Abstract
Biomedical natural language processing (BioNLP) is a useful technique that unlocks valuable information stored in textual data for practice and/or research. Syntactic parsing is a critical component of BioNLP applications that rely on correctly determining the sentence and phrase structure of free text. In addition to dealing with the vast amount of domain-specific terms, a robust biomedical parser needs to model the semantic grammar to obtain viable syntactic structures. With either a rule-based or corpus-based approach, the grammar engineering process requires substantial time and knowledge from experts, and does not always yield a semantically transferable grammar. To reduce the human effort and to promote semantic transferability, we propose an automated method for deriving a probabilistic grammar based on a training corpus consisting of concept strings and semantic classes from the Unified Medical Language System (UMLS), a comprehensive terminology resource widely used by the community. The grammar is designed to specify noun phrases only due to the nominal nature of the majority of biomedical terminological concepts. Evaluated on manually parsed clinical notes, the derived grammar achieved a recall of 0.644, precision of 0.737, and average cross-bracketing of 0.61, which demonstrated better performance than a control grammar with the semantic information removed. Error analysis revealed shortcomings that could be addressed to improve performance. The results indicated the feasibility of an approach which automatically incorporates terminology semantics in the building of an operational grammar. Although the current performance of the unsupervised solution does not adequately replace manual engineering, we believe once the performance issues are addressed, it could serve as an aide in a semi-supervised solution.
Keywords: Natural language processing, Biomedical terminology, Semantic grammar, Probabilistic parsing
1. Introduction
Biomedical natural language processing (BioNLP) is a sub-discipline of biomedical informatics that aims at automatically processing natural language text such as clinical notes and research literature. It has been reported that BioNLP contributes in various applications: automated medical coding, de-identification, syndromic surveillance, literature-based hypothesis generation, etc. For comprehensive reviews, readers may refer to Zweigenbaum et al.[1] and Meystre et al.[2]. Among the core functions of many NLP systems, syntactic parsing (or simply referred as parsing) is important because it helps analyze/reconstruct the underlying hierarchical structure of free text and therefore benefits downstream processes such as determining semantic roles of the identified entities [3]. For example, given the text “sore throat and congestion of nose”, it would be desirable for a parser to generate the phrase structure so that the modifiers of the conjoined noun phrase are distributed correctly as shown in Figure 1. Note that according to Figure 1, “sore” does not modify “congestion” and “of nose” does not modify “throat”. Chung [4] showed that parsing contributed in determining correct coordinate conjunctions for extracting alternative interventions from randomized controlled trial reports. Parsing has also been highly emphasized in biological text mining. A comparative study by Miyao et al. [5] reported that parsing improved accuracy in the task of extracting protein-protein interactions.
Figure 1. Example of a correct syntactic parse of a conjoined phrase.

NP = noun phrase, PP = prepositional phrase.
Due to domain specificity, developing biomedical parsers is very challenging. A biomedical parser needs to handle not only the huge lexical space of professional terms (e.g., there exist more than 7 million concept names in the Unified Medical Language System [6]) but also the grammatical characteristics of the biomedical language, particularly when handling complex terms consisting of prepositional phrases and conjunctions. Correct distribution of modifiers in those complex phrase structures is critical for correct interpretation, but it is difficult for a syntactic parser to generate a correct parse for such structures without additional semantic information. We believe that the additional modeling of domain-specific semantic regularity should help parse those phrases correctly. Taking Figure 1 as an example, knowing that “throat” and “nose” belong to a semantic class anatomy and that “sore” and “congestion” belong to the class symptom would help determine the correct phrase structure, because the syntacto-semantic pattern ((symptomadjective anatomynoun) and (symptomnoun of anatomynoun)) is more viable than ((symptomadjective (anatomynoun and symptomnoun)) of anatomynoun) or (symptomadjective ((anatomynoun and symptomnoun) of anatomynoun)). Note that in certain context the word “sore” can also be a noun instead of an adjective, but in this example we are assuming that the correct syntactic part of speech has already been determined (e.g., by an automated tagger introduced in the subsection 4.3 of Methods). Interestingly we can see there are still several possible parses given the correct syntactic labeling of adjective and nouns, and it is the additional semantic information that resolves the structural ambiguity. State-of-art biomedical parsers address the semantic requirement via two major approaches, i.e., by using a rule-based or a probabilistic grammar. In either approach, a set of semantic classes representative of the domain have to be defined and associated with lexical terms that appear in the text to be parsed. In the rule-based approach, considerable effort is needed to induce (generally a few hundred) grammar rules using the pre-defined semantic classes. On the other hand, the probabilistic approach requires knowledgeable annotators to semantically parse a large training corpus (with at least a few hundred sentences for performance). Therefore, the cost for developing a biomedical parser is high, and usually re-investment of the effort is inevitable when the target text changes even just from one medical specialty to another.
To address the high cost and limited transferability of semantic engineering in building biomedical parsers, we propose a solution that automatically incorporates the terms and semantic classes of a publicly available, integrated biomedical terminology resource to derive an operational grammar. The underlying hypothesis of this approach is that incorporation of domain semantics should improve the parsing performance, particularly for complex noun phrases. By using comprehensive and sharable semantic classes, we expect this approach could benefit the community with transferable/reusable semantic grammar rules and parses. Most importantly, our method does not require manual creation of a semantic grammar or manual annotation of a training corpus. We focused on parsing noun phrases in clinical notes and evaluating correctness of the parsed syntactic structures. The results showed the grammar achieved recall 0.644 and precision 0.737 in bracketing the phrase structures, with average cross-bracketing of 0.61 per sentence (see section 3.8 for explanation of the metrics). Error analysis indicated more considerations that need to be addressed in order to refine and use the proposed solution.
In the following two sections we review background and related work on parsing natural language, biomedical terminology, and the relation between biomedical terminology and biomedical parsers.
2. Background
2.1 Natural language parsing
Parsing, usually referring to syntactic parsing in NLP, determines the syntactic structure of free text. For example, Figure 1 shows the syntactic parse of a noun phrase. The nested structure of syntactic constituents in Figure 1 represents a major syntactic paradigm of parsing, referred as constituency grammar. A common implementation of constituency grammar is a context-free grammar (CFG), which consists of a set of production rules in the form of LHS ➜ RHS, where LHS (left-hand side) is a single non-terminal and the RHS (right-hand side) can be a mixed sequence of terminals and/or non-terminals. In a pure syntactic CFG for English, the non-terminals are high-level syntactic constructs such as NP (noun phrase) and PP (prepositional phrase), while the terminals are syntactic parts of speech (POS), such as JJ (adjective) and NN (singular noun). For example, the first column of Table 1 displays some production rules of a syntactic CFG. Given a CFG and a POS-tagged text, there are considerable choices of existing parsing algorithms (e.g., the CYK algorithm [7-8]) that can be used to obtain the phrase structure shown in Figure 1. Detail of parsing algorithms can be found in references such as Jurafsky & Martin [9].
Table 1.
Example rules of CFG, lexicalized CFG, and semantically augmented CFG
| CFG | lexicalized CFG | semantically augmented CFG |
|---|---|---|
| NP ➜ NN | NP~throat ➜ NN~throat | NP~anatomy~throat ➜ NN~anatomy~throat |
| NP ➜ JJ NN | NP~throat ➜ JJ~sore *NN~throat |
NP~symptom~throat ➜ JJ~ symptom~sore *NN~anatomy~throat |
▪ Denotations: NP = noun phrase, PP = prepositional phrase, NN = singular noun, and JJ = adjective.
▪ The head node in each RHS is preceded by an asterisk * symbol.
With sufficient manually parsed text for training, a CFG can be augmented into a PCFG (probabilistic CFG) by estimating the probability of each rule. An advantage of PCFG is that it enables selection of the most probable parse. Following Collins [10], the probability of a parse T for a given input text S, denoted as P(T, S), can be computed with Eq.(1), where T applies n production rules LHSi ➜ RHSi and individual conditional probabilities can be estimated from a manually parsed corpus by using Eq.(2). In the end of parsing, the specific T which yields the highest P(T, S) is considered the most probable parse.
| (1) |
| (2) |
However, a pure syntactic CFG does not allow for modeling of specific lexical patterns, and a lexicalization technique was proposed accordingly, which incorporates lexemes (words in text) into the grammar. For example, instead of representing rules as the first column of Table 1, the syntactic tag can be paired with the lexical token to represent each node (see the second column of Table 1). Note that lexicalization is usually implemented with a “heading” technique, in which the LHS inherits the head word of its RHS. For example, in the second row, second column of Table 1, “throat” is inherited as the head word into the LHS node. A CFG with the head information modeled is usually called a head-driven CFG, and in this article we mark the head node in the RHS with an asterisk * symbol. Lexicalization benefits a PCFG by enriching the probability estimation with lexical information. Using the example in Figure 1, the phrases “sore throat” and “congestion of nose” should prevail in a training corpus representative of biomedical language and therefore boost the probabilities of the associated rules. On the other hand, an alternative parse with the incorrect coordination [NP sore [NP throat and congestion]] would be associated with low probability because “congestion” is rarely modified by “sore” in biomedical text. To avoid the issue of sparse training patterns in a lexicalized PCFG, the probability estimation is usually implemented by decomposing the generation of the RHS into smaller terms with independence assumption. We followed Collins’ approach [10], by which a rule of a head-driven PCFG can be written as Eq.(3).
| (3) |
In the abstract rule, W(h) stands for the LHS node with h being the head word inherited from the RHS. In the center of the RHS, H(h) stands for the head node, flanked by its left and right adjuncts (Li(li) and Ri(ri) respectively). Accordingly, the probability for a specific rule P(rhs | lhs) can then be computed by using Eq.(4), which is the product of the probability of generating the head node given the LHS node (including h) and the probabilities of generating individual left (and right) adjuncts given the head node and the LHS node.
| (4) |
Note that in Eq.(4) Ln+1(ln+1) = Rm+1(rm+1) = STOP, and they are used for modeling the outer boundary condition in generating the adjuncts. Either L1(l1) or R1(r1) could be empty. Taking the second rule in the center column of Table 1 as example, the probability P(rhs | lhs) is computed as:
where the rule’s RHS JJ~“sore” *NN~“throat” has no right adjuncts and therefore R1(r1) is immediately the STOP boundary.
To further model the semantic constraints for parsing domain-specific languages, Ge & Mooney [11] augmented Collins’ lexicalized PCFG by associating each node with a semantic label in addition to the head word and the syntactic tag. For example, NP~ballOwner~“player” represents such a triplex with ballOwner being the semantic class. They reported improved performance on symbolic learning tasks that involved parsing two domain-specific languages (one for coaching robots and the other for querying geography database). In this study we adapted Ge & Mooney’s techniques to the biomedical language. Our motivation is that the semantics of many relevant terms in the biomedical domain are well-defined (e.g., anatomy, symptom, procedure, medication, etc.) and sensible semantic sequences can be specified. Therefore, including the semantic classes in the PCFG should also help. For example, in the simple phrase “swelling in arm and legs”, the correct parse would specify that “arms and legs” are conjoined because they both involve the semantic class anatomy whereas the parse consisting of “swelling in arm” conjoined to “legs” is incorrect because it would not be appropriate to conjoin a symptom phrase with an anatomy phrase. In the third column of Table 1, we show how a lexicalized CFG can be further augmented with semantic information. Each node in the grammar is expanded into a triplex of syntacticTag~semanticTag~headLexeme. For example, the second rule in the third column contains a syntacto-semantic pattern indicating that a symptom NP can be composed of a symptom adjective modifying an anatomy head noun. After adding the semantic tag, the probability estimation in Eq.(4) is expanded into Eq.(5), with the subscripts syn and sem denoting the syntactic and semantic components respectively.
| (5) |
Taking the second rule in the third column of Table 1 as example, the probability estimation P(rhs | lhs) is computed as:
From the example above we can see that a semantic PCFG has the advantage of incorporating semantic information for modeling biomedical grammar. However, training such a syntacto-semantic grammar requires a corpus annotated with both syntactic and semantic information, which is a substantial amount of effort additional to pure syntactic annotation. To address the issue, it is desirable to develop automated methods to generate a semantically annotated training corpus.
2.2 Biomedical terminologies and related tools
A biomedical terminology generally consists of comprehensive biomedical concepts along with their lexical synonyms, which can assist NLP programs in term recognition and standardization. In some terminologies, concepts are also assigned semantic classes that model the categorical knowledge about a domain. The Unified Medical Language System (UMLS) [6] is an integrated biomedical terminology resource maintained by the National Library of Medicine (NLM). The UMLS has three main components: the Metathesaurus, the Semantic Network, and the Specialist LEXICON. The Metathesaurus integrates about 150 biomedical terminologies (ranging from clinical to molecular) into a concept-centered terminology (~2 million concepts) and associates each distinct concept with a Concept Unique Identifier (CUI). Each CUI subsumes a set of synonyms (e.g., “facial edema” and “face oedema”) representing the concept. The Semantic Network includes 135 semantic types and serves as a semantic framework for the UMLS concepts. Each concept in the Metathesaurus is assigned at least one semantic type. For example, the concept Nose Congestion (CUI: C0027424) is assigned the semantic type T184 Sign or Symptom. The Specialist LEXICON contains comprehensive syntactic, morphological, and orthographic information of biomedical terms. Based on the LEXICON, the NLM has also developed a set of tools [12] for performing tasks such as normalizing and deriving lexical variants (e.g., simple conversion between “nose” and “noses”), which conveniently support the development of various NLP applications.
3. Related work
3.1 Augmenting general parsers with biomedical terminologies
A common approach to developing a biomedical parser is by augmenting a general English parser with lexical knowledge from a domain-specific terminology. Aimed at processing emergency department notes, Szolovits [13] proposed using the UMLS Specialist LEXICON to augment the default lexicon of the Link Grammar (LG) parser [14]. Huang et al.[15] reported that augmenting the Stanford parser [16] with the Specialist LEXICON improved performance in a task of noun phrase identification from radiology reports. Lease & Charniak [17] also reported lexical augmenting techniques to be helpful in adapting a general PCFG [18] for parsing biology text, where they forced lexical collocations learned from the UMLS Specialist LEXICON to be unbreakable units. Although augmenting knowledge from biomedical terminologies has been found to help adapt general English parsers to the sublanguage, the existing solutions remain basically at lexical preprocessing and do not involve deeper customization of the grammar. The pure lexical adaption approach has limitations of 1) assuming that biomedical sublanguage does not syntactically diverge from general English, and 2) not integrating the useful semantic patterns between biomedical concepts into grammar development. The lexical collocation from the UMLS used by Lease & Charniak [17] was a closer step towards lexical semantics, but still not a formal modeling of a grammar that includes domain semantic classes.
3.2 Biomedical parsers involving proprietary semantic classes
In addition to only lexical adaption, many dedicated biomedical parsers have focused on engineering (in a broad sense) semantic grammars specifically for biomedical language. The Medical Language Processor (MLP) [19] from NYU’s Linguistic String Project (LSP) used manually derived semantic constraints to rule out incorrect syntactic parses, where the semantic classes were based on a homemade semantic lexicon of the LSP. The MedLEE [20] system implemented a syntacto-semantic grammar, also with homemade semantic classes. Sample rules of MedLEE are provided below (the square-bracketed nodes are optional) to give readers a sense about such syntacto-semantic grammars:
Analyzing radiology texts with help from physicians, Schröder [21] built an ontology with concepts, types, and their relations and used it to verify the semantic viability of candidate syntactic parses. With inspiration from field theory of physics, Taira et al. [22] developed a probabilistic parser, with a homemade semantic lexicon containing a few hundred of semantic classes for parsing radiology reports. The MEDSYNDIKATE [23] system associates lexical terms with semantic features from specially engineered medical ontologies and performs semantic constraint-checking in the parsing process. The NLUS [24] and MPLUS [25] systems implemented hierarchical Bayesian Networks [26] with specially customized semantic classes to guide on top of syntactic parsing.
The biomedical parsers involving proprietary semantic classes mostly focused on a specific specialty (e.g., radiology) and used domain-specific terminologies to increase lexical coverage. Many of the semantically customized parsers achieved precision higher than 0.85. Not surprisingly, the performance resulted from time-consuming and knowledge-intensive grammar engineering and/or corpus annotation. For example, it was estimated [27] that 6 person-months were spent in creating 350 grammar rules of the MedLEE system. Moreover, the generality and interoperability of these parsers’ input/output can be limited because of the locally optimized semantic classes.
3.3 Preparing semantic infrastructure sharable by parsers
An endeavor with potentially broader influence has been on preparing a sharable infrastructure that uses semantic classes of public terminologies. Two of the major approaches are preparing sharable semantic lexica and preparing sharable semantic grammar rules:
To create a semantic lexicon especially for processing discharge summaries, Johnson [28] proposed associating the Specialist lexemes via Metathesaurus concepts to appropriate UMLS semantic types. Similarly, Verspoor [29] created a semantic lexicon for processing biological literature.
- As early as late 60s, Pratt & Pacak [30] already proposed intricate syntacto-semantic grammars that incorporated the semantic classes (Etiology, Function, General, Morphology, and Topography) of the Systematized Nomenclature of Pathology (SNOP) [31], the precursor of the SNOMED [32]. Decades later, Do Amaral Marcio & Satomura [33] picked up the idea again and incorporated SNOMED semantic classes into a syntacto-semantic grammar. They manually examined case reports in the New England Journal of Medicine and derived rules such as:
The pioneering studies in preparing sharable semantic lexica and grammars laid the foundation towards pursuing semantic interoperability in developing biomedical parsers. The previous work had also demonstrated inspiring techniques such as the use of hybrid syntacto-semantic representation. However, none of the early work has been reported to be part of a full functioning parser. There still appears to be a gap from creating a semantic lexicon to formulizing a broad-coverage semantic grammar, and the main obstacle lies in that the current methods for engineering semantic grammars are costly and not scalable.
4. Methods
4.1 Overview
To parse noun phrases in biomedical text, we developed a PCFG with each non-terminal node represented as a triplex of syntacticTag~semanticTag~headLexeme. Figure 2 illustrates the steps applied in generating the grammar, and each step is elaborated in subsections 4.2 to 4.6. Integration of our semantic PCFG with a bottom-up parsing algorithm is described in 4.7, and evaluation is described in 4.8.
Figure 2.
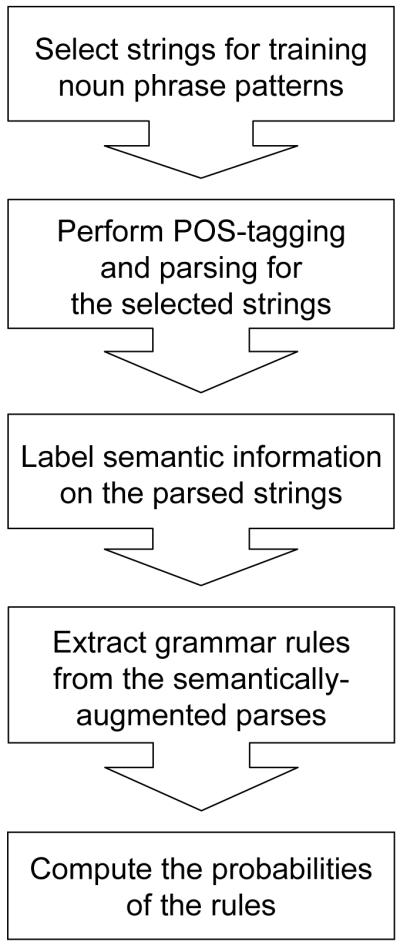
Flow diagram of the steps involved in generating the semantic PCFG
4.2 Select strings for training noun phrase patterns
The UMLS Metathesaurus main table MRCONSO (2010AA) was used as the source of our training strings. Strings containing special characters such as “:” and “@” were excluded, for they are less likely to appear in natural language. For example, the colon-delimited string “Zidovudine:Susc:Pt:Isolate:OrdQn” in LOINC is not useful for training noun phrases. Artificial tokens such as “NEC” and “NOS” were also removed.
4.3 Perform POS-tagging and parsing for the selected strings
After the filtering in 4.2, the MedPost tagger [34] was used to label POS tags on the selected Metathesaurus strings. For example, the string “ulcer of eyelid” was tagged as “ulcer/NN of/IN eyelid/NN”, where “IN” denotes preposition and “NN” denotes singular noun. The Stanford parser [16] was then used to parse the tagged strings. We configured the parser to mark the head nodes in its output so that we could use Eq.(5) to model our probabilistic grammar based on the syntactic, semantic, and lexical information of the head words. For example, the above tagged string was parsed as shown in Figure 3. Note that we selectively used the output of the Stanford parser, which was not originally designed to handle domain-specific syntax. The selection criteria are described in 4.5 below.
Figure 3. Example of a headed parse by the Stanford parser.
The square brackets mark the head words and their part-of-speeches (NN = singular noun, IN = preposition). Head roles are marked specially by “=H”. For example, the second level NP that contains only “ulcer” is marked as the head of the whole NP.
4.4 Label semantic information on the parsed strings
A critical step in creating a semantic grammar is attaching semantic information (e.g., the UMLS semantic types in this study) to the grammar rules. Our approach was labeling the semantic types on the parsed training strings, followed by extracting the semantically augmented rules automatically (see 4.5). Since some of the semantic types such as T170 Intellectual Product and T089 Regulation or Law were not useful for capturing information we were interested in when parsing biomedical noun phrases, we excluded them when labeling the training instances (see Supplementary data 1 for the final list of semantic types selected into our grammar). The labeling of semantic information involved three steps:
Label the semantic type of the entire concept string if it was parsed as an NP by the Stanford parser. The process was straightforward attaching the semantic type(s) of the CUI to the entire NP.
Based on the UMLS, semantic type(s) of the CUI associated with each lexeme in the NP was labeled. A “NULL” type was labeled if the lexeme was not a string of any CUI or was a function word (e.g., the preposition “of”).
Copy the semantic type(s) of the head noun of a PP as the semantic type(s) of the entire PP, so that the semantic head of the PP can be exposed for modeling semantic patterns in rules such as NP ➜ NP PP.
Figure 4 illustrates the same phrase in Figure 3 after the above 3-step semantic labeling. Note that in more complicated cases a lexeme might be ambiguously associated with multiple semantic types, e.g., “dilatation” could be labeled T046 Pathologic Function or T061 Therapeutic or Preventive Procedure. The solution we devised involved using the semantic group [35] of the entire NP to disambiguate the semantic type of the head noun. For example, the “dilatation” in “endoscopic dilatation” was labeled T061 because it belonged to the same semantic group Procedures as the concept represented by the entire NP. Once the head was disambiguated, its syntactic dependents (e.g., adjective modifiers) were disambiguated by using the semantic types of the head and the whole concept.
Figure 4. Example of a semantically labeled parse.
The same phrase from Figure 3 after semantic labeling. Each labeled triplex is represented as syntacticClass~semanticClass~headLexeme, where the head nodes (identified by Stanford parser) are marked by an asterisk * symbol. Semantic types: T047 Disease or Syndrome, T046 Pathologic Function, T023 Body Part, Organ, or Organ Component.
4.5 Extract grammar rules from the semantically-augmented parses
For extracting grammar rules from the semantically labeled parses, we used a program (count-trees.c) by Mark Johnson [36] that can automatically pull out context-free grammar from Treebank [37] style parses. To avoid potentially erroneous patterns (e.g., incorrect distribution of modifiers) induced by pure syntactic parsing we used the simple training instances and discarded complex training instances that contained a mixture of coordination and prepositions such as the example in Figure 1. Instances containing nested coordination or prepositions (e.g., “increase of pain in joint”) were also excluded from training. Figure 5 shows some grammar rules extracted from the labeled parse in Figure 4. To increase recall of the grammar, we also used the Specialist LEXICON’s LVG program [12] to derive rules with inflected forms of the lexemes. For example, the rule NNS~T023~eyelids ➜ eyelids was derived from NN~T023~eyelid ➜ eyelid.
Figure 5. Example of grammar rules extracted from a semantically labeled parse.
Grammar rules automatically extracted from the labeled parse in Figure 4. Except for the RHS of the lexical rules, each grammar node is represented as syntacticClass~semanticClass~headLexeme, with head nodes marked by asterisk * symbol. Semantic types: T047 Disease or Syndrome, T046 Pathologic Function, T023 Body Part, Organ, or Organ Component.
4.6 Compute the probabilities of the rules
After obtaining the rules, we computed the typical probability estimation P(rhs | lhs) for PCFG rules. To model potentially unseen lexemes in test data, we substituted lexemes occurring less than 3 times in the training data for the UNKNOWN token. We also followed Taira et al.’s [38] idea of mixing lexicalized/unlexicalized nodes to reduce rule specificity and increase grammar recall. For example, the unlexicalized LHS of a lexical rule became a less strict syntacto-semantic node: posTag~semanticType ➜ lexeme/posTag (if the semanticType is not “NULL”). Probability of the smoothed rule was estimated by using Eq.(6), where Wsem stands for the semantic type of the LHS (the entire concept), and Count is frequency function.
| (6) |
Note that due to more specific conditioning, rules with lexicalized LHS generally have higher probability. As a result, lexicalized rules are favored in the ranked parsing and should help make more specific global parses ranked higher. Adapting Ge & Mooney’s probability estimation for semantically-augmented PCFG, the P(rhs | lhs) in Eq.(5) was estimated as the product of the three terms described as follows:
-
P(Hsyn, Hsem | Wsyn, Wsem, h)
-
∏i=1…m+1 P(Rsyn:i, Rsem:i, ri | Rsyn:i-1, Rsem:i-1, ri-1, Hsyn, Hsem, h, Wsyn, Wsem)
Probability of the right adjuncts given each preceding right adjunct, the syntactic class and semantic type of the head, and the LHS triplex. The estimation is symmetric to that for the left adjuncts, which is explained in details below.
-
∏i=1…n+1 P(Lsyn:i, Lsem:i, li | Lsyn:i+1, Lsem:i+1, li+1, Hsyn, Hsem, h, Wsyn, Wsem)
Probability of the left adjuncts given each preceding left adjunct, the syntactic class and semantic type of the head, and the LHS triplex. Different from the 0th order assumption in Collins’ [10] original implementation, we used a 1st order Markov model and had Li(li) depend on Li+1(li+1). Note that Ln+1(ln+1) = STOP, which models the left boundary of the RHS. Each conditional probability in the chain of product was estimated using Eq.(9):(9)
4.7 Implement an NP parser using the semantic PCFG
We adapted a Cocke-Younger-Kasami (CYK) parser freely distributed by Sarkar [39] for executing our semantic PCFG. Before parsing, the input text was POS-tagged by the MedPost tagger [34]. To contain the huge search space resulting from the lexicalized semantic grammar, for each LHS only the top 3 rules (ranked according to probability) were used in parsing. In the end, the parser determined the longest spanning NPs that were covered by the grammar and output the syntactic parse with the highest probability for each identified NP. Post-processing was performed to convert the parse granularity to a coarser level that conformed to our annotation guideline (see Supplementary data 2).
4.8 Evaluate the NP parser
To create a gold standard set, the second author (CF) manually annotated 10 de-identified (by DeID [40]) clinical notes by following our NP bracketing guideline (Supplementary data 2). In this study, we focused on evaluating only the correctness of the syntactic structure. The evaluation notes were randomly selected from dictated discharge summaries at University of Pittsburgh Medical Center Hospitals during 2008. We preprocessed the notes by removing the XML tags and retaining the scrubbing tags in DeID output, e.g., “Jul_33_2019/DATE”. In addition, we compared the semantic PCFG to a non-semantic, lexicalized version by making the semantic component of every grammar node into an identical dummy NULL. The semantic-null model was smoothed by including unlexicalized nodes such as IN~NULL, which literally backed off to a pure syntactic node.
NLP community-agreed metrics (bracketing recall, bracketing precision, and average cross-bracketing) for the phrase-structure syntactic parse were computed by using the Evalb program [41]. The bracketing recall is the number of correctly parsed (bracketed) noun phrase constituents over the gold standard bracketing. The bracketing precision is the number of correctly parsed (bracketed) noun phrase constituents over all the bracketing by the parser. Average cross-bracketing is the number of non-aligned bracketing between the parser and the gold standard per sentence. For example, [… {… ] …} is considered a cross, where the square and curly bracketing stand for the parser output and the gold standard respectively. The lower the average cross-bracketing the better a parser performs.
5. Results
5.1 The derived semantic PCFG
The derived semantic PCFG contains 5,498,365 rules with associated probabilities (available as Supplementary data 3). The grammar contains essential information that is ready to be loaded into an operational PCFG parser. A sample rule in the grammar is shown below:
−4.48017369581341 NP~T047~aneurysm JJ~T029~abdominal NN~T190~aneurysm
The log-likelihood of rule probability is followed by a tab, then the LHS node, and a sequence of RHS nodes delimited by space. The example rule states that a T047 Disease or Syndrome NP with head word “aneurysm” can be re-written as a T029 Body Location or Region adjective “abdominal” modifying a T190 Anatomical Abnormality head noun “aneurysm”.
5.2 Evaluation using the manually annotated noun phrases
From the 10 discharge summaries, 212 sentences were annotated and used in evaluation. Table 2 shows the performance statistics of two versions of the PCFG that were compared: one is the fully-implemented semantic PCFG, and the other is the PCFG with semantic components removed. We can see that the semantically-augmented grammar resulted in higher bracketing precision (with a difference of 0.161) and lower cross-bracketing rate (0.61 versus 1.27). The lower recall by the semantic-null model appears counter-intuitive, but it actually reflects less precise parsing from another perspective, i.e., some of the incorrect parses had simpler structures (therefore fewer brackets). For example, Figure 6 compares the parses for the phrase “left arm and left leg numbness” by the semantic-null model (6a) and by the fully implemented model (6b).
Table 2.
Performance statistics of the two compared PCFG implementations
| Bracketing recall |
Bracketing precision |
Average cross-bracketing |
|
|---|---|---|---|
| Semantic PCFG | 0.644 | 0.737 | 0.61 |
| PCFG w/o semantics | 0.601 | 0.576 | 1.27 |
▪ Bracketing recall = (# of correct bracketing) / (gold standard bracketing)
▪ Bracketing precision = (# of correct bracketing) / (parser bracketing)
▪ Average cross-bracketing = (# of crossing brackets between parser and gold standard) / (# of sentences)
Figure 6. Comparing the parses without/with using semantic information.
Figure 6a (top) shows an incorrect parse for “left arm and left leg numbness” by the PCFG with all semantic labels made null. According to this parse, “left” modifies both “arm” and “left leg” so that “left arm” and “left left leg” modify “numbness”.
Figure 6b (bottom) shows the correct parse for the same phrase by the fully implemented semantic PCFG. In this parse, “left arm” and “left leg” are parsed correctly and modify “numbness”.
Figure 6b agrees with our manually annotated gold standard, while Figure 6a has the following errors:
The extra NP bracketing for “arm and left leg” is not correct.
The NP bracketing for “left arm”, “left leg”, and their coordination are missing.
The adjective “left” of “arm” was incorrectly distributed, resulting in a cross-bracketing with the gold standard, i.e., crossed the NP “left arm”.
5.3 Error analysis
We performed manual inspection into arbitrarily sampled parses and summarized the errors we could find explanations for into several categories (with estimated percentages):
1. Mis-tagging by the POS tagger (20%)
In some cases, incorrect POS tagging had prevented correct parsing from the first place. For example, in one sentence “history of significant left lower quadrant pain…” the “left” was mis-tagged by MedPost as NN (singular noun) and “lower” was mis-tagged as VBP (non-3rd person singular present verb), making the phrase parsed into two nonsensical segments “history of significant left” and “quadrant pain”.
2. Failure in separating contiguous NPs that belong to disjoint sentence-level structures (20%)
The grammar was created for parsing longest-spanning NPs (and only NPs), which sometimes lumped together contiguous NPs that should belong to different sub-trees of a full parse. For example, the NP “the original CT scan 3 days” was parsed out from the sentence “…reviewed the original CT scan 3 days earlier…”. Apparently “3 days earlier” adverbially modifies “reviewed” and should have been separated as an ADVP (adverb phrase).
3. Incorrect scoping of pre-modifiers (4%)
Certain pre-modifiers (especially determiners) were not correctly scoped even with the help of semantic parsing. For example, “no rhonchi, rales, or wheezes” was parsed as ((no rhonchi), (rubs), or (gallops)), in which the three nouns should have been coordinated first. As most determiners (e.g., “the” and “no”) were not associated with any clinically relevant semantic types included in building our grammar (e.g., “no” belongs to the general T078 Idea or Concept), the grammar had limited effect in determining their correct scope because the determiner’s semantic label was NULL. The appropriate scoping of negation is a critical but difficult task, and it is likely that it would require additional semantic annotation/training for accuracy.
4. Inadequate coverage of the UMLS as a semantic lexicon (6%)
Although the UMLS was considered a comprehensive terminology resource, there were still lexemes not covered by the Metathesaurus and therefore no semantic types could be associated through the automatic labeling. For example, “rub” and “gallop” were not UMLS synonyms of their corresponding concepts C0232267 Pericardial rub and C0232278 Protodiastolic gallop (both belonging to T033 Finding). The under-labeling of semantic information also had implication on parsing of coordination. For example, had “rub” and “gallop” been associated with T033 but not NULL, the phrase “no murmurs, rubs, or gallops” (the UMLS had no trouble associating “murmurs” to T033 via the concept C0018808 Heart murmur) could have been parsed correctly provided that the three nouns belonged to the same semantic type and were of higher probability to be coordinated first.
5. Limited coverage of NP patterns learned from the UMLS strings (45%)
The NP structures in the UMLS strings were diverse, but they did not align perfectly with NP patterns occurring in natural language. For example, some NPs such as “a sodium of 152” (i.e., PP attachment containing only a quantifier) have phrase structures that are prevalent in clinical notes but are not well covered in terminology strings. As a result, those clinical NP patterns were not modeled into the UMLS-derived grammar and were missed in the parsing.
6. Incorrect labeling of semantic types to the grammar rules (5%)
Nonsensical semantic patterns were observed in the parses. For example, one parse contained a constituent by the rule: NP~T046 ➜ NP~T061 PP~T023~of, which states that a Pathologic Function can be semantically composed as a Therapeutic or Preventive Procedure of a Body Part, Organ, or Organ Component. By tracing the source of the rule, we found it resulted from the string “Percutaneous transluminal cutting balloon angioplasty of popliteal vein using fluoroscopic guidance” of C2315793, which was mistakenly assigned as T046 Pathologic Function (SNOMED-CT labeled the concept as a procedure). After correcting the source error, the derived rule should be NP~T061 ➜ NP~T061 PP~T023~of. We also observed nonsensical semantic patterns that appeared to result from errors in the automated labeling process itself (e.g., from incorrect semantic disambiguation).
Note that the above categories of errors were those we could give an explanation. Due to the nature of automatic rule-ranking in probabilistic parsing, it was difficult to trace in many cases why the correct parse did not receive the highest probability (even if it actually existed in the grammar space).
6. Discussion
6.1 Comments on the unsupervised semantic grammar derivation
We demonstrated the feasibility of a method that incorporated terminology semantic classes into building an operational PCFG for parsing biomedical NPs. The solution is basically unsupervised, with the goal of reducing human effort in grammar engineering. The UMLS was chosen for it was public, comprehensive, and appeared promising in terms of semantic transferability. In addition, we assumed that domain terminologies were composed dominantly of nominal concepts suitable as training instances for noun phrases. Although the performance did not result in a solution that was adequate for deriving a grammar with comparable quality of that by human, our controlled experiment indicated that even unsupervised semantic augmentation could benefit syntactic parsing. The error analysis revealed critical and yet general issues to be addressed in order to improve the method:
Terminology concepts can be a rich base training set but do not include all types of phrase structures and semantically-annotated lexemes that occur in natural language. In addition, the rule frequencies derived from a terminology may not quantitatively reflect the usage in a real corpus. To bridge the gap, we project that a certain amount of manually annotated texts and/or a semantic lexicon already learned from such a domain-specific corpus will be required. Associating certain semi-semantic function words (e.g., negation determiner “no”) with special semantic type could also help model their appropriate scoping over structures with coordination and PP attachment. Other than enriching with additional semantic types, it should be interesting to investigate the effects of using coarser-granularity semantic classes (e.g., McCray’s semantic groups) for building the grammar, which could also significantly reduce the size and complexity.
The over-spanning of noun phrase boundaries is intrinsic to a partial grammar that models only noun phrases and does not model full-sentence parsing because when a prepositional phrase follows a noun, it is always associated with the noun phrase and never with a remote verb phrase. For example, when parsing only noun phrases in “She was admitted to the ER on Monday”, the prepositional phrase “on Monday” will be attached to “the ER” and not to the verb “was admitted”. To overcome the limitation, we project several possible solutions: a) search for an optimal cutoff value to trim off any sub-tree parse that has a probability below the cutoff, b) fuse the domain-specific semantic grammar with a general grammar that also models full-sentence parsing; c) apply the semantic grammar on the noun phrases output by another high-performance phrase chunker that can handle phrase boundaries correctly, i.e., use shallow parsing to determine the phrase boundaries and then the semantic PCFG as a post-processor to determine internal phrase structure.
Some incorrect semantic grammar rules were found to result from errors in our automated semantic labeling, which was based on UMLS semantic categorization. Although no formal evaluation was performed to verify whether every semantic type in the parses was appropriate, arbitrary inspections indicated that nonsensical semantic constituents were prevalent. In some parses, even when the correct phrase structure was obtained, the underlying semantic constituents did not match the lexical meaning. For example, in one case “squamous cell” was parsed with NP~T033 ➜ JJ~T080 NN~T082, which states that a Finding is a Spatial Concept modified by a Qualitative Concept. We project that still more systematic analysis is needed to identify the exact sources of the errors, but it is likely that some of the less well-defined UMLS semantic types (e.g., T033, T080, and T082) complicated the automated labeling process.
We used the MedPost tagger without customization and did not formally evaluate its performance on the clinical notes. The tagger was trained on PubMed abstract sentences and was not expected to be optimal for clinical notes, which is a different genre. We believe POS tagging is critical to accurate parsing and will improve/evaluate it as an independent module. As the NLP community is putting considerable effort in annotating clinical notes, we anticipate that a more appropriate POS tagger will be developed in the near future. In the meanwhile, it would also be possible to identify common tagging errors for words such as ‘left’ and ‘lower’, which frequently occur as adjectives in clinical notes.
In summary, we believe existing computational models for implementing a domain-specific semantic PCFG are already well-studied and available to the community. The missing part remains designing efficient methods to obtain quality training data. Our methods demonstrated one approach to this challenge and can be improved by addressing the observed issues. We consider some of our methods are general and useful to other biomedical genres and to different grammar paradigms. For example, it is possible to apply the methods described in 4.5-4.6 to derive a semantic PCFG from a semantically annotated and parsed corpus (e.g., the Genia Corpus [42]). Before satisfactory performance could be achieved with a completely unsupervised solution, we suggest that a semi-supervised approach would be more practical to benefit the semantic engineering of biomedical grammars. A domain expert can review/validate the common unlexicalized syntacto-semantic patterns in the derived grammar, and the deliverable will be a collection of sharable syntacto-semantic grammar rules similar to the work by Pratt & Pacak [30] and Do Amaral Marcio & Satomura [33] described in subsection 3.3.
6.2 Limitations
We recognize the study design was limited in term of the following aspects:
There was only one annotator (the second author) in generating the gold standard.
Our control grammar by normalizing the semantic labels to NULL was literally a straightforward single-factor control and might not represent other lexicalized, semantic-null, biomedical NP parsers that were developed/obtained in different ways.
We preserved the de-identification tags (e.g., DATE) of DeID into the parsing pipeline, which avoided some false positives and therefore favored precision. For example, “had/VBD arthroplasty/NN on/IN 07-27-2009/DATE” directly prevents the “07-27-2009” from being attached to “arthroplasty” because DATE is not a legitimate POS tag in the grammar.
We did not perform an independent evaluation for each component in the pipeline.
7. Conclusion
To facilitate the efficiency of semantic grammar engineering and the transferability of the grammar, we developed a complete pipeline with automated methods to derive a probabilistic PCFG by using selected UMLS concept strings and semantic types for training. This study focused on implementing a grammar for parsing biomedical noun phrases and evaluating it on clinical notes. The derived grammar achieved recall 0.644, precision 0.737, and average cross-bracketing 0.61, compared to a semantic-null control grammar, which achieved recall 0.601, precision 0.576, and average cross-bracketing 1.27. The results indicated that, with an unsupervised approach, incorporation of terminology semantics into a PCFG benefited the parsing of biomedical noun phrases. We believe the methods can be applied to developing grammars for other biomedical genres and can be further improved by addressing the issues discovered in our error analysis.
Supplementary Material
Supplementary data 1 – Semantic types used in our grammar The UMLS semantic types considered relevant for parsing biomedical noun phrases.
Supplementary data 2 – NP annotation guideline Phrase-structure bracketing instructions for NPs defined in this study.
Supplementary data 3 – The semantic PCFG derived in this study Format: [log-likelihood]<tab>[LHS]<space>[RHS node1]<space>[RHS node2]… [RHS noden]
Acknowledgements
We thank Dr. Wendy Chapman for help with access to the University of Pittsburgh NLP Repository. We thank Drs. Noémie Elhadad, Yang Huang, Herbert Chase, Chintan Patel, and Francis Morrison for their intellectual input in discussing the research ideas. This study was performed during the first author’s Ph.D. training in the Department of Biomedical Informatics, Columbia University, and was supported by Grant LM008635 from the National Library of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of biomedical text mining: current progress. Brief Bioinform. 2007;8(5):358–75. doi: 10.1093/bib/bbm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–44. [PubMed] [Google Scholar]
- 3.Punyakanok V, Roth D, Yih W. The importance of syntactic parsing and inference in semantic role labeling. Comput Linguist. 2008;34(2):257–87. [Google Scholar]
- 4.Chung GY. Towards identifying intervention arms in randomized controlled trials: extracting coordinating constructions. J Biomed Inform. 2009;42(5):790–800. doi: 10.1016/j.jbi.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Miyao Y, Sagae K, Saetre R, Matsuzaki T, Tsujii J. Evaluating contributions of natural language parsers to protein-protein interaction extraction. Bioinformatics. 2009;25(3):394–400. doi: 10.1093/bioinformatics/btn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Younger DH. Recognition and parsing of context-free languages in time n3. Inf Control. 1967;10(2):189–208. [Google Scholar]
- 8.Kasami J. An efficient recognition and syntax analysis algorithm for context-free languages. Technical Report. Air Force Cambridge Research Laboratory; Bedford, MA: 1965. Report No.: AFCRL-65-758. [Google Scholar]
- 9.Jurafsky D, Martin JH. Speech and language processing: an introduction to natural language processing, computational linguistics, and speech recognition. 2nd ed Pearson Prentice Hall; Upper Saddle River, N.J.: 2009. [Google Scholar]
- 10.Collins M. Head-driven statistical models for natural language parsing. Comput Linguist. 2003;29(4):589–637. [Google Scholar]
- 11.Ge R, Mooney RJ. A statistical semantic parser that integrates syntax and semantics. Proceedings of the Ninth Conference on Computational Natural Language Learning; Ann Arbor, Michigan: Association for Computational Linguistics; 2005. pp. 9–16. [Google Scholar]
- 12.Browne AC, Divita G, Aronson AR, McCray AT. UMLS language and vocabulary tools. AMIA Annu Symp Proc; Washington, DC, USA: American Medical Informatics Association; 2003. p. 798. [PMC free article] [PubMed] [Google Scholar]
- 13.Szolovits P. Adding a medical lexicon to an English parser. AMIA Annu Symp Proc; Washington, DC, USA: American Medical Informatics Association; 2003. pp. 639–43. [PMC free article] [PubMed] [Google Scholar]
- 14.Sleator D, Temperly D. Parsing English with a link grammar. Third International Workshop on Parsing Technologies; Tilburg, The Netherlands: SIGPARSE, Association for Computational Linguistics; 1993. [Google Scholar]
- 15.Huang Y, Lowe HJ, Klein D, Cucina RJ. Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the UMLS specialist lexicon. J Am Med Inform Assoc. 2005;12(3):275–85. doi: 10.1197/jamia.M1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein D, Manning CD. Advances in Neural Information Processing Systems 15 (NIPS 2002) MIT Press; Cambridge, MA: 2003. Fast exact inference with a factorized model for natural language processing; pp. 3–10. [Google Scholar]
- 17.Lease M, Charniak E. Parsing biomedical literature. 2nd International Joint Conference of Natural Language Processing; Jeju Island, Korea: Springer; 2005. pp. 58–69. [Google Scholar]
- 18.Eugene C. A maximum-entropy-inspired parser. Proceedings of the 1st North American chapter of the Association for Computational Linguistics conference; Seattle, Washington, USA: Association for Computational Linguistics; 2000. pp. 132–9. [Google Scholar]
- 19.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994;1(2):142–60. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schröder M. Knowledge-based processing of medical language: a language engineering approach. Proceedings of the 16th German Conference on Artificial Intellegence: Advances in Artificial Intelligence; London, UK: Springer-Verlag; 1992. pp. 221–34. [Google Scholar]
- 22.Taira RK, Bashyam V, Kangarloo H. A field theoretical approach to medical natural language processing. IEEE Trans Inf Technol Biomed. 2007 Jul;11(4):364–75. doi: 10.1109/titb.2006.884368. [DOI] [PubMed] [Google Scholar]
- 23.Hahn U, Romacker M, Schulz S. MEDSYNDIKATE--a natural language system for the extraction of medical information from findings reports. Int J Med Inform. 2002;67(1-3):63–74. doi: 10.1016/s1386-5056(02)00053-9. [DOI] [PubMed] [Google Scholar]
- 24.Haug P, Koehler S, Lau LM, Wang P, Rocha R, Huff S. A natural language understanding system combining syntactic and semantic techniques. Proc Annu Symp Comput Appl Med Care. 1994:247–51. [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen LM, Haug PJ, Fiszman M. MPLUS: a probabilistic medical language understanding system. Proceedings of the ACL-02 workshop on Natural language processing in the biomedical domain; Phildadelphia, Pennsylvania, USA: Association for Computational Linguistics; 2002. pp. 29–36. [Google Scholar]
- 26.Pearl J. Probabilistic reasoning in intelligent systems: networks of plausible inference. Morgan Kaufmann Publishers; San Mateo, Calif.: 1988. [Google Scholar]
- 27.Spyns P. Natural language processing in medicine: an overview. Methods Inf Med. 1996;35(4-5):285–301. [PubMed] [Google Scholar]
- 28.Johnson SB. A semantic lexicon for medical language processing. J Am Med Inform Assoc. 1999;6(3):205–18. doi: 10.1136/jamia.1999.0060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verspoor K. Towards a semantic lexicon for biological language processing. Comp Funct Genomics. 2005;6(1-2):61–6. doi: 10.1002/cfg.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratt AW, Pacak MG. Automated processing of medical English. Proceedings of the 1969 conference on Computational linguistics; Sang-Saby, Sweden: Association for Computational Linguistics; 1969. [Google Scholar]
- 31.Systematized Nomenclature of Pathology. College of American Pathologists; Chicago, Illinois: 1965. [Google Scholar]
- 32.Systematized Nomenclature of Medicine-Clinical Terms [database on the Internet] International Health Terminology Standards Development Organization; [cited May 14, 2009]. Available from: http://www.ihtsdo.org/snomed-ct/ [Google Scholar]
- 33.Do Amaral Marcio B, Satomura Y. Medinfo. International Medical Informatics Association; Vancouver, BC, Canada: 1995. Associating semantic grammars with the SNOMED: processing medical language and representing clinical facts into a language-independent frame; pp. 18–22. [PubMed] [Google Scholar]
- 34.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics. 2004;20(14):2320–1. doi: 10.1093/bioinformatics/bth227. [DOI] [PubMed] [Google Scholar]
- 35.McCray AT, Burgun A, Bodenreider O. Medinfo. International Medical Informatics Association; London, UK: 2001. Aggregating UMLS semantic types for reducing conceptual complexity; pp. 216–20. [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson M. [Accessed on: September 8, 2009];A C implementation of PCFG parser with utility programs. 2006 Available from: http://web.science.mq.edu.au/~mjohnson/code/cky.tbz.
- 37.The Penn Treebank Project [cited September 10, 2009];1999 Available from: http://www.cis.upenn.edu/~treebank/
- 38.Taira RK, Soderland SG. A statistical natural language processor for medical reports. AMIA Annu Symp Proc; Washington, DC, USA: American Medical Informatics Association; 1999. pp. 970–4. [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar A. [Accessed on: July 17, 2008];Simple Perl implementation of a CKY recognizer and parser for probabilistic Context Free Grammars (CFGs) 2004 Available from: http://www.cs.sfu.ca/~anoop/distrib/ckycfg/readme.html.
- 40.Neamatullah I, Douglass M, Lehman L-w, Reisner A, Villarroel M, Long W, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak. 2008;8(1):32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekine S, Collins MJ, Brroks D, Ellis D, Blaheta D. [Accessed on: Feb 6, 2010];Evalb: a bracket scoring program for Treebank-style syntactic parses. 2008 Available from: http://nlp.cs.nyu.edu/evalb/
- 42.Kim J-D, Ohta T, Tateisi Y, Tsujii J. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(suppl 1):i180–i2. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data 1 – Semantic types used in our grammar The UMLS semantic types considered relevant for parsing biomedical noun phrases.
Supplementary data 2 – NP annotation guideline Phrase-structure bracketing instructions for NPs defined in this study.
Supplementary data 3 – The semantic PCFG derived in this study Format: [log-likelihood]<tab>[LHS]<space>[RHS node1]<space>[RHS node2]… [RHS noden]







