Abstract
Proliferating Cell Nuclear Antigen (PCNA) ubiquitylation plays a crucial role in maintaining genomic stability during DNA replication. DNA damage stalling the DNA replication fork induces PCNA ubiquitylation that activates DNA damage bypass to prevent the collapse of DNA replication forks that could potentially produce double strand breaks and chromosomal rearrangements. PCNA ubiquitylation dictates the mode of bypass depending on the level of ubiquitiylation; monoubiquitylation and polyubiquitylation activate error-prone translesion synthesis and error-free template switching, respectively. Due to the error-prone nature of DNA damage bypass, PCNA ubiquitylation needs to be tightly regulated. Here, we review the molecular mechanisms to remove ubiquitin from PCNA including the emerging role of USP1 and ELG1 in this fascinating process.
Keywords: Ubiquitylation, Deubiquitylation, PCNA, USP1, ELG1
Introduction
Protein ubiquitylation is a reversible, 76 amino acid post-translational modification carried out by the coordinated activites of an E1 ubiquitin-activating enzyme, an E2 ubiquitin conjugase, and an E3 ubiquitin ligase. In the first step of the ubiquitylation reaction, ubiquitin is activated in an ATP-dependent process in which a ubiquitin-adenylate intermediate is formed and ubiquitin is transferred to a cysteine residue in the E1 active site, resulting in a thioester linkage between the C-terminal carboxyl group of ubiquitin and the E1 cysteine sulfydral group. Ubiquitin is then transferred to an E2 via a trans(thio)esterification reaction. Finally, an E3 binds to both the ubiquitin-carrying E2 and the substrate, and catalyzes the formation of an isopeptide bond between a lysine of the target protein and the C-terminal glycine of ubiquitin. Protein ubiquitylation plays an important role in the regulation of many biological processes including protein stability, cell cycle progression, apoptosis, growth signal transduction, transcriptional regulation, endocytosis, vesicle trafficking, and the DNA damage response [1]. The functional consequence of a particular ubiquitin moiety is dictated by the length of the ubiquitin chain as well as the linkage type. For example, lysine 48 (K48)-linked polyubiquitin chains mainly mark proteins for proteolysis, whereas lysine 164 (K164)-linked monoubiquitylation triggers different cellular processes including DNA repair [2].
Opposing the activities of the E1/E2/E3 ubiquitylating enzymes are the deubiquitylating enzymes (DUBs). These cysteine or metallo-proteases cleave ubiquitin from linear ubiquitin polypeptides or from specific mono- or polyubiquitylated substrates and are responsible for processing inactive ubiquitin precursors, proofreading ubiquitin-protein conjugates, and removing and recycling ubiquitin from cellular adducts [3]. Cysteine protease DUBs are organized into four subclasses based on their ubiquitin-protease domains: ubiquitin-specific protease (USP), ubiquitin C-terminal hydrolase (UCH), Otubain protease (OTU), and Machado-Joseph disease protease (MJD). The metallo-protease DUBs belong to the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain superfamily.
To date, numerous cellular targets of ubiquitin have been identified [4–10]. In this review, we focus on the ubiquitylation and deubiquitylation of Proliferating Cell Nuclear Antigen (PCNA), a homotrimeric ring that functions as a DNA polymerase sliding clamp accessory protein as well as a scaffold for numerous proteins involved in DNA replication, repair, cell cycle regulation, and chromatin assembly. We review the importance of PCNA ubiquitylation and deubiquitylation in the DNA damage response, with special emphasis on DNA damage bypass following replication stress. Finally, we will discuss the coordinated activities of ubiquitin specific protease 1 (USP1) and the newly identified function of Enhanced Level of Genome Instability Gene 1 (ELG1) as a regulator of PCNA ubiquitylation.
The Role of PCNA Ubiquitylation in DNA Damage Bypass
DNA Damage Bypass
Replication stress can arise from both endogenous metabolic processes and exogenous DNA damaging agents and can interfere with the progression of the DNA replication fork. For example, UV light promotes the formation of cyclobutane pyrimidine dimers that alter the structure of DNA and consequently inhibit DNA polymerases and hydroxyurea depletes nucleotide pools thereby halting DNA synthesis. Failure to relieve such DNA replication stress can have dire consequences for the cell, as stalled replication forks are prone to collapse and could potentially lead to double-strand breaks (DSBs) that result in cell death, or to gross chromosomal rearrangements that have a close link to tumorigenesis. To prevent such a situation, cells have evolved a mode of DNA damage bypass termed post-replication repair (PRR). Although PRR does not remove the actual lesion from DNA, it does enable the cell cycle to safely progress from the S to G2 phase, where damage can then be repaired by appropriate DNA repair pathways including base excision repair, nucleotide excision repair, or homologous recombination repair. PRR can be divided into two distinct pathways: translesion synthesis (TLS) and template switching. Whereas the latter bypass is thought to utilize the genetic information encoded by the newly synthesized, undamaged sister chromatid to carry out a recombination-mediated mechanism, TLS employs specialized, DNA damage-tolerant polymerases that can synthesize DNA directly across the damaged template. As discussed below, the post-translational modification of PCNA by ubiquitin and the small ubiquitin-like modifier (SUMO) plays a key role in deciding which of these pathways is used for the processing of DNA lesions that arise during replication.
PCNA Monoubiquitylation
Following DNA damage that stalls the progression of the DNA replication fork, the highly conserved K164 of PCNA is monoubiquitylated by the E2 Rad6 and the E3 Rad18 (Figure 1) [11]. PCNA monoubiquitylation has been reported in a wide variety of organisms including yeast, Xenopus, chicken, and mammals. The trigger for DNA damage-induced PCNA monoubiquitylation is believed to result from the uncoupling of the stalled replicative polymerase and the MCM helicase, which results in production of single-stranded DNA exposed in the vicinity of the stalled fork. This single-stranded DNA is coated by Replication Protein A (RPA), which in turn interacts with Rad18 and directs the Rad6/Rad18 complex to the site of DNA damage [12,13]. Consistent with this model, there are data demonstrating that Rad18 and RPA physically interact, the siRNA-mediated depletion of RPA2 in human cells results in a moderate reduction in damage-induced PCNA ubiquitylation, and a RPA-coated immobilized oligonucleotide can recruit Rad6/Rad18 [13–15]. In vitro reactions, RPA appears to be dispensable for PCNA modification, although this could be explained by the high concentration of conjugation factors present in the reaction [14,16]. The direct binding of Rad18 to DNA appears to be important for PCNA monoubiquitylation [13]. However, since the in vitro affinity of the Rad6/Rad18 complex for single-standed DNA is highly dependent on salt concentration, the physiological relevance of the direct binding of Rad18 to DNA is still controversial [14].
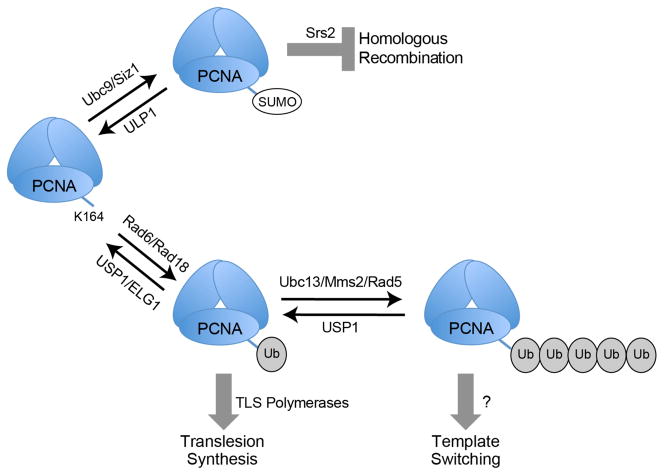
Figure 1.
PCNA modifications occur following DNA replication stress. In response to DNA damage stalling the DNA replication fork, K164 of PCNA can either be SUMOylated (SUMO) by Ubc9/Siz1 or monoubiquitylated (Ub) by Rad6/Rad18. The former modification inhibits inappropriate homologous recombination by recruiting the helicase Srs2, and the latter attracts specialized polymerases that carry out translesion synthesis. K164 monoubiquitylation can also be further extended by Ubc13/Mms2/Rad5 to initiate template switching by a currently unknown mechanism. PCNA SUMOylation and ubiquitylation are both reversible – SUMO is removed by ULP1 and ubiquitin is removed by USP1/ELG1.
Since RPA-bound single-stranded DNA is thought to initiate the ATR-Chk1 DNA damage checkpoint pathway [17], several studies have been conducted to investigate the relationship between PCNA monoubiquitylation and the activation of the S phase checkpoint. Although one such study demonstrated that the siRNA-mediated knockdown of ATR and the overexpression of an inactive Chk1 significantly reduced PCNA ubiquitylation upon exposure to the DNA adduct benzo[a]pyrene dihydrodiol epoxide (BDPE) [18], the results from several other experiments indicate that PCNA ubiquitylation occurs independently of checkpoint activation following DNA damage [13,19–22]. One notable exception is PCNA monoubiquitylation at K107, which was recently shown to be a prerequisite for checkpoint activation in DNA ligase I-deficient cells [23].
Once monoubiquitylated at K164, PCNA recruits members of the Y-family TLS polymerases (Polη, Polκ, Polι, and REV1) as well as the B-family TLS polymerase Polζ to the site of DNA damage (Figure 1). The TLS polymerases interact with monoubiquitylated PCNA through their UBM or UBZ ubiquitin-binding domains as well as through their PCNA interacting peptide (PIP) motifs. The preferential binding of the TLS polymerases to monoubiquitylated PCNA enables the TLS polymerases to replace the stalled replicative DNA polymerase (Polδ or Polε) at the blocked sites of the DNA replication fork [24–29]. In contrast to the replicative polymerases, the TLS polymerases have large open active sites that can accommodate bulky DNA lesions. Consequently, the TLS polymerases have the ability to bypass DNA adducts without removing the actual lesion [30]. Whereas some TLS polymerases, such as Polη, can bypass DNA damage in a fairly error-free fashion [31–33], others, such as Polζ, exhibit low fidelity when bypassing DNA damage and result in elevated mutagenesis [34]. The physiological importance of TLS is highlighted by the discovery that a mutation in the human POLH gene encoding Polη is responsible for the variant form of the skin cancer-prone syndrome Xeroderma Pigmentosum [31,32].
PCNA Polyubiquitylation
In many organisms including budding yeast, S. cerevisiae, K164 monoubiquitylation can be further polyubiquitylated by the E2/E2 variant Ubc13/Mms2 and the E3 Rad5 (Figure 1). Ubc13/Mms2 is recruited from the cytoplasm to the site of the DNA lesion through the RING finger domain of Rad5, and Rad5 is recruited to the stalled fork through interaction with Rad18 [11,35]. The coordinated enzymatic activities of Ubc13/Mms2 and Rad5 generate a polyubiquitylated chain linked through the K63 residue of ubiquitin [36,37]. The addition of this chain activates the template switching mechanism of DNA damage bypass (Figure 1) by dislodging TLS polymerases from PCNA, preventing the interaction of the TLS polymerses with PCNA, or recruiting factors that carry out template switching processes [38]. Unlike TLS, template switching seems to be error-free. In fact, epistatic analyses in yeast revealed that loss of the template switching mechanism results in increased mutagenesis, presumably due to an increased reliance on the more error-prone TLS for lesion bypass [39–42]. The mechanism of template switching is still poorly understood due to the transient nature of the intermediates formed and the current inability to identify many of the genes directly involved in the process but it is known to require the helicase activity of Rad5 [43]. Although still the subject of debate, it has been hypothesized that template switching proceeds by a “chicken foot” mechanism. In this model, Rad5 unwinds and anneals the nascent and template strands, and subsequent fork regression activity produces a four-way junction intermediate resembling a “chicken foot.” DNA polymerase then extends the 3′ end of the leading nascent strand by copying from the nascent lagging strand, and the regression of the four-way junction completes error-free replication through the DNA damage. In a second model, a homologous recombination-like mechanism occurs whereby the sister duplex is invaded by a single-stranded gap, forming a D-loop [11,41].
There is increasing evidence that PCNA polyubiquitylation also exists in mammalian systems, although it occurs at much lower levels than monoubiquitylation and is only readily detectible after over-expression of the relevant E3s [41,44]. RAD18 and UBC13 are required for PCNA polyubiquitylation in human cells, but MMS2 appears to be dispensable [45]. Recently, HLTF and SHPRH were identified as putative mammalian homologues of yeast Rad5. Both proteins interact with RAD6/RAD18 and UBC13/MMS2 and promote the polyubiquitylation of PCNA [44,46–48]. However, hltf/shprh double mutant mouse embryonic fibroblasts (MEFs) are still able to polyubiquitylate PCNA, suggesting the existence of an alternative E3 ligase [49].
So what triggers the switch from PCNA monoubiquitylation to polyubiquitylation (and by extension the switch from TLS to template switching)? Although the answer to this question is not totally clear, the level of DNA damage is thought to play a role, since polyubiquitylation is observed following treatment with increasing amounts of DNA damaging agents [48] and with increasing amounts of RPA-coated single-stranded DNA [21]. It has been suggested that an elevation in the amount of RPA-coated single-stranded DNA results in a concomitant increase in RPA-bound Rad18, and a subsequent decrease in Rad18 homodimerization. Since Rad5 interacts with Rad18 at the same site at which homodimerization occurs, Rad5 would then be able to better compete for binding to Rad18 and would thus promote polyubiquitylation of PCNA [38].
PCNA SUMOylation
In addition to ubiquitin, PCNA can be covalently modified by SUMO following replication stress or during S phase in the absence of DNA damage. PCNA SUMOylation occurs in an enzymatic cascade that is analogous to that involved in ubiquitylation. PCNA is SUMOylated at K164 and K127 by the E2 Ubc9 and the E3 Siz1 [11,50]. In S. cerevisiae, SUMOylated PCNA recruits the helicase Srs2 through a conserved SUMO-interaction motif in the carboxy-terminus of Srs2 [51]. Recruitment of Srs2 disrupts Rad51 single-stranded filaments [52], thereby preventing inappropriate homologous recombination [53]. When this occurs at a stalled replication fork, the inhibition of recombination by Srs2 allows for the processing of lesions by ubiquitin-dependent TLS or template switching [16,54–56] (Figure 1). PCNA SUMOylation also assists recombination-dependent gross chromosomal rearrangement [57]. The SUMOylation of PCNA is a reversible modification that can be removed by the SUMO protease Ubl-specific protease 1 (ULP1) (Figure 1).
In addition to S.cerevisiae, PCNA SUMOylation has also been shown to occur in X. laevis egg extracts and chicken DT40 cells [58–60]. Given that none of the Srs2-like helicases in X. laevis egg extracts and chicken DT40 cells are known to exert an effect on the Rad6 pathway, it is likely that PCNA SUMOylation in these organisms functions in a different manner than PCNA SUMOylation in yeast [14]. Modification of PCNA by SUMO has yet to be reported in human cells [49].
The Deubiquitylation of PCNA as a Safeguard Against Error-Prone TLS
USP1 was identified in a siRNA screen as a DUB responsible for the deubiquitylation of PCNA following DNA damage bypass [61]. USP1 is also involved in the deubiquitylation of FANCD2, a Fanconi anemia effector protein that functions in the repair of DNA interstrand crosslinks [62]. Considering the opposing roles of the E1/E2/E3 ubiquitinating enzymes and the DUBs, it is not surprising that exposure to UV light, which promotes PCNA monoubiquitylation, also decreases USP1 protein levels. The mechanism for this decrease involves an autocleavage event followed by proteasomal degredation of the cleaved products. The ubiquitylation of PCNA following replication stress is not always accompanied by the disappearance of USP1, though. For example, treatment with hydroxyurea (HU) results in no detectable change in USP1. To explain this phenomenon, it has been suggested that HU disrupts the interaction between USP1 and its activating partner protein UAF1 (USP1-associated factor 1) [12,61].
The importance of PCNA deubiquitylation is highlighted by the finding that depletion of USP1 increases the level of mutagenesis in the cell [61]. It is thought that persistent PCNA ubiquitylation results in the overuse and subsequent disregulation of the TLS polymerases and allows the error-prone TLS polymerases to replicate undamaged DNA [61]. Even Polη, which can faithfully replicate past UV-induced cyclobutane pyrimidine dimers, exhibits much lower fidelity compared to replicative polymerases when copying undamaged DNA [63], and would thus contribute to increased mutagenesis if used inappropriately. Such mutations could ultimately disrupt basic cellular processes and/or cause uncontrolled proliferation, the latter of which is a hallmark of cancer. In addition to being more error-prone than the replicative DNA polymerases, the TLS polymerases also catalyze DNA synthesis much more slowly and thus could potentially cause replication fork collapse. As discussed above, collapsed replication forks are particularly dangerous to cells, as they often result in genomic instability.
Insights into the possible physiological consequences of defects in PCNA deubiquitylation come from a USP1 transgenic mouse model. Usp1−/− mice displayed an increase in both FANCD2 and PCNA monoubiquitylation, and exhibited a high rate of perinatal lethality, depletion of male germ cells, infertility, hypersensitivity to the crosslinking agent mitomycin C, and chromosome instability. Usp1−/− mice were also much smaller in size than their wildtype littermates [64]. It remains to be determined whether these mice exhibit increased point mutation frequency and increased cancer incidence as would be expected given the in vitro data described above.
ELG1 and its Role in the Deubiquitylation of PCNA
ELG1 and the RFC complex
The loading and unloading of PCNA on DNA is carried out by an ATP-dependent Replication Factor C (RFC) complex. The canonical RFC complex consists of 5 subunits, RFC 1-5. There also exist three alternative RFC complexes in which RFC1 is replaced by either CTF18, RAD17, or ELG1. The canonical RFC loads PCNA onto DNA during general DNA replication [65]. CTF18-RFC unloads PCNA from DNA where sister chromatids are held by a cohesion complex [66], and has recently been implicated in the replication fork bypass of lesions that arise from triplet repeats [67] and in S-phase checkpoint activation [68,69]. RAD17-RFC loads the PCNA-like 9-1-1 complex (consisting of RAD9, RAD1, and HUS1) onto damaged DNA for activation of the DNA damage checkpoint [70].
Initially, the role of the ELG1-RFC complex was not clearly understood, even though it was known to play an important part in the suppression of gross chromosomal rearrangements and the maintenance of genomic stability during normal cell growth [71–75]. However, studies in yeast soon brought forth several possibilities [76]. The first was that ELG1-RFC functions as a clamp loader during DNA replication. Although this hypothesis was supported by ELG1’s interaction with PCNA and the flap endonuclease Rad27 (yeast FEN1) [73], PCNA loading or unloading by ELG1-RFC could not be detected in vitro. ELG1’s interaction with PCNA and its colocalization with Polη also raised the possibility that ELG1-RFC could serve as a platform for polymerase switching during TLS. Alternatively, because ELG1 and template switching both suppress gross chromosomal rearrangements, it was easy to envision a role for ELG1 in the error-free mode of DNA damage bypass. Finally, it was suggested that ELG1-RFC functions in chromatin assembly through PCNA interaction. Evidence for this came from the synthetic lethality between elg1 and the htb1 or bre11 mutants that create defects in histone levels and histone modification [77].
ELG1 as a Regulator of PCNA Ubiquitylation
Last year, great strides were made in understanding the functional significance of ELG1 when it was discovered that the human homolog of ELG1 (which is also called ATAD5) not only interacts with PCNA and colocalizes with the sliding clamp at stalled replication forks with distinct foci structures in the nucleus, but also associates with the USP1-UAF1 complex. This observation led to the hypothesis that ELG1 affects the deubiquitylation of PCNA (Figure 2). Indeed, the siRNA-mediated knockdown of ELG1 resulted in an increase in PCNA monoubiquitylation that could be rescued by ectopic expression of siRNA-resistant ELG1. The regulation of PCNA ubiquitylation was specific to ELG1 and vice versa, as the knockdown of CTF18 and RAD17 did not increase PCNA ubiquitylation and the knockdown of ELG1 did not affect the ubiquitylation of USP1’s other target protein FANCD2. The knockdown of RFC1 or RFC4 also did not increase PCNA ubiquitylation, suggesting that this function of ELG1 is independent of its role as an alternative RFC complex [78].
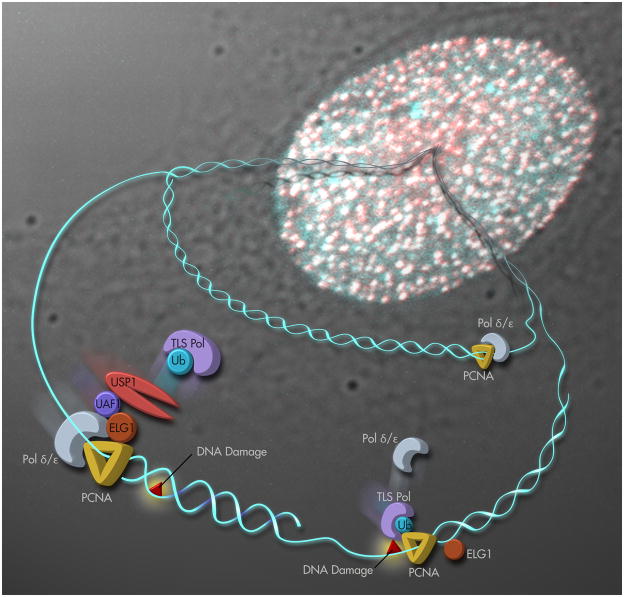
Figure 2.
A model for the role of ELG1 in PCNA deubiquitylation. Upon encountering DNA damage, the replication machinery becomes stalled. PCNA is then monoubiquitylated, and a TLS polymerase is recruited to the damage site to replace the replicative polymerase (Polδ/ε) and bypass the lesion. Once bypass has occurred, ELG1, which becomes concentrated at stalled forks with distinct foci structures in the nucleus, binds to PCNA and recruits the USP1/UAF1 complex. USP1 deubiquitylates PCNA, displacing the TLS polymerase and allowing Polδ/ε to resume normal replication.
How does ELG1 down-regulate PCNA ubiquitylation? One possibility is that ELG1 modulates the expression or stability of USP1. However, no detectable changes in USP1 protein levels were observed following ELG1 knockdown. The second possibility is that ELG1 enhances or stabilizes the interaction between USP1 and its activating factor UAF1. This scenario is also unlikely, considering that the ability of USP1 to interact with UAF1 remained unchanged in ELG1-knockdown cells. Finally, ELG1 could regulate USP1 activity or recruit USP1-UAF1 to monoubiquitylated PCNA. In support of this theory, a synergistic reduction in UV-induced PCNA monoubiquitylation was observed following the overexpression of both ELG1 and USP1. Additionally, the overexpression of USP1 alone had no effect on PCNA monoubiquitylation in the absence of ELG1, indicating that ELG1 is required for the deubiquitylation of PCNA by USP1. The recruitment of USP1-UAF1 appears to be transient because ELG1 knockdown did not affect the levels of USP1 bound to chromatin, and in contrast to ELG1, neither USP1 nor UAF1 formed foci following DNA damage [78].
Unlike the observation in human cells, the knockout of the ELG1 gene in S. cerevisiae caused a higher level of chromatin-bound SUMOylated PCNA [79]. The SUMO-interacting domain of yeast Elg1 appears to be important for the interaction between Elg1 and PCNA and for regulating chromatin-bound SUMOylated PCNA. Since SUMOylated PCNA inhibits homologous recombination [55,56], and elg1 mutation is synthetic lethal with genes involved in homologous recombination [77], yeast Elg1 might function to reduce the level of SUMOylated PCNA in chromatin. However, the fact that elg1 mutation generates a hyper-recombination phenotype [72,80], hints further complexity regarding the genetic interaction between ELG1 and SUMOylated PCNA. It is also still unclear whether Elg1-RFC actively unloads SUMOylated PCNA from chromatin or recruits the deSUMOylating enzyme Ulp1 to chromatin to reduce the level of SUMOylated PCNA. Due to the absence of SUMOylated PCNA (at least in published literature), it is unclear if there is a similar mechanism conserved in mammalian systems [49].
As discussed above, PCNA deubiquitylation plays an important role in preventing TLS polymerases with low fidelity and processivity from replicating undamaged DNA, a process that could result in mutagenesis, DSBs, and gross chromosomal rearrangements. Consistent with the proposed role for ELG1 in the USP1-UAF1-mediated deubiquitylation of PCNA, ELG1 knockdown cells display a significant increase in mutation frequency [78], a higher incidence of chromosome end-to-end fusions, inversions and aneuploidy [81], and an increase in the foci formation of the DSB markers γH2AX, 53BP1, and phospho-ATM [81]. Elg1+/− MEFs, which display an increase in the levels of chromatin-bound PCNA compared to wildtype MEFs, also exhibit genomic instability and spontaneous DNA damage. Furthermore, Elg1+/− mice develop a wide variety of tumors displaying genomic instability and die between 11 and 18 months of age (Myung lab, unpublished results).
Conclusion
Much progress has been made in understanding the regulation of PCNA ubiquitylation and its role in DNA damage bypass and the maintenance of genomic integrity. We now know that K164 monoubiquitylation by RAD6/RAD18 functions to recruit the Y-family DNA polymerases and activate TLS, whereas further extension of this modification by UBC13/MMS2/RAD5 initiates an error-free template switching mechanism. PCNA SUMOylation can also occur at the same residue as ubiquitylation and serves to suppress homologous recombination at the site of the stalled replication fork through the recruitment of Srs2. Equally important as PCNA ubiquitylation is PCNA deubiquitylation, which is carried out by the USP1-UAF1 complex in conjunction with ELG1. ELG1 specifically directs the USP1-UAF1 to PCNA at the damage site, and in doing so initiates the switch from the error-prone and poorly processive TLS polymerases to the faithful replicative polymerases Polδ and Polε. Such a switch is necessary to reduce the likelihood of mutagenic effects by the TLS polymerases following DNA damage bypass.
Despite the significant advancements made, many questions still remain. For example, exactly when does DNA damage bypass occur? What are the conditions under which the template switching mode of PRR is activated in favor of TLS? What is the mechanism of template switching? And what regulates USP1 activity when the protein is not actively degraded following DNA damage? Future studies addressing these issues will provide more detailed insight into the pathways in which PCNA ubiquitylation/deubiquitylation function, and further highlight the importance of these modifications in the maintenance of genomic integrity.
Highlights.
In this review, we went through post-translational modifications of PCNA.
PCNA ubiquitylation allows cells to bypass DNA damage by translesion synthesis.
PCNA deubiquitylation after DNA damage bypass is catalyzed by USP1-UAF1.
PCNA deubiquitylation is promoted by ELG1-RFC in mammals.
There are alternative post-translational modifications of PCNA.
Acknowledgments
We thank A. D’Andrea (Dana Faber Cancer Institute), J. McCulley (NHGRI, NIH) and R. Woodgate (NICHD, NIH) for helpful discussions and comments on the manuscript; K.M. especially thanks E. Cho. This research was supported by the intramural research program of the NHGRI, NIH to K.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg EC. Reversible monoubiquitination of PCNA: A novel slant on regulating translesion DNA synthesis. Mol Cell. 2006;22:150–2. doi: 10.1016/j.molcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–42. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Campanero MR, Flemington EK. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci U S A. 1997;94:2221–6. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem. 2003;278:38315–24. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- 6.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–51. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 7.Isobe T, Uchida C, Hattori T, Kitagawa K, Oda T, Kitagawa M. Ubiquitin-dependent degradation of adenovirus E1A protein is inhibited by BS69. Biochem Biophys Res Commun. 2006;339:367–74. doi: 10.1016/j.bbrc.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki H, Chiba T, Kobayashi M, Takeuchi M, Furuichi K, Tanaka K. In vivo and in vitro recruitment of an IkappaBalpha-ubiquitin ligase to IkappaBalpha phosphorylated by IKK, leading to ubiquitination. Biochem Biophys Res Commun. 1999;256:121–6. doi: 10.1006/bbrc.1999.0296. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc Natl Acad Sci U S A. 2001;98:8662–7. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uchida C, et al. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 2005;24:160–9. doi: 10.1038/sj.emboj.7600486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 12.Brown S, Niimi A, Lehmann AR. Ubiquitination and deubiquitination of PCNA in response to stalling of the replication fork. Cell Cycle. 2009;8:689–92. doi: 10.4161/cc.8.5.7707. [DOI] [PubMed] [Google Scholar]
- 13.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–36. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8:461–9. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Huttner D, Ulrich HD. Cooperation of replication protein A with the ubiquitin ligase Rad18 in DNA damage bypass. Cell Cycle. 2008;7:3629–33. doi: 10.4161/cc.7.23.7166. [DOI] [PubMed] [Google Scholar]
- 16.Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:4267–74. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–85. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 18.Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–40. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, Green CM, Lehmann AR. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16125–30. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brun J, Chiu RK, Wouters BG, Gray DA. Regulation of PCNA polyubiquitination in human cells. BMC Res Notes. 2010;3:85. doi: 10.1186/1756-0500-3-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem. 2006;281:32081–8. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 22.Frampton J, et al. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:2976–85. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das-Bradoo S, Nguyen HD, Wood JL, Ricke RM, Haworth JC, Bielinsky AK. Defects in DNA ligase I trigger PCNA ubiquitylation at Lys 107. Nat Cell Biol. 2010;12:74–9. doi: 10.1038/ncb2007. sup pp 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bienko M, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–4. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 25.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 26.Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3:1011–3. [PubMed] [Google Scholar]
- 27.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 28.Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006;25:2847–55. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–96. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci U S A. 2007;104:15591–8. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–4. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 32.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–4. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 34.Lin W, Wu X, Wang Z. A full-length cDNA of hREV3 is predicted to encode DNA polymerase zeta for damage-induced mutagenesis in humans. Mutat Res. 1999;433:89–98. doi: 10.1016/s0921-8777(98)00065-2. [DOI] [PubMed] [Google Scholar]
- 35.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–97. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brusky J, Zhu Y, Xiao W. UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr Genet. 2000;37:168–74. doi: 10.1007/s002940050515. [DOI] [PubMed] [Google Scholar]
- 37.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–53. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 38.Chang DJ, Cimprich KA. DNA damage tolerance: when it’s OK to make mistakes. Nat Chem Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc Natl Acad Sci U S A. 1998;95:5678–83. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broomfield S, Hryciw T, Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res. 2001;486:167–84. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 41.Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao W, Chow BL, Broomfield S, Hanna M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000;155:1633–41. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–75. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol. 2006;175:703–8. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brun J, Chiu R, Lockhart K, Xiao W, Wouters BG, Gray DA. hMMS2 serves a redundant role in human PCNA polyubiquitination. BMC Mol Biol. 2008;9:24. doi: 10.1186/1471-2199-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unk I, Hajdu I, Fatyol K, Hurwitz J, Yoon JH, Prakash L, Prakash S, Haracska L. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc Natl Acad Sci U S A. 2008;105:3768–73. doi: 10.1073/pnas.0800563105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unk I, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2006;103:18107–12. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Motegi A, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci U S A. 2008;105:12411–6. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krijger PH, et al. HLTF and SHPRH are not essential for PCNA polyubiquitination, survival and somatic hypermutation: Existence of an alternative E3 ligase. DNA Repair (Amst) 2011 doi: 10.1016/j.dnarep.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–91. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 51.Kerscher O. SUMO junction-what’s your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–5. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Branzei D, Foiani M. RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev. 2007;21:3019–26. doi: 10.1101/gad.1624707. [DOI] [PubMed] [Google Scholar]
- 53.Branzei D, et al. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–22. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 54.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–20. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 55.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–33. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–33. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 57.Motegi A, Kuntz K, Majeed A, Smith S, Myung K. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:1424–33. doi: 10.1128/MCB.26.4.1424-1433.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arakawa H, Moldovan GL, Saribasak H, Saribasak NN, Jentsch S, Buerstedde JM. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leach CA, Michael WM. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J Cell Biol. 2005;171:947–54. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gohler T, Munoz IM, Rouse J, Blow JJ. PTIP/Swift is required for efficient PCNA ubiquitination in response to DNA damage. DNA Repair (Amst) 2008;7:775–87. doi: 10.1016/j.dnarep.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 61.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 62.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–9. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 63.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404:1011–3. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 64.Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D’Andrea AD. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16:314–20. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–51. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 66.Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol. 2005;25:5445–55. doi: 10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gellon L, Razidlo DF, Gleeson O, Verra L, Schulz D, Lahue RS, Freudenreich CH. New Functions of Ctf18-RFC in Preserving Genome Stability outside Its Role in Sister Chromatid Cohesion. PLoS Genet. 2011;7:e1001298. doi: 10.1371/journal.pgen.1001298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kubota T, Hiraga SI, Yamada K, Lamond AI, Donaldson AD. Quantitative proteomic analysis of chromatin reveals that Ctf18 acts in the DNA replication checkpoint. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M110.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Naiki T, Kondo T, Nakada D, Matsumoto K, Sugimoto K. Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol Cell Biol. 2001;21:5838–45. doi: 10.1128/MCB.21.17.5838-5845.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci U S A. 2001;98:11236–41. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bellaoui M, Chang M, Ou J, Xu H, Boone C, Brown GW. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 2003;22:4304–13. doi: 10.1093/emboj/cdg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci U S A. 2003;100:9906–11. doi: 10.1073/pnas.1633757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol. 2003;13:1583–95. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- 74.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:9039–44. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Banerjee S, Myung K. Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot Cell. 2004;3:1557–66. doi: 10.1128/EC.3.6.1557-1566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Banerjee S, Sikdar N, Myung K. Suppression of gross chromosomal rearrangements by a new alternative replication factor C complex. Biochem Biophys Res Commun. 2007;362:546–9. doi: 10.1016/j.bbrc.2007.07.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aroya SB, Kupiec M. The Elg1 replication factor C-like complex: a novel guardian of genome stability. DNA Repair (Amst) 2005;4:409–17. doi: 10.1016/j.dnarep.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 78.Lee KY, Yang K, Cohn MA, Sikdar N, D’Andrea AD, Myung K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J Biol Chem. 2010;285:10362–9. doi: 10.1074/jbc.M109.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parnas O, Zipin-Roitman A, Pfander B, Liefshitz B, Mazor Y, Ben-Aroya S, Jentsch S, Kupiec M. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J. 2010;29:2611–22. doi: 10.1038/emboj.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ogiwara H, Ui A, Enomoto T, Seki M. Role of Elg1 protein in double strand break repair. Nucleic Acids Res. 2007;35:353–62. doi: 10.1093/nar/gkl1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sikdar N, et al. DNA damage responses by human ELG1 in S phase are important to maintain genomic integrity. Cell Cycle. 2009;8:3199–207. doi: 10.4161/cc.8.19.9752. [DOI] [PMC free article] [PubMed] [Google Scholar]