Abstract
One of the first steps in neurogenesis is the diversification of cells along the dorsoventral axis. In Drosophila the central nervous system develops from three longitudinal columns of cells: ventral cells that express the vnd/nk2 homeobox gene, intermediate cells, and dorsal cells that express the msh homeobox gene. Here we describe a new Drosophila homeobox gene, intermediate neuroblasts defective (ind), which is expressed specifically in the intermediate column cells. ind is essential for intermediate column development: Null mutants have a transformation of intermediate to dorsal column neuroectoderm fate, and only 10% of the intermediate column neuroblasts develop. The establishment of dorsoventral column identity involves negative regulation: Vnd represses ind in the ventral column, whereas ind represses msh in the intermediate column. Vertebrate genes closely related to vnd (Nkx2.1 and Nkx2.2), ind (Gsh1 and Gsh2), and msh (Msx1 and Msx3) are expressed in corresponding ventral, intermediate, and dorsal domains during vertebrate neurogenesis, raising the possibility that dorsoventral patterning within the central nervous system is evolutionarily conserved.
Keywords: vnd, NK2, tinman, neuroectoderm, central nervous system
The Drosophila central nervous system (CNS) develops from a bilateral neuroectoderm that lies to each side of a narrow strip of ventral midline cells. Single neuroectodermal cells delaminate from the surface epithelium and move into the interior of the embryo to form neural precursor cells called neuroblasts. The early neuroblasts form an orthogonal grid of four rows (1, 3, 5, and 7) along the anterior-posterior (AP) axis and three columns (ventral, intermediate, and dorsal) along the dorsoventral (DV) axis. Subsequently, each neuroblast expresses a characteristic combination of genes and contributes a stereotyped family of neurons and glia to the CNS. Thus the earliest steps in patterning the CNS are the formation and specification of neuroblasts.
Neuroblast formation is regulated by two phenotypically opposite classes of genes: Proneural genes promote neuroblast formation, whereas the neurogenic genes inhibit neuroblast formation. Proneural genes encode a family of basic helix–loop–helix transcription factors that are expressed in 4–6 cell clusters at specific positions within the neuroectoderm. Embryos lacking the proneural genes achaete/scute or lethal of scute have a reduced number of neuroblasts (for review, see Skeath and Carroll 1994). Conversely, neurogenic genes are expressed uniformly in the neuroectoderm, and embryos that lack any one neurogenic gene function, such as Notch or Delta, develop an excess number of neuroblasts (for review, see Campos-Ortega 1995).
The generation of neuronal diversity begins with the specification of unique neuroblast identities along both the AP and DV axes. The wingless, hedgehog, gooseberry, and engrailed genes are expressed in stripes of neuroectoderm that subdivide the AP axis. They are required for establishing AP row identity within the neuroectoderm and neuroblasts (Chu-LaGraff and Doe 1993; Zhang et al. 1994; Skeath et al. 1995; Bhat 1996; Matsuzaki and Saigo 1996; Bhat and Schedl 1997; McDonald and Doe 1997). For example, gooseberry is expressed in row 5 neuroectoderm. Embryos lacking gooseberry function have a transformation of row 5 into row 3 neuroectoderm and neuroblast identity, whereas misexpression of gooseberry results in the converse row 3 to row 5 transformation (Zhang et al. 1994; Skeath et al. 1995). Similarly, wingless encodes a protein secreted from row 5 and required for specifying the fate of the adjacent rows 4 and 6 neuroectoderm and neuroblasts (Chu-LaGraff and Doe 1993). Although we have learned a great deal about how the Drosophila CNS is patterned along the AP axis recently, relatively little is known about patterning along the DV axis.
Two genes are expressed in restricted domains along the DV axis within the neuroectoderm: ventral nervous system defective (vnd) is an NK2 class homeobox gene expressed in the ventral column neuroectoderm (Jimenez et al. 1995; Mellerick and Nirenberg 1995) and muscle segment homeobox (msh) is a homeobox gene expressed in the dorsal column neuroectoderm and neuroblasts (D’Alessio and Frasch 1996; Isshiki et al. 1997). Mutations in vnd cause defects in neuroblast formation and lead to severe defects later in neurogenesis (White et al. 1983; Skeath et al. 1994), but the role of vnd in patterning the neuroectoderm and neuroblasts along the DV axis has not been determined. Mutations in msh result in a partial transformation of dorsal neuroblasts into a more ventral or intermediate column identity, without affecting neuroblast formation (Isshiki et al. 1997). Signaling via the EGF receptor is required to establish ventral and/or intermediate column fates in the neuroectoderm (Rutledge et al. 1992; Raz and Shilo 1993; Schweitzer et al. 1995). Although vnd and msh are candidate genes for establishing ventral and dorsal column fates within the CNS, no genes are known currently to be expressed specifically in the intermediate column of the CNS.
In this paper we describe the identification and genetic characterization of a new homeobox gene, intermediate neuroblasts defective (ind) (GenBank accession no. AF095926), that is the first gene known to be expressed specifically in the intermediate column of neuroectoderm and neuroblasts. We show that ind function is required for the establishment of intermediate column identity in the neuroectoderm, and for the formation of intermediate column neuroblasts. In this paper and in McDonald et al. (1998), we examine the regulatory interactions between vnd, ind, and msh, and show that there is a hierarchical cascade of transcriptional repression. Vnd represses ind expression to establish the ventral boundary of ind transcription, and ind represses msh to establish the ventral boundary of msh transcription. The homeobox genes expressed in columns within the Drosophila neuroectoderm—vnd, ind, and msh—each have gene homologs expressed in corresponding domains along the DV axis of the vertebrate neural ectoderm. On this basis it appears that fundamental molecular mechanisms of DV patterning may be similar in Drosophila and vertebrates.
Results
A screen for genes regulated by Tinman class homeodomain proteins
The tinman gene encodes a homeodomain protein required for heart and visceral mesoderm development (Azpiazu and Frasch 1993; Bodmer 1993). To discover how tinman directs mesodermal cell fates, we performed a screen to identify genes that are regulated by tinman. The screen relies on genetic selection in yeast for a protein–DNA interaction (Liu et al. 1993; Mastick et al. 1995). In brief, yeast expressing a gene encoding the Tinman homeodomain fused to the yeast GAL4 activation domain were transformed with a library of Drosophila genomic DNA fragments inserted upstream of a selectable marker in a reporter plasmid. When the fusion protein recognizes a Drosophila genomic DNA fragment it will activate transcription of the selectable marker. Genomic DNA fragments identified in the screen are therefore presumptive binding sites for the Tinman homeodomain.
Transcribed regions flanking the genomic DNA fragments isolated in the screen were identified and used as probes for whole-mount in situ hybridization to embryos. Several of the transcripts found in this way are produced in the mesoderm in patterns consistent with Tinman regulation, and will be described elsewhere (J.B. Weiss and M.P. Scott, in prep.). One of the identified genes, ind, located in polytene interval 71A, is transcribed in the CNS but not in the mesoderm. In this paper we describe the expression, regulation, and function of ind in patterning the DV axis of the CNS.
ind is a homeobox gene related to two homeobox genes expressed in the developing vertebrate CNS
The genomic DNA (Fig. 1A) adjacent to the putative Vnd binding site was employed as a hybridization probe to isolate full-length cDNA clones of ind. The predicted protein sequence shows that ind encodes a protein containing a homeodomain that is most closely related to the vertebrate Gsh1 and Gsh2 homeodomain proteins. There is 85% amino acid identity between Ind and either of the Gsh homeodomains (Fig. 1B). Ind, Gsh1, and Gsh2 also share a short amino-terminal region of homology (Fig. 1B). As is shown below, ind is expressed in two longitudinal stripes in sharply defined DV domains of the Drosophila CNS. This shows striking similarity to the expression of Gsh1 and Gsh2 in the developing murine CNS (Fig. 7 below, and Valerius et al. 1995; Szucsik et al. 1997).
Figure 1.
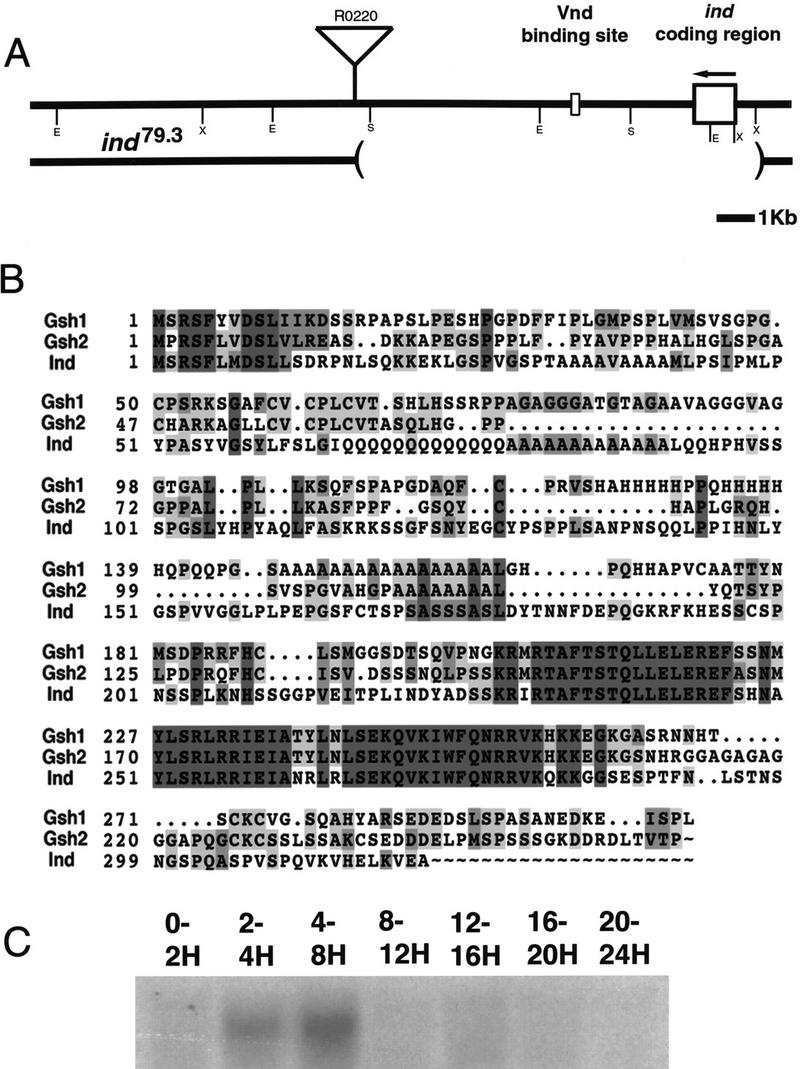
The structure of the ind genomic region, amino acid sequence of Ind, and developmental expression of ind. (A) The structure of the ind gene in wild-type and a mutant allele, ind79.3. (Top) The structure of the wild-type allele. The coding sequence for ind is contained in a single exon, shown as a large box. ind is transcribed in the direction indicated by the arrow. The binding site for Vnd, marked by a smaller box, lies 3 kb 3′ to the coding sequence. The insertion site of a P element, R0220, in relation to the gene is shown. Restriction endonuclease sites are marked with single letters: (E) EcoRI, (S) SalI, (X) XhoI. (Bottom) The extent of a deletion that was generated by mobilization of the R0220 P element. The deletion is 7 kb long and removes the coding sequence for ind. (B) The predicted amino acid sequence of Ind aligned with two closely related mouse genes, Gsh1 and Gsh2. The alignment shows conservation primarily within the homeodomain (amino acids 227–286 in Ind) though there is also a short region of homology at the amino terminus. Identities are marked in dark gray, similarities in light gray. Numbers at the left indicate the amino acid positions in the respective proteins. (C) An RNA blot of staged embryos hybridized with a radiolabeled ind cDNA clone. The times in hours postfertilization of the embryo collections are marked above the lanes. All lanes were loaded with 20 μg of total RNA.
Figure 7.
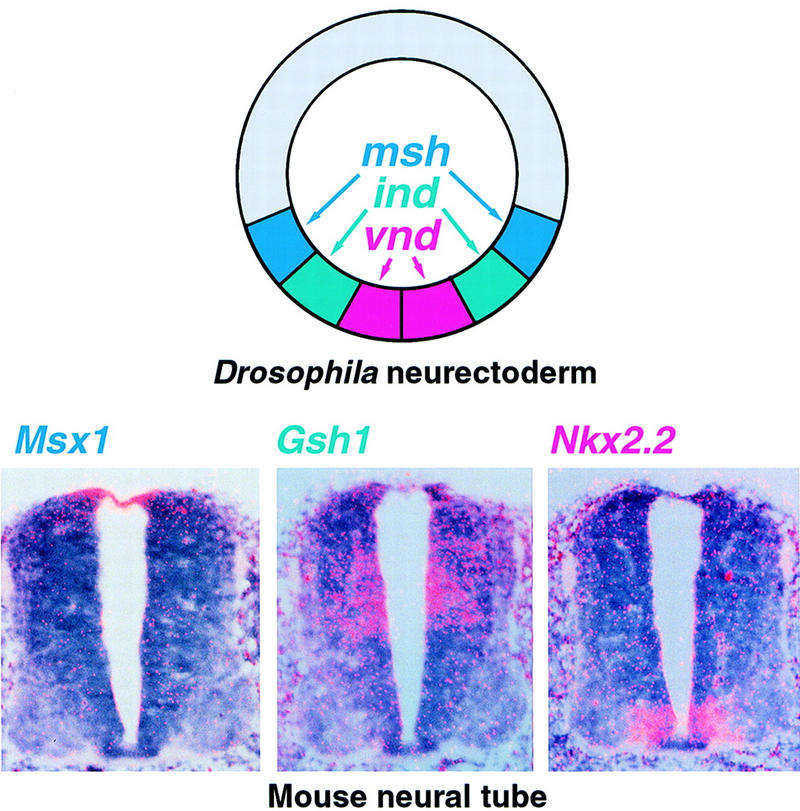
DV domains of expression of Drosophila and related mouse homeobox genes in the developing CNS. On the left a schematic cross section of a stage 7 Drosophila embryo showing the expression of msh (blue), ind (green), and vnd (red) in symmetric columns of neurectoderm. On the right, three panels show in situ hybridization with three related mouse genes performed on serial cross sections of the neural tube at the midthoracic level of an 11.5 day mouse embryo. Dorsal is up, ventral is down.
ind is transiently expressed in the intermediate column neuroectoderm and neuroblasts
The temporal and spatial pattern of ind RNA accumulation was determined using RNA blots (Fig. 1C) and whole-mount in situ hybridization (Fig. 2). A single 1.3-kb ind transcript appears at 2–4 hr of development with peak accumulation at 4–8 hr of development; no transcripts were detected later in embryogenesis (Fig. 1C). Whole-mount in situ hybridization first detects ind expression in two parallel, lateral columns in stage 5 cellular blastoderm embryos (Fig. 2). At this stage each column is about five cells wide and runs from the procephalic region to the most posterior region of the embryo (Fig. 2). During gastrulation, the ind expression domain narrows until each column is two cells wide. The position of each of the two symmetric ind columns is just dorsal to the domain of vnd transcription (Fig. 4, below), in cells that will become the intermediate column of neuroectoderm. When neuroblast formation begins at late stage 8, ind mRNA is expressed in both the intermediate column neuroectoderm and in the S1 neuroblasts (Nbs) derived from the intermediate column (Nbs 3-2 and 5-3). By stage 9, ind mRNA is absent from the neuroectoderm, but is detectable in all of the neuroblasts in the intermediate column (Nbs 3-2, 4-2, 5-3, 6-2, and 7-2). ind is not expressed in any ventral or dorsal column neuroectoderm or neuroblasts. By stage 11, ind mRNA is detectable in just a single intermediate column neuroblast, Nb 6-2. After stage 11, ind mRNA was not detected anywhere in the embryo.
Figure 2.
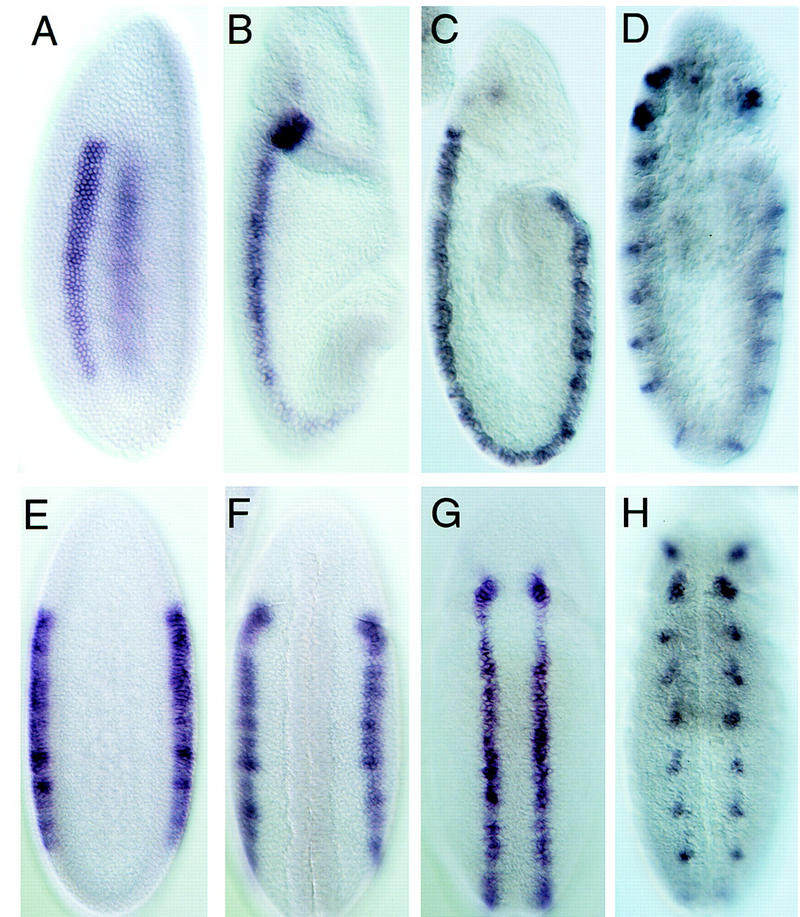
ind expression in wild-type embryos. Shown are lateral and ventral views of whole-mount in situ hybridizations with the ind cDNA to a developmental series of wild-type embryos. The embryos on the top (A–D) are shown in lateral views with anterior up and ventral to the left. The embryos on the bottom (E–H) are ventral views again with anterior up. The embryos are approximately stage matched and represent a developmental series starting at the left. (A,E) Stage 5, cellular blastoderm, embryos in which ind is expressed in two longitudinal stripes five cells wide in the intermediate neurectoderm. (B,F) Stage 7 embryos, following ventral furrow formation and ventral migration of the neurectoderm. ind expression in these embryos is narrowed to longitudinal stripes three cells wide. (C,G) Early stage 9 embryos in which ind is expressed in the intermediate neurectoderm and S1, intermediate column neuroblasts. (D,H) Late stage 9 to early stage 10 embryos. At this stage ind mRNA is progressively restricted to neuroblasts 6-2 and 7-2.
Figure 4.
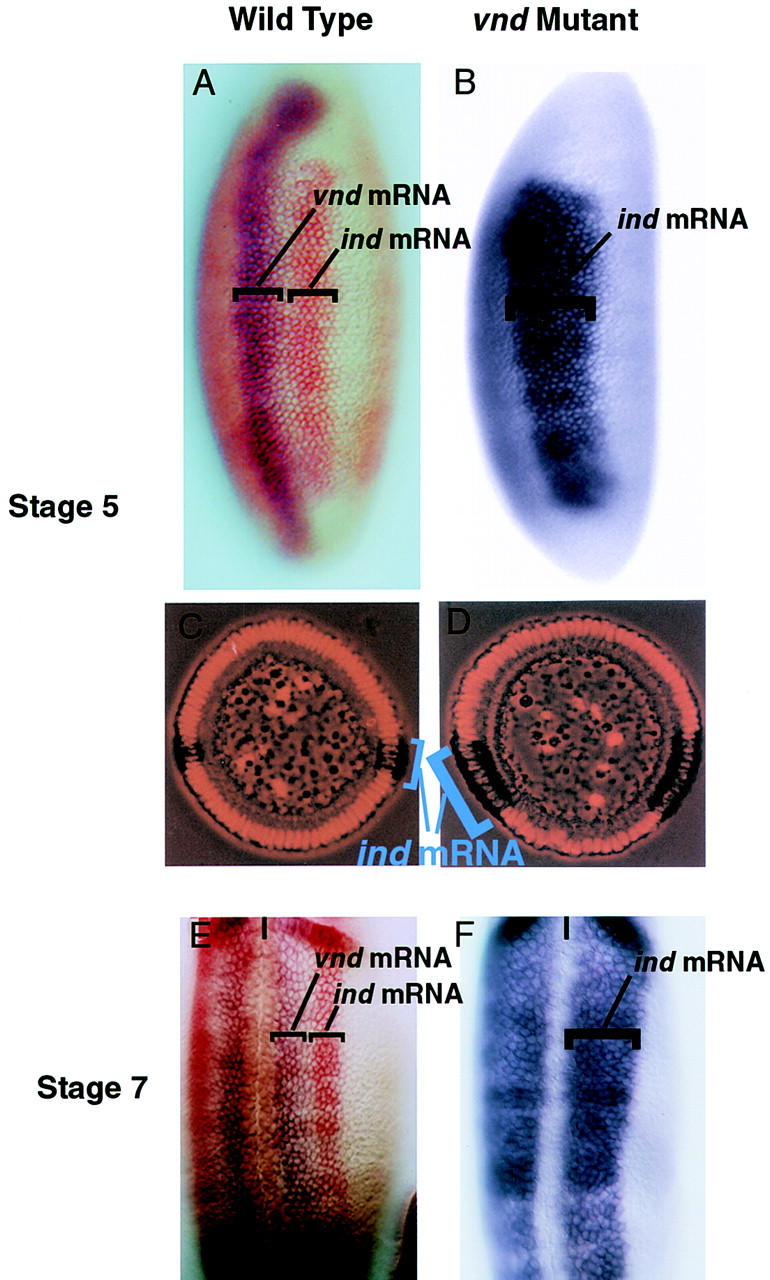
ind and vnd expression in wild-type embryos. Ventral derepression of ind in vnd mutant embryos. (A–D) Lateral (A,B) and cross sectional (C,D) views of wild-type (A,C) and vnd mutant (B,D) stage 5 embryos. Anterior is up, ventral to the left in A and B. In C and D ventral is down. (A) ind (red) is dorsal and adjacent to vnd (blue) in wild-type embryos. (B) In vnd mutant embryos ind (blue) expression is broadened ventrally to encompass the vnd expression domain. (C,D) Cross sectional views of wild-type (C) and vnd mutant (D) stage 5 embryos stained with ind (blue-black) and propidium iodide to mark nuclei. The ventral expansion of ind in vnd mutants is seen clearly in these cellular blastoderm embryos. (E,F) Ventral views of wild-type and vnd mutant stage 7 embryos. Anterior is up, the ventral midline is marked with a black line. (E) As above ind (red) is dorsal to vnd (blue). (F) In the vnd mutant ind (blue) expands ventrally to encompass the neurectoderm and the vnd expression domain.
Monoclonal and polyclonal antibodies against the Ind protein were used to determine the subcellular localization of ind protein, assay the extent of ind posttranscriptional regulation, and confirm the identity of ind-expressing cells using double labels with other antibody markers. The Ind protein is detected in the nucleus at all stages assayed, as expected for a homeodomain protein (Fig. 3). We see no evidence for post-transcriptional regulation: ind mRNA and Ind protein patterns are virtually identical, except that the protein persists slightly longer than the mRNA (Figs. 2 and 3). The antibody staining confirms that Ind is restricted to the intermediate column neuroectoderm and neuroblasts. The Ind protein-containing cells are immediately adjacent but nonoverlapping with Vnd-containing cells of the ventral column neuroectoderm (see McDonald et al. 1998). Double labeling with various neuroblast markers shows that Ind is detected in the same intermediate column neuroblasts as ind mRNA (Figs. 2 and 3). In contrast to ind mRNA, Ind protein is detectable in all intermediate column neuroblasts at stage 13, after the mRNA is no longer detectable in most of these neuroblasts. Ind protein is not detectable after stage 11 of embryogenesis.
Figure 3.
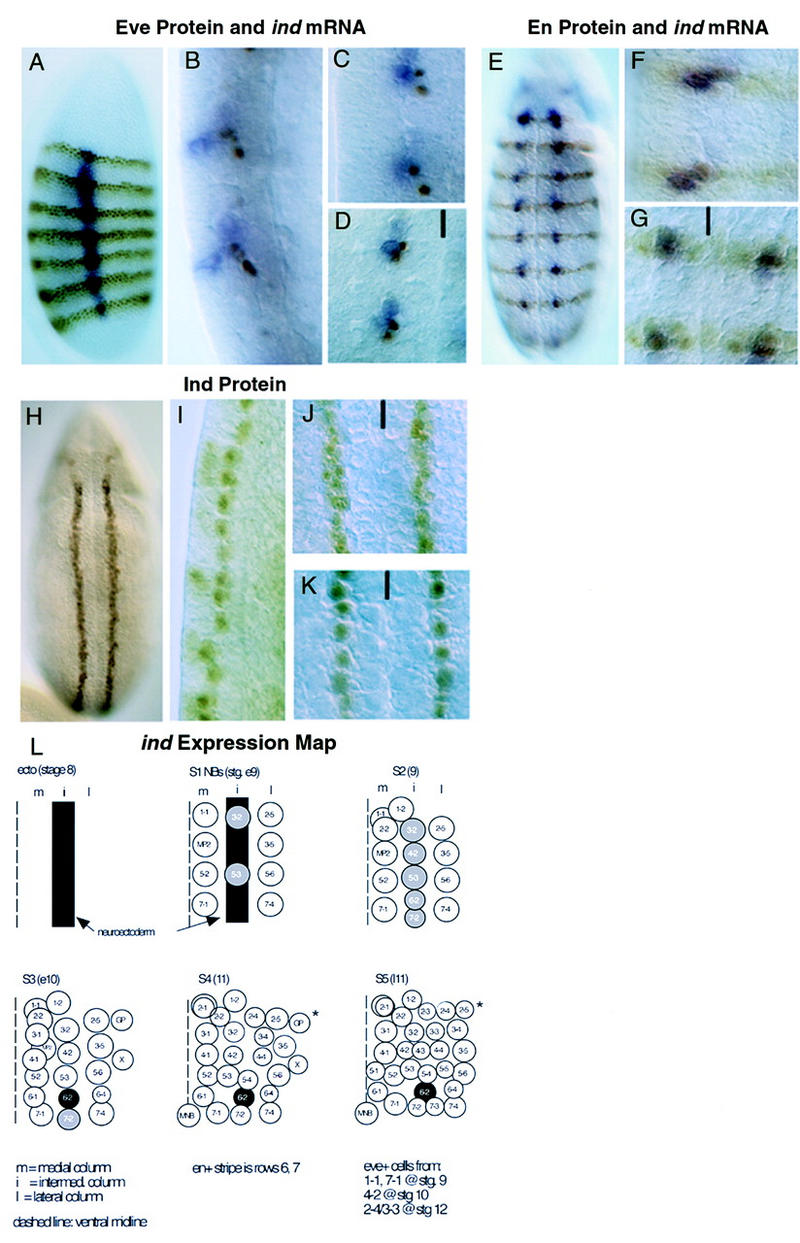
ind expression monitored with antibodies. (A–D) DV localization of ind-expressing Nbs. Anti-Eve antibody staining is brown, hybridization to ind mRNA is blue. Anterior is up, ventral to the left. (A) Lateral view of a stage 5, cellular blastoderm embryo showing ind expression in two parallel bands in intermediate neurectoderm, orthogonal to Eve. (B) Higher magnification, lateral view of late stage 9 embryos showing delamination of the Eve positive medial Nbs 1-1 and 7-1 prior to delamination of the ind-expressing intermediate Nbs 6-2 and 7-2. (C) Lateral view of midstage 10 embryo showing delamination of the ind expressing Nbs 6-2 and 7-2. (D) Ventral view of midstage 10 embryo showing medial Eve positive Nbs 1-1 and 7-1 adjacent to intermediate ind-expressing Nbs 6-2 and 7-2. The ventral midline is marked with a black line. (E–G) AP localization of ind-expressing Nbs. Anti-Engrailed (En) antibody staining is in brown, hybridization to ind mRNA is in blue. All embryos are at stage 10. (E) Whole embryo showing expression of ind in register with En in the AP axis. (F,G) Lateral and ventral views, respectively, of delaminated ind expressing Nbs showing the identity of En- and Ind-positive intermediate Nbs. The ventral midline is marked with a black line. (H–K) Ind protein expression in the neurectoderm and intermediate Nbs. (H) Ventral view of stage 8 embryo showing two parallel stripes of neurectodermal Ind protein. (I) Higher magnification, lateral view of late stage 9 embryo showing persistent accumulation of Ind protein in all delaminated intermediate column NBs. (J) Higher magnification, ventral view of stage 8 embryo showing intermediate neurectoderm presence of Ind. (K) Higher magnification ventral view of stage 9 embryo showing Ind in all intermediate-column Nbs. The ventral midline is marked with a black line. (L) ind expression map diagram of neuroblast formation in one hemisegment stages 8–11. Ventral view, anterior is up, the midline is at the left of each panel. ind expression is shown in gray and black. Neuroblasts are designated by their final row number–column number.
ind is regulated by Vnd/Nk2 in the central nervous system
Although ind was identified in a screen for Tinman transcriptional targets, ind and Tinman are expressed in nonoverlapping, nonadjacent regions of the CNS and mesoderm, so it is unlikely that Tinman regulates ind directly. Furthermore, tinman mutant embryos have no change in ind expression (data not shown). We therefore tested the hypothesis that ind is transcriptionally regulated by Vnd, a homeodomain protein related closely to Tinman. Vnd is produced in the ventral neuroectoderm immediately adjacent to the ind-expression domain.
Genetic and molecular data demonstrate that ind is transcriptionally repressed by Vnd (Figs. 4, 5). In wild-type embryos the two genes are expressed in adjacent but nonoverlapping portions of the neuroectoderm. vnd is expressed in the ventral column, whereas ind is expressed in the intermediate column. In vnd mutant embryos, ind expression is broader and encompasses what would normally be the vnd-expression domain. This can be observed clearly in lateral views of whole-mount embryos as well as in embryo cross sections (Fig. 4). These genetic experiments show that vnd is required to repress ind expression within the ventral column neuroectoderm.
Figure 5.
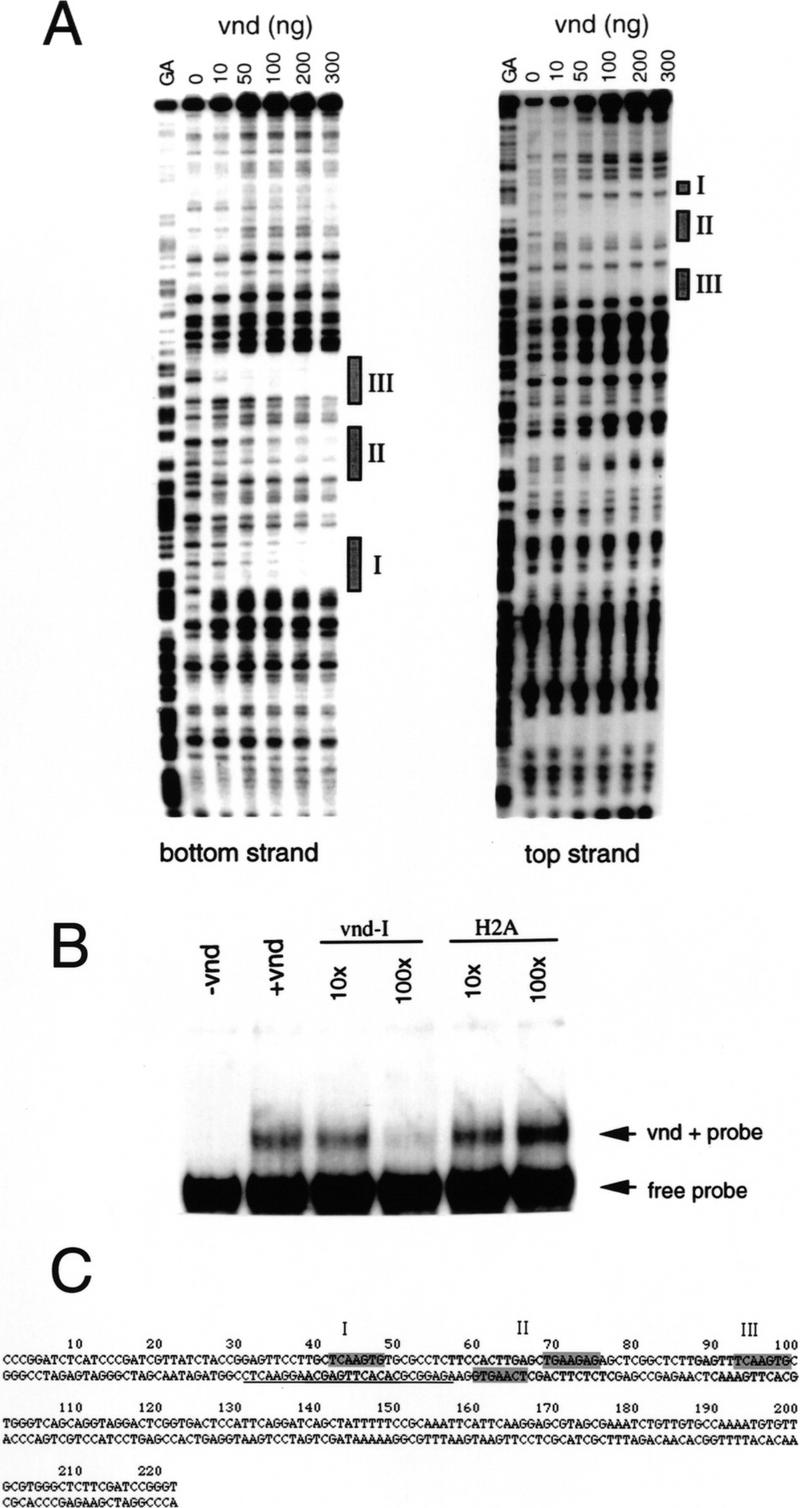
Binding of the Vnd protein to ind sequences. (A) DNase I footprints of both strands of the ind DNA. Three major protected regions become visible as the amount of recombinant Vnd homeodomain is increased. (B) Electrophoresis mobility-shift assay using ind DNA nucleotides 5–135, which includes all three Vnd-binding regions. A DNA/protein complex is observed upon addition of Vnd. This complex can be competed out with increasing amounts of the Vnd-I oligonucleotide but not with an irrelevant oligonucleotide (H2A). (C) Sequence showing the Vnd-binding regions as determined by DNase I footprinting. The protected regions are shaded in light gray, whereas a candidate consensus sequence is shaded in dark gray. The Vnd-I oligonucleotide sequence is underlined.
To determine whether Vnd regulates ind transcription directly, we used bacterially expressed Vnd protein to perform electrophoretic mobility-shift and footprinting assays with the genomic ind DNA fragment identified in our initial screen (GenBank accession no. AF096297). Vnd specifically binds the fragment of ind genomic DNA isolated in the screen (Fig. 5A). Three specific binding sites of roughly equal affinity can be identified using footprinting assays (Fig. 5B). The three sites protected in the footprinting assay each contain one copy of the sequence GTGAACT, which has been found to be a recognition sequence for both Vnd and the Tinman-related Nkx2.5 vertebrate protein (Chen and Schwartz 1995; Gruschus et al. 1997).
ind is required for the specification of intermediate-column neuroectoderm and neuroblasts
ind null mutants were generated in two different screens. We identified a single ind allele (indRR108) in an ethyl methane sulfonate mutagenesis screen for altered even-skipped (eve) expression in the CNS (J. Skeath and C.Q. Doe, unpubl.). In addition, three ind alleles (ind16.2, ind79.3, and ind4.1) were obtained by mobilizing a P element located next to the ind locus. All four alleles appear to be null mutations in ind for the following reasons: (1) one allele, ind79.3, has a deletion of the ind-coding region (Fig. 1); (2) ind16.2 and indRR108 homozygous embryos have no detectable Ind protein during neurogenesis (data not shown); (3) ind16.2 in trans with a large deficiency shows the same phenotype as the ind16.2 homozygotes (data not shown); and (4) all four ind alleles have the same phenotype when homozygous (data not shown). We believe that the ind phenotype described below is caused specifically by the loss of ind function because the ind79.3 mutation is a small deficiency that deletes the ind-coding region and 6 kb of flanking DNA, but not any other detectable gene (Fig. 1).
The earliest ind phenotype is observed in stage 7 embryonic neuroectoderm, when msh expression occurs both in its normal locations in the dorsal columns and in the adjacent intermediate columns (Fig. 6A,B). Thus ind represses transcription of msh directly or indirectly within intermediate column neuroectoderm. Normally the ind and msh expression domains are adjacent but nonoverlapping, consistent with negative regulation of msh by ind. During the earliest stage of neurogenesis (stage 8), wild-type embryos have expression of the proneural gene achaete in rows 3 and 7 of the neuroectoderm, with expression restricted to the ventral and dorsal columns and excluded from the intermediate column (Fig. 6G). ind expression in the intermediate column abuts these clusters of achaete-expressing cells precisely without overlapping them. In ind mutant embryos, derepression of achaete expression is observed within the intermediate column of neuroectoderm in rows 3 and 7 (Fig. 6H). This is consistent with a transformation of intermediate to dorsal neuroectoderm, as is seen at stage 7 with the msh marker. We conclude that ind represses msh and achaete gene expression directly or indirectly, and that ind is necessary for establishing proper intermediate-column identity within the neuroectoderm.
Figure 6.
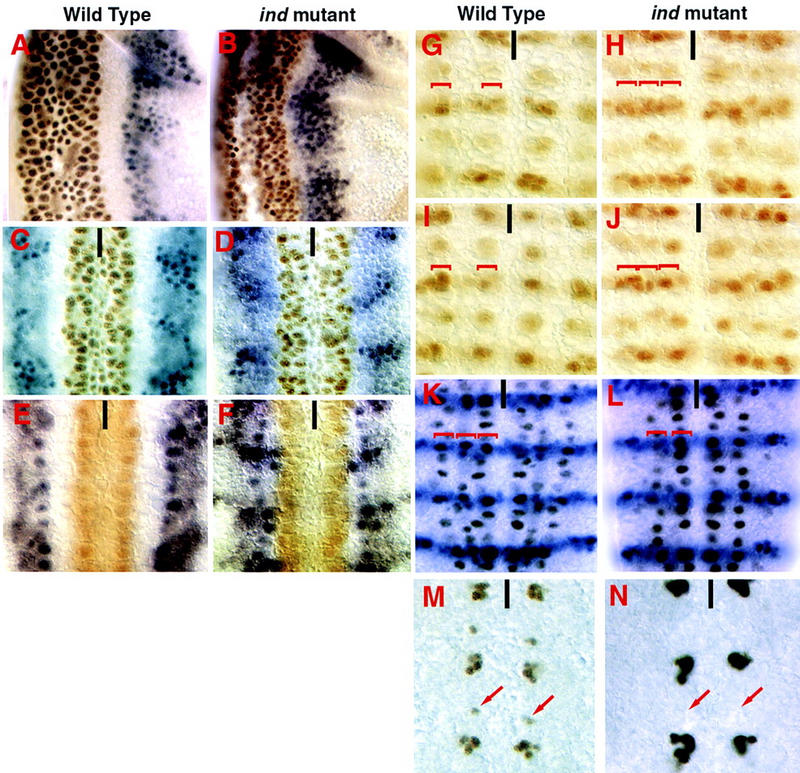
Loss of ind function results in loss of intermediate-column neuroblasts and intermediate-column-derived neurons. All views are ventral except A and B which are lateral. Anterior is up, ventral to the left in A and B. The ventral midline is marked with a black line. (A–F) Embryos doubly labeled for msh protein (blue) and Vnd protein (brown). (A,C,E) Wild-type embryos, blank space delineates the domain of Ind expression. (B,D,F). ind16.2 mutant embryos show Msh is expanded in the neurectoderm at stage 7 (B). This persists until stage 9 when the neuroblasts delaminate (D,F). (G–J) Achaete (Ac) is repressed in the neurectoderm and neuroblasts by Ind. In wild-type embryos Ac is expressed in four clusters per hemisegment in the neurectoderm (G) and in four neuroblasts per hemisegment (I). In ind mutant embryos Ac expression is derepressed in the intermediate column (H). When a neuroblast forms at these positions it expresses Ac inappropriately (J). (K,L) Hunchback, En double labels showing loss of intermediate-column neuroblasts in ind mutant embryos. Pan neural expression of Hunchback in wild-type late stage 9 embryos shows there are three columns of neuroblasts (K). In ind16.2 embryos only two columns of S1 neuroblasts are observed. Ind is therefore required for formation of intermediate-column neuroblasts (L). (M,N) Ind is required for neurons derived from intermediate-column neuroblasts. Even-skipped staining is shown in brown. The embryos are stage 13 and two types of Even-skipped positive cells are seen per hemisegment in this focal plane. The larger clusters are neurons derived from medial column neuroblasts. They are unaffected in ind mutants (N). The smaller clusters, highlighted by arrows, are RP2/RP2sib neurons that are derived from intermediate-column neuroblasts. These neurons are absent in the ind mutants (N).
In addition to regulating gene expression in the neuroectoderm, ind has an essential role in neuroblast formation and specification. In wild-type embryos, Hunchback staining reveals three columns of neuroblasts (ventral, intermediate, and dorsal; Fig. 6K). In ind mutant embryos, only 10% of the intermediate-column neuroblasts develop, although adjacent ventral and dorsal column neuroblasts form normally (Fig. 6L). The 10% of neuroblasts that develop in the intermediate column always express achaete, which is never detected in wild-type intermediate column neuroblasts (Fig. 6I,J).
We stained the infrequent neuroblasts that form in the intermediate column of ind mutant embryos with anti-Msh and anti-Vnd antibodies to determine if there is a transformation to either dorsal or ventral cell fates in these mutants. A total of 80 hemisegments were scored. In a wild-type embryo 400 intermediate neuroblasts would be expected. In the ind mutant embryo 32 were observed. Eleven expressed Vnd, 12 expressed Msh, and 9 expressed neither marker. Thus, ind is essential for the formation of intermediate column neuroblasts, and for the repression of ventral- and dorsal-specific genes within these neuroblasts.
To test whether intermediate-column neuroblasts that form in ind mutant embryos have the potential to generate cell lineages characteristic of intermediate column neuroblasts, we assayed development of the Eve-containing RP2 neuron, which is a motor neuron derived from the intermediate-column neuroblast 4-2 (Doe 1992; Chu-LaGraff et al. 1995). The pattern of Eve is a sensitive indicator for normal cell fates within neuroblast cell lineages (Duffy et al. 1991; Chu-LaGraff and Doe 1993; Yang et al. 1993, 1997; Zhang et al. 1994; Chu-LaGraff et al. 1995). In wild-type embryos, an Eve-protein-containing RP2 is found in every hemisegment (100/100), whereas in ind mutant embryos the Eve-containing RP2 is never detected (0/96) (Fig. 6M,N). This defect is caused in part by a failure in NB 4-2 formation, but is also likely to be caused by defects in NB 4-2 specification or cell division in the 10% of the NB 4-2s that appear to form. These data suggest that ind regulates intermediate column neuroblast-cell lineages.
Discussion
The Drosophila CNS develops from ventral, intermediate, and dorsal columns of neuroectoderm and neuroblasts. Each column runs longitudinally along the embryo, and each gives rise to a specific set of neurons and glia (Bossing et al. 1996; Schmidt et al. 1997). The ventral neuroectoderm and neuroblasts express the vnd/nk2 homeobox gene (Jimenez et al. 1995; Mellerick and Nirenberg 1995), whereas the dorsal neuroectoderm and neuroblasts express the msh homeobox gene (D’Alessio and Frasch 1996; Isshiki et al. 1997). ind appears to be the missing regulator, a gene that precisely marks and controls the development of the intermediate column of neuroectoderm and neuroblasts. The DV control genes vnd, ind, and msh—together with the AP-patterning genes—constitute a Cartesian cell-fate determination system for the developing CNS.
Identification and expression of ind
tinman encodes a homeodomain protein that is required for heart and visceral mesoderm development, and is transcribed only in the tissues it affects. ind is not expressed in the mesoderm, yet was identified in a yeast one-hybrid screen for transcriptional targets of Tinman. This paradox was resolved with the cloning of vnd (Jimenez et al. 1995) and the recognition that Vnd and Tinman proteins contain closely related homeodomains. Both genetic and molecular data demonstrate that ind is repressed by Vnd protein. DNA-binding assays show that Vnd protein binds to a sequence adjacent to the ind transcript unit; this interaction is likely to be relevant biologically because Vnd is necessary to repress ind transcription in the ventral column of neuroectoderm and neuroblasts. Other transcription units identified in the yeast Tinman target gene screen have expression patterns consistent with regulation by Tinman, and not Vnd, and will be described elsewhere.
ind is required for repression of dorsal-column neuroectodermal markers within the intermediate column
Embryos lacking ind function have profound defects in the development of the intermediate column of the CNS. The defects include a derepression of dorsal column markers within the intermediate-column neuroectoderm. The msh gene is expressed normally in the dorsal column of the neuroectoderm, but ind mutant embryos show ectopic msh transcription in the intermediate column. Similarly, achaete is expressed normally in the ventral and dorsal regions of the neuroectoderm, but ind mutant embryos show ectopic achaete in the intermediate column of neuroectoderm. Whether ind acts directly or indirectly to repress msh and achaete expression is unknown. Taken together, the msh and achaete data suggest that one function of ind is to prevent transcription of dorsal column genes within the intermediate column of the neuroectoderm.
ind promotes intermediate-column neuroblast formation
In stage 9 wild-type embryos, five neuroblasts constitute the intermediate column in each hemisegment. In ind mutant embryos, at most one intermediate-column neuroblast is observed in each hemisegment, whereas the normal number of ventral and dorsal column neuroblasts form. Why do ind mutants have reduced neuroblast formation? One possibility is that ind activates proneural gene expression in the intermediate column of neuroblasts. The only proneural gene known to be expressed in this domain is lethal of scute, which we have not assayed. However, the ectopic expression of the proneural gene achaete that we observe in the intermediate-column neuroectoderm should promote—not reduce—neuroblast formation. Alternative explanations for the failure to generate intermediate column neuroblasts in ind mutant embryos include the following. (1) Proneural clusters of the dorsal column expand to include cells of the intermediate column, but still produce a single dorsal column neuroblast per row. This is consistent with the achaete expression in the intermediate-column neuroectoderm. (2) Intermediate-column neuroectoderm assumes a novel cell fate that is incompatible with neuroblast formation. This hypothesis is supported by data showing that alterations in neuroectoderm cell fate along the AP axis can lead to reduced neuroblast formation without affecting proneural gene expression (Chu-LaGraff and Doe 1993).
Our results suggest that ind may act in parallel to the known proneural genes to promote neuroblast formation in the intermediate column. Similarly, vnd is thought to promote neuroblast formation by proneural-dependent and proneural-independent pathways (Jimenez et. al. 1995; McDonald et al. 1998). Vnd and Ind could promote neuroblast formation by transcriptionally activating known or novel proneural genes, by transcriptionally repressing neurogenic genes (e.g., Notch), or by regulating genes currently unlinked to the proneural or neurogenic pathways.
Regulation of ind expression along the DV axis of the CNS
The three longitudinal columns of cells along the DV axis are defined by the ventral expression and function of vnd; the intermediate expression and function of ind; and the dorsal expression and function of msh. In this paper and in McDonald et al. (1998) we show that vnd function is required to establish the ventral border of ind: In the absence of vnd, expression of ind is observed in the ventral-column neuroectoderm and neuroblasts. How the dorsal border of ind expression is established is unknown. The absence of msh function from the dorsal column has no effect on ind expression. The activating regulators for ind expression are also unknown, although candidates include the Dorsal protein, which regulates DV patterning of the cellular blastoderm (for review, see Rusch and Levine 1996), and the epidermal growth factor (EGF)-receptor pathway, which regulates DV cell fate in the neuroectoderm (for review, see Schweitzer and Shilo 1997).
Parallels with vertebrate neural patterning
The vertebrate CNS develops from a dorsal neuroepithelium that is similar in many respects to the Drosophila ventral neuroectoderm. For example, in Xenopus the neural primordium is established by Chordin-mediated repression of BMP4 activity, whereas in Drosophila, the Chordin homolog Sog inhibits the BMP4 homolog Dpp to establish the neuroectoderm (for review, see Ferguson 1996; Bier 1997). In addition vertebrate orthologs of vnd and msh have been identified: Nkx2.1 and Nkx2.2 and Msx1 and Msx3, respectively (Holland 1991; Barth and Wilson 1995). Like vnd expression, Nkx2.2 expression is restricted to ventral regions of the nerve cord in the regions that give rise to motor neurons (Fig. 7). Similarly msh expression in the dorsal neuroectoderm is mirrored by Msx1 transcription in the dorsal neural tube (D’Alessio and Frasch 1996; Isshiki et al. 1997). The similar spatial distribution of Nkx2.2 and Msx1 and Msx3 expression to that of vnd and msh, respectively suggests a conserved mechanism of DV patterning across species (D’Alessio and Frasch 1996; Wang et al. 1996). Consistent with this, a putative ind ortholog, Gsh1, is expressed in symmetric lateral domains in the mouse neural tube (Fig. 7; Valerius et al. 1995). In Figure 7 the DV order of the Drosophila and mouse genes are clearly the same. It is also apparent that there are gaps between the expression domains of the three vertebrate genes that we have compared. We speculate that other related vertebrate genes will fill in these gaps. If the overall parallel is indicative of an evolutionary relationship, it will be of great importance to investigate what target genes might be regulated in common by the fly and vertebrate genes. Common targets might be revealing about ancient cell type diversification during neurogenesis.
Materials and methods
Yeast screen for genomic DNA bound by Tinman
The screen was performed according to the protocol of Liu et al. (1993). Amino acids 221–416 of the Tinman protein were cloned into the yeast vector pBM2463, the activator plasmid. This construct was transformed into yeast strain YM4271 by standard methods (Ausubel et al. 1994). The transformed yeast strain was subsequently transformed with a library of Drosophila melanogaster (DM) genomic DNA fragments cloned into the yeast vector pHR307a, the reporter plasmid (Mastick et al. 1995). The library construction is described in Mastick et al. (1995) and the library was provided by Javier Lopez. The library represents ∼15% of the genome. Transformants (5 × 106) were screened for growth on medium lacking histidine. Secondary screening was performed for maintenance of the Ade5 gene contained in the activator plasmid by a sectoring assay. Nonsectoring colonies were patched to medium lacking histidine and tested by replica plating for growth on medium lacking histidine in the presence of 5-fluoro-orotic acid (5-FOA). Growth on medium lacking histidine in the presence of 5-FOA acid implies that the activator plasmid coding for the Tinman fusion protein is not necessary for histidine auxotrophy. Positives were scored as yeast colonies that were nonsectoring on medium lacking histidine and unable to grow on medium lacking histidine in the presence of 5-FOA. Twenty-one independent positives were isolated from the screen. The reporter plasmids containing the Tinman binding sites were isolated from the yeast and the genomic DNA inserts subcloned and sequenced (Ausubel et al. 1994). Sequence analysis demonstrated that the 21 isolates represented 7 different genomic sites. Hybridization of these fragments to DM genomic DNA and polytene chromosomes confirmed that they are single-copy sequences of DM DNA (Pardue 1994).
Gene discovery
The genomic DNA inserts from the screen were employed as probes in hybridization screening for λ phage genomic clones by standard protocols (Sambrook et al. 1989). The library screened was provided by John Tamkun (Tamkun et al. 1992). Transcribed regions within the genomic λ were detected by hybridization to radiolabeled cDNA essentially as in Frei et al. (1986). Restriction digests of DM genomic λ phage that contained the Tinman binding sites were size fractionated in 0.8% agarose gel and transferred to a nylon filter (Sambrook et al. 1989). The filters were hybridized with radiolabeled cDNA synthesized by random priming of poly(A)-enriched RNA extracted from 0- to 24-hr embryos. Total embryonic RNA was extracted with guanidine HCl and organic solvents as described in Sambrook et al. (1989). Total RNA was enriched for poly(A)+ RNA by selection on oligo-dT cellulose according to the manufacturer’s directions (Pharmacia, mRNA Purification Kit). Radioactive cDNA was synthesized according to the protocol in Sambrook et al. (1989) with the modification that SuperScript Reverse Transcriptase and Buffers were used (GIBCO-BRL). Hybridization of radioactive cDNA to genomic phage DNA was performed in Church buffer (Church and Gilbert 1984). Fragments of genomic DNA that hybridized to radiolabeled cDNA were employed as probes for whole-mount in situ hybridization to embryos and for hybridization screening of cDNA libraries. Full-length cDNA clones were isolated by standard hybridization screening from the staged embryonic cDNA library of N. Brown (Brown and Kafatos 1988). The sequence of both strands was determined by the cycle sequencing reaction and an automated ABI sequencer (ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit, Perkin Elmer). Homology to known genes was determined by BLAST search of the NCBI database (Altschul et al. 1997). The cytogenetic location of the gene was determined by hybridization to squashes of salivary polytene chromosomes by standard methods (Pardue 1994).
In situ hybridization
Whole-mount in situ hybridization to Drosophila embryos with both DNA and RNA digoxigenin-labeled probes was performed according to the protocol of Lehmann and Tautz (1994). For RNA probes proteinase K digestion was omitted. For double-label in situ hybridization both probes were RNA, one was labeled with fluorescein and the other was labeled with digoxigenin (Boehringer Mannheim). Following hybridization with both probes the embryos were incubated with alkaline-phosphatase-conjugated anti-fluorescein antibody. The first color reaction was developed with Fast-Red alkaline-phosphatase substrate (Boehringer Mannheim). After washing, the embryos were incubated in 0.1 m glycine HCl (pH 2.2), 0.1% Tween, for 10 min to remove the anti-fluorescein antibody (S. Small, pers. comm.). The embryos were washed again and the anti-digoxigenin antibody was added and the usual protocol for whole-mount in situ hybridization followed. For double-label experiments with whole-mount in situ hybridization and antibody staining the in situ hybridization was performed first without proteinase K digestion. Following development of the alkaline-phosphatase reaction antibody staining was performed as described below. For sectioning embryos were postfixed in 2% formaldehyde in PBS. Then 10-μm cryostat sections were made following embedding in gelatin according to Mastick et al. (1997). Briefly, following sequential infiltration in 5% sucrose and then 15% sucrose in 0.1 m phosphate buffer (pH 7.4), embryos were embedded in 7.5% gelatin, 15% sucrose in 0.1 m phosphate buffer. Cryosections were counterstained with propidium iodide and photographed.
In situ hybridization with radiolabeled probes to mouse embryos was performed according to the protocol of Angerer and Angerer (1992).
Isolation of mutants
A P-element insertion obtained from the Drosophila Genome Project was mapped near the coding sequence of ind (Fig. 1). This element was mobilized by a dysgenic cross and progeny tested for noncomplementation of a deficiency that uncovers ind. Six hundred lines from the dysgenic cross were tested for noncomplementation; 10 lines failed to complement the deficiency. Of these one was found to lack Ind protein by antibody staining (ind16.2). Additional complementation testing placed two other mutant lines in the same complementation group (ind79.3 and ind4.2). All three showed the same CNS phenotype when homozygous or in combination with the deficiency. The indRR108 allele was identified in a large scale EMS mutagenesis of the third chromosome by assaying for changes in the number or pattern of Eve-protein-positive neurons (J. Skeath and C.Q. Doe, unpubl.).
The extent of the deficiency in ind79.3 was determined by Southern analysis (Sambrook et al. 1989). The ends of the deletion were mapped by probes at both ends that recognize a common polymorphic fragment that is not recognized by fragments in the intervening interval.
DNase I footprinting and gel-shift analysis
The amino acid residues 563–723 containing the homeodomain of Vnd were inserted into the vector pRSET (InVitrogen) to generate a poly-histidine-tagged bacterial fusion construct. After induction, the bacteria were lysed in 6 m guanidine-HCl and the Vnd fusion protein was purified by nickel affinity chromatography. The protein was eluted in 8 m urea/sodium phosphate (pH 4.0). The purified Vnd fusion protein was dialyzed stepwise into 4, 2, and 1 m urea and finally into 0.5× Z-buffer [10% glycerol, 12.5 mm HEPES (pH 7.8), 6 mm MgCl2, 0.5 mm DTT, 50 mm KCl, 0.05% NP-40]. The final concentration of the Vnd fusion protein stock was 500 μg/ml.
DNase I footprinting was done essentially as described by Ryan et al. (1995). The ind repressor region was labeled using Klenow polymerase fragment to fill in the NotI or HindIII sites in the polylinker, for the bottom and top strands, respectively. The GA tract was generated by formic acid modification and piperidine cleavage (Maxam and Gilbert 1980). Approximately 20,000 dpm of DNA was added to increasing amounts of Vnd fusion protein in 0.5× Z-buffer with 1 μg of poly[d(I-C)]. After incubation for 15 min at room temperature, 0.2 unit of DNase I (Boehringer) was added and incubation proceeded for another 2 min. The reactions were stopped by adding 0.1 ml of STOP mix [50 mm Tris (pH 8.0), 100 mm NaCl, 1% SDS, 10 mm EDTA, 5 μg salmon sperm DNA, 50 μg protease-K] and the samples were digested overnight at 50°C. After phenol/chloroform extraction and ethanol precipitation, the samples were electrophoresed on 6% denaturing polyacrylamide gels.
Electrophoresis mobility-shift experiments used a 130-bp Sau3A fragment corresponding to nucleotides 5–135 of the ind repressor region and labeled by the Klenow fill-in reaction. Approximately 20,000 dpm of probe was incubated with 1 ng of Vnd fusion protein and a molar excess of specific and nonspecific competitor oligonucleotides. The double-stranded oligonucleotide corresponding to Vnd-binding site I, nucleotides 31–57, was used as a specific competitor, whereas the paired-domain binding site H2A (Adams et al. 1992) was used as a nonspecific control oligonucleotide. The binding reactions contained 0.5× Z-buffer and 1 mg of poly[d(I-C)]. After 15 min at room temperature, binding reactions were loaded onto 6% neutral acrylamide gels and run in 0.5× TBE.
Antibody generation and immunocytochemistry
Rat serum and monoclonal antibodies were raised against an Ind peptide conjugated to bovine serum albumin (BSA). After the monoclonal fusion, hybridoma supernatants were screened by ELISA against a peptide-KLH conjugate. Positive hybridoma lines were tested by immunohistochemistry. Embryo fixation, antibody staining, and identification of homozygous mutant embryos were performed essentially as by McDonald and Doe (1997). Embryos were staged according to Campos-Ortega and Hartenstein (1997). Primary antibodies were used at the following dilutions: mouse anti-Achaete monoclonal (990E5F1; 1:10; Skeath and Carroll 1992), rabbit anti-β galactosidase serum (1:3000; Cappel), rat anti-β-galactosidase monoclonal (1:10; Srinivasan et al. 1998), mouse anti-Eve [2B8; 1:10; (Patel 1994)], rabbit anti-Msh serum (1:500; T. Isshiki and A. Nose, unpubl.), rat anti-Ind serum (1:500), rat anti-IndN1 monoclonal (1:1), and rabbit anti-Vnd serum (1:10). Secondary antibodies of the appropriate species conjugated to biotin (Vector Labs) or alkaline phosphatase (Southern Biotechnology Associates) were used at a dilution of 1:400. Histochemical detection of primary antibodies was done with the HRP Vectastain Elite kit (Vector Labs) in conjunction with the Renaissance TSA Indirect kit (NEN Life Sciences) for some antibodies. After staining, embryos were dissected, mounted in 70% or 85% glycerol and examined on a Zeiss Axioplan microscope. Images were captured with a Sony DKC-5000 digital camera.
Acknowledgments
We thank Javier Lopez for advice on the yeast screen and for libraries of Drosophila DNA, John Tamkun and Nick Brown for libraries, Takako Isshiki for the Msh antibody, Steve Potter for probes, Lisa Goodrich and Tony Oro for probes and help with mouse in situs, and Matt Fish for polytene mapping. This work was supported by a KO8 award from the National Heart, Lung, and Blood Institute to J.B.W., by the National Science Foundation (D.M.), by the National Institutes of Health (C.Q.D.), and by the Howard Hughes Medical Institute (M.P.S. and C.Q.D.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL scott@cmgm.stanford.edu; FAX (650) 723-9878.
E-MAIL cdoe@uoneuro.uoregon.edu; FAX (541) 346-4736.
References
- Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes & Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer L, Angerer R. In situ hybridization to cellular RNA with radiolabelled RNA probes. In: Wilkinson DG, editor. In situ hybridization: A practical approach. Oxford, UK: Oxford University Press; 1992. pp. 15–32. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley & Sons; 1994. [Google Scholar]
- Azpiazu N, Frasch M. tinman and bagpipe: Two homeobox genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes & Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Barth KA, Wilson SW. Expression of zebrafish nk2.2 is influenced by sonic hedgehog/vertebrate hedgehog-1 and demarcates a zone of neuronal differentiation in the embryonic forebrain. Development. 1995;121:1755–1768. doi: 10.1242/dev.121.6.1755. [DOI] [PubMed] [Google Scholar]
- Bhat KM. The patched signaling pathway mediates repression of gooseberry allowing neuroblast specification by wingless during Drosophila neurogenesis. Development. 1996;122:2921–2932. doi: 10.1242/dev.122.9.2921. [DOI] [PubMed] [Google Scholar]
- Bhat KM, Schedl P. Requirement for engrailed and invected genes reveals novel regulatory interactions between engrailed/invected, patched, gooseberry and wingless during Drosophila neurogenesis. Development. 1997;124:1675–1688. doi: 10.1242/dev.124.9.1675. [DOI] [PubMed] [Google Scholar]
- Bier E. Anti-neural-inhibition a conserved mechanism for neural induction. Cell. 1997;89:681–684. doi: 10.1016/s0092-8674(00)80250-0. [DOI] [PubMed] [Google Scholar]
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA. Genetic mechanisms of early neurogenesis in Drosophila melanogaster. Mol Neurobiol. 1995;10:75–89. doi: 10.1007/BF02740668. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. 2nd ed. Berlin, Germany: Springer-Verlag; 1997. [Google Scholar]
- Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- Chu-LaGraff Q, Doe CQ. Neuroblast specification and formation regulated by wingless in the Drosophila CNS. Science. 1993;261:1594–1597. doi: 10.1126/science.8372355. [DOI] [PubMed] [Google Scholar]
- Chu-LaGraff Q, Schmid A, Leidel J, Bronner G, Jackle H, Doe CQ. huckebein specifies aspects of CNS precursor identity required for motoneuron axon pathfinding. Neuron. 1995;15:1041–1051. doi: 10.1016/0896-6273(95)90093-4. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio M, Frasch M. msh may play a conserved role in dorsoventral patterning of the neuroectoderm and mesoderm. Mech Dev. 1996;58:217–231. doi: 10.1016/s0925-4773(96)00583-7. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Ferguson EL. Conservation of dorsal-ventral patterning in arthropods and chordates. Curr Opin Genet Dev. 1996;6:424–431. doi: 10.1016/s0959-437x(96)80063-3. [DOI] [PubMed] [Google Scholar]
- Frei E, Baumgartner S, Edström J-E, Noll M. Cloning of the extra sex combs gene of Drosophila and its identification by P-element-mediated gene transfer. EMBO J. 1986;4:979–987. doi: 10.1002/j.1460-2075.1985.tb03727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruschus JM, Tsao DH, Wang LH, Nirenberg M, Ferretti JA. Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: Basis of binding specificity. Biochemistry. 1997;36:5372–5380. doi: 10.1021/bi9620060. [DOI] [PubMed] [Google Scholar]
- Holland PW. Cloning and evolutionary analysis of msh-like homeobox genes from mouse, zebrafish and ascidian. Gene. 1991;98:253–257. doi: 10.1016/0378-1119(91)90182-b. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Takeichi M, Nose A. The role of the msh homeobox gene during Drosophila neurogenesis: Implication for the dorsoventral specification of the neuroectoderm. Development. 1997;124:3099–3109. doi: 10.1242/dev.124.16.3099. [DOI] [PubMed] [Google Scholar]
- Jimenez F, Martin-Morris LE, Velasco L, Chu H, Sierra J, Rosen DR, White K. vnd, a gene required for early neurogenesis of Drosophila, encodes a homeodomain protein. EMBO J. 1995;14:3487–3495. doi: 10.1002/j.1460-2075.1995.tb07355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In situ hybridization to RNA. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical uses in cell and molecular biology. San Diego, CA: Academic Press; 1994. pp. 575–598. [DOI] [PubMed] [Google Scholar]
- Liu J, Wilson TE, Milbrandt J, Johnston M. Identifying DNA-binding sites and analyzing DNA-binding domains using a yeast selection system. Methods: A Companion to Meth Enzymol. 1993;5:125–137. [Google Scholar]
- Mastick GS, McKay R, Oligino T, Donovan K, Lopez AJ. Identification of target genes regulated by homeotic proteins in Drosophila melanogaster through genetic selection of Ultrabithorax protein-binding sites in yeast. Genetics. 1995;139:349–363. doi: 10.1093/genetics/139.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastick GS, Davis NM, Andrew GL, Easter SS., Jr Pax-6 functions in boundary formation and axon guidance in the embryonic mouse forebrain. Development. 1997;124:1985–1997. doi: 10.1242/dev.124.10.1985. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Saigo K. hedgehog signaling independent of engrailed and wingless required for post-S1 neuroblast formation in Drosophila CNS. Development. 1996;122:3567–3575. doi: 10.1242/dev.122.11.3567. [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Meth Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Doe CQ. Establishing neuroblast-specific gene expression in the Drosophila CNS: Huckebein is activated by Wingless and Hedgehog and repressed by Engrailed and Gooseberry. Development. 1997;124:1079–1087. doi: 10.1242/dev.124.5.1079. [DOI] [PubMed] [Google Scholar]
- McDonald, J.A., S. Holbrook, T. Isshiki, J. Weiss, C.Q. Doe, and D.M. Mellerick. 1998. Dorsoventral patterning in the Drosophila CNS: The vnd homeobox gene specifies ventral column identity. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Mellerick DM, Nirenberg M. Dorsal-ventral patterning genes restrict NK-2 homeobox gene expression to the ventral half of the central nervous system of Drosophila embryos. Dev Biol. 1995;171:306–316. doi: 10.1006/dbio.1995.1283. [DOI] [PubMed] [Google Scholar]
- Pardue M-L. Looking at polytene chromosomes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical uses in cell and molecular biology. San Diego, CA: Academic Press; 1994. pp. 333–351. [Google Scholar]
- Patel NH. Imagining neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical uses in cell and molecular biology. San Diego, CA: Academic Press; 1994. pp. 445–487. [DOI] [PubMed] [Google Scholar]
- Raz E, Shilo BZ. Establishment of ventral cell fates in the Drosophila embryonic ectoderm requires DER, the EGF receptor homolog. Genes & Dev. 1993;7:1937–1948. doi: 10.1101/gad.7.10.1937. [DOI] [PubMed] [Google Scholar]
- Rutledge BJ, Zhang K, Bier E, Jan YN, Perrimon N. The Drosophila spitz gene encodes a putative EGF-like growth factor involved in dorsal-ventral axis formation and neurogenesis. Genes & Dev. 1992;6:1503–1517. doi: 10.1101/gad.6.8.1503. [DOI] [PubMed] [Google Scholar]
- Rusch J, Levine M. Threshold responses to the dorsal regulatory gradient and the subdivision of primary tissue territories in the Drosophila embryo. Curr Opin Genet Dev. 1996;6:416–423. doi: 10.1016/s0959-437x(96)80062-1. [DOI] [PubMed] [Google Scholar]
- Ryan G, Steele-Perkins V, Morris JF, Rauscher FJR, Dressler GR. Repression of Pax-2 by WT1 during normal kidney development. Development. 1995;121:867–875. doi: 10.1242/dev.121.3.867. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes & Dev. 1995;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shilo BZ. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 1997;13:191–196. doi: 10.1016/s0168-9525(97)01091-3. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Carroll SB. Regulation of proneural gene expression and cell fate during neuroblast segregation in the Drosophila embryo. Development. 1992;114:939–946. doi: 10.1242/dev.114.4.939. [DOI] [PubMed] [Google Scholar]
- ————— The achaete–scute complex: Generation of cellular pattern and fate within the Drosophila nervous system. FASEB J. 1994;8:714–721. doi: 10.1096/fasebj.8.10.8050670. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Panganiban GF, Carroll SB. The ventral nervous system defective gene controls proneural gene expression at two distinct steps during neuroblast formation in Drosophila. Development. 1994;120:1517–1524. doi: 10.1242/dev.120.6.1517. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Zhang Y, Holmgren R, Carroll SB, Doe CQ. Specification of neuroblast identity in the Drosophila embryonic central nervous system by gooseberry-distal. Nature. 1995;376:427–430. doi: 10.1038/376427a0. [DOI] [PubMed] [Google Scholar]
- Srinivasan, S., C.-Y. Peng, S. Nair, J.B. Skeath, E.P. Spana, and C.Q. Doe. 1998. Biochemical analysis of Prospero protein during asymmetric cell division: Cortical Prospero is highly phosphorylated relative to neclear Prospero. Dev. Biol. (in press). [DOI] [PubMed]
- Szucsik JC, Witte DP, Li H, Pixley SK, Small KM, Potter SS. Altered forebrain and hindbrain development in mice mutant for the Gsh2 homeobox gene. Dev Biol. 1997;191:230–242. doi: 10.1006/dbio.1997.8733. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Valerius MT, Li H, Stock JL, Weinstein M, Kaur S, Singh G, Potter SS. Gsh1: A novel murine homeobox gene expressed in the central nervous system. Dev Dyn. 1995;203:337–351. doi: 10.1002/aja.1002030306. [DOI] [PubMed] [Google Scholar]
- Wang W, Chen X, Xu H, Lufkin T. Msx3: A novel murine homolog of the Drosophila msh homeobox gene restricted to the dorsal embryonic central nervous system. Mech Dev. 1996;58:203–215. doi: 10.1016/s0925-4773(96)00562-x. [DOI] [PubMed] [Google Scholar]
- White K, DeCelles NL, Enlow TC. Genetic and developmental analysis of the locus vnd in Drosophila melanogaster. Genetics. 1983;104:433–448. doi: 10.1093/genetics/104.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bahri S, Klein T, Chia W. Klumpfuss, a putative Drosophila zinc finger transcription factor, acts to differentiate between the identities of two secondary precursor cells within one neuroblast lineage. Genes & Dev. 1997;11:1396–1408. doi: 10.1101/gad.11.11.1396. [DOI] [PubMed] [Google Scholar]
- Yang X, Yeo S, Dick T, Chia W. The role of a Drosophila POU homeo domain gene in the specification of neural precursor cell identity in the developing embryonic central nervous system. Genes & Dev. 1993;7:504–516. doi: 10.1101/gad.7.3.504. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ungar A, Fresquez C, Holmgren R. Ectopic expression of either the Drosophila gooseberry-distal or proximal gene causes alterations of cell fate in the epidermis and central nervous system. Development. 1994;120:1151–1161. doi: 10.1242/dev.120.5.1151. [DOI] [PubMed] [Google Scholar]


