Abstract
During Drosophila neural development, neuroblasts delaminate from the neuroectoderm of each hemisegment in a stereotypic orthogonal array of five rows and three columns (ventral, intermediate, and dorsal). Prevailing evidence indicates that the individual neuroblast fate is determined by the domain-specific expression of genes along the dorsoventral and anteroposterior axis. Here, we analyze the role of Vnd, a NK-2 homeodomain protein, expressed initially in the ventral neuroectoderm adjacent to the ventral midline, in the dorsoventral patterning of the neuroectoderm and the neuroblasts. We show that in vnd null mutants most ventral neuroblasts do not form and the few that form do not develop ventral fates, but instead develop intermediate-like fates. Furthermore, we demonstrate that Vnd influences the gene expression patterns in the ventral proneural clusters and neuroectoderm, and that its action in neuroblast formation includes, but is not exclusive to the activation of proneural AS-C genes. Through the use of GAL4/UAS gene-expression system we show that ectopic Vnd expression can promote ventral-like fates in intermediate and dorsal neuroblasts and can suppress certain normal characteristics of the intermediate and dorsal neuroectoderm. Our results are discussed in the context of the current evidence in dorsoventral patterning in the Drosophila neuroectoderm.
Keywords: Neurogenesis, NK-2 homeodomain, ventral nervous system defective, cell fate, dorsoventral axis patterning
The ventral nerve cord, a major component of the Drosophila central nervous system (CNS), is formed mainly by the fusion of thoracic and abdominal neuromeres. Each bilateral hemineuromere consists of ∼380 different postmitotic cells that derive from a set of 30 different neuroblasts (Nbs), the progenitor stem cells (Bossing et al. 1996; Schmidt et al. 1997). Nbs are first observed as individual cells that enlarge and then delaminate from the neuroectoderm (NE) in an orderly spatiotemporal sequence (S1–S5 waves) to form a stereotypical subepidermal array composed of seven anteroposterior (AP) rows and three columns along the dorsoventral (DV) axis: ventral (also called medial), intermediate, and dorsal (also called lateral) (for review, see Campos-Ortega 1993). Patterned expression of proneural genes in the NE within small groups of cells, a proneural cell cluster, precedes the enlargement and delamination of Nbs (Martín-Bermudo et al. 1991; Skeath and Carroll 1992). A competitive process involving the Notch signaling pathway allows singling out of an individual cell to become an Nb, from within the equipotential cells of each proneural cluster (for review, see Campos-Ortega 1993).
Careful lineage studies with individual Nbs have demonstrated that each Nb has a unique identity at birth, which is reflected by the timing of its divisions, the size of its lineage, and the specific neurons and glia that it produces (Bossing et al. 1996; Schmidt et al. 1997). Evidence using several molecular-identity markers illustrate amply molecular distinctions between delaminating Nbs (Doe 1992; Broadus et al. 1995). Moreover, in some cases such distinctions are manifested already in the proneural cluster from which the Nb derives (Martín-Bermudo et al. 1991; Skeath and Carroll 1992; Chu-LaGraff and Doe 1993; Skeath et al. 1995). Therefore, the question of how Nb identities are specified can be posed as to how the two-dimensional neuroectodermal layer is partitioned into discrete domains of specific gene expression.
Several studies have provided convincing evidence that determination of NE and Nb identity along the AP axis of the Drosophila embryo is under the control of segment polarity genes. Specific examples include wingless, gooseberry-distal, and hedgehog (Patel et al. 1989a; Chu-LaGraff and Doe 1993; Skeath et al. 1995; Matsuzaki and Saigo 1996), which are expressed as transverse stripes within each segment at the time when Nbs are specified. However, little is known about how the NE is partitioned along the DV axis. Heterotopic cell transplantation experiments within the NE of early gastrula embryos have shown that V–D transplanted cells are firmly committed to a ventral fate, producing cell lineages typical of ventral Nbs, whereas D–V transplanted cells adopt ventral cell fates (Udolph et al. 1995). Recently, the Drosophila gene muscle segment homeobox (msh) which, like its vertebrate homologs (Msx1-3), is expressed initially in the dorsal portion of the NE (D’Alessio and Frasch 1996; Isshiki et al. 1997), has been shown to be required for proper development of dorsal Nbs, but not for their formation (Isshiki et al. 1997). From these data, it has been proposed that msh may control dorsal identities of the NE and Nbs.
Several lines of genetic and molecular evidence demonstrate that the ventral nervous system defective (vnd) gene influences the delamination of Nbs in the ventral NE. Early analysis of mutants revealed a severely affected ventral nerve cord (White et al. 1983; Jiménez and Campos-Ortega 1987). Subsequent analysis demonstrated a pronounced effect on the delamination of Nbs in the ventral NE (Jiménez and Campos-Ortega 1990; Skeath et al. 1994); however, specific Nb assignments were not possible in these studies. Furthermore, Skeath et al. (1994) found that one function of vnd is to activate expression of the achaete-scute gene complex (AS-C) in specific proneural clusters in the ventral NE, which might explain the defective Nb formation in vnd mutants to some extent. Recent molecular studies have demonstrated that vnd encodes the prototype of the NK-2 homeobox gene family (Kim and Nirenberg 1989; Jiménez et al. 1995). vnd is transcribed initially in the ventral-most portion of the NE as two bilateral longitudinal stripes that overlap in time and space with the formation of proneural clusters from which S1 and S2 Nbs of the ventral column delaminate (Jiménez et al. 1995; Mellerick and Nirenberg 1995). Expression of vnd is also observed in these Nbs and some of their progeny. This early expression pattern is compatible with the role of vnd in formation and fate specification of early ventral Nbs.
In this paper we address two issues: First, what is the function of vnd in Nb formation and does it act exclusively by regulating proneural AS-C function? Second, does vnd specify the identity of the ventral NE and Nbs? Our results indicate an almost absolute requirement for vnd in the formation of early ventral Nbs that is not mediated simply through AS-C activation. Additionally, we demonstrate that vnd is essential in the specification of the identity of the ventral NE and Nbs, which in its absence, adopt intermediate identities, and that ectopic expression of vnd can promote ventral fates in intermediate and dorsal Nbs.
Results
Generation of a new vnd mutation
Previous detailed analyses of the vnd mutant phenotype utilized the lethal allele vnd6 (Jiménez and Campos-Ortega 1990; Skeath et al. 1994). Whether vnd6 is a true null is not known. Recently we generated a new null allele of vnd, using a lacZ enhancer trap (rF124) (a gift of Christian Klämbt, University of Münster, Germany) inserted ∼9 kb, 5′ to the vnd transcription unit (data not shown). Excisions of the rosy+ marked P element were induced and chromosomal deletions that failed to complement a vnd lethal allele were isolated (see Materials and Methods). Analysis of an excision chromosome, vndD38, indicated that the deletion included only one other known gene, the adjacent Appl gene, but not the flanking lethal loci. Inclusion of Appl is not likely to influence this analysis as Appl is nonessential and there is no evidence that Appl has any role in neurogenesis (Martin-Morris and White 1990; Luo et al. 1992). Polymerase chain reaction analysis of DNA from mutant embryos demonstrated that a genomic fragment from the 3′ coding sequence of the vnd gene was absent from the vnd mutant, verifying that this was a vnd null mutation (data not shown).
Expression of vnd during early neurogenesis
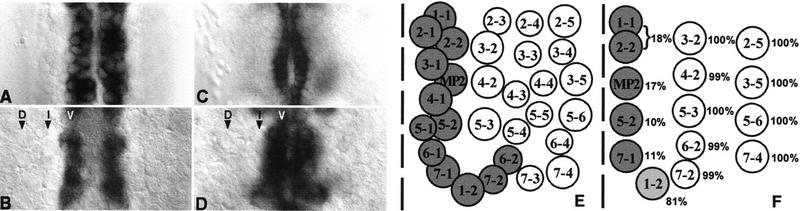
We focus here on particular aspects of the vnd expression pattern that are related temporally and spatially to the process of Nb formation. Previous work has shown that initial expression of vnd in the NE occurs at the blastoderm stage as two longitudinal stripes flanking the presumptive ventral midline, preceding the formation of the S1 proneural clusters (Jiménez et al. 1995; Mellerick and Nirenberg 1995). Position of these clusters, defined by the expression of AS-C genes subdivides the NE into three columns: ventral, intermediate, and dorsal (Martín-Bermudo et al. 1991; Skeath and Carroll 1992). The DV extent of the vnd stripe appears to match that of the ventral column (Jiménez et al. 1995; Mellerick and Nirenberg 1995). After S1 Nb delamination (stage 8), the ventral-most NE continues expressing vnd (Fig. 1A). Only ventral Nbs appear to transcribe vnd after S2 Nb delamination (stage 9, Fig. 1B). Neuroectodermal expression of vnd is narrowed progressively and, by completion of Nb formation (late stage 11), it is restricted to the ventral-most cell row (Fig. 1C). At this stage, vnd transcripts are detected in all ventral Nbs and in the three intermediate S2 Nbs located in the posterior compartment of the hemisegment (Nbs 1-2, 6-2, and 7-2, defined by engrailed expression) (Fig. 1D; Doe 1992) even though these intermediate Nbs did not express vnd initially (Fig. 1B). Strong vnd transcription is also found at stage 11 in the neural progeny underlying vnd-positive Nbs (data not shown). A schema of Nbs that transcribe vnd at any given time is shown in Fig. 1E.
Figure 1.
Early vnd transcription and Nb phenotype of vnd null embryos. Transcription of vnd during early Nb segregation detected by in situ hybridization (A–D); each panel depicts a single segment, anterior is up. (A) After delamination of the S1 Nbs, vnd transcript is in a two-cell–wide ventral stripe that runs anteroposterior in the ventral-most NE. (B) After delamination of the S2 Nbs, expression in the Nb layer is restricted to the ventral column (v); no expression is apparent in Nbs of the intermediate (i) and dorsal (d) columns. (C) After completion of Nb delamination (stage 11), transcripts in the NE are still detected in a single-cell wide stripe. (D) In the Nb layer of the same embryo as in (C), vnd is transcribed in ventral Nbs, as well as in three intermediate Nbs (6-2, 7-2, and 1-2), in the posterior part of the segment that delaminated in the S2 phase. (E) Diagram of the wild-type Nb map where vnd-expressing Nbs are indicated in gray; ventral midline, dotted line. (F) Diagram of S1 + S2 Nbs with percentage of presence of each Nb in vnd null embryos indicated. Affected Nbs are in gray.
Defective formation of early ventral Nbs in the absence of vnd
The formation of S1 and S2 Nbs in vndD38 embryos (hereafter called vnd embryos) was analyzed. Morphological features such as size and position, as well as anti-Hunchback (Hb) antibodies, a general Nb marker, were used to assess the presence of Nbs as described (Jiménez and Campos-Ortega 1990). Additionally, resolution of mapping was improved by the use of two lacZ enhancer trap lines in genes svp (H162) and hkb (5953), each expressing in a different subset of Nbs in a precise temporal sequence (Fig. 2C–F, I–L; Doe 1992). These markers greatly aid mapping of lacZ-expressing Nbs as well as those surrounding them, despite possible displacements of their position after delamination.
Figure 2.
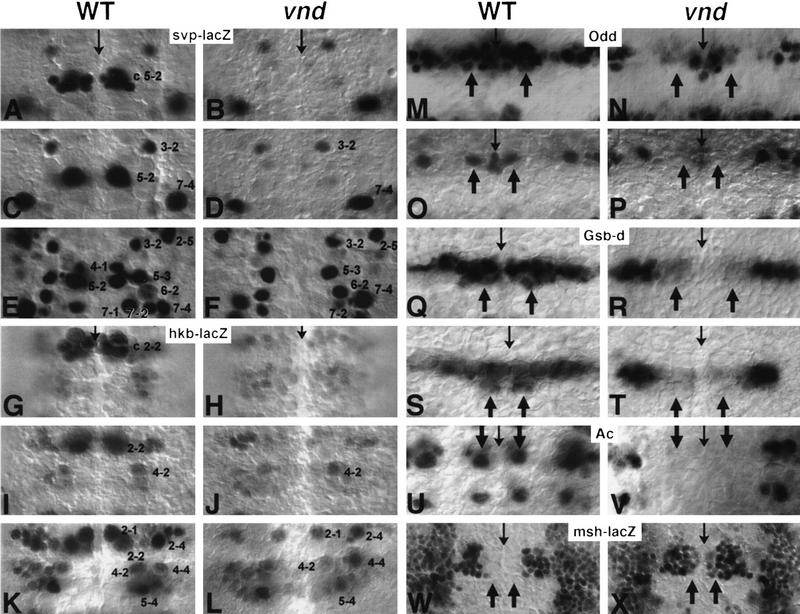
Gene expression in the NE and Nb of the wild type and the vnd mutant. Molecular markers to analyze the formation and specification of Nbs were β-galactosidase from the enhancer trap insertions H162 (svp–lacZ; A–F), 5953 (hkb–lacZ; G–L), rH96 (msh–lacZ; W,X), and proteins Odd (M–P), Gsb-d (Q–T), Ac (U,V). Each panel set depicts a single wild-type (left) and mutant (right) segment; thin arrows mark the ventral midline; anterior is up. At stage 9, normal svp expression is lacking in the mutant in the ventral proneural cluster c5-2 (A,B), also lacking is a svp-expressing Nb 5-2 (C,D); at stage 10, all the svp-expressing ventral Nbs 4-1, 5-2, and 7-1 are missing (E,F). Note a ventral shift of Nbs 3-2 and 7-4 in the mutant (D). Likewise, the mutant lacks hkb expression in ventral proneural cluster c2-2 (G,H) and in Nb 2-2 (I,J). However, the hkb-expressing S3 ventral Nb 2-1 (stage 11) is formed normally in the mutant (K,L). Note that the intermediate Nb 4-2 is shifted ventrally in the right mutant hemisegment shown in (L). At stage 8, expression of Odd in ventral proneural cluster c1-1 is abolished in the mutant (M,N). Also, at stage 9, a normal Odd-positive Nb 1-1 (arrows in O) is never found in the mutant (P). Wild-type Gsb-d expression (Q) is localized to the entire row 5 and to the ventral column of row 6/7; arrows indicate ventral column. All ventral expression is abolished in the mutant NE (R). In the Nb layer at stage 9, Gsb-d is expressed normally in all row 5 Nbs (5-2, 5-3, and 5-6) and in row 7 ventral Nb 7-1 (S); arrows indicate ventral column. In the mutant (T), Gsb-d positive ventral Nbs 5-2 and 7-1 are not observed; expression in intermediate Nb 5-3 is weaker than normal. (U) In wild-type, alternate row ventral Nbs (MP2 and 7-1) and dorsal Nbs (3-5 and 7-4) (stage 9) express Ac; arrows indicate ventral column. No Ac-positive ventral Nbs are found in the mutant (V). (W) During stage 10, msh–lacZ expression in the wild-type NE is detected in the entire dorsal column and part of the intermediate column, but not in the ventral column (arrows). In the mutant (X), ventral expansion of msh–lacZ expression is observed.
Figure 1F shows a schema of the S1 + S2 Nbs map and the frequency at which each Nb is found in vnd embryos. In these mutant embryos, formation of all ventral Nbs is affected dramatically, but Nbs of the intermediate and dorsal columns form normally. However, a few residual Nbs are still observed in the ventral column. To test if the possible maternal expression of vnd was responsible for the residual ventral Nbs, homozygous vnd germ-line clones were generated, and mutant embryos devoid of any maternal vnd product were analyzed. Residual S1 + S2 ventral Nbs were still observed at a similar frequency (data not shown). Although not studied in detail, formation of S3-S5 Nbs seems to be only partially affected. Thus, instead of the three ventral Nbs (MP2, 3-1, and 4-1) present in rows 3/4 at the S3 stage in the wild type, only one Nb at most could be found at the same position in vnd embryos, and this happened in only 33% of cases. On the contrary, the S3 Nb 6-1 (identified by normal expression of ming–lacZ and Gsb-d) was detected in >90% of cases (not shown).
Lack of vnd function alters the identity of ventral Nbs
In the vnd embryos some early Nbs were still observed in the ventral column; thus we questioned their developmental potential. Virtual absence of several progeny of S1 ventral Nbs determined by using different specific markers, suggests that the residual Nbs do not possess ventral identities (Table 1). These absences include (1) Eve- and FasII-positive aCC/pCC neurons (Fig. 4, cf. D, G, and J with E, H, and K, below) and Repo-positive SPG glial cells, progeny of Nb 1-1, (2) 22C10-positive dMP2/vMP2 neurons, progeny of MP2, and (3) Eve-positive CQ neurons and the Eve- and Fas II-positive U motoneurons (Fig. 4E,H, below), progeny of Nb 7-1. Collectively, these data indicate that early ventral Nb lineages were affected in the vnd embryos.
Table 1.
vnd affects the formation and specification of neuroblast lineages
| NB lineagea
|
Genotypeb
|
Eliminationc (%)
|
Formation
|
Duplication
|
|---|---|---|---|---|
| 1-1 | vnd (127) | 97.8 | 2.2 | N.A. |
| sca–GAL4; UAS–vnd (105) | N.A. | 100 | 17.4/2d | |
| MP2 | vnd (143) | 100 | 0 | N.A. |
| sca–GAL4; UAS–vnd (91) | 0 | 100 | 0 | |
| 4-2 | vnd (127) | 13 | 87 | 19.6 |
| sca–GAL4; UAS–vnd (105) | 16.7 | 83.4 | 47.6 | |
| 7-1 | vnd (127) | 100 | 0 | N.A. |
| sca–GAL4; UAS–vnd (93) | 0 | 100 | 11 |
The following progeny are analyzed for each NB: NB 1-1, aCC/pCC; MP2, dMP2/vMP2; NB 4-2, RP-2; NB 7-1, GMC 7-1a.
Number of hemisegments scored indicated in parentheses.
Percent elimination, formation, or duplication in the observed hemisegments. (N.A.) Data not available.
Triplication of aCC/pCC neurons.
Figure 4.
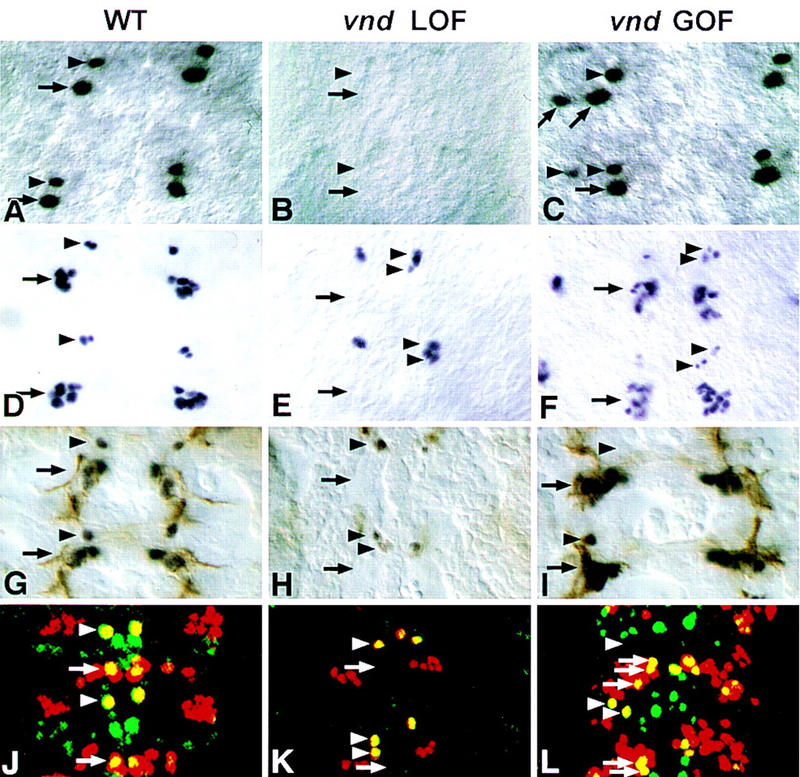
Nb lineages are altered in vnd mutants. Partial lineages (GMCs and neurons) of Nbs 1-1, 4-2, and 7-1 are traced with cell-specific markers in different backgrounds (wild type, vnd loss-of-function, and vnd gain-of-function). Two segments are shown in each case. In the mutant at stage 10, the Eve-positive cells GMC1-1a and GMC7-1a are never detected (compare B and A), whereas, in sca–GAL4; UAS–vnd embryos, both GMC1-1a and GMC7-1a can be duplicated (C). Normally, Eve staining reveals, at stage 11-12, GMC 4-2a, its daughters RP2/RP2-sib (arrowhead), and a cluster of neurons containing aCC/pCC, and 2-3 Us (arrow) (D). In the mutant, Eve-positive Nb 4-2 progeny is duplicated, at both GMC and neuron level (arrowheads), but the Eve-positive neuronal cluster is not detected (E); but in sca–GAL4; UAS–vnd embryos, the Nb 4-2 lineage can be either duplicated (arrowheads) or eliminated and more cells are observed in the Eve-positive cluster (F). (G–I) Eve (dark brown) and FasII (light brown) double-stained stage 13 embryos. In the wild type, RP2 (arrowhead) and a cluster of five neurons aCC/pCC and three Us (arrow) coexpress Eve and FasII (G). In the vnd mutant, RP2 can be duplicated and the cluster of five neurons is never detected (H), but in Kr–GAL4; UAS–vnd embryos, RP2 is most frequently absent, or its position displaced (arrowhead), and aCC/pCC and Us appear to be duplicated (arrow) (I). (J–L) Eve (red) and Zfh-1 (green) double staining in stage 15 embryos. In the wild type, Eve and Zfh-1 are coexpressed in aCC (arrow) and RP-2 (arrowhead) (J). In the mutant aCC is eliminated, but the RP-2 neurons are duplicated (K). In sca–GAL4; UAS–vnd embryos aCC is duplicated; RP-2 can be either duplicated or eliminated (L).
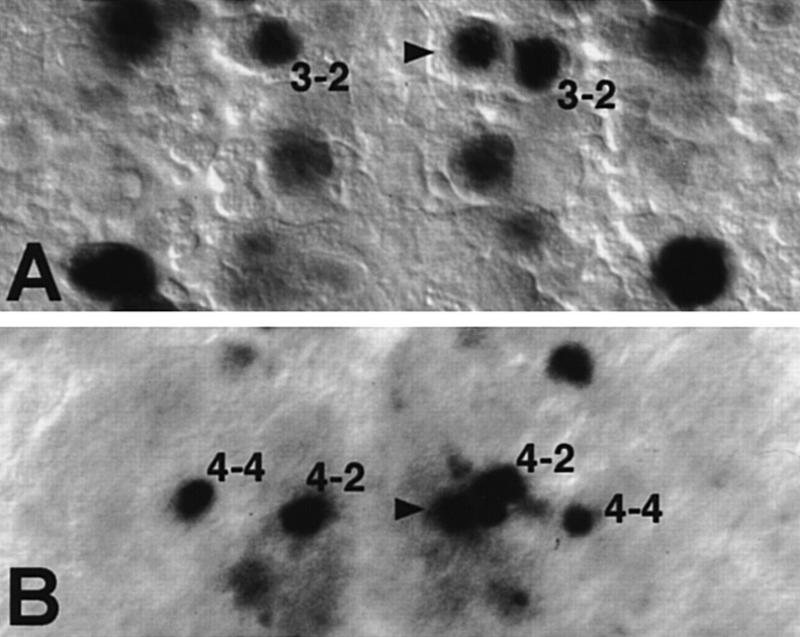
Absence of specific lineages could mean that the Nb identities are modified, or that the residual Nbs do not develop further, or that vnd is also required for the formation of the neurons in question. Several observations indicate that residual early ventral Nbs have altered identities in vnd embryos: (1) The Nb occasionally found at the 1-1/2-2 position in late stage 9 never expressed Odd, a Nb 1-1 marker (Fig. 2O,P), or hkb–lacZ (Nbs 1-1 and 2-2 marker) (not shown). On the contrary, in 38% (n = 29) of cases in which an Nb formed at the 1-1/2-2 position, it expressed svp–lacZ which is a distinctive characteristic of the neighboring intermediate Nb 3-2 at late stage 9 (Fig. 3A). These results suggest that Nb 1-1/2-2 has adopted a Nb 3-2 fate. (2) When an Nb was found at the MP2 position in stage 8, it failed to express Ac (Fig. 2U,V), nuclear Pros and Ftz (not shown), characteristic markers of the wild-type MP2. (3) In 27% (n = 37) of cases in which an Nb was observed at the 3-1/4-1/MP2 position in stage 10 (see above), it expressed Hkb (Fig. 3B), whereas expression of Hkb in rows 3/4 in stage 10 is restricted to the intermediate Nb 4-2 normally (Fig. 3A; Chu-LaGraff et al. 1995). This indicates that NB 3-1/4-1/MP2 has transformed to a NB 4-2-like identity. Indeed, an ectopic neuron expressing Eve and Zfh-1, characteristic markers of the RP2 neuron derived from Nb 4-2, was found slightly anterior and ventral to RP2 in 20% mutant hemisegments (n = 127) (Fig. 4, cf. G and J with H and K). Moreover, we were able to ascertain an Eve-positive ectopic GMC4-2a-like cell ventral to GMC4-2a (cf. Fig. 4, D and E). (4) In early stage 9, svp–lacZ expression was never observed at ventral position 5-2 (Fig. 2C,D). (5) In stage 8/9, Gsb-d expression, observed normally in S1 ventral Nbs 5-2 and 7-1 (Gutjahr et al. 1993), is lacking at those positions in vnd embryos (Fig. 2S,T); however, at later stages, Gsb-d is expressed normally in mutant S3 ventral Nb 6-1 (95%, n = 128). In addition, normal expression of svp–lacZ in position 7-1 in stage 9 was never detected in the mutant (Fig. 2E,F). Taken together, these data show that the residual S1 + S2 ventral Nbs formed in vnd embryos, and probably other late ventral Nbs as suggested for Nb 3-1/4-1, are transformed to fates characteristic of adjacent, intermediate neural precursors.
Figure 3.
Residual ventral Nbs acquire intermediate characteristics in vnd embryos. (A) Ventral expression of svp–lacZ at late stage 9 in row 2/3 is restricted normally to the intermediate Nb 3-2, as seen in the mutant left hemisegment (also see Fig. 2E,F). When a residual Nb at position 1-1/2-2 forms (arrowhead), as seen in the right hemisegment, it may express svp–lacZ abnormally, like its neighbor Nb 3-2. (B) At stage 11, the left hemisegment of a vnd mutant shows normal row 4 Hkb expression in intermediate Nb 4-2 (shifted ventrally) and dorsal Nb 4-4. As seen in the right hemisegment, when a residual ventral Nb forms in row 4 (arrowhead), it may express Hkb abnormally, like its more dorsal neighbors Nbs 4-2 and 4-4.
Absence of vnd modifies gene expression in ventral proneural clusters and NE
Loss of AS-C expression in the proneural clusters of S1 ventral Nbs MP2 and 7-1 in vnd mutant embryos was noted by Skeath et al. (1994). Because vnd expression in the ventral NE precedes Nb formation, we investigated if loss of vnd also influenced other distinctive identity features of the ventral proneural clusters. Indeed, proneural clusters of Nb 1-1, Nb 2-2, and Nb 5-2 failed to express Odd (Fig. 2M,N), hkb–lacZ (Fig. 2G,H), and svp–lacZ (Fig. 2A,B) respectively in vnd embryos. The neuroectodermal expression of Gsb-d in the ventral column is also reduced strongly (Fig. 2Q,R). Therefore, vnd seems to control Nb fates already at the proneural cluster stage.
msh, a homeobox gene implicated in the control of dorsal Nb fate (Isshiki et al. 1997), is not expressed in the ventral NE of the wild type. Initially, msh expression is restricted to the dorsal NE (Fig. 5D; D’Alessio and Frasch 1996; Isshiki et al. 1997); at stage 10, it is also expressed in the putative proneural clusters of some intermediate Nbs (Fig. 2W; Isshiki et al. 1997). In stage 10 vnd embryos, the msh expression domain, determined with the enhancer trap insertion rH96 (Isshiki et al. 1997), is expanded into part of the ventral NE (Fig. 2X).
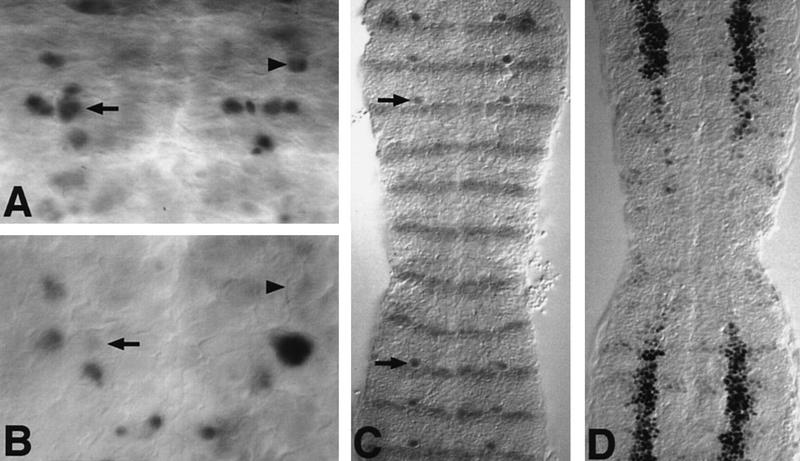
Figure 5.
Ectopic expression of vnd suppresses the identity of intermediate and dorsal NE and Nbs. (A,B) Vnd suppresses Hkb expression in the intermediate and dorsal neuroblasts. (A) In stage 11 wild-type embryos, Hkb protein is strongly expressed by Nb 2-4 (arrowhead), Nb 4-2 (arrow), Nb 4-4, and Nb 5-4. (B) In sca–GAL4; UAS–vnd embryos, the Hkb-positive Nb4-2 and Nb2-4 are missing. Other Hkb-positive Nbs are out of focus and are weakly stained. (C) Kr–GAL4; UAS–vnd embryos double stained for Lbe and Engrailed. Outside the Kr domain, Nb 5-6 expressing Lbe (arrow) is detected just above the Engrailed stripe. Within the Kr domain, Lbe expression in Nb 5-6 is abolished. (D) Kr–GAL4; UAS–vnd embryos carrying a msh–lacZ insertion and stained for β-gal to detect msh expression. Early msh expression in the dorsal column of the NE is abolished within the Kr domain.
Targeted expression of Vnd in the NE rescues early ventral Nbs in vnd embryos
To target Vnd expression using the UAS/GAL4 system (Brand and Perrimon 1993), we generated transformant lines that carry UAS–vnd transgenes using cDNA. First, we verified if UAS–vnd transgene could rescue the ventral Nb formation in the vnd embryos, using Kr–GAL4, which drives expression in T2-A4 parasegments (Kr domain).
In vndD38; Kr–GAL4/+; UAS–vnd/+ embryos, ventral S1 + S2 Nb formation was restored fully within the Kr domain (not shown). In particular, the Nb rescued at the MP2 position always localized Pros in the nucleus, a unique feature of the wild-type MP2 (Spana and Doe 1995); also rescued were the sibling dMP2/vMP2 neurons, as identified with mAb 22C10 (not shown). The complementation of vnd function in the above-mentioned embryos demonstrates that the protein encoded by the known vnd cDNA is sufficient for the ventral Nb formation and likely, as found for MP2, for specification. Two additional drivers that express in the entire NE, but at lower levels (rho–GAL4) or later (sca–GAL4) than Kr–GAL4 were tried. Rescue of Nb formation with rho–GAL4 was variable from embryo to embryo (50%–100%), whereas no rescue was observed with sca–GAL4.
Proneural gene expression is insufficient to promote Nb formation in the absence of vnd
In vnd6 embryos AS-C gene expression in proneural clusters corresponding to ventral Nbs 1-1 and 5-2 is unaffected (Skeath et al. 1994). Nevertheless, these two Nbs fail to delaminate frequently (Fig. 1F), suggesting an additional requirement for vnd in Nb formation, which is distinct from its capacity to control AS-C expression. Because Kr-GAL4-driven vnd expression restores Nb formation, we chose it to drive AS-C expression in vnd embryos. Kr-GAL4-driven expression of either UAS–l′sc or UAS–ac + UAS–sc was insufficient to induce formation of S1 ventral Nbs above the levels found in vnd embryos alone (not shown). On the contrary, the Kr–GAL4/UAS–ac combination could rescue formation of S1 ventral Nbs fully in Df(1)sc19 (ac− sc− l′sc−) embryos (not shown). Likewise, the rho–GAL4/UAS–ac combination, which rescued MP2 formation in Df(1)y3PLsc8R (ac− sc−) embryos very efficiently (Parras et al. 1996), was unable to do so in vnd embryos (not shown). These results confirm that the role of vnd in Nb formation is not solely through the activation of proneural AS-C gene expression.
Targeted ectopic expression of Vnd promotes ventral-like fates in intermediate and dorsal Nbs and in their progeny
To assess if Vnd expression can modify the fate of intermediate and dorsal Nbs, UAS–vnd/Kr–GAL4 or sca–GAL4 embryos were analyzed. Cell-specific staining data collected from these embryos at the earliest stages indicate that in the gain-of-function situation, intermediate and dorsal Nbs exhibit two kinds of changes: (1) acquisition of specific features of ventral Nbs, and (2) suppression of some of their normal characteristics. Furthermore, intermediate and dorsal NE exhibited certain ventral-like features. As a rule, the penetrance of the transformations is not complete, affecting the intermediate Nbs more frequently than the dorsal ones. Also, it is unlikely that the transformations are perfect, although in our analysis transformations of intermediate Nbs in every row were observed and the transformed intermediate Nb progeny exhibited all identity markers tested.
Transformations in Nbs fates in rows 1 and 7 were discerned with the markers Odd and Gsb-d, respectively (Fig. 6A,B,K,L). When Vnd is expressed in the sca domain, the row 1 intermediate Nb 3-2 is Odd-positive (22%, n = 124; Fig. 6B), whereas only Nbs 1-1 (ventral) and 2-5 (dorsal) are normally Odd-positive in row 1 (Fig. 6A). Likewise, the row 7 intermediate Nb 7-2 is Gsb-d-positive (24%, n = 124; Fig. 6L), whereas only the ventral Nb 7-1 is normally Gsb-d-positive in row 7 (Fig. 6K). No ectopic Gsb-d was, however, detected in the dorsal Nb 7-4. Several observations using markers of the Nb 1-1 and 7-1 lineages also indicated dorsal-to-ventral transformations. Eve-staining of stage 10 sca–GAL4; UAS–vnd embryos revealed duplications, and at a much lower frequency triplications, of GMC 1-1a-like and GMC 7-1a-like cells (Fig. 4C). Later, at stage 15, duplications (17.4%, n = 92) and triplications (2%; n = 92) of aCC/pCC neurons were observed using Eve, Zfh-1, and FasII antibodies (Table 1; Fig. 4F,L). Furthermore, Fas II antibodies in Kr–GAL4; UAS–vnd embryos in stage 13 revealed duplications of U motor neurons, normal progeny of Nb 7-1 (Fig. 4I). The Nb that adopts the 1-1 fate could be its neighbor in the intermediate column, Nb 3-2, as it fails to express its characteristic marker svp–lacZ (Fig. 6I,J). Similarly, the Nb acquiring 7-1-like fate could be its neighbor in the intermediate column, Nb 7-2, because ectopic Vnd induces both Ac and Gsb-d expression in a Nb at position 7-2 (cf. Fig. 6E,F and 7K,L; see also below).
When Vnd is ectopically expressed, normal ac expression during the S1 phase in ventral and dorsal NE/Nbs of rows 3 and 7 expands into the intermediate column (Fig. 6C–F). The expansion effect with Kr driver is more pronounced in row 3 (NE: 91%, Nb: 60%) than in row 7 (NE: 70%, Nb: 35%), as determined by ac in situ hybridization.
In line with the preceding result, nuclear Pros localization, a distinctive MP2 feature, can be also seen in stage 9 in row 3/4 in the intermediate Nb 4-2 (24%, n = 170) and at a much lower frequency in the dorsal Nb 3-5 (4%, n = 170), using the Kr–GAL4 driver (Fig. 6G,H). These values are roughly twice as much if scored only in segments A1–A4, where expression of the driver is stronger. However, whereas MP2 expresses nuclear Pros until it divides at the end of stage 10, the fraction of Nbs at 4-2 and 3-5 positions that still expressed nuclear Pros during stage 10 decayed considerably and no 22C10-positive MP2-like neurons could be detected later as progeny of those two Nbs. These results indicate that the expressivity of dorsal-to-ventral transformation of Nb fates in row 3/4 was incomplete.
In Kr–GAL4; UAS–vnd embryos, svp–lacZ is expressed precociously at early stage 9 in intermediate Nb 5-3 (50%) and dorsal Nb 5-6 (10%) compared to the wild type (Fig. 6J). Note that in row 5 only the ventral Nb 5-2 expresses svp at that stage in the wild type (Fig. 6I). This early induction is not seen using sca–GAL4, suggesting that with this driver appropriate Vnd levels to effect the transformation are either not attained or attained too late.
Whereas ectopic Vnd can activate ventral-marker expression in the intermediate Nbs, it can also suppress or delay the normal marker expression in intermediate and dorsal Nbs, like that of Hkb in Nb 4-2 (13%, n = 66) and in Nb 2-4 ( 11%, n = 66) in the sca–GAL4; UAS–vnd embryos (Fig. 5, cf. A and B).
Normal expression of the homeobox gene ladybird early (lbe) in the dorsal S1 Nb 5-6 and its lineage (Jagla et al. 1997; J. Urban, pers. comm.) was abolished by ectopic Vnd expression (Kr driver 100%, n = 240; sca driver 90%, n = 130) (Fig. 5C), although formation and division of a Nb at the 5-6 position was detected by Hb antibody staining (not shown).
Expression of the homeobox gene msh was assayed using the enhancer trap msh–lacZ. Early expression of msh–lacZ in the dorsal NE (Fig. 5D) and subsequently in a region of the intermediate NE (not shown) were both totally suppressed in the Kr domain of Kr–GAL4; UAS–vnd embryos. Therefore, some of the suppression of dorsal specific features may be through suppression of msh.
Figure 6.
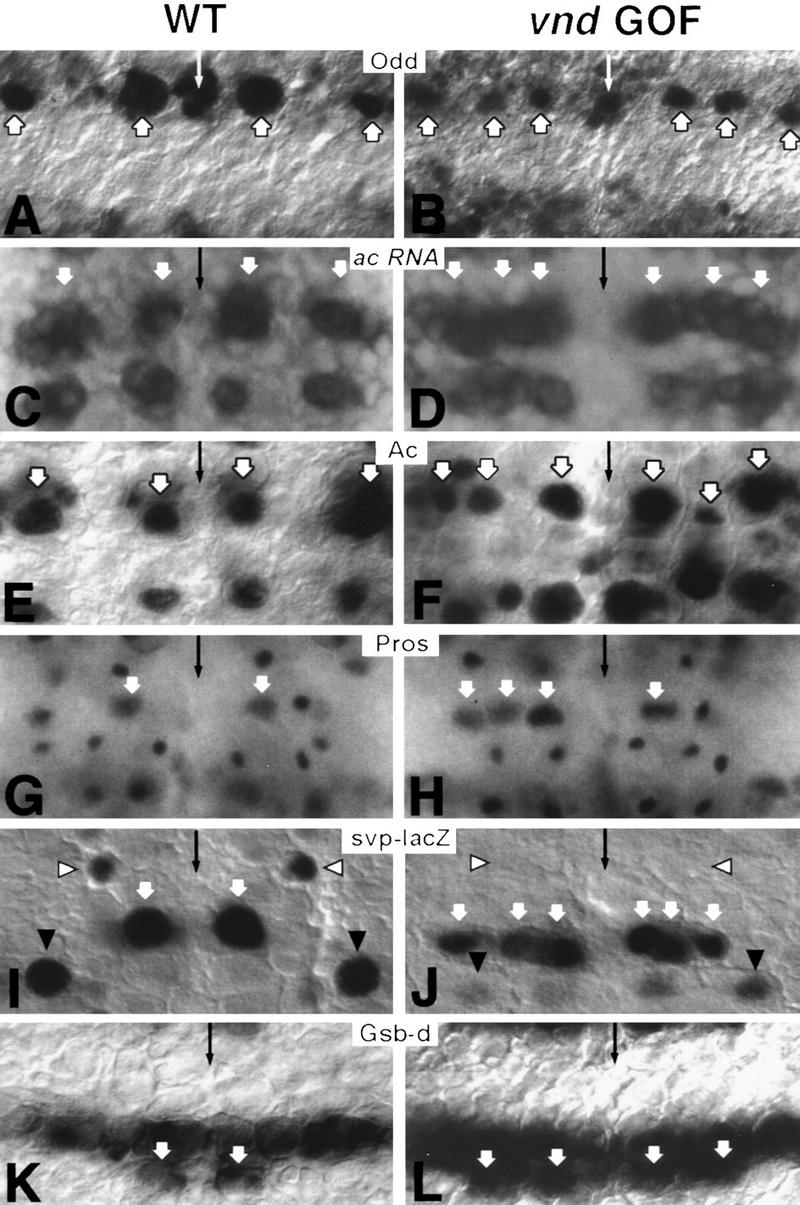
Ectopic expression of vnd promotes ventral identities in NE and Nbs. Molecular markers to analyze the specification of the NE and Nbs include the protein products of odd (A,B), ac (E,F), pros (G,H), gsb-d (K,L), enhancer trap H162 (svp–lacZ; I,J), and ac RNA (C,D). Micrographs from sca–GAL4; UAS–vnd embryos (B,F,L) and Kr–GAL4; UAS–vnd embryos (D,J,H) depict gain-of-function phenotype. A single segment is shown in each case; thin arrows mark the ventral midline; anterior is up. (A) At stage 9, Odd protein is detected normally only at row 1/2 in ventral Nb 1-1 and dorsal Nb 2-5 (thick arrows). (B) Under ectopic Vnd, the intermediate Nb at position 3-2 is also Odd-positive. (C) At stage 8, neuroectodermal ac expression is detected normally in ventral and dorsal proneural clusters of rows 3 and 7, those corresponding to Nbs MP2, 3-5, 7-1, and 7-4, respectively. (E) After Nb delamination, Ac is detected in those four Nbs. White arrows (C,E) point to ventral and dorsal columns. (D) Under ectopic Vnd, ac is expressed additionally in the intermediate column at the level of rows 3 and 7, resulting in a continuous stripe of ac expression in both rows. (F) After Nb delamination, the intermediate Nb in rows 3 and 7 is also Ac-positive. White arrows (D,F) point to ventral, intermediate and dorsal columns. (G) Nuclear accumulation of Pros protein in Nbs is detected normally only in MP2 (white arrow); other Pros-positive smaller nuclei observed in the picture correspond to GMCs. (H) Under ectopic Vnd, nuclear Pros can also be found in intermediate and dorsal Nbs in the same row as MP2, as seen in the left hemisegment (white arrows). (I) In early stage 9 wild-type embryos, svp–lacZ expression is detected in only three Nbs: 3-2 (white arrowhead), 5-2 (white arrow), and 7-4 (black arrowhead). (J) Under ectopic Vnd at the same stage, svp–lacZ expression is abolished in the intermediate Nb 3-2 (white arrowhead), and much reduced in the dorsal Nb 7-4 (black arrowhead). Conversely, svp–lacZ is expressed precociously in intermediate and dorsal row 5 Nbs (5-3 and 5-6), resembling their neighbor ventral Nb 5-2 (white arrows). (K) At stage 9, Gsb-d is expressed by row 7 ventral Nb 7-1 (white arrow) as well as the entire row 5 Nbs (5-2, 5-3, and 5-6). (L) Under ectopic Vnd, two row 7 Nbs (white arrows) are Gsb-d-positive: one in the intermediate column in addition to Nb 7-1.
In addition to the examples that illustrate the acquisition of ventral-like fates and the suppression of normal fates, when Vnd is expressed in the intermediate and dorsal NE, situations that defy simple interpretation were also observed. This is not unexpected, as the positional informational matrix depends on the interplay of complex regulatory networks in which both the concentration and timing of the ectopic expression will influence the final outcome. This point is illustrated in our analysis of the Nb 4-2-like lineage. In stage 15 sca–GAL4; UAS–vnd embryos, RP2 deletions were observed in 16% hemisegments (n = 105; Fig. 4F,L). Interestingly, in these same embryos, ∼47% of the hemisegments exhibited two Eve- and Zfh-1-positive RP2 neurons (Fig. 4F,L). Furthermore, in stage 11 embryos, Eve-positive GMC4-2a-like cell was observed in the same position as the ectopic RP-2-like cell (Fig. 4C). With the Kr driver, the suppression of RP2 is more pronounced and no RP2 duplications were detected (Fig. 4I). Our current interpretation is that high and/or early levels of Vnd suppress Nb 4-2 fate by directly imposing a ventral fate, but somewhat lower and/or later levels of Vnd could cause Nb 4-2 duplications through the suppression of msh or other dorsal regulatory genes. Indeed, duplication of Nb 4-2 is seen in msh loss-of-function phenotype (Isshiki et al. 1997).
Discussion
It is generally accepted that an individual Nb acquires a singular fate based on when and where it forms. The genetic program that distinguishes Nbs is already evident in specific gene-expression patterns in the proneural clusters. Here we discuss our findings that indicate that vnd is indispensable for the formation and specification of the ventral column of early Nbs and relate them to the overall emerging picture on how the Nb field is patterned.
Vnd promotes the formation of early ventral Nbs
A prominent feature of the early Nb map in vnd mutant embryos is the specific and near complete absence of the S1 + S2 ventral Nbs (Fig. 1F). This phenotype indicates an almost absolute requirement for Vnd in the formation of ventral Nbs and correlates nicely with the early-expression domain. Two probable reasons contributed to the underestimation of Nb loss in an earlier study (Jiménez and Campos-Ortega 1990): (1) inclusion of later forming Nbs, and (2) lack of specific markers available at that time.
Results presented in this paper suggest that role of vnd in the formation of vnd- and AS-C-dependent Nbs MP2 and 7-1 is not solely through the activation of AS-C, as restoration of AS-C expression in vnd null embryos provided no rescue. To explain the absence of Nbs 1-1 and 5-2 that are largely AS-C-independent, as they are present in AS-C-deficient embryos (H. Chu et al., unpubl.), and their proneural clusters still express l′sc in vnd mutant embryos (Skeath et al. 1994), we propose a second proneural activity under vnd regulation. That Vnd itself will provide proneural activity is unlikely, because its ectopic expression in imaginal wing discs is unable to promote the formation of extrasensory organs (H. Chu et al., unpubl.), a property common to all known proneural genes (Rodríguez et al. 1990; Jarman et al. 1993; Hinz et al. 1994).
With respect to post-S2 ventral Nbs, we only examined those that delaminate at stage S3 and found that formation of Nbs 3-1 and 4-1, but not that of Nb 6-1, is affected appreciably in the mutant. This incomplete requirement is unexplained currently. Formation of S1 + S2 intermediate and dorsal Nbs is clearly vnd independent and presumably the same applies to later forming Nbs in those two columns, as vnd expression is never observed in the corresponding neuroectodermal domains.
vnd is essential in fate specification of ventral Nbs
Our analysis shows that vnd begins to exert its influence on Nb specification as early as the proneural cluster stage as, in addition to activation of AS-C (Skeath et al. 1994), it also controls activation of two other genes, hkb and svp, in the proneural clusters. In vnd mutant embryos, the residual S1 + S2 ventral Nbs not only did not produce the expected progeny, but exhibited features typical of intermediate Nbs, indicating an absolute requirement of vnd for fate specification of ventral Nbs. A diagram that summarizes altered patterns of gene expression in S1 + S2 Nbs in vnd loss-of-function conditions is shown in Figure 7A. The ventral-to-intermediate Nb fate transformations in the absence of vnd are elicited most likely by derepression in the ventral column of intermediate neuroblasts defective (ind), a gene that positively regulates formation and specification of intermediate Nbs (Weiss et al. 1998).
Figure 7.
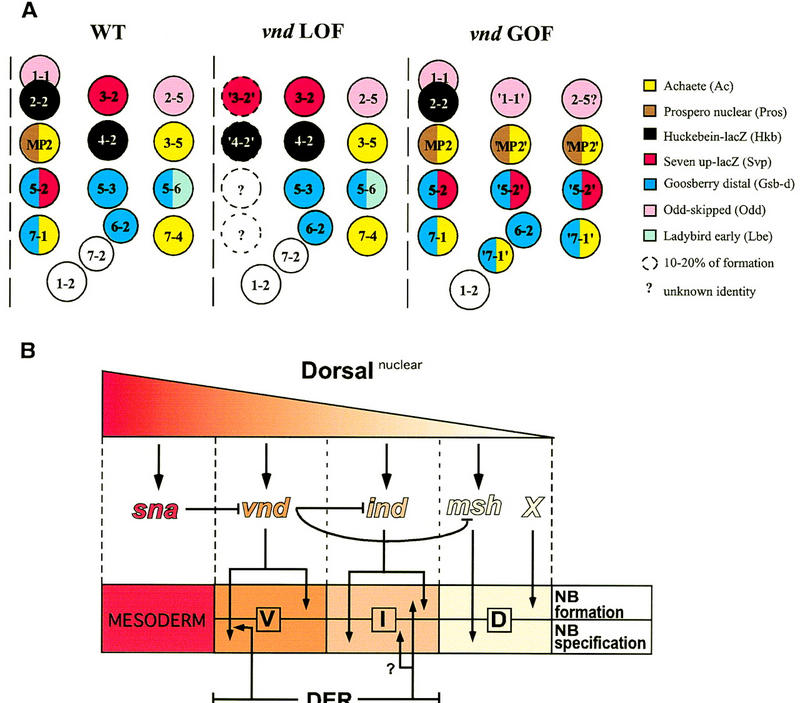
(A) Summary of neuroblast identities according to the panel of molecular markers tested in the wild type, in the vnd null mutant (LOF), and under ectopic expression conditions for vnd (GOF). (B) A model for neuroectodermal patterning along the DV axis. Activation of relevant genes by different nuclear concentrations of the maternal morphogen Dorsal, the inhibitory relationships between those genes, and their action upon formation and/or fate specification of Nbs are indicated. (See text for further details.)
Ectopic expression of Vnd can promote ventral-like transformations
When vnd was expressed along the entire DV extent of the NE, severe perturbations in Nb specification pattern were observed, with the exception of ventral Nbs (Fig. 7A). Early ectopic induction of Vnd resulted in S1 + S2 Nbs in both the intermediate and dorsal columns to adopt gene-expression patterns specific of ventral Nbs (Fig. 6). Instances indicative of dorsal-to-intermediate transformation were also observed. Furthermore, transformations for the intermediate Nbs were observed in all four rows, indicating that ectopic Vnd can effect the dorsal-to-ventral fate switch along the entire A/P axis. These results, together with the loss-of-function phenotypes, indicate that vnd plays a key instructive role in the specification of ventral Nb fates.
Because our criteria for transformations are limited to the expression of cell-type-specific markers in the Nb itself or in its early progeny, it is difficult to judge the full authenticity of the transformation. Only in the case of the MP2 fate, characterized by a single symmetric division, dMP2/vMP2 neurons were not detected at intermediate and dorsal domains of row 3, even though the corresponding Nbs did express several MP2 markers. Incomplete penetrance and expressivity of Nb fate transformation seen in ectopic expression studies may partially lie in the ectopic Vnd delivery system. Thus, transformation frequencies were about twofold higher for the Kr–GAL4 driver compared to the sca–GAL4 driver, suggesting that they could still be improved with an optimal system.
The difference in the efficacy of ventral-fate activation, in the intermediate and dorsal column with similar level of Vnd expression, suggests that regulatory proteins expressed normally in a specific Nb could interfere with the action of ectopic Vnd, and/or that Vnd may not be able to activate the ventral specification pathway fully and additional regulatory factors are necessary. Nevertheless, at least for the intermediate row, sufficient Vnd concentrations can beguile an Nb into a ventral fate.
Drosophila EGF receptor has also been implicated in the fate of early ventral Nbs (Skeath 1998; Udolph et al. 1998), raising two possibilities: Vnd and DER could act in the same pathway or in parallel, but independent pathways. Because DER signals through the conserved RAS pathway (Seger and Krebs 1995), we have coexpressed Vnd and an activated form of Drosophila RAS1 (Queenan et al. 1997) using the Kr–GAL4 driver, in the expectation of improving the dorsal-to-ventral fate transformation. However, using the MP2 identity as paradigm, the frequency of intermediate and dorsal row 3 Nbs that expressed nuclear Pros did not increase with respect to the ectopic expression of Vnd alone (not shown). This lack of an additive effect suggests that Vnd and DER operate in the same pathway and is compatible with the proposal that DER controls ventral Nb fates by modulating Vnd activity through the MAP kinase pathway (Skeath 1998).
Dorsoventral fate specification in the NE
A major insight provided by the cell transplantation studies is that neural precursors from the dorsal ectoderm are more plastic and will assume ventral fates when transplanted to the ventral NE; in contrast, ventral neural precursors tend to retain their ventral fates in dorsal locations (Udolph et al. 1995). Thus, with respect to the ventrally transplanted dorsal neural precursors that generate a ventral-neural clone, we speculate that these cells must respond to cues in the ventral NE that activate vnd transcription. Further, it suggest that the mechanism of vnd activation can be, and perhaps is, non-cell autonomous.
In some respects, msh serves a role similar to vnd in the dorsal NE; its early expression temporally parallels that of vnd and it is necessary for the specification of dorsal Nbs. Moreover, ectopic msh expression disrupts DV pattern specification but, whether it promotes dorsal fates, is not completely clear (Isshiki et al. 1997). In support of this view, we observed that ectopic msh induces lbe expression in intermediate Nb 5-3, in addition to its normal expression in the adjacent dorsal Nb 5-6 (H. Chu et al., unpubl.). Nevertheless, the plasticity of dorsal neural precursors suggest that, unlike Vnd, Msh does not commit cells irreversibly to dorsal fate. In vnd mutants msh expression domain is expanded (Fig. 2X); moreover, ectopic expression of vnd suppresses msh expression, indicating that vnd suppresses msh (Fig. 5D) either directly or indirectly. Thus, with respect to the nonplastic behavior of the dorsally transplanted ventral cells (Udolph et al. 1995), we speculate that vnd expression has committed them to the ventral fates already, and/or it inhibits msh expression that might be activated eventually by nonautonomous signaling mechanism in the dorsal NE.
How is the NE initially patterned along the DV axis? The model we offer is based on several recent studies that have illuminated key players, at or near the top of the regulatory cascades, that controls early Nb formation and/or specification (Isshiki et al. 1997; McDonald et al. 1998; Skeath 1998; Weiss et al. 1998) and DV patterning mechanisms that initially subdivide the NE. The two key presuppositions for this model are that the dorsal encoded morphogen activates at different thresholds a series of genes in consecutive domains in the form of DV stripes, and that expression of these genes is negatively regulated, at least in part, by the more ventrally activated genes. These genes, expressed in nonoverlapping but contiguous DV stripes are vnd in the ventral domain (this work; Jiménez et al. 1995; Mellerick and Nirenberg 1995), rhomboid and vein through their effect upon localized DER activation in the ventral and intermediate domains (Ip et al. 1992; Golembo et al. 1996; Schnepp et al. 1996), ind in the intermediate domain (Weiss et al. 1998) and msh in the dorsal domain (Isshiki et al. 1997), all affecting Nb formation and/or specification. Several pieces of data support the supposition that repression by a more ventrally expressing gene maintains the sharp domain boundaries. Indeed, mutant analysis indicates that msh is repressed ventrally by DER (D’Alessio and Frasch 1996; Skeath 1998), by ind (Weiss et al. 1998), and by vnd (this work); ind is repressed by vnd (McDonald et al. 1998), and vnd and rho are repressed by snail (Ip et al. 1992; Mellerick and Nirenberg 1995), a mesoderm master gene that is expressed in the ventral-most region of the blastoderm. We have not considered in this model the presumptive mesectoderm, a bilateral single cell row placed between mesoderm and ventral NE that gives rise to the ventral midline, because there are discrepancies as to the expression of vnd in it at early stages (Jiménez et al. 1995; Mellerick and Nirenberg 1995). Finally, the existence of an unknown gene has been postulated to control Nb formation in the dorsal NE, as dorsal Nbs form normally in msh mutants.
Evolutionary conservation of NK-2 homoeodomin protein function
Expression domains of the NK2 gene family members from different phyla strongly suggests that their role in the DV patterning of the NE is evolutionary conserved (García-Fernández et al. 1991; Price et al. 1992; Nardelli-Haefliger and Shankland 1993; Saha et al. 1993). Two mouse genes, Nkx2.1 and Nkx2.2, are implicated in the ventral brain development in mouse. Nkx2.1 knockout mice display strong defects in the ventral forebrain (Kimura et al. 1996) consistent with its expression as a longitudinal stripe adjacent to the ventral midline/floor plate (Price et al. 1992; Shimamura et al. 1995), and it has been suggested that it participates in the control of neuronal (ventral motoneurons) identity at caudal levels of the CNS (Ericson et al. 1997).
Materials and methods
Generation and detection of a null vnd mutation
A ry+ transposon, rF124, inserted 5′ to the vnd gene was used to generate independent excisions using standard methods (Robertson et al. 1988), by crossing virgin females carrying the transposon to +/Y; Sb D2-3 ry+/TM6, Tb males. Excisions were screened in the progeny of Fm7a; ry females crossed to dysgenic males of the genotype rF124/Y; Sb D2-3 ry+/TM6. Independent excision chromosomes were tested for complementation to vnd by crossing to vnd5/Balancer chromosome and those that failed to complement were further mapped with respect to the flanking lethal markers elav and l(1)Bg. To test complementation with the nonvital gene Appl, flies of the Appld/excision genotypes were assayed for APPL protein as described previously (Luo et al. 1992).
Ectopic expression by GAL4–UAS system
The GAL4–UAS system (Brand and Perrimon 1993) was used for the ectopic expression of vnd. To generate UAS–vnd construct, the XhoI–XbaI fragment of a vnd cDNA containing the entire open reading frame was inserted into the XhoI–XbaI cut pUAST vector. This construct was introduced into y,w; +/+; Kipp,D2-3/+ embryos by germ-line transformation according standard protocol (Rubin and Spradling 1982). Two independent inserts, UAS–vnd 11 and 21 lines, mapping to X and the second chromosome, respectively, were used. For the rescue of the mutant phenotype of vnd embryos, UAS–vnd 21 and different GAL4 driver lines were used. Ectopic expression of vnd was achieved by crossing either sca–GAL4 or Kr–GAL4 driver lines with the UAS–vnd 11 or 21 line indistinctly.
Immunohistochemistry and in situ hybridization
Antibody staining and dissection of embryos were performed as described (Martín-Bermudo et al. 1991; Patel 1994). For Ac and Hkb antibodies, the RENAISSANCE tyramide-indirect kit (NEN Life Science) was used to amplify the signal following manufacturer’s protocol. The following primary antibodies were used: rat anti-Hb [from P. MacDonald (Jiménez and Campos-Ortega 1990)], rat anti-Hkb (McDonald and Doe 1997), rabbit anti-Eve (Frasch et al. 1987), rat anti-Gsb-d (Zhang et al. 1994), rat anti-Zfh1 (Lai et al. 1991), rabbit anti-Odd (E. Ward and D. Coulter, unpubl.), rabbit anti-βGal (Cappel), and the following mouse monoclonal antibodies: anti-Ac (Skeath and Carroll 1992), anti-Eve 2B8 (Patel et al. 1994), anti-Pros (Spana and Doe 1995), anti-FasII 1D4 (Grenningloh et al. 1991), anti-Lbe (Jagla et al. 1997), anti-En 4D9 (Patel et al. 1989b), 22C10 (Fujita et al. 1982). Mapping of Nbs labeled with some of the above markers was aided by simultaneous staining of the posterior segmental compartment with anti-En antibody.
In situ hybridization to embryos was performed with an ac cDNA probe (Campuzano et al. 1985) or a vnd riboprobe (Jiménez et al. 1995) according to the protocol of Tautz and Pfeifle (1989).
Acknowledgments
We thank J.A. McDonald, J. Urban, J.B. Weiss, and J. Skeath for communicating results before publication. We also thank F.P. Tam, who helped generate vnd line; S. Carrol, E. Coulter, C. Doe, C. Goodman, R. Holmgren, C. Klämbt, T. Orenic, and N. Patel, for flies and antibodies; and E. Dougherty, for help in imaging. C.P. was supported by a predoctoral fellowship from Spanish Dirección General de Enseñanza Superior e Investigación Científica (D.G.E.S.I.C.). This work was supported by grant PB96-0904 from Spanish D.G.E.S.I.C. to F.J., an institutional grant from Fundación Ramón Areces to the Centro Biología Molecular ‘Severo Ochoa’, and a National Institutes of Health grant GM-22350 to K.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL fjimenez@mussol.umh.es; FAX 34-96-5919424.
References
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster .1. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau GM, Doe CQ. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech Dev. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA. Early neurogenesis in Drosophila melanogaster. In: Bate M, Martínez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 1091–1129. [Google Scholar]
- Campuzano S, Carramolino L, Cabrera CV, Ruiz-Gómez M, Villares R, Boronat A, Modolell J. Molecular genetics of the achaete–scute gene complex of D. melanogaster. Cell. 1985;40:327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- Chu-LaGraff Q, Doe CQ. Neuroblast specification and formation regulated by wingless in the Drosophila CNS. Science. 1993;261:1594–1597. doi: 10.1126/science.8372355. [DOI] [PubMed] [Google Scholar]
- Chu-LaGraff Q, Schmid A, Leidel J, Brönner G, Jäckle H, Doe CQ. huckebein specifies aspects of CNS precursor identity required for motoneuron axon pathfinding. Neuron. 1995;15:1041–1051. doi: 10.1016/0896-6273(95)90093-4. [DOI] [PubMed] [Google Scholar]
- D’Alessio M, Frasch M. msh may play a conserved role in dorsoventral patterning of the neuroectoderm and mesoderm. Mech Dev. 1996;58:217–231. doi: 10.1016/s0925-4773(96)00583-7. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita SC, Zipurski SL, Benzer S, Ferrús A, Shotwell SL. Monoclonal antibodies against the Drosophila nervous system. Proc Nat Acad Sci. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Fernández J, Baguñá J, Saló E. Planarian homeobox genes: Cloning, sequence analysis, and expression. Proc Natl Acad Sci. 1991;88:7338–7342. doi: 10.1073/pnas.88.16.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembo M, Raz E, Shilo BZ. The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development. 1996;122:3363–3370. doi: 10.1242/dev.122.11.3363. [DOI] [PubMed] [Google Scholar]
- Grenningloh G, Rehm EJ, Goodman CS. Genetic analysis of growth cone guidance in Drosophila: Fasciclin II functions as a neuronal recognition molecule. Cell. 1991;67:45–57. doi: 10.1016/0092-8674(91)90571-f. [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Patel NH, Li X, Goodman CS, Noll M. Analysis of the goosberry locus in Drosophila embryos: goosberry determines the cuticular pattern and activates goosberry neuro. Development. 1993;118:21–31. doi: 10.1242/dev.118.1.21. [DOI] [PubMed] [Google Scholar]
- Hinz U, Giebel B, Campos-Ortega JA. The basic-helix-loop-helix of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes & Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Takeichi M, Nose A. The role of the msh homeobox gene during Drosophila neurogenesis: Implication for the dorsoventral specification of the neuroectoderm. Development. 1997;124:3099–3109. doi: 10.1242/dev.124.16.3099. [DOI] [PubMed] [Google Scholar]
- Jagla K, Jagla T, Heitzler P, Dretzen G, Bellard F, Bellard M. ladybird, a tandem of homeobox genes that maintain late wingless expression in terminal and dorsal epidermis of the Drosophila embryo. Development. 1997;124:91–100. doi: 10.1242/dev.124.1.91. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jiménez F, Campos-Ortega JA. Genes of the subdivision 1B of the genome of Drosophila melanogaster and their participation in neural development. J Neurogenet. 1987;4:179–200. [PubMed] [Google Scholar]
- Jiménez F, Campos-Ortega JA. Defective neuroblast commitment in mutants of the achaete–scute complex and adjacent genes of D. melanogaster. Neuron. 1990;5:81–89. doi: 10.1016/0896-6273(90)90036-f. [DOI] [PubMed] [Google Scholar]
- Jiménez F, Martin-Morris LE, Velasco L, Chu H, Rosen SJ, DR, White K. vnd, a gene required for early neurogenesis of Drosophila, encodes a homeodomain protein. EMBO J. 1995;14:3487–3495. doi: 10.1002/j.1460-2075.1995.tb07355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Nirenberg M. Drosophila NK-homeobox genes. Proc Natl Acad Sci. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: Thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes & Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Lai Z, Fortini ME, Rubin GM. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev. 1991;34:123–134. doi: 10.1016/0925-4773(91)90049-c. [DOI] [PubMed] [Google Scholar]
- Luo L, Tully T, White K. Human Amyloid precursor protein ameliorates behavioral deficit of flies deleted for Appl gene. Neuron. 1992;9:595–605. doi: 10.1016/0896-6273(92)90024-8. [DOI] [PubMed] [Google Scholar]
- Martín-Bermudo MD, Martínez C, Rodríguez A, Jiménez F. Distribution and function of the lethal of scute gene product during early neurogenesis in Drosophila. Development. 1991;113:445–454. doi: 10.1242/dev.113.2.445. [DOI] [PubMed] [Google Scholar]
- Martin-Morris LE, White K. The Drosophila transcript encoded by the β-amyloid protein precursor-like gene is restricted to the nervous system. Development. 1990;110:185–195. doi: 10.1242/dev.110.1.185. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Saigo K. hedgehog signaling independent of engrailed and wingless required for post-S1 neuroblast formation in Drosophila CNS. Development. 1996;122:3567–3575. doi: 10.1242/dev.122.11.3567. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Doe CQ. Establishing neuroblast-specific gene expression in the Drosophila CNS: Huckebein is activated by Wingless and Hedgehog and repressed by Engrailed and Gooseberry. Development. 1997;124:1079–1087. doi: 10.1242/dev.124.5.1079. [DOI] [PubMed] [Google Scholar]
- McDonald, J.A., S. Holbrook, T. Isshiki, J.B. Weiss, C.Q. Doe, and D. Mellerick. 1998. Dorso-ventral patterning in the Drosophila CNS: The vnd homeobox gene specifies ventral column identity. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Mellerick DM, Nirenberg M. Dorsal-ventral patterning genes restrict NK-2 homeobox gene expression to the ventral half of the central nervous system of Drosophila embryos. Dev Biol. 1995;171:306–316. doi: 10.1006/dbio.1995.1283. [DOI] [PubMed] [Google Scholar]
- Nardelli-Haefliger D, Shankland M. Lox10, a member of the NK-2 homeobox gene class, is expressed in a segmental pattern in the endoderm and in the cephalic nervous system of the leech Helobdella. Development. 1993;118:877–892. doi: 10.1242/dev.118.3.877. [DOI] [PubMed] [Google Scholar]
- Parras C, García-Alonso L, Rodríguez I, Jiménez F. Control of neural precursor specification by proneural genes in the CNS of Drosophila. EMBO J. 1996;15:6394–6399. [PMC free article] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: Practical uses in cell and molecular biology. New York, NY: Academic Press; 1994. pp. 445–487. [DOI] [PubMed] [Google Scholar]
- Patel NH, Schafer B, Goodman CS, Holmgren R. The role of segment polarity genes during Drosophila neurogenesis. Genes & Dev. 1989a;3:890–904. doi: 10.1101/gad.3.6.890. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martín-Blanco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of engrailed proteins in arthropods, annelids, and chordates. Cell. 1989b;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Patel NH, Condron BG, Zinn K. Pair-rule expression patterns of even-skipped are found in both short- and long-germ beetles. Nature. 1994;367:429–434. doi: 10.1038/367429a0. [DOI] [PubMed] [Google Scholar]
- Price M, Lazzaro D, Pohl T, Mattei M-G, Rüther U, Olivo J-C, Duboule D, Di Lauro R. Regional expression of the homeobox gene Nkx-2.2 in the developing mammalian forebrain. Neuron. 1992;8:241–255. doi: 10.1016/0896-6273(92)90291-k. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Ghabrial A, Schüpach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124:3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez I, Hernández R, Modolell J, Ruiz-Gómez M. Competence to develop sensory organs is temporally and spatially regulated in Drosophila epidermal primordia. EMBO J. 1990;9:3583–3592. doi: 10.1002/j.1460-2075.1990.tb07569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Saha SM, Michel RB, Gulding KM, Grainger RM. A Xenopus homeobox gene defines dorsal-ventral domains in the developing brain. Development. 1993;118:193–202. doi: 10.1242/dev.118.1.193. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. 2. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Schnepp B, Grumbling G, Donaldson T, Simcox A. Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes & Dev. 1996;10:2302–2313. doi: 10.1101/gad.10.18.2302. [DOI] [PubMed] [Google Scholar]
- Seger R, Krebs EG. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Skeath JB. The Drosophila EGF receptor controls the formation and specification of neuroblasts along the dorsal-ventral axis of the Drosophila embryo. Development. 1998;17:3301–3312. doi: 10.1242/dev.125.17.3301. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Carroll SB. Regulation of proneural gene expression and cell fate during neuroblast segregation in the Drosophila embryo. Development. 1992;114:939–946. doi: 10.1242/dev.114.4.939. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Panganiban GF, Carroll SB. The ventral nervous system defective gene controls proneural gene expression at two distinct steps during neuroblast formation in Drosophila. Development. 1994;120:1517–1524. doi: 10.1242/dev.120.6.1517. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Zhang Y, Holmgren R, Carroll SB, Doe CQ. Specification of neuroblast identity in the Drosophila embryonic central nervous system by goosberry-distal. Nature. 1995;376:427–430. doi: 10.1038/376427a0. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asimetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Udolph G, Luer K, Bossing T, Technau GM. Commitment of CNS progenitors along the dorsoventral axis of Drosophila neuroectoderm. Science. 1995;269:1278–1281. doi: 10.1126/science.7652576. [DOI] [PubMed] [Google Scholar]
- Udolph G, Urban J, Rüsing G, Lüer K, Technau GM. Differential effects of EGF receptor signalling on neuroblast lineages along the dorsoventral axis of the Drosophila CNS. Development. 1998;17:3291–3299. doi: 10.1242/dev.125.17.3291. [DOI] [PubMed] [Google Scholar]
- Weiss, J.B., T. Von Ohlen, D. Mellerick, G. Dressler, C.Q. Doe, and M.P. Scott. 1998. Dorsal-ventral patterning in the Drosophila central nervous system: The intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- White K, DeCelles NL, Enlow TC. Genetic and developmental analysis of the locus vnd in Drosophila melanogaster. Genetics. 1983;104:433–448. doi: 10.1093/genetics/104.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ungar A, Fresquez C, Holgrem R. Ectopic expression of either Drosophila gooseberry-distal or proximal gene causes alterations of cell fate in the epidermis and central nervous system. Development. 1994;120:1151–1161. doi: 10.1242/dev.120.5.1151. [DOI] [PubMed] [Google Scholar]