Abstract
The ubiquitination pathway is a highly dynamic and coordinated process that regulates degradation as well as numerous processes of proteins within a cell. The p53 tumor suppressor and several factors in the pathway are regulated by ubiquitin as well as ubiquitin-like proteins. These modifications are critical for the function of p53 and control both the degradation of the protein as well as localization and activity. Importantly, more recent studies have identified deubiquitination enzymes that can specifically remove ubiquitin moieties from p53 or other factors in the pathway, and the reversible nature of this process adds yet another layer of regulatory control of p53. This review highlights the recent advances in our knowledge of ubiquitin and the p53 pathway.
Keywords: p53, Mdm2, antirepression, destabilization, ubiquitination, transcriptional activation and stability
Introduction
The tumor suppressor p53 is a complex, multi-functional sequence specific transcription factor that has critical regulatory functions in a cell. The seemingly endless discovery of additional factors that have a regulatory effect on p53 underscores its significance in the control of normal cellular growth and function. While initial in vitro and cell-based assays have provided a detailed understanding of p53 function, more recent and complex in vivo modeling has identified some interesting and unexpected mechanisms of p53 control.
Post-translational modifications are key processes that control how p53 protein functions. Acetylation, methylation, phosphorylation, neddylation, sumoylation, and ubiquitination all have important downstream effects on how p53 is stabilized and activated as a transcription factor. In addition, the spatial and temporal overlap of these reactions can have a profound impact on the outcome of cellular fate. Since p53 has important control of cell cycle arrest and apoptosis in response to cellular stress, these modifications are critical for the stability and transcriptional activity of p53 as the protein activates downstream target genes that dictate the cellular response.
Ubiquitination of p53 and the functions of the ubiquitin-proteosome pathway have a marked impact on p53 protein levels and turnover. p53 was first shown to be regulated by this pathway with the discovery that the human papilloma virus (HPV) E6 protein could induce degradation [1]. Since then, a greater understanding of the importance of this process in controlling p53 activity has uncovered several E3 ubiquitin ligases and other associated factors that directly affect the p53 levels, sub-cellular localization, and activity. In addition, it has been found that the process is reversible by the activity of deubiquitination enzymes, and other ubiquitin-like (UBL) proteins have also been shown to have an impact on p53 control. Taken together, the process of ubiquitination has profound effects on p53 activity, and the cell has a number of factors that are orchestrated together to ensure that p53 remains under strict regulatory control. This review highlights our current understanding of p53 ubiquitination and summarizes recent findings of p53 regulation by ubiquitin.
Regulation of p53 through ubiquitination
The ubiquitin pathway provides an efficient, structured, and reversible mechanism for controlling a number of cellular processes, including signaling, protein degradation, trafficking, DNA repair, and apoptosis [2,3]. The process of ubiquitination is an enzymatically orchestrated event that involves E1 activating enzymes, E2 conjugating enzymes, and E3 ubiquitin ligases [4]. Together, these enzymes function in a cascading event to attach an ubiquitin molecule, which is an evolutionarily conserved protein that consists of 76 amino acids, to a lysine residue on a target substrate. Ubiquitin is linked to lysine residues of the target substrate directly (monoubiquitination) or to another ubiquitin protein (polyubiquitination) though a covalent isopeptide bond. Monoubiquitination acts as an important signaling event for the regulation of a number of proteins, whereas polyubiquitination of at least 4 ubiquitins serves as a signal for degradation by the 26S proteosome.
An important control mechanism for p53 regulation is through ubiquitination. It has been shown that p53 ubiquitination is highly dynamic and reversible, with monoubiquitination and polyubiquitation playing important and distinct roles in the functions of p53. The first indication that p53 was regulated by the ubiquitination pathway was by the identification of the human papilloma virus (HPV) E6-associated cellular protein E6AP [5]. The HPV E6 protein commandeers E6AP to reduce p53 levels as a mechanism for replicating in the host cell. Shortly thereafter, Mdm2 was identified as a cellular factor that ubiquitinates and degrades p53 in the absence of exogenous factors [6–8]. A number of studies have shown that Mdm2 is the predominant and critical E3 ubiquitin ligase for p53 and mediates p53 ubiquitination through a RING domain. Together, p53 and Mdm2 function in a negative feedback loop, with p53 driving the transcription of mdm2 during times of normal homeostasis and maintaining low levels of p53 protein. However, upon DNA damage or other type of cellular stress event, p53 protein levels rise due to both a disruption of p53-mediated mdm2 transcription and the post-translational inhibition of Mdm2 function. The direct importance of these interactions was highlighted with the generation of mdm2 null mice, which exhibit embryonic lethality at day E6.5. Interestingly, the lethality is completely rescued in a double knockout of mdm2 and p53 [9–12].
A number of mechanisms have been described that inhibit the p53-Mdm2 interaction during cellular stress, suggesting that this interplay is critical for regulating the balance of p53 protein at any given time in the cell. Phosphorylation of p53 at Ser15 and Ser20 in response to DNA damage and other types of cellular stress by ATM, ATR, DNA-PK, Chk1, and Chk2 is thought to abrogate the Mdm2-p53 protein-protein interaction and thereby stabilize p53 [13]. Moreover, the acetylation of p53, which is a process that is critically important for transcriptional activity, occurs on the same C-terminal lysine residues as ubiquitination, and therefore this enzymatic process can compete with and block ubiquitination to induce p53 stabilization. Indirect mechanisms of p53 stabilization exist as well. The tumor suppressor p14ARF can stabilize p53 by binding to and preventing Mdm2 from physically interacting with p53 [11,12]. Although p14ARF is known to be a nucleolar protein, the Mdm2 sequestration and subsequent p53 activation has been shown to occur both inside and outside of the nucleolus [14,15]. It has also been shown that p14ARF can block Mdm2 in response to aberrant oncogenes, which provides the cell with a response mechanism for the activation of p53 to this type of cellular stress [12].
Mdm2 ubiquitinates p53 at six key lysine residues located at the C-terminus of the protein, including K370, K372, K373, K381, K382, and K386 (Figure 1) [16,17]. Knock-in studies, where the lysines are replaced with arginines (the so-called p53-6KR mutant) in vivo, have shown that p53 expression levels are not dramatically altered, suggesting that while these lysines are important for the regulation and function of p53, they are not sufficient for degradation [18,19]. The in vivo half-life of the p53-7KR mutant, which is the murine equivalent of the p53-6KR mammalian mutant, was also shown to be similar to wild-type p53 [19]. These findings suggested that alternative sites on p53 may be important for stability. More recently, it was shown that p53 can also be ubiquitinated in vitro within the DNA binding domain as well [20]. When this domain was removed, the overall ubiquitination and stability of p53 decreased, though these sites we also not sufficient for complete p53 degradation.
Figure 1.
Overview of major ubiquitination, neddylation, and sumoylation sites of p53. The major sites are shown with their corresponding modifying enzymes. Ub, Ubiquitination; Ne, Neddylation; Su, Sumoylation.
Another structurally related protein to Mdm2, MdmX, adds a layer of complexity to p53 regulation. Although MdmX has sequence homology to Mdm2 and possesses a RING domain, it does not have E3 ligase activity for p53. Instead, MdmX was shown to repress p53-mediated transcriptional activation [21]. Interestingly, mdmx null mice are embryonic lethal, but can be rescued by crossing with p53 null mice [22,23]. More recently, animal studies have aimed to address the intricate functions of Mdm2 and MdmX on p53. In one study, mutant knockout mice were generated that lacked p53 together with either mdm2 or mdmx [24–27]. A temperature sensitive p53 mutant was then reintroduced into the mice, which allowed for a detailed in vivo analysis of p53 in the selective absence of Mdm2 or MdmX. Interestingly, a differential effect was observed, where loss of mdm2 promoted p53-dependent activation of apoptosis-related genes, while loss of MdmX promoted p53-dependent activation of cell cycle arrest genes. This study provided the first in vivo analysis on the specific effects of these proteins on p53 activity, and suggests distinct functions for these two proteins. MdmX and Mdm2 both bind to the promoters of p53-responsive genes and form a 3 protein complex with p53 by interacting with the transactivation domain [28–30]. This interaction has been shown to inhibit p53-mediated transcription of some p53 target genes [30]. In addition, although both Mdm2 and MdmX have important effects on p53 function, in vivo evidence suggests that only Mdm2 has an effect on p53 protein levels [31,32]. Nevertheless, additional in vivo studies of MdmX in relation to Mdm2 and p53 will help identify the true physiological mechanisms at play in p53 regulation.
Mdm2-independent ubiquitination of p53
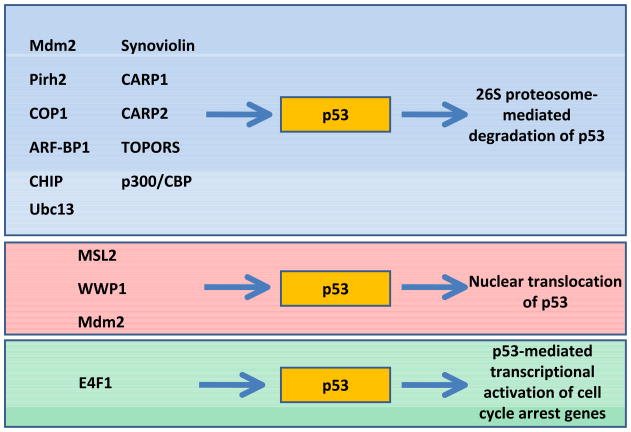
Although Mdm2 has been shown to be the predominant E3 ubiquitin ligase for p53, and mdm2 null mice exhibit embryonic lethality, later studies have shown that p53 is still degraded in vivo in mdm2 deficient mice [33]. Using a switchable endogenous p53 model, mice were raised to adulthood possessing a p53 deficiency, crossed with mdm2 null mice, and then acutely restored with p53 function, which allowed for an in vivo analysis of homeostatic p53 stability in the absence of Mdm2. Remarkably, p53 was still degraded in the absence of Mdm2, suggesting the presence of other mechanisms for p53 degradation. Indeed, a number of E3 ligases have been described for p53, including, Pirh2, COP1, ARF-BP1, WWP1, E4F1, and Ubc13 (Figure 2) [34]. Both Pirh2 and COP1 can ubiquitinate p53 independently of Mdm2, which leads to degradation mediated by the 26S proteosome. Topors have been shown to both ubiquitinate and sumoylate p53, though the physiological effects of these modifications are unclear [35,36]. CARP1 and CARP2 ubiquitinate both unmodified p53 and phosphorylated p53 at S20 in stressed cells [37]. Moreover, the reduction of CARP2 by siRNA increases the G1/S ratio, indicating an accumulation of p53. ARF-BP1 was purified as an ARF associated factor from p53 null cells and associates with ARF in vivo [38,39]. Interestingly, ARF-BP1 can directly ubiquitinate p53, and this effect is inhibited by ARF. These findings suggest that ARF-BP1 is a critical regulator of both p53-dependent and p53-indpendent tumor suppression functions of ARF.
Figure 2.
E3 ubiquitin ligases that target p53. E3 and E4 ubiquitin ligases have diverse effects on p53, including 26S proteosome-mediated degradation, nuclear export, and transcriptional activation.
The type of ubiquitination occurring on a substrate can have very different downstream consequences, with polyubiquitination acting as a signal for degradation by the 26S proteosome and monoubiquitination leading to several degradation-independent processes, including endocytosis and transcriptional regulation [40]. Mdm2 has the ability to catalyze both mono- and polyubiquitination of p53. Interestingly, monoubiquitination of p53 induces nuclear export in an Mdm2 dose-dependent manner [16,41]. Therefore, maintenance of Mdm2 levels may provide another mechanism for controlling p53 activity, since during times of non-stressed homeostasis the low levels of Mdm2 would induce p53 monoubiquitination and drive the protein out of the nucleus and away from transcriptional targets. During cellular stress, when p53 levels are quickly up-regulated, the increased expression of Mdm2 would provide a mechanism for polyubiquitinating and degrading p53 to subsequently reduce levels after repair has occurred and the protein is no longer needed. These interesting findings will require further investigation; however, they do suggest an added mechanism for regulating p53 function.
The movement of p53 into the cytoplasm allows for transcription-independent activities, including apoptosis and autophagy [42–44]. Once in the cytoplasm, p53 can interact with members of the Bcl family, such as Bcl-XL and Bcl-2, and promote the oligomerization of proapoptotic factors Bak and Bax at the outer mitochondrial membrane. The formation of a pore in the membrane allows for the release of cytochrome c and other apoptotic factors into the cytoplasm [45–47]. CBP and p300 have been described as cytoplasmic E4 ubiquitin ligases for p53, and are required for the endogenous polyubiquitination of p53 [48,49]. Another protein, synoviolin, a protein that is located in the endoplasmic reticulum (ER), mediates ER-associated degradation (ERAD) of p53 by sequestering and polyubiquitinating the protein in the ER [50]. ERAD was shown to be a mechanism for the transport of polyubiquitinated p53 to the 26S proteosome. However, the specific roles of p53 with the ER are as yet unclear. The chaperone-associated ubiquitin ligase CHIP is also able to polyubiquitinate p53 and induces degradation. CHIP ubiquitin ligase functions are dependent on an association with the molecular chaperones Hsc70 and Hsp90.
Ubiquitination and Acetylation
Post-translational modifications of p53 are intricately complex and often times occur at the same residues of p53. While the ubiquitination of p53 is critical for maintaining appropriate protein levels at all times and under all conditions, acetylation of p53 is an equally important step for quickly stabilizing and activating the protein. Acetylation of p53 occurs at a number of key residues throughout the protein, but predominantly occurs at the C-terminus. Within this region, it has been shown that acetylation at key C-terminal lysine residues can inhibit Mdm2-mediated ubiquitination [51]. Moreover, purified acetylated p53 cannot be ubiquitinated in vitro, and the level of ubiquitination has been shown to be reduced after the induction of acetylation [52,53]. Acetylation of eight specific lysine residues of p53 also blocks interaction with Mdm2 [30]. Therefore, the stepwise process of quickly stabilizing and activating p53 to a level that is sufficient to induce cell growth arrest or apoptosis requires the interplay and balance between ubiquitination inhibition and acetylation.
The histone acetyltransferase (HAT) CBP/p300, which, as mentioned above, possesses intrinsic E4 ligase function, is also a major HAT for p53 and Mdm2. Whereas CBP/p300-mediated p53 acetylation is critical for activity as a transcription factor, the acetylation of Mdm2 within the RING domain has been shown to prevent interaction with p53 and promotes p53 stabilization [54]. Another HAT that has activity on Lys320 of p53, PCAF, has also recently been shown to have intrinsic ubiquitination function and can directly ubiquitinate Mdm2 [55]. Both HATs therefore have dual mechanisms for inhibiting the p53-Mdm2 interaction and activating p53 transcriptional activity, showing that some factors in the p53 pathway have multiple roles for the quick stabilization and activation of p53.
Non-degradation effects of p53 ubiquitination
Although one of the predominant functions of the ubiquitin-proteosome pathway is for the efficient and expeditious degradation and removal of proteins from the cell, it has become more apparent in recent years that ubiquitin is also an important signaling factor in several processes. These unique non-degradation functions of ubiquitin have also been identified in the p53 pathway, and several E3 ubiquitin ligases can induce proteosome-independent ubiquitination of p53. Msl2 was identified as an E3 ubiquitin ligase that can induce polyubiquitination of p53 to promote nuclear export [56]. Although Msl2 has a RING domain, is does not affect p53 protein levels or Mdm2-mediated ubiquitination. In addition, Msl2 ubiquitinates K351 and K357 of p53, which are not sites of Mdm2 ubiquitination. Similar to Msl2, WWP1 can also ubiquitinate p53 and induce nuclear export of a fraction of the protein [57]. Interestingly, WWP1-mediated ubiquitination of p53 seems to stabilize the protein and ubiquitination is not dependent on Mdm2. p53 also reduces WWP1 expression, suggesting another intricate feedback loop. The E2-ubiquitin conjugating enzyme Ubc13 induces K63-dependent ubiquitination of p53, which reduces Mdm2-mediated p53 ubiquitination and promotes cytoplasmic translocation. Ubc13 also increases p53 stability, but it was shown to reduce p53 transcriptional activity and can increase the pool of monomeric p53, thereby reducing the availability of p53 monomers for tetramerization.
The transcription factor E4F1 was also recently shown to ubiquitinate p53, though it does not have an effect on p53 stability [58]. E4F1 is an atypical ubiquitin ligase because it does possess a commonly associated ubiquitin ligase domain (RING or HECT), though the enzymatic activity was narrowed to amino acids 41–85, which resemble the core enzymatic domain of the SUMO E3 ligase RanBP2 [59,60]. E4F1-mediated ubiquitination of p53 increases the fraction of protein associated with chromatin, and this association specifically coincides with p53-mediated transcriptional activation of genes involved with cell cycle arrest, but not apoptosis. These findings are interesting, as the mechanisms the allow p53 to selectively induce a cell cycle arrest or apoptosis response due to DNA damage or cell stress has remained elusive. Interestingly, E4F1 ubiquitinates K320 of p53, which is also a PCAF acetylation residue, and therefore these two enzymes compete for modification of the same site. The effect of PCAF acetylation on p53function remains unclear, though it has been hypothesized that by blocking PCAF-mediated acetylation of p53 at K320, E4F1 ubiquitination at this position may promote a particular p53-mediated response. Future studies of the consequences of these interactions may add clarity to these findings and mechanisms.
p53 sumoylation and neddylation
Other ubiquitin-like (UBL) proteins can be covalently linked to p53 through enzymatic processes similar to ubiquitination and have an impact on the function of the protein (Table 1). Mdm2 has been shown to neddylate K370, K372, and K373, which inhibit p53-mediated transcriptional activation [61]. Moreover, Fbxo11 seems to have a similar effect on p53 function by neddylating K320 and K31 [62]. Interestingly, these sites coincide with Mdm2 ubiquitination sites, and since Mdm2 is capable of both ubiquitinating and neddylating p53, it is as yet unclear what mechanism allows for Mdm2 to selectively induce one reaction over the other. More recently, it has been shown that NUB1, a non-covalent interacting protein of Nedd8, decreases p53 neddylation and stimulates ubiquitination [63]. NUB1 promotes the cytoplasmic localization of p53, suggesting the p53 neddylation may block nuclear export.
Table 1.
Deubiquitinases and ubiquitin-like proteins that affect the p53 pathway.
| Deubiquitinase | Target | Effect |
|---|---|---|
| HAUSP | p53 Mdm2 MdmX FOXO4 |
Stabilization Stabilization Stabilization Inhibits transcriptional activity |
| USP2a | Mdm2 | Stabilization |
| USP10 | p53 | Stabilization |
| Sumo ligase | Target | Effect |
| PIAS | p53 | Promotes p53 transcriptional activity and cytoplasmic translocation |
| Topors | p53 | Increases p53 levels |
| Nedd8 ligase/interacting proteins | Target | Effect |
| Mdm2 | p53 | Inhibits p53 transcriptional activation |
| NUB1 | Nedd8 | Promotes p53 cytoplasmic translocation |
| FBXO11 | p53 | Inhibits p53 transcriptional activity |
Sumoylation of p53 occurs predominantly at K386, and the major cellular factors that mediate this reaction are members of the PIAS family [64]. The function of sumoylated p53 is not clear, with some studies showing the sumoylation promotes p53 transcriptional activity and others showing that the process promotes cytoplasmic translocation [65,66]. Since the lysines of p53 that have been identified for sumoylation and neddylation are also ubiquitinated, and in some cases acetylated, the detailed mechanisms and in vivo effects of these modification remains to be elucidated.
Deubiquitination
Deubiquitination enzymes (DUBs) have important and diverse functions in a number of cellular processes [2,67,68]. The ability to reverse ubiquitination is particularly important when ubiquitin acts as a signal for non-proteosomal processes [69]. DUBs have been shown to have activity on several factors in the p53-Mdm2 pathway (Table 1). The first DUB shown to function in this pathway was USP7, also called Herpes-Specific Ubiquitin Specific Protease (HAUSP), which was found to directly deubiquitinate and stabilize p53 [51]. Interestingly, it was also found that a somatic knockout of hausp in HCT116 cells caused a dramatic increase in p53 protein levels, suggesting that the effects of HAUSP on p53 were more complex than a single downstream function [70]. Indeed, HAUSP can also deubiquitinate Mdm2, which has auto-ubiquitination activity and is inherently unstable [71]. The somatic loss of HAUSP destabilizes and reduces the levels of Mdm2, which results in the indirect stabilization of Mdm2. HAUSP can also deubiquitinate and stabilize MdmX and has a role in DNA-damaged induced degradation of MdmX [72]. Although the effects of HAUSP on p53, Mdm2, and MdmX presumably occurs in the nucleus, HAUSP localizes to both the nucleus and cytoplasm, and mitochondrial HAUSP has been shown to deubiquitinate a cytoplasmic pool of monoubiquitinated p53 upon arriving at the mitochondria through a stress-induced p53-HAUSP complex, which thereby creates a sub-fraction of non-ubiquitinated p53 that can induce transcription-independent apoptosis [43]. These findings suggest an interesting mechanism that allows for monoubiquitinated p53, which is exported into the cytoplasm, to be functionally active in mitochondria though deubiquitination by HAUSP.
The effects of HAUSP on p53, Mdm2, and MdmX are complex and clearly do not occur as isolated interactions. In addition, HAUSP has been shown to have other important effects on cellular function, including the negative regulation of FOXO4, and has been shown to exhibit some tumor suppression characteristics in a human colon carcinoma xenogaft model [73,74]. Inactivation of hausp in vivo causes early embryonic lethality between E6.5 and E7.5 due to a severe reduction of cell proliferation from both p53-dependent and –independent mechanisms [75]. Interestingly, the cross between hausp and p53 null mice did not rescue this defect, suggesting a complex network of HAUSP-mediated effects that are both p53-dependent and p53-indendent. It is also possible that HAUSP-mediated p53 effects could be tissue specific, and more recently it was shown that neural cell-specific inactivation of HAUSP in mice caused p53-dependent hypoplasia and brain development deficiencies [76]. Together, these findings suggest a complex role of HAUSP in the deubiquitination of p53, Mdm2, and MdmX, and additional in vivo studies may further refine the roles of this important DUB in the context of the p53 pathway.
Other DUBs also impact the functions of p53 and other members of this pathway. USP2a was shown to associate with and specifically deubiquitinate Mdm2, but not p53, and promote Mdm2-dependent p53 degradation [77]. The endogenous inhibition of USP2a also induced the stabilization and activation of p53, suggesting a clear role in the regulation of Mdm2. More recently, USP10, a cytoplasmic DUB, was found to directly deubiquitinate p53 [78]. Upon induction by DNA damage, a fraction of USP10 is phosphorylated by ATM and translocates to the nucleus where it deubiquitinates p53. This action directly opposed Mdm2-mediated nuclear export of p53, which provides yet another mechanism for the nuclear accumulation of p53 and subsequent transcriptional activation.
The p53 pathway – a therapeutic target?
Given the global cellular impact of p53 in cellular regulation and the occurrence of mutations in over 50% of all human tumors, p53 is a logical cellular factor to consider for therapeutic intervention. However, the fact that p53 is a potent tumor suppressor and is generally deleted or mutated in a vast majority of tumors, one has to ask, is p53 a druggable target [79]? In tumors that possess an inactive form of p53, the reintroduction of a wild-type copy of p53 would potentially render the tumor susceptible to the introduced gene. Animal studies have shown that the reactivation of p53 in p53 null tumors induces tumor regression [80–82]. Moreover, gene therapy using p53 expressed in an adenovirus has shown some promise in clinical trials [83]. As an alternative, gene therapy has been used to re-introduce an oncolytic adenovirus containing p53, which selectively replicates in tumor cells that are p53 negative and induces cell death [84]. The therapy, ONYX-015, was recently assessed in patients with advanced carcinoma or sarcomas and showed both efficacy and safety [85,86]. Gene therapy is currently being tested for several conditions, and the reintroduction of p53 into p53 negative tumors represents one possible therapeutic intervention that will require additional trial analysis.
For tumors that retain wild-type p53, which represent approximately 25% of human cancers, an alternative therapeutic strategy is to directly target upstream regulators of p53 as an indirect method for activating p53. One such agent, called reactivation of p53 and induction of tumor cell apoptosis (RITA), is a small molecule that induces p53-mediated apoptosis in several cancer lines by blocking the p53-Mdm2 interaction [87]. Another class of small molecules, called Nutlins, are potent inhibitors of Mdm2 and induce non-genotoxic activation of p53 by binding Mdm2 in the p53 binding pocket in nanomolar range [88]. Nutlins readily penetrate several tumor cell types and potently activate p53, and Nutlin-3a was shown to be orally bioavailable in human tumor xenografts. Two other compounds, HIL98 and MI-219, inhibit the E3 ubiquitin ligase activity or p53-Mdm2 interaction, respectively [89,90]. In particular, MI-219 was shown to have 10,000-fold higher selectivity for Mdm2 than MdmX.
In addition to targeting Mdm2, inhibitors for other p53 regulators are also being developed. Tenovins are small molecule inhibitors of SIRT1, which is an NAD-dependent histone deacetylase for p53 [91]. The inhibition of p53 acetylation prevents transcription, and therefore by blocking SIRT1 activity, Tenovins have been shown to potently activate p53 transcription at single-digit micromolar concentrations [91]. Other SIRT1 inhibitors, such as sirtonol, cambinol, and EX-527 have also shown promise as p53 activators [92–94]. More recently, it has been shown that a small molecule inhibitor of HAUSP, HBX41108, stabilizes p53 and induces p53-dependent apoptosis in cancer cell lines that retain wild-type p53 [95]. Importantly, HBX41108 was shown to inhibit cancer cell growth without inducing genotoxic stress.
Conclusions
The reversible process of ubiquitination provides a critical regulatory control in the p53 pathway. Although the maintenance of p53 at low protein levels by polyubiquitination and subsequent degradation by the 26S proteosome is imperative for normal cellular growth and homeostasis, the monoubiquitination of p53 has more recently been shown to have important signaling effects of p53 as well. The transcriptional activity of nuclear p53 is required for efficient and effective activation of downstream signaling events in response to cellular stress, but cytoplasmic p53 has also been shown to have important transcription-independent functions, and ubiquitin is a key factor for mediating these effects. In addition, the number of E3 ligases that have specificity for p53 and the redundancy of their functions indicate the importance of this process for maintaining p53 at low levels when not needed. Still, a number of unanswered questions remain, including how the various E3 ligases network for effective p53 ubiquitination and how DUBs are regulated. Future studies, and in particular creative in vivo studies, will help elucidate the complexity of p53 ubiquitination.
Research Highlights.
The ubiquitination pathway is a highly dynamic and coordinated process
The p53 tumor suppressor and several factors in the pathway are regulated by ubiquitin as well as ubiquitin-like proteins
Ubiquitination of p53 leads to both 26S proteosome-dependent and independent effects
Deubiquitination enzymes can specifically remove ubiquitin moieties from p53 or other factors in the pathway
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–30. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–5. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 5.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 6.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 7.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–7. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 8.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 9.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–8. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 10.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 11.Sharpless NE, DePinho RA. Telomeres, stem cells, senescence, and cancer. J Clin Invest. 2004;113:160–8. doi: 10.1172/JCI20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–7. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 13.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25:1258–71. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH. C-terminal ubiquitination of p53 contributes to nuclear export. Mol Cell Biol. 2001;21:8521–32. doi: 10.1128/MCB.21.24.8521-8532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol. 2000;20:8458–67. doi: 10.1128/mcb.20.22.8458-8467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–95. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci U S A. 2005;102:10188–93. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan WM, Mak MC, Fung TK, Lau A, Siu WY, Poon RY. Ubiquitination of p53 at multiple sites in the DNA-binding domain. Mol Cancer Res. 2006;4:15–25. doi: 10.1158/1541-7786.MCR-05-0097. [DOI] [PubMed] [Google Scholar]
- 21.Marine JC, Jochemsen AG. Mdmx as an essential regulator of p53 activity. Biochem Biophys Res Commun. 2005;331:750–60. doi: 10.1016/j.bbrc.2005.03.151. [DOI] [PubMed] [Google Scholar]
- 22.Migliorini D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–38. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–5. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 24.Barboza JA, Iwakuma T, Terzian T, El-Naggar AK, Lozano G. Mdm2 and Mdm4 loss regulates distinct p53 activities. Mol Cancer Res. 2008;6:947–54. doi: 10.1158/1541-7786.MCR-07-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwakuma T, Lozano G. Crippling p53 activities via knock-in mutations in mouse models. Oncogene. 2007;26:2177–84. doi: 10.1038/sj.onc.1210278. [DOI] [PubMed] [Google Scholar]
- 26.Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–34. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 27.Wahl GM. Mouse bites dogma: how mouse models are changing our views of how P53 is regulated in vivo. Cell Death Differ. 2006;13:973–83. doi: 10.1038/sj.cdd.4401911. [DOI] [PubMed] [Google Scholar]
- 28.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–9. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 29.Ohkubo S, Tanaka T, Taya Y, Kitazato K, Prives C. Excess HDM2 impacts cell cycle and apoptosis and has a selective effect on p53-dependent transcription. J Biol Chem. 2006;281:16943–50. doi: 10.1074/jbc.M601388200. [DOI] [PubMed] [Google Scholar]
- 30.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–26. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci U S A. 2006;103:3232–7. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci U S A. 2006;103:3226–31. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–14. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajendra R, et al. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem. 2004;279:36440–4. doi: 10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 36.Weger S, Hammer E, Heilbronn R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005;579:5007–12. doi: 10.1016/j.febslet.2005.07.088. [DOI] [PubMed] [Google Scholar]
- 37.Yang W, et al. CARPs are ubiquitin ligases that promote MDM2-independent p53 and phospho-p53ser20 degradation. J Biol Chem. 2007;282:3273–81. doi: 10.1074/jbc.M610793200. [DOI] [PubMed] [Google Scholar]
- 38.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–83. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 39.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–95. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–72. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 41.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–5. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 42.Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 2007;6:1718–23. doi: 10.4161/cc.6.14.4503. [DOI] [PubMed] [Google Scholar]
- 43.Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–34. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–4. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 46.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–90. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 47.Tomita Y, et al. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem. 2006;281:8600–6. doi: 10.1074/jbc.M507611200. [DOI] [PubMed] [Google Scholar]
- 48.Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–4. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 49.Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci U S A. 2009;106:16275–80. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamasaki S, et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 2007;26:113–22. doi: 10.1038/sj.emboj.7601490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–53. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 52.Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–40. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–11. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 54.Wang X, Taplick J, Geva N, Oren M. Inhibition of p53 degradation by Mdm2 acetylation. FEBS Lett. 2004;561:195–201. doi: 10.1016/S0014-5793(04)00168-1. [DOI] [PubMed] [Google Scholar]
- 55.Linares LK, et al. Intrinsic ubiquitination activity of PCAF controls the stability of the oncoprotein Hdm2. Nat Cell Biol. 2007;9:331–8. doi: 10.1038/ncb1545. [DOI] [PubMed] [Google Scholar]
- 56.Kruse JP, Gu W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J Biol Chem. 2009;284:3250–63. doi: 10.1074/jbc.M805658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laine A, Ronai Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene. 2007;26:1477–83. doi: 10.1038/sj.onc.1209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Cam L, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775–88. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 59.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–20. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 60.Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat Struct Mol Biol. 2004;11:984–91. doi: 10.1038/nsmb834. [DOI] [PubMed] [Google Scholar]
- 61.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 62.Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu G, Xirodimas DP. NUB1 promotes cytoplasmic localization of p53 through cooperation of the NEDD8 and ubiquitin pathways. Oncogene. 29:2252–61. doi: 10.1038/onc.2009.494. [DOI] [PubMed] [Google Scholar]
- 64.Stehmeier P, Muller S. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair (Amst) 2009;8:491–8. doi: 10.1016/j.dnarep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 65.Bischof O, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol Cell. 2006;22:783–94. doi: 10.1016/j.molcel.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 66.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–35. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 67.Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–30. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 68.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 69.Katz EJ, Isasa M, Crosas B. A new map to understand deubiquitination. Biochem Soc Trans. 38:21–8. doi: 10.1042/BST0380021. [DOI] [PubMed] [Google Scholar]
- 70.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. following 486. [DOI] [PubMed] [Google Scholar]
- 71.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–86. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 72.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–76. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 73.Becker K, Marchenko ND, Palacios G, Moll UM. A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle. 2008;7:1205–13. doi: 10.4161/cc.7.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 75.Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 29:1270–9. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kon N, Zhong J, Kobayashi Y, Li M, Szabolcs M, Ludwig T, Canoll PD, Gu W. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell Death Differ. doi: 10.1038/cdd.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–86. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 140:384–96. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen F, Wang W, El-Deiry WS. Current strategies to target p53 in cancer. Biochem Pharmacol. 80:724–30. doi: 10.1016/j.bcp.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 80.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 81.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 82.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang PI, Chang JF, Kirn DH, Liu TC. Targeted genetic and viral therapy for advanced head and neck cancers. Drug Discov Today. 2009;14:570–8. doi: 10.1016/j.drudis.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 84.Lane DP, Cheok CF, Lain S. p53-based cancer therapy. Cold Spring Harb Perspect Biol. 2:a001222. doi: 10.1101/cshperspect.a001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Galanis E, et al. Phase I-II trial of ONYX-015 in combination with MAP chemotherapy in patients with advanced sarcomas. Gene Ther. 2005;12:437–45. doi: 10.1038/sj.gt.3302436. [DOI] [PubMed] [Google Scholar]
- 86.Nemunaitis J, Senzer N, Sarmiento S, Zhang YA, Arzaga R, Sands B, Maples P, Tong AW. A phase I trial of intravenous infusion of ONYX-015 and enbrel in solid tumor patients. Cancer Gene Ther. 2007;14:885–93. doi: 10.1038/sj.cgt.7701080. [DOI] [PubMed] [Google Scholar]
- 87.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–8. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 88.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 89.Shangary S, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–8. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang Y, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–59. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 91.Lain S, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:454–63. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heltweg B, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–77. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 93.Nayagam VM, et al. SIRT1 modulating compounds from high-throughput screening as anti-inflammatory and insulin-sensitizing agents. J Biomol Screen. 2006;11:959–67. doi: 10.1177/1087057106294710. [DOI] [PubMed] [Google Scholar]
- 94.Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Colland F, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–95. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]