Abstract
The human breast cancer resistance protein (BCRP/ABCG2) confers multidrug resistance and mediates the active efflux of drugs and xenobiotics. BCRP contains one nucleotide-binding domain (NBD) followed by one membrane-spanning domain (MSD). We investigated whether prolines in or near the transmembrane helices are essential for BCRP function. Six proline residues were substituted with alanine individually, and the mutants were stably expressed in Flp-In™-293 cells at levels comparable to wild-type BCRP and predominantly localized on the plasma membrane of the cells. While P392A showed a significant reduction in the efflux of mitoxantrone, BODIPY-prazosin, and Hoechst33342 by 35–50%, P485A exhibited a significant decrease by approximately 70% in the efflux of only BODIPY-prazosin. Other mutants had no significant changes in efflux of these substrates. Drug resistance profiles of the cells expressing the mutants correlated well with the efflux data. ATPase activity was not substantially affected for P392A or P485A compared to wild-type BCRP. These results strongly suggest Pro392 and Pro485 are important in determining the overall transport activity and substrate selectivity of BCRP, respectively. Prazosin differentially affected the binding of 5D3, a conformation-sensitive antibody, to wild-type BCRP, P392A or P485A in a concentration-dependent manner. In contrast, mitoxantrone had no significant effect on 5D3 binding. Homology modeling indicates that Pro392 may play an important role in the communication between the MSD and NBD as it is predicted to be located at the interface between the two functional domains, and Pro485 induces flexible hinges that may be essential for the broad substrate specificity of BCRP.
The human breast cancer resistance protein (BCRP, gene symbol ABCG2) is an ATP-binding cassette (ABC) efflux transporter that confers resistance to chemotherapeutic agents such as mitoxantrone and SN-38 (1–4). BCRP also actively transports a large variety of non-chemotherapeutic drugs and xenobiotics (1, 5). BCRP is highly expressed in stem cells (6), the liver canalicular membrane (7), the apical membrane of the small intestine and colon epithelium (7), the brain microvessel endothelium (8), and the apical membrane of placental syncytiotrophoblasts (7). Therefore, BCRP plays an important role in drug disposition and tissue protection (1–2).
BCRP is a half transporter comprising one N-terminal nucleotide-binding domain (NBD) followed by one membrane-spanning domain (MSD) (1, 5). BCRP is believed to form a homodimer or homooligomer, and its function depends on homodimerization (9–12). The MSD of BCRP contains six transmembrane (TM) α-helices (13). To date, little is known about the structure of BCRP and the mechanism of its ability to transport a broad range of substrates. Recently, we have determined the projection structures of BCRP at moderate resolutions (5 – 7 Å) and demonstrated that BCRP undergoes significant conformational changes upon mitoxantrone binding (14). Site-directed mutagenesis studies from various laboratories have shown that some charged or polar residues within or near TM helices are important in determining overall transport activity and substrate specificity of BCRP (15–20). However, how exactly BCRP recognizes and transports a wide spectrum of structurally unrelated substrates remains largely unknown.
Proline residues are found more frequently in TM α-helices of polytopic membrane proteins than in α-helices of soluble proteins and have been suggested to play structural and/or dynamic roles in membrane proteins (21–22). Proline cannot form normal backbone hydrogen bonds in the helix as proline lacks an amide hydrogen. The bulky proline may also cause a steric constraint on the conformation of adjacent residues. These two factors can potentially distort the regular structure of TM helices by introducing a kink between the segments adjacent to the proline. This can induce flexibility of the region by introducing a molecular switch or hinge (23). Thus, proline residues in or near TM helices can have important structural (e.g., facilitating helix packing (24)) and/or functional (e.g., involved in ligand binding (25), conformational changes (26), or substrate selectivity (27–28)) roles in membrane proteins including transport proteins.
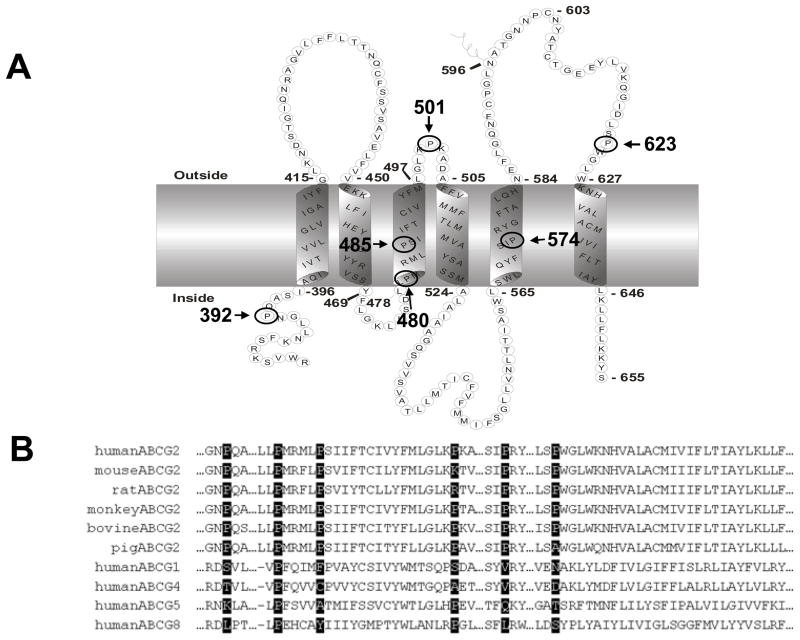
There are six proline residues in or near TM helices of BCRP, namely Pro392, Pro480, Pro485, Pro501, Pro574 and Pro623 (Figure 1), which are the focus of this study. Pro602 is located in the middle of the large extracellular loop connecting TM5 and TM6 and is far away from the TM boundaries. In analysis of previous studies for P-gp and MRP1, we noted that mutation of proline residues in the intracellular or extracellular loops connecting TM helices had either no effect (such as those in P-gp (27)) or moderate effect (such as some of those in MRP1 (29–30)) on transport activity, with only one exception of Pro1150 in MRP1 which is located in the intracellular loop connecting TM15 and TM16 (28, 31). Mutation of Pro1150 of MRP1 substantially increased transport of 17beta-estradiol-17-beta-(D-glucuronide) and methotrexate, but had no effect on or reduced transport of other organic anions (28), possibly due to alteration in the role of the intracellular loop as a coupling helix between the MSD and the NBD of MRP1, which is not the case for Pro602 in BCRP (31). We therefore did not included Pro602 in this study. We investigated the functional importance of the six prolines in or near TM helices in BCRP activity. These prolines were replaced with alanine individually by site-directed mutagenesis. The BCRP mutants were expressed in Flp-In™-293 cells by stable transfection and characterized with respect to their ability to transport drugs, drug resistance, basal ATPase activity, and the binding affinity for the 5D3 monoclonal antibody in the presence of substrates. Homology model of BCRP was used to assess potential structural mechanism.
Figure 1. Human BCRP topology (A) and sequence alignment (B).
A) The boundary of TM helices is based on the experimentally determined membrane topology of BCRP and only the MSD is shown. Proline residues analyzed in this study are circled and indicated with amino acid positions. B) Human BCRP amino acid sequence in the TM helices was aligned with the corresponding sequences of BCRP homologs and orthologs using Clustal W. Proline residues analyzed in this study are highlighted.
EXPERIMENTAL PROCEDURES
Materials
Fumitremorgin C (FTC) was obtained from National Cancer Institute (Bethesda, MD). BODIPY FL-prazosin was from Molecular Probes (Eugene, OR). Mitoxantrone hydrochloride (MX), doxorubicin hydrochloride (Dox), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) were from Sigma (St. Louis, MO). SN-38 was from TOCRIS Bioscience (Ellisville, MO). Rhodamine 123 (Rho123) was from MP Biomedicals (Irvine, CA). HPLC-grade DMSO was from Fisher Scientific (Waltham, MA). Fetal bovine serum (FBS) was from Atlanta Biologicals (Lawrenceville, GA). PBS, DMEM media, 4′,6-diamidino-2-phenylindole (DAPI), Hoechst33342, and Alexa-Fluor 488-conjugated goat anti-mouse IgG (H + L) (Fab′)2 fragment were from Invitrogen (Carlsbad, CA). FuGENE HD transfection reagent and protease inhibitor cocktail tablets were from Roche Applied Science (Indianapolis, IN). All restriction enzymes were from New England Biolabs (Beverly, MA). Primers used for PCR were synthesized by Operon (Huntsville, AL). The BCRP-specific mouse monoclonal antibody (mAb) BXP-21 was from Kamiya Biomedical Co. (Seattle, WA). The monoclonal goat anti-mouse HRP-conjugated antibody was from Bio-Rad (Hercules, CA). The phycoerythrin-conjugated anti-BCRP mAb 5D3 (catalog number 12-8888-73) and phycoerythrin-conjugated negative control antibody IgG2b were from eBioscience (San Diego, CA). Fluoromount G was from Southern Biotech (Birmingham, Alabama). The pcDNA3.1 plasmid containing full-length wild-type human BCRP cDNA was kindly provided by Dr. Susan E. Bates (National Cancer Institute, Bethesda, MD). Flp-In™-293 cells, the pcDNA5/FRT and pOG44 plasmids, zeocin, and hygromycin B were from Invitrogen. All other reagents and chemicals were of the highest purity available commercially.
Site-directed mutagenesis
The full-length wild-type human BCRP cDNA in pcDNA3.1 was subcloned into pcDNA5/FRT (15). The pcDNA5/FRT plasmid containing full-length human BCRP cDNA was used as a template to generate all the BCRP mutants using PCR mutagenesis as previously described (15). The forward primers used for mutagenesis were as follows: P392A (5′-AAC TTG CTG GGT AAT GCC CAG GCC TCT ATA GCT C-3′), P480A (5′-CTG TTA TCT GAT TTA TTA GCC ATG AGG ATG TTA CCA AG-3′), P485A (5′-CCC ATG AGG ATG TTA GCA AGT ATT ATA TTT ACC TGT ATA G-3′), P501A (5′-CAT GTT AGG ATT GAA GGC AAA GGC AGA TGC CTT C-3′), P574A (5′-CAG TAC TTC AGC ATT GCA CGA TAT GGA TTT ACG-3′), and P623A (5′-GGC ATC GAT CTC TCA GCC TGG GGC TTG TGG AAG AAT-3′). The reverse primers were complementary to the respective forward primers. All the mutants were confirmed by DNA sequencing, and the entire BCRP cDNA in each construct was also sequenced to make sure that no additional mutations were introduced during PCR.
Generation of Flp-In™-293 cells stably expressing wild-type BCRP and proline mutants
One μg of the pcDNA5/FRT plasmid containing wild-type or mutant BCRP cDNA or the empty vector were transfected into Flp-In™-293 cells in combination with 2 μg of pOG44 using the FuGENE HD transfection, according to the manufacturer’s instruction. After recovery, cells were selected in 125 μg/ml of hygromycin for approximately 3 – 4 weeks. The cells expressing wild-type or mutant BCRP were then maintained in complete DMEM medium containing 10% FBS, L-glutamine, and 125 μg/ml of hygromycin for subsequent experiments.
Immunoblotting, confocal microscopy, and cell surface expression of BCRP
Immunoblotting, confocal microscopy, and detection of cell surface expression of BCRP were performed exactly the same as previously described (15, 17). For immunoblotting assay and confocal microscopy study, the BCRP-specific mAb BXP-21 was used. The 5D3 mAb was used for cell surface detection of BCRP (20 μl of the 5D3 antibody incubated with approximately 5 × 105 cells). Human β-actin was detected as an internal standard in immunoblotting of whole cell lysates. Relative protein expression levels of wild-type and mutant BCRP were determined by densitometric analysis and normalization to β-actin as previously described (15, 17).
Plasma membrane preparation
Plasma membranes were prepared as previously described (15, 17). BCRP expression in the plasma membrane samples was determined by immunoblotting using the mAb BXP-21. The human plasma membrane protein Na+/K+-ATPase was detected as an internal standard. Relative protein expression levels of wild-type and mutant BCRP were determined by densitometric analysis and normalization to Na+/K+-ATPase as previously described (15, 17).
Flow cytometric efflux assay
Flow cytometric efflux assays were performed essentially the same as previously described (15). Briefly, cells were first incubated with 10 μM MX, 500 nM BODIPY-prazosin, or 0.05 μg/ml Hoechst33342, in the presence and absence of 10 μM FTC for 30 min at 37°C. Cells were then washed and resuspended in 1 ml of fluorescent compound-free incubation buffer with or without 10 μM FTC, and the incubation was continued for 1 h at 37°C. Cells were washed and intracellular fluorescence was measured as previously described (15). Cells in medium containing a fluorescent compound alone or in medium containing the fluorescent compound and FTC generated the efflux and FTC/efflux histograms, respectively. The difference (ΔF) in median fluorescence between the FTC/efflux histogram and the respective efflux histogram was used as a measure of FTC-inhibitable efflux activity of wild-type and mutant BCRP. Statistical significance of the differences in efflux activity between wild-type and mutant BCRP was analyzed using the Student’s t-test. A difference with a p value of < 0.05 was considered statistically significant.
Cytotoxicity assay
Drug resistance profiles of Flp-In™-293 cells expressing wild-type and mutant BCRP and the vector control cells were determined using the MTT microtiter plate assay as previously described (15).
Vanadate-sensitive ATPase activity
Vanadate-sensitive ATPase activities of wild-type and mutant BCRP were determined by measuring inorganic phosphate liberation from MgATP as previously described (15).
Concentration-dependent effects of BCRP substrates on 5D3 binding to wild-type and mutant BCRP
The effects of BCRP substrates, prazosin and MX, on 5D3 binding to wild-type and mutant BCRP were determined as previously described (15). Briefly, ~5 × 105 cells were incubated in the presence or absence of various concentrations of prazosin (0 – 40 μM) or MX (0 – 80 μM) for 5 min at 37°C followed by the addition of the 5D3 or IgG2b antibody (20 μl each), and incubation was continued for 30 min at 37 °C. Cells were then washed and analyzed using flow cytometry as previously described (15). The concentration-dependent effects of substrates on 5D3 binding to BCRP were determined by the differences (ΔF/F0) in phycoerythrin fluorescence between the cells incubated with 5D3 in the presence of a substrate (prozosin or MX) and 5D3 alone (F0). In preliminary experiments, we found that MX alone up to 80 μM did not have any interference on phycoerythrin fluorescence of phycoerythrin-conjugated antibodies.
Homology modeling
We have previously built a homology model of BCRP based on the recently published crystal structure of mouse P-gp, which represents the substrate-bound and nucleotide-free inward-facing form of BCRP (14). We based on this model to assess potential structural roles of proline resides in BCRP. To explore the potential conformational flexibility of TM3 which contains Pro480 and Pro485 in a PXXXXP helix motif, we tested several kink conformations in our homology model, based on X-ray crystal structures of TM helices that contain prolines at both and positions from a TM helix database (22). Two PXXXXP TM helix motifs, PAFVAP (PDB code: 1EHK1) and PHAAVP (PDB code: 1EHK2), found in the PDB entry 1EHK (32) were selected for modeling of the potential alternate helix conformations of TM3. We also tested a similar GXXXXP motif found in the PDB entry 2RCR (33). The BCRP models with alternate TM3 conformations were generated as follows. First, 3D coordinates of the proline-containing TM helices were cut from the original crystal structures; Second, these TM helices were structurally aligned to the N-terminal half of TM3 in the original BCRP model as templates; Finally, new models of BCRP were built using its own homology model (14) as a template except for the TM3 region where the kink conformations of the selected TM helices were used as the structural templates. The BCRP models were built using the Modeller 9.8 software (34) exactly the same as previously described (14).
RESULTS
Expression of wild-type BCRP and proline mutants in Flp-In™-293 cells
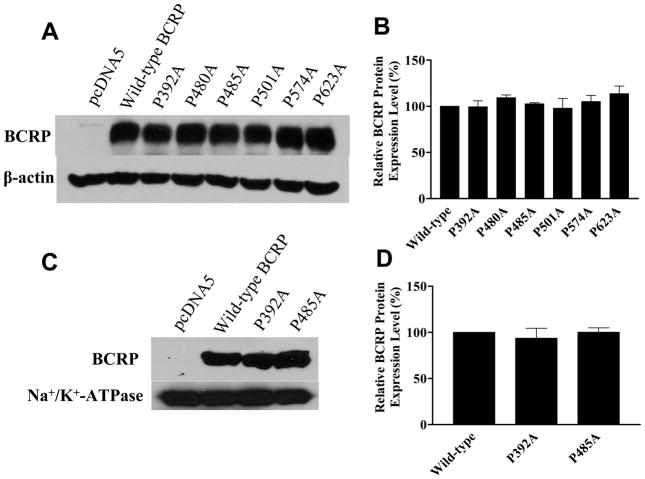
According to the membrane topology of BCRP we determined (13), six proline residues, namely Pro392, Pro480, Pro485, Pro501, Pro574 and Pro623, exist in the TM helices or no more than 5 amino acid residues outside of the TM boundaries (Figure 1A). These proline residues are highly conserved in the human, mouse, rat, monkey, bovine, and porcine orthologs of BCRP (Figure 1B). To investigate the importance of the six proline residues in BCRP transport function, we substituted the six proline residues with alanine individually. Alanine was used because it has been shown to possess the highest helix propensity compared to other amino acids (35), and therefore alanine substitutions of proline may result in mutants with potentially minimal effects on BCRP stability and structural integrity. Also, substitution with alanine or any other amino acids would similarly eliminate the molecular hinges potentially introduced by a proline in TM helices. Flp-In™-293 cell lines stably expressing wild-type BCRP and the proline mutants were generated by co-transfection of pcDNA5/FRT expression vectors and pOG44 and subsequent selection with hygromycin. The resultant cell lines expressed the mutants at levels comparable to wild-type BCRP as determined by immunoblotting of whole cell lysates using β-actin as an internal standard (Figure 2A and 2B).
Figure 2. Immunoblotting analysis of the expression levels of wild-type and mutant BCRP.
The protein expression levels of wild-type and mutant BCRP in stably transfected Flp-In™-293 cells were determined by immunoblotting, densitometric analysis of the blots, and normalization to internal standard. A) Immunoblot of whole cell lysates for wild-type and mutant BCRP. Each lane was loaded with 20 μg of protein. B) The protein expression levels of the mutants relative to wild-type BCRP (100%) in whole cell lysates. Shown are means ± SD of three experiments. C) Immunoblot of plasma membrane preparations for wild-type and mutant BCRP. Each lane was loaded with 5 μg of protein. D) The protein expression levels of P392A and P485A relative to wild-type BCRP (100%) in the plasma membrane samples. Shown are means ± SD of three experiments.
Plasma membrane localization and cell surface expression of wild-type and mutant BCRP in Flp-In™-293 cells
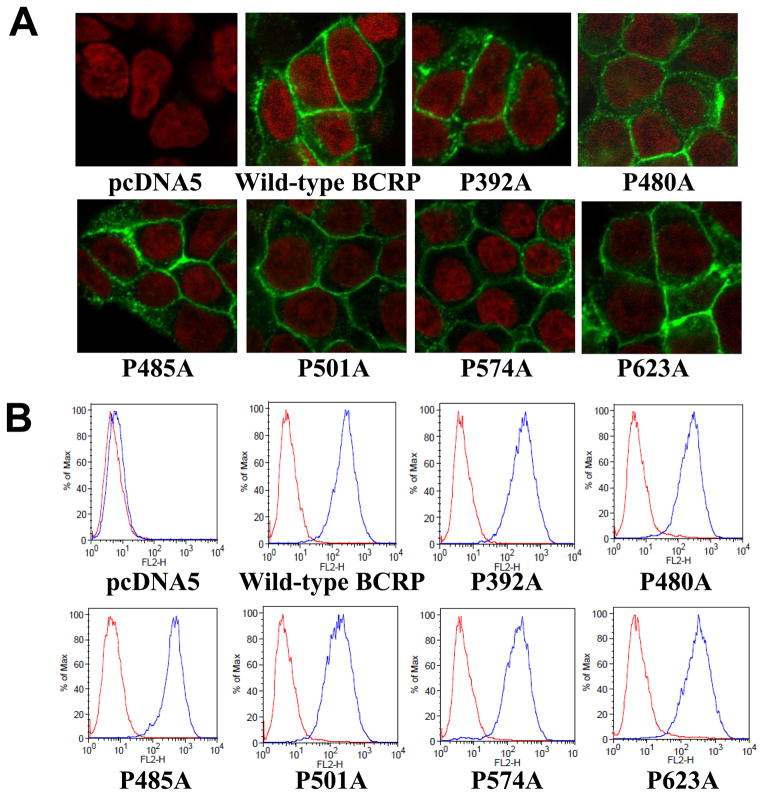
To determine if the proline mutations affect proper folding and trafficking of BCRP to the plasma membrane, Flp-In™-293 cells stably expressing wild-type and mutant BCRP were analyzed by immunofluorescent confocal microscopy with the BCRP-specific mAb BXP-21. Wild-type BCRP and all the six proline mutants were observed to be predominantly localized on the plasma membrane (Figure 3A), suggesting that the mutants retained proper folding and maturation as wild-type BCRP. To further verify cell surface expression of these mutants, cells were incubated with the phycoerythrin-conjugated BCRP-specific mAb 5D3 or the IgG2b negative control antibody. Cell surface expression of BCRP was demonstrated by determining the differences in cell-associated phycoerythrin fluorescence between the 5D3 and IgG2b incubations using flow cytometry. As expected, the pcDNA5 vector control cells did not exhibit any cell surface expression of BCRP (Figure 3B). Wild-type and mutant BCRP all showed significant cell surface expression. The estimated relative cell surface expression of wild-type and mutant BCRP based on the differences in median fluorescence between 5D3 and IgG2b control incubations was 100 ± 22%, 108 ± 4%, 89 ± 1%, 139 ± 32%, 75 ± 16%, 76 ± 9%, 114 ± 0.7% of three independent determinations for wild-type BCRP, P392A, P480A, P485A, P501A, P574A, and P623A, respectively. Overall, these results were consistent with the immunoblotting data (Figure 2) with generally less than 25% of difference between wild-type and mutant BCRP. The 5D3 fluorescence associated with P485A was increased by 39% compared to wild-type BCRP, but the difference was not statistically significant (p > 0.05 by the Student’s t-test). The variation in 5D3 fluorescence of the mutants could be caused by experimental errors and/or reflect the change in BCRP conformation caused by the mutations.
Figure 3. Confocal microscopy analysis and cell surface expression of Flp-In™-293 cells stably expressing wild-type and mutant BCRP.
A) The cellular localization of wild-type and mutant BCRP in Flp-In™-293 cells (shown in green) was determined by immunofluorescent confocal microscopy using the BCRP-specific mAb BXP-21. Cell nuclei were stained with DAPI and are shown in red. B) Expression of wild-type and mutant BCRP on cell surface of stably transfected Flp-In™-293 cells was detected using the 5D3 monoclonal antibody. Representative flow cytometry histograms showing cell surface expression of wild-type and mutant BCRP are presented. The red and blue peaks represent the phycoerythrin fluorescence associated with cells treated with the IgG2b negative control and the 5D3 antibodies, respectively. No surface expression of BCRP was detected in the pcDNA5 vector control cells.
FTC-inhibitable efflux activities of wild-type and mutant BCRP
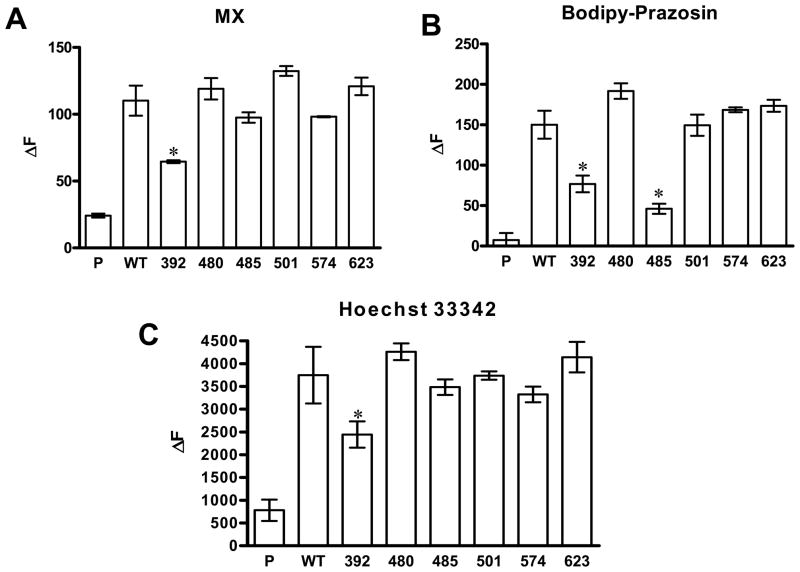
After validation of BCRP expression in Flp-In™-293 cells, we examined the efflux activity of wild-type and mutant BCRP using a well established flow cytometric efflux assay with three fluorescent substrates, MX, BODIPY-prazosin, and Hoechst 33342 (15, 18, 36–37). MX, BODIPY-prazosin, and Hoechst 33342 have been widely used and well characterized as BCRP substrates (15, 18, 36–37). They do not share any chemical and structural similarity, and therefore, the use of these compounds may help us determine BCRP-substrate interactions. Cells expressing wild-type BCRP showed efflux activities for all the three substrates tested (Figure 4). The mutation of Pro392 caused a significant reduction in the efflux of all the three substrates by 35–50% (Figure 4). In contrast, the mutation of Pro485 only significantly decreased the efflux of BODIPY-prozasin by ~70% (Figure 4B), but had no significant effect on the efflux of MX (Figure 4A) or Hoechst33342 (Figure 4C). Thus, the mutation of Pro485 selectively affected BCRP transport activity. Mutations of other prolines had no significant effect on the efflux of the three substrates (Figure 4).
Figure 4. FTC-inhibitable efflux activities of Flp-In™-293 cells stably expressing wild-type and mutant BCRP.
Three substrates, mitoxantrone (A), BODIPY-prazosin (B), and Hoechst33342 (C) were used. The efflux activities were expressed as the differences in median fluorescence (ΔF) between the FTC/efflux and efflux histograms. Shown are means ± S.D. of three experiments. Differences in efflux activities between wild-type and mutant BCRP are statistically significant: * p < 0.05 by the Student’s t-test. The pcDNA5 vector control, wild-type BCRP, P392A, P480A, P485A, P501A, P574A and P623A are indicated by P, WT, 392, 480, 485, 501, 574 and 623, respectively. Since the expression levels of wild-type and mutant BCRP were comparable, we did not normalize the efflux activity data to BCRP protein expression.
Drug resistance conferred by wild-type and mutant BCRP
To further test if the mutations affect drug resistance conferred by BCRP, we determined resistance of Flp-In™-293 cells expressing wild-type and mutant BCRP to MX, SN-38, Dox, and Rho123. As shown in Table 1, only P392A exhibited a significantly lower resistance to MX compared to wild-type BCRP, which is consistent with the efflux activity data. On the other hand, both P392A and P485A showed significantly decreased resistance to SN-38 as compared to wild-type, thus providing additional evidence that alanine substitution of Pro485 changed substrate specificity of BCRP. Mutations of other prolines had no significant effect on resistance to MX or SN-38. We also tested resistance to Rho123 and Dox conferred by the mutants to determine if there is so-called “gain-of-function” because neither drug can be transported by wild-type BCRP. All the mutants did not exhibit any significant resistance to either Dox or Rho123.
Table 1.
Drug resistance profile of wild-type and mutant BCRP
| MX | SN-38 | Dox | Rho123 | |||||
|---|---|---|---|---|---|---|---|---|
| IC50 (nM) | RR | IC50 (nM) | RR | IC50 (nM) | RR | IC50 (nM) | RR | |
| pcDNA5/FRT | 5.6 ± 0.8 | - | 1.7 ± 0.1 | - | 33.1 ± 2.2 | - | 4218 ± 400.0 | - |
| Wild-type BCRP | 51.5 ± 8.5 | 9.2 | 30.3 ± 2.0 | 17.8 | 40.9 ± 6.7 | 1.2 | 4446 ± 261.5 | 1.1 |
| P392A | 24.6 ± 3.8 | 4.4* | 13.1 ± 1.0 | 7.7* | 34.8 ± 1.7 | 1.1 | 5326 ± 414.1 | 1.3 |
| P480A | 69.4 ± 10.5 | 12.4 | 37.8 ± 1.5 | 22.2 | 29.0 ± 4.0 | 0.9 | 5107 ± 272.7 | 1.2 |
| P485A | 38.5 ± 7.0 | 6.9 | 15.9 ± 2.6 | 9.4* | 41.8 ± 5.3 | 1.3 | 3237 ± 199.5 | 0.8 |
| P501A | 40.9 ± 14.6 | 7.3 | 27.5 ± 2.4 | 16.2 | 23.2 ± 3.8 | 0.7 | 4480 ± 816.4 | 1.1 |
| P574A | 49.6 ± 12.5 | 8.9 | 32.5 ± 2.6 | 19.1 | 29.6 ± 3.5 | 0.9 | 5320 ± 419.8 | 1.3 |
| P623A | 68.4 ± 14.4 | 12.2 | 36.8 ± 2.8 | 21.6 | 37.0 ± 8.8 | 1.1 | 3302 ± 388.1 | 0.8 |
The relative resistance (RR) values represent the relative levels of resistance of the mutants compared to wild-type BCRP, and were calculated by dividing the IC50 values of wild-type and mutant BCRP by the IC50 values of the vector control. Wild-type and mutant BCRP did not confer resistance to Dox or Rho123. The IC50 values shown are means ± S.D. of three experiments. Differences in IC50 values between wild-type and mutant BCRP are statistically significant:
p < 0.05 by the Student’s t- test. Since the expression levels of wild-type and mutant BCRP were comparable, we did not normalize the resistance data to BCRP protein expression.
ATPase activities of wild-type and mutant BCRP
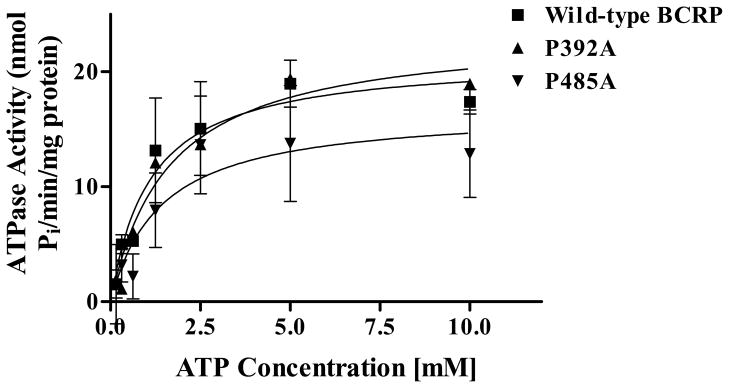
To examine if the changes in efflux activity of mutant BCRP are caused by alterations in its ability to hydrolyze ATP, we measured vanadate-sensitive ATPase activities of wild-type BCRP, P392A and P485A in plasma membrane preparations. The protein expression levels of wild-type BCRP, P392A, and P485A in plasma membranes were determined by immunoblotting using the mAb BXP-21 and found to be comparable after normalization to the internal plasma membrane control Na+/K+-ATPase (Figure 2C and 2D). Vanadate-sensitive ATPase activities attributable to wild-type and mutant BCRP were calculated by subtracting the background activities associated with the plasma membranes isolated from the vector control cells. P392A exhibited similar ATPase activities to wild-type BCRP. P485A showed a modest decrease in ATPase activities compared to wild-type BCRP; however, the differences were not statistically significant (Figure 5). Overall, the mutations of Pro392 and Pro485 did not substantially change ATPase activity, suggesting that the alterations in transport activity of the two proline mutants are not primarily due to the effect of the mutations on ATP hydrolytic capability of BCRP.
Figure 5. Venadate-sensitive ATPase of plasma membrane preparations containing wild-type and mutant BCRP.
ATPase activities were measured using plasma membrane preparations isolated from Flp-In™-293 cells stably expressing wild-type and mutant BCRP over a range of MgATP concentration (0 – 10 mM). Shown are mean ± S.D. of three experiments. Since the expression levels of wild-type and mutant BCRP were comparable, we did not normalize the ATPase activity data to BCRP protein expression.
Concentration-dependent effects of prazosin and MX on 5D3 binding to wild-type and mutant BCRP
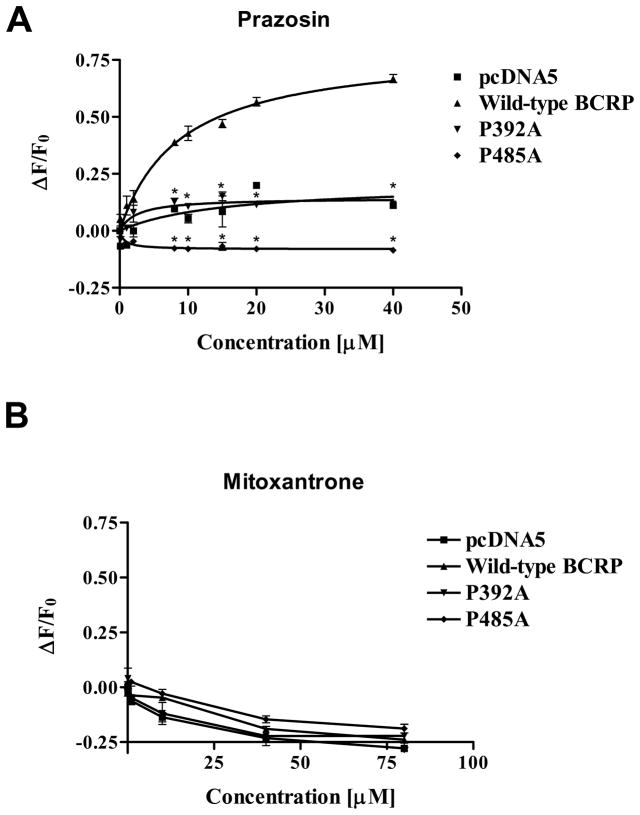
Previous studies have demonstrated that the mAb 5D3 is conformation-sensitive and can be used to monitor potential conformational changes in BCRP associated with ATP binding/hydrolysis, formation of a catalytic intermediate, or substrate and inhibitor binding (38–40). Therefore, 5D3 binding to BCRP can be measured to reflect conformational changes induced by BCRP-ligand interactions. We have previously shown that prazosin binding-induced conformational changes as monitored by measuring 5D3 binding to BCRP could be affected by single mutations, thereby leading to altered transport activity (15). To investigate whether the mutations of Pro392 and Pro485 cause conformational changes in BCRP that affect transport activity, we measured 5D3 binding to wild-type BCRP, P392A or P485A in the presence and absence of varying concentrations of the substrate prazosin (0 – 40 μM) or MX (0 – 80 μM). When cells expressing wild-type BCRP were incubated with the control phycoerythrin-conjugated IgG2b, neither prazosin at up to 40 μM nor MX at up to 80 μM had any effect on phycoerythrin fluorescence (data not shown), suggesting that these drugs themselves had no contribution to fluorescence detection. Also, there was no significant prazosin concentration-dependent change in 5D3-phycoerythrin fluorescence for the vector control cells (Figure 6A). In the presence of prazosin, wild-type BCRP was associated with a concentration-dependent increase in 5D3-phycoerythrin fluorescence by up to 70%; however, there was only a slight increase in 5D3-phycoerythrin fluorescence for P392A and a slight decrease in 5D3-phycoerythrin fluorescence for P485A (Figure 6A). These results suggest that the mutations of Pro392 and Pro485 dramatically and differentially affect prazosin binding-induced conformational changes in BCRP. The patterns of 5D3-phycoerythrin fluorescence with MX for wild-type BCRP, P392A or P485A were similar, but different from those with prazosin (Figure 6B). This observation implies that BCRP has differential response to prazosin and MX binding and the mutations of Pro392 and Pro485 do not seem to affect the interaction of BCRP with MX.
Figure 6. Concentration-dependent effects of prazosin and MX on 5D3 binding to wild-type BCRP, P392A, and P485A.
The effects of prazosin (A) and MX (B) on 5D3 binding to wild-type and mutant BCRP over a concentration range of 0 – 40 μM and 0 – 80 μM, respectively, were determined using flow cytometry. Shown are means ± S.D. of three experiments. * indicates statistically significant differences between wild-type BCRP and P392A or P485A (p < 0.05 by the Student’s t-test) at various prazosin concentrations.
A potential molecular hinge in TM3 introduced by Pro485
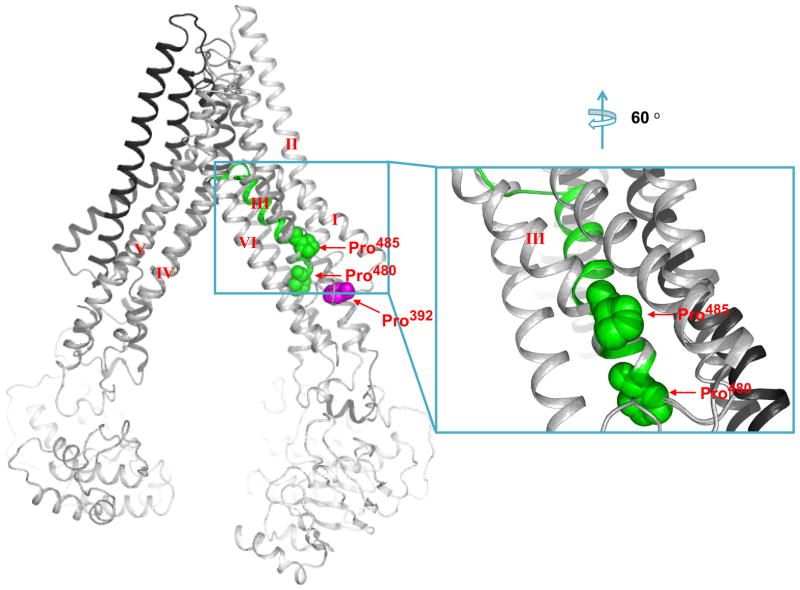
According to the homology model of BCRP (Figure 7), Pro485 of TM3 is located in the large drug-binding cavity. In contrast to α-helices in soluble proteins, proline residues often occur within TM helices where they may induce the formation of molecular hinges (22). Accordingly, Pro485 being 5 residues away from Pro480 at the N-terminus of TM3 may introduce a flexible hinge in BCRP. While it is difficult to obtain atomic details of such dynamic movements of TM helices experimentally, one may explore possible TM conformations allowed by structural features of the protein using homology modeling. We tested three different conformations of TM3 in our BCRP homology model. By doing this, if the alternate TM3 conformation fits into the overall structure of the original BCRP model, other parts of the original model should remain unchanged. We found that the model based on the GXXXXP motif was distorted, suggesting that the GXXXXP helix conformation cannot exist in BCRP. The model based on the 1EHK1 helix was very similar to the original BCRP model, while the model based on the 1EHK2 helix resulted in an alternative TM3 conformation compared to the original model (Figure 7, shown in green). The TM3 with a flexible hinge is oriented towards the interior of the drug-binding cavity, thus providing a potential conformation of the drug-binding cavity that coordinately brings residues involved in drug binding to optimal positions for interactions with certain substrates. This hinge would allow rearrangement of the substrates-binding sites of BCRP, depending on the nature and geometry of substrates (e.g., size and charge) (Supplementary Figure 1). It is important to note that the model shown in Figure 7 just provides one possible alternative TM3 conformation that is allowed by the overall domain arrangement of BCRP; however, more precise and atomic analysis of this presumed hinge will only be possible by determining high resolution structures of BCRP experimentally.
Figure 7. Schematic illustration of a molecular hinge in TM3 introduced by Pro485.
A side view of a three-dimensional model of BCRP in the nucleotide-free closed apo inward-facing form is presented. The TM helices are labeled with roman numerals, I, II, III, IV, V and VI. The residues Pro392, Pro480 and Pro485 are shown in magenta, green and green, respectively, and pointed by arrows. Also shown on the right is an expanded view of TM3 and neighboring helices indicating a potential hinge (shown in green) introduced by Pro485. The C-terminal half of TM3 is kinked between TM1 and TM6 in the hinged conformation and rotated closer to the interior of the drug-binding cavity of BCRP. The undistorted TM3 in the original BCRP model is shown in grey.
DISCUSSION
In the present study, we have analyzed the functional importance of six proline residues within or near TM helices of BCRP for drug transport (Figure 1). The prolines were substituted with alanine individually. All the mutants were well expressed and properly targeted to the plasma membrane (Figures 2 and 3), suggesting that none of the mutations had a significant impact on proper folding and maturation of BCRP.
We found that alanine substitution of Pro485 had a profound impact on substrate selectivity of BCRP. P485A showed a significant decrease in the efflux of BODIPY-prazosin, but not of MX and Hoechst33342, and conferred lower resistance to SN-38, but not to MX (Figure 4 and Table 1). Prior to this study, mutations of Arg482 were shown to significantly affect substrate specificity of BCRP (16, 18, 36, 41). This study provides another example that mutation of a single residue in BCRP can change its substrate specificity. On the other hand, alanine substitution of Pro392 caused a significant decrease in transport of all the substrates tested (Figure 4 and Table 1). Such changes in transport activity of P392A and P485A do not seem to be related to alterations in ATP hydrolytic capability (Figure 5). No significant changes in efflux of or resistance to these substrates were observed for alanine substitutions of Pro480, Pro501, Pro574 and Pro623, suggesting that these prolines are likely not functionally important for BCRP.
To assess potential mechanisms by which the mutations of Pro392 and Pro485 alter transport activity and substrate specificity of BCRP, we utilized the homology model of BCRP we previously constructed based on the X-ray structure of mouse P-gp (14). The homology model suggests that Pro485 is located inside the putative drug-binding cavity (Figure 7). Cordes et al. analyzed sequences of 199 TM helices of membrane proteins and found that prolines are preferentially located in a terminal or central position in TM helices (22). In the terminal position, prolines may serve as helix breaking residues. In a central region, prolines may have a possible structural and/or functional role by introducing a hinge (a region of possible flexibility) in the middle of a TM helix (21–22, 42–43). This is exactly what we see in TM3 of BCRP. Pro485 is located in the central region of TM3 which is 5 residues away from the starting residue Pro480 (Figure 1A). Pro485 therefore may introduce a structurally flexible hinge in the middle of TM3. From the structural point of view, such a hinge may bring in a curved structure of TM3 which could make the drug binding cavity structurally more dynamic. We have constructed a homology model of the hinge to illustrate this concept (Figure 7). The motion of the flexible hinge may be dependent on the chemical or physical nature of substrates such as size or charge. Indeed, the chemical structure and molecular volume of the substrates tested in this study are quite diverse (Supplementary Figure 1). We believe that this could be one of the reasons why BCRP can accommodate and transport structurally and chemically diverse substrates. Thus, Pro485 may play an important role in drug binding by introducing a flexible hinge to make the drug-binding cavity optimal for interactions with certain substrates. Alanine substitution of Pro485 would eliminate the helical distortion of TM3 and the flexibility and influence the helical packing with neighboring TM helices, thereby affecting transport activity for some substrates. The results of this study suggest that alanine substitution of Pro485 does not affect binding/transport for MX and Hoechst33342, but deteriorates binding/transport for prazosin and SN-38.
Clark et al. performed equilibrium and kinetic drug binding studies for BCRP and showed that MX and Hoechst33322 may share similar binding sites, which were distinct from that of prazosin (44). The results of the present study appear to be consistent with the direct drug binding data based upon the assumption that the molecular hinge introduced by Pro485 may differentially affect interaction of BCRP with some substrates (e.g., prazosin and SN-38), but not others (e.g., MX and Hoechst33342) possibly due to different binding sites or different binding regions within the same site. Although it is generally believed that the MSD participates in drug binding, the exact location of drug binding sites in BCRP remains to be determined. Functional importance of Pro223 and Pro866 in P-gp has also been demonstrated (27). Pro223 and Pro866 are located in the central region of TM4 and TM10 of P-gp, respectively. Likewise, alanine substitution of Pro223 or Pro866 significantly decreased the efflux of colchicine, but had no effect on efflux of vinblastine, suggesting that colchicine and vinblastine likely bind to different sites or different regions within the same binding site.
Regarding Pro392, our homology model suggests that it is located in the linker region between the MSD and the NBD (Figure 7). This led us to hypothesize that the mutation of Pro392 affects the communication between the two domains which is essential for BCRP function. This is consistent with previous studies showing the functional importance of the linker region. For example, Asn387 and Gln398 in the linker region were replaced with alanine, leading to a significant decrease in BCRP activity (15).
Based on the membrane topology of BCRP we recently determined (13), Pro480 is predicted to be at the N-terminal end of TM3 and thus may play a role in helix breaking. Pro574 is predicted to be in the middle of TM5. However, mutations of either proline showed no significant effect on BCRP function. It is important to note that Pro485 is only 3 residues away from Arg482 (both residues are within TM3), of which mutations have been shown to significantly alter substrate specificity of BCRP (16, 18, 36, 41). Arg482 is possibly part of the molecular hinge introduced by Pro485 and both residues may contribute to the broad substrate specificity of BCRP. However, how exactly Arg482 and Pro485 work together to determine substrate specificity is not clear. Collectively, TM3 is very likely to be involved in drug binding and transport, and the role of TM3 in determining transport activity and substrate specificity of BCRP warrants further investigation.
We have previously demonstrated substrate binding-induced conformational changes in BCRP by analyzing 2-dimensional crystals with and without MX (14). Such conformational changes can be monitored by measuring 5D3 binding. The mAb 5D3 binds to a highly specific epitope in an extracellular loop of BCRP and is conformation-sensitive. Therefore, to further illustrate if mutations of Pro392 and Pro485 affect BCRP-substrate interactions by perturbing substrate binding-induced conformational changes and thus transport activity and substrate specificity, we analyzed 5D3 binding to wild-type and mutant BCRP in the presence and absence of prazosin or MX. First, wild-type BCRP responded very differently to 5D3 binding in the presence of prazosin or MX (Figure 6), again suggesting differential interactions between BCRP and the two substrates. Second, the response of wild-type BCRP, P392A or P485A to 5D3 binding in the presence of prazosin was significantly different from each other (Figure 6A). However, there was no difference between wild-type BCRP, P392A and P485A in response to 5D3 binding in the presence of MX (Figure 6B). This further suggests that the mutations of Pro392 and Pro485 induce conformational changes in BCRP that differentially affect BCRP-substrate interactions and thus the two prolines play different functional roles in BCRP activity. Such effects are substrate-dependent (i.e., due to different binding sites).
In summary, we have demonstrated the importance of Pro392 in overall transport activity and Pro485 in substrate selectivity of BCRP. We also provided functional evidence that Pro485 in TM3 may introduce flexible hinges that allow BCRP to accommodate a wide spectrum of substrates, whereas Pro392 may play a role in the communication between the MSD and the NBD during the transport cycle.
Supplementary Material
Acknowledgments
We thank Dr. Susan E. Bates (National Cancer Institute, Bethesda, MD) for providing the pcDNA3.1 expression vector containing full-length human wild-type BCRP cDNA. We also acknowledge the Drug Synthesis & Chemistry Branch, National Cancer Institute (Bethesda, MD) for providing FTC. We greatly acknowledge Drs. Haichuan Duan and Joanne Wang (Department of Pharmaceutics, University of Washington) for providing the pcDNA5/FRT expression vector.
Abbreviations
- BCRP
breast cancer resistance protein
- P-gp
P-glycoprotein
- ABC
ATP-binding cassette
- ABCG2
ATP-binding cassette G2
- NBD
nucleotide-binding domain
- MSD
membrane-spanning domain
- TM
transmembrane
- WT
wild-type
- MX
mitoxantrone
- Dox
doxorubicin
- Rho123
rhodamine 123
- FTC
fumitremorgin C
- DMSO
dimethyl sulfoxide
- DMEM
Dulbecco’s modified Eagle minimum essential medium
- FBS
fetal bovine serum
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- LB
Luria-Bertani
- SDS
sodium dodecyl sulfate
- mAb
monoclonal antibody
- DAPI
4′, 6-diamidino-2-phenylindole
- SN-38
7-ethyl-10-hydroxycamptothecin
- BODIPY
boron-dipyrromethene
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide
- PDB code
Protein Data Bank ID code
Footnotes
This work was supported by a grant from the National Institutes of Health, GM073715.
The chemical structure, molecular volume, and cLogP value of the substrates used in this study are shown in Supplementary Figure 1. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Robey RW, To KK, Polgar O, Dohse M, Fetsch P, Dean M, Bates SE. ABCG2: a perspective. Adv Drug Deliv Rev. 2009;61:3–13. doi: 10.1016/j.addr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao Q, Unadkat JD. Role of the breast cancer resistance protein (ABCG2) in drug transport. Aaps J. 2005;7:E118–133. doi: 10.1208/aapsj070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarkadi B, Homolya L, Szakacs G, Varadi A. Human multidrug resistance ABCB and ABCG transporters: participation in a chemoimmunity defense system. Physiol Rev. 2006;86:1179–1236. doi: 10.1152/physrev.00037.2005. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa T, Nakagawa H. Human ABC transporter ABCG2 in cancer chemotherapy and pharmacogenomics. J Exp Ther Oncol. 2009;8:5–24. [PubMed] [Google Scholar]
- 5.Ni Z, Bikadi Z, Rosenberg MF, Mao Q. Structure and function of the human breast cancer resistance protein (BCRP/ABCG2) Curr Drug Metab. 2010;11:603–617. doi: 10.2174/138920010792927325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou S, Morris JJ, Barnes Y, Lan L, Schuetz JD, Sorrentino BP. Bcrp1 gene expression is required for normal numbers of side population stem cells in mice, and confers relative protection to mitoxantrone in hematopoietic cells in vivo. Proc Natl Acad Sci U S A. 2002;99:12339–12344. doi: 10.1073/pnas.192276999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maliepaard M, Scheffer GL, Faneyte IF, van Gastelen MA, Pijnenborg AC, Schinkel AH, van De Vijver MJ, Scheper RJ, Schellens JH. Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Res. 2001;61:3458–3464. [PubMed] [Google Scholar]
- 8.Cooray HC, Blackmore CG, Maskell L, Barrand MA. Localisation of breast cancer resistance protein in microvessel endothelium of human brain. Neuroreport. 2002;13:2059–2063. doi: 10.1097/00001756-200211150-00014. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Peng H, Chen Q, Liu Y, Dong Z, Zhang JT. Oligomerization domain of the multidrug resistance-associated transporter ABCG2 and its dominant inhibitory activity. Cancer Res. 2007;67:4373–4381. doi: 10.1158/0008-5472.CAN-06-3169. [DOI] [PubMed] [Google Scholar]
- 10.Ni Z, Mark ME, Cai X, Mao Q. Fluorescence resonance energy transfer (FRET) analysis demonstrates dimer/oligomer formation of the human breast cancer resistance protein (BCRP/ABCG2) in intact cells. Int J Biochem Mol Biol. 2010;1:1–11. [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Yang Y, Qi J, Peng H, Zhang JT. Effect of cysteine mutagenesis on the function and disulfide bond formation of human ABCG2. J Pharmacol Exp Ther. 2008;326:33–40. doi: 10.1124/jpet.108.138115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDevitt CA, Collins RF, Conway M, Modok S, Storm J, Kerr ID, Ford RC, Callaghan R. Purification and 3D structural analysis of oligomeric human multidrug transporter ABCG2. Structure. 2006;14:1623–1632. doi: 10.1016/j.str.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Lee EW, Cai X, Ni Z, Zhou L, Mao Q. Membrane topology of the human breast cancer resistance protein (BCRP/ABCG2) determined by epitope insertion and immunofluorescence. Biochemistry. 2008;47:13778–13787. doi: 10.1021/bi801644v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenberg MF, Bikadi Z, Chan J, Liu X, Ni Z, Cai X, Ford RC, Mao Q. The human breast cancer resistance protein (BCRP/ABCG2) shows conformational changes with mitoxantrone. Structure. 2010;18:482–493. doi: 10.1016/j.str.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni Z, Bikadi Z, Cai X, Rosenberg MF, Mao Q. Transmembrane helices 1 and 6 of the human breast cancer resistance protein (BCRP/ABCG2): identification of polar residues important for drug transport. Am J Physiol Cell Physiol. 2010;299:C1100–1109. doi: 10.1152/ajpcell.00160.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honjo Y, Hrycyna CA, Yan QW, Medina-Perez WY, Robey RW, van de Laar A, Litman T, Dean M, Bates SE. Acquired mutations in the MXR/BCRP/ABCP gene alter substrate specificity in MXR/BCRP/ABCP-overexpressing cells. Cancer Res. 2001;61:6635–6639. [PubMed] [Google Scholar]
- 17.Cai X, Bikadi Z, Ni Z, Lee EW, Wang H, Rosenberg MF, Mao Q. Role of basic residues within or near the predicted transmembrane helix 2 of the human breast cancer resistance protein in drug transport. J Pharmacol Exp Ther. 2010;333:670–681. doi: 10.1124/jpet.109.163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robey RW, Honjo Y, Morisaki K, Nadjem TA, Runge S, Risbood M, Poruchynsky MS, Bates SE. Mutations at amino-acid 482 in the ABCG2 gene affect substrate and antagonist specificity. Br J Cancer. 2003;89:1971–1978. doi: 10.1038/sj.bjc.6601370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen ZS, Robey RW, Belinsky MG, Shchaveleva I, Ren XQ, Sugimoto Y, Ross DD, Bates SE, Kruh GD. Transport of methotrexate, methotrexate polyglutamates, and 17beta-estradiol 17-(beta-D-glucuronide) by ABCG2: effects of acquired mutations at R482 on methotrexate transport. Cancer Res. 2003;63:4048–4054. [PubMed] [Google Scholar]
- 20.Polgar O, Ierano C, Tamaki A, Stanley B, Ward Y, Xia D, Tarasova N, Robey RW, Bates SE. Mutational analysis of threonine 402 adjacent to the GXXXG dimerization motif in transmembrane segment 1 of ABCG2. Biochemistry. 2010;49:2235–2245. doi: 10.1021/bi902085q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams KA, Deber CM. Proline residues in transmembrane helices: structural or dynamic role? Biochemistry. 1991;30:8919–8923. doi: 10.1021/bi00101a001. [DOI] [PubMed] [Google Scholar]
- 22.Cordes FS, Bright JN, Sansom MS. Proline-induced distortions of transmembrane helices. J Mol Biol. 2002;323:951–960. doi: 10.1016/s0022-2836(02)01006-9. [DOI] [PubMed] [Google Scholar]
- 23.von Heijne G. Proline kinks in transmembrane alpha-helices. J Mol Biol. 1991;218:499–503. doi: 10.1016/0022-2836(91)90695-3. [DOI] [PubMed] [Google Scholar]
- 24.Woolfson DN, Williams DH. The influence of proline residues on alpha-helical structure. FEBS Lett. 1990;277:185–188. doi: 10.1016/0014-5793(90)80839-b. [DOI] [PubMed] [Google Scholar]
- 25.Vilsen B, Andersen JP, Clarke DM, MacLennan DH. Functional consequences of proline mutations in the cytoplasmic and transmembrane sectors of the Ca2(+)-ATPase of sarcoplasmic reticulum. J Biol Chem. 1989;264:21024–21030. [PubMed] [Google Scholar]
- 26.Rothschild KJ, He YW, Mogi T, Marti T, Stern LJ, Khorana HG. Vibrational spectroscopy of bacteriorhodopsin mutants: evidence for the interaction of proline-186 with the retinylidene chromophore. Biochemistry. 1990;29:5954–5960. doi: 10.1021/bi00477a011. [DOI] [PubMed] [Google Scholar]
- 27.Loo TW, Clarke DM. Functional consequences of proline mutations in the predicted transmembrane domain of P-glycoprotein. J Biol Chem. 1993;268:3143–3149. [PubMed] [Google Scholar]
- 28.Koike K, Conseil G, Leslie EM, Deeley RG, Cole SP. Identification of proline residues in the core cytoplasmic and transmembrane regions of multidrug resistance protein 1 (MRP1/ABCC1) important for transport function, substrate specificity, and nucleotide interactions. J Biol Chem. 2004;279:12325–12336. doi: 10.1074/jbc.M311435200. [DOI] [PubMed] [Google Scholar]
- 29.Ito K, Weigl KE, Deeley RG, Cole SP. Mutation of proline residues in the NH(2)-terminal region of the multidrug resistance protein, MRP1 (ABCC1): effects on protein expression, membrane localization, and transport function. Biochim Biophys Acta. 2003;1615:103–114. doi: 10.1016/s0005-2736(03)00228-1. [DOI] [PubMed] [Google Scholar]
- 30.Koike K, Conseil G, Leslie EM, Deeley RG, Cole SP. Identification of proline residues in the core cytoplasmic and transmembrane regions of multidrug resistance protein 1 (MRP1/ABCC1) important for transport function, substrate specificity, and nucleotide interactions. J Biol Chem. 2004;279:12325–12336. doi: 10.1074/jbc.M311435200. [DOI] [PubMed] [Google Scholar]
- 31.Letourneau IJ, Nakajima A, Deeley RG, Cole SP. Role of proline 1150 in functional interactions between the membrane spanning domains and nucleotide binding domains of the MRP1 (ABCC1) transporter. Biochem Pharmacol. 2008;75:1659–1669. doi: 10.1016/j.bcp.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang CH, el-Kabbani O, Tiede D, Norris J, Schiffer M. Structure of the membrane-bound protein photosynthetic reaction center from Rhodobacter sphaeroides. Biochemistry. 1991;30:5352–5360. doi: 10.1021/bi00236a005. [DOI] [PubMed] [Google Scholar]
- 34.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 35.Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozvegy-Laczka C, Koblos G, Sarkadi B, Varadi A. Single amino acid (482) variants of the ABCG2 multidrug transporter: major differences in transport capacity and substrate recognition. Biochim Biophys Acta. 2005;1668:53–63. doi: 10.1016/j.bbamem.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Robey RW, Honjo Y, van de Laar A, Miyake K, Regis JT, Litman T, Bates SE. A functional assay for detection of the mitoxantrone resistance protein, MXR (ABCG2) Biochim Biophys Acta. 2001;1512:171–182. doi: 10.1016/s0005-2736(01)00308-x. [DOI] [PubMed] [Google Scholar]
- 38.Ozvegy-Laczka C, Varady G, Koblos G, Ujhelly O, Cervenak J, Schuetz JD, Sorrentino BP, Koomen GJ, Varadi A, Nemet K, Sarkadi B. Function-dependent conformational changes of the ABCG2 multidrug transporter modify its interaction with a monoclonal antibody on the cell surface. J Biol Chem. 2005;280:4219–4227. doi: 10.1074/jbc.M411338200. [DOI] [PubMed] [Google Scholar]
- 39.Shukla S, Robey RW, Bates SE, Ambudkar SV. Sunitinib (Sutent, SU11248), a small-molecule receptor tyrosine kinase inhibitor, blocks function of the ATP-binding cassette (ABC) transporters P-glycoprotein (ABCB1) and ABCG2. Drug Metab Dispos. 2009;37:359–365. doi: 10.1124/dmd.108.024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozvegy-Laczka C, Laczko R, Hegedus C, Litman T, Varady G, Goda K, Hegedus T, Dokholyan NV, Sorrentino BP, Varadi A, Sarkadi B. Interaction with the 5D3 monoclonal antibody is regulated by intramolecular rearrangements but not by covalent dimer formation of the human ABCG2 multidrug transporter. J Biol Chem. 2008;283:26059–26070. doi: 10.1074/jbc.M803230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miwa M, Tsukahara S, Ishikawa E, Asada S, Imai Y, Sugimoto Y. Single amino acid substitutions in the transmembrane domains of breast cancer resistance protein (BCRP) alter cross resistance patterns in transfectants. Int J Cancer. 2003;107:757–763. doi: 10.1002/ijc.11484. [DOI] [PubMed] [Google Scholar]
- 42.Govaerts C, Blanpain C, Deupi X, Ballet S, Ballesteros JA, Wodak SJ, Vassart G, Pardo L, Parmentier M. The TXP motif in the second transmembrane helix of CCR5. A structural determinant of chemokine-induced activation. J Biol Chem. 2001;276:13217–13225. doi: 10.1074/jbc.M011670200. [DOI] [PubMed] [Google Scholar]
- 43.Sansom MS, Weinstein H. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol Sci. 2000;21:445–451. doi: 10.1016/s0165-6147(00)01553-4. [DOI] [PubMed] [Google Scholar]
- 44.Clark R, Kerr ID, Callaghan R. Multiple drugbinding sites on the R482G isoform of the ABCG2 transporter. Br J Pharmacol. 2006;149:506–515. doi: 10.1038/sj.bjp.0706904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.