, Li DF. Lipid cubic phase as a membrane mimetic for integral membrane protein enzymes. P Natl Acad Sci USA. 2011;108:8639–8644. doi: 10.1073/pnas.1101815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramer JK, Bell RM. Expression of the Phospholipid-Dependent Escherichia-Coli Sn-1,2-Diacylglycerol Kinase in Cos Cells Perturbs Cellular Lipid-Composition. Journal of Biological Chemistry. 1990;265:16478–16483. [PubMed] [Google Scholar]
- 82.Pilot JD, East JM, Lee AG. Effects of bilayer thickness on the activity of diacylglycerol kinase of Escherichia coli. Biochemistry-Us. 2001;40:8188–8195. doi: 10.1021/bi0103258. [DOI] [PubMed] [Google Scholar]
- 83.Pilot JD, East JM, Lee AG. Effects of phospholipid headgroup and phase on the activity of diacylglycerol kinase of Escherichia coli. Biochemistry-Us. 2001;40:14891–14897. doi: 10.1021/bi011333r. [DOI] [PubMed] [Google Scholar]
- 84.Russ E, Kaiser U, Sandermann H., Jr Lipid-dependent membrane enzymes. Purification to homogeneity and further characterization of diacylglycerol kinase from Escherichia coli. Eur J Biochem. 1988;171:335–342. doi: 10.1111/j.1432-1033.1988.tb13795.x. [DOI] [PubMed] [Google Scholar]
- 85.Berger C, Ho JT, Kimura T, Hess S, Gawrisch K, Yeliseev A. Preparation of stable isotope-labeled peripheral cannabinoid receptor CB2 by bacterial fermentation. Protein Expr Purif. 2010;70:236–247. doi: 10.1016/j.pep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Breyer RM, Strosberg AD, Guillet JG. Mutational analysis of ligand binding activity of beta 2 adrenergic receptor expressed in Escherichia coli. EMBO J. 1990;9:2679–2684. doi: 10.1002/j.1460-2075.1990.tb07453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Furukawa H, Haga T. Expression of functional M2 muscarinic acetylcholine receptor in Escherichia coli. J Biochem. 2000;127:151–161. doi: 10.1093/oxfordjournals.jbchem.a022577. [DOI] [PubMed] [Google Scholar]
- 88.Grisshammer R, White JF, Trinh LB, Shiloach J. Large-scale expression and purification of a G-protein-coupled receptor for structure determination -- an overview. J Struct Funct Genomics. 2005;6:159–163. doi: 10.1007/s10969-005-1917-6. [DOI] [PubMed] [Google Scholar]
- 89.Stanasila L, Massotte D, Kieffer BL, Pattus F. Expression of delta, kappa and mu human opioid receptors in Escherichia coli and reconstitution of the high-affinity state for agonist with heterotrimeric G proteins. Eur J Biochem. 1999;260:430–438. doi: 10.1046/j.1432-1327.1999.00187.x. [DOI] [PubMed] [Google Scholar]
- 90.Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping P, Abramson J. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci U S A. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hite RK, Gonen T, Harrison SC, Walz T. Interactions of lipids with aquaporin-0 and other membrane proteins. Pflugers Arch. 2008;456:651–661. doi: 10.1007/s00424-007-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]