Abstract
Salinity and drought have a huge impact on agriculture since there are few areas free of these abiotic stresses and the problem continues to increase. In tomato, the most important horticultural crop worldwide, there are accessions of wild-related species with a high degree of tolerance to salinity and drought. Thus, the finding of insertional mutants with other tolerance levels could lead to the identification and tagging of key genes responsible for abiotic stress tolerance. To this end, we are performing an insertional mutagenesis programme with an enhancer trap in the tomato wild-related species Solanum pennellii. First, we developed an efficient transformation method which has allowed us to generate more than 2,000 T-DNA lines. Next, the collection of S. pennelli T0 lines has been screened in saline or drought conditions and several presumptive mutants have been selected for their salt and drought sensitivity. Moreover, T-DNA lines with expression of the reporter uidA gene in specific organs, such as vascular bundles, trichomes and stomata, which may play key roles in processes related to abiotic stress tolerance, have been identified. Finally, the growth of T-DNA lines in control conditions allowed us the identification of different development mutants. Taking into account that progenies from the lines are being obtained and that the collection of T-DNA lines is going to enlarge progressively due to the high transformation efficiency achieved, there are great possibilities for identifying key genes involved in different tolerance mechanisms to salinity and drought.
Keywords: Insertional mutagenesis, Solanum pennellii, Enhancer trap, Salinity, Drought, Gene tagging
Introduction
Abiotic stress, which includes numerous effects caused by diverse environmental conditions, e.g. drought, salinity and low/high temperatures, have a huge impact on agriculture since there are few areas free of abiotic stresses in which crops can reach their maximum potential. Thus, the maintenance of crop production depends to a large extent on irrigation, but water availability for agricultural purposes is decreasing daily. Today, the scarcity of water does not only affect arid lands, which can only be used for subsistence agriculture, but also affect wide agricultural areas which were highly productive not so long ago. Attempting to maintain the productivity in the latter areas, the farmers are sometimes forced to use low quality water (e.g. with excessive concentrations of Na+, Cl− and other toxic ions) which in the long term leads to a secondary salinization process. Some authors consider that drought affects 45% of the total cultivated surface (Bot et al. 2000) and losses in agricultural production by secondary salinity processes, mainly generated by the use of low quality water and excessive fertilization, are estimated to be around 20% (Ashraf and Harris 2005). Therefore, it is not surprising that great efforts are currently being invested with the aim of increasing the degree of abiotic stress tolerance in species essential for human consumption. However, despite the great efforts to improve the degree of abiotic stress tolerance in species of agronomic interest, the results attained through conventional breeding methods have been rather scarce (Flowers 2004; Cuartero et al. 2006; Ashraf et al. 2008). Moreover, the attempt to overcome this challenge through a transgenic approach has not been as successful as initially expected despite the great number of genes, most of them isolated from model species, which have been transferred and expressed in crop plants (Cuartero et al. 2010). The present situation advices to reconsider what do we actually know on the genetic basis of adaptive mechanisms leading to salinity and drought tolerance and raises the questions of which would it be the most adequate plant material to perform the genetic dissection of these complex traits.
On the basis of studies performed to compare gene expression patterns in salt-tolerant- and salt-sensitive-related species (e.g. Thellungiella halophila and Arabidopsis thaliana), it has been hypothesized that differences in salt tolerance mechanisms between salt-sensitive glycophytes and salt-tolerant halophytes may result from changes in regulation of the same basic set of genes rather than in the presence or absence of particular genes (Gong et al. 2005; Kant et al. 2006; Wong et al. 2006). Flowers and Colmer (2008), after reviewing the tolerance mechanisms in different halophytes, proposed that the future research should be focused on a limited number of ‘model’ halophytes that are representative of the different mechanisms engaged in tolerance. This is in our opinion a sensible proposal, as it would help to unveil some of the adaptive physiological mechanisms leading to abiotic stress tolerance and the genetic basis underlying those mechanisms in tolerant plants. However, as these halophytes are evolutionarily very remote from the main crop species it is unclear if this approach would be useful for breeding purposes. For example, provided that it was possible to identify the subset of genes responsible for the high level of salt tolerance in Thellungiella halophyla it is conceivable that upon transference of those genes we were able to increase the salt tolerance level in its close relative species A. thaliana. However, it is not so clear that the same subset of genes would operate in the same way in tomato, corn, rice or any other crop species. In the same way it is hard to see that the extreme physiological adaptations acquired by the common ice plant Mesembryanthemum crystallinum to thrive in extreme drought conditions could work equally well in a crop species. Actually the problem from a breeding point of view is that the objective is not only to increase the level of tolerance to an abiotic stress but also the quality and yield of the original cultivar are maintained. For all these reasons we think that if the final objective is the genetic improvement of a crop it would be better to take advantage from the genetic variation existing in wild-related species.
The tomato is the main horticultural crop from an economic point of view, but at the same time it could be an excellent model in this sense. Tomato cultivars are usually considered as moderately salt-tolerant (Cuartero et al. 2006) but their level of tolerance is not sufficient to keep yield in areas affected by secondary salinization processes and, furthermore, they do not display a noticeable level of water-deficit stress tolerance. By contrast, there are accessions of wild-related species, both red-fruited (e.g. Solanum cheesmaniae, Solanum pimpinellifolium) and green-fruited (e.g. Solanum pennellii), with a high degree of tolerance to either salinity or drought (Bolarin et al. 1991, 2001); Cano et al. 1996). Moreover, molecular maps derived from interspecific crosses between S. lycopersicum and S. pennellii, S. habrochaites, S. pimpinellifollium, and other tomato wild-related species are also available (Foolad 2007). The comparative study of tomato and wild species can help us to identify key genetic factors involved in domestication, which will benefit breeding programs, as well as genes involved in abiotic stress tolerance.
The priority in this research field continues to be the identification of genes truly involved in the tolerance process (Cuartero et al. 2010). Research on mutants has successfully been applied in model species for identifying genes putatively involved in abiotic stress response mechanisms (Medina et al. 2005; Mowla et al. 2006; An et al. 2007; Indorf et al. 2007; Galbiati et al. 2008; Magnan et al. 2008; Sakamoto et al. 2008). For example, through the identification in Arabidopsis of the salt overly sensitive (sos) mutants and the cloning and characterization of the SOS genes, an important and novel signalling pathway called the SOS pathway, which mediates ion homeostasis, was discovered (Wu et al. 1996; Zhu et al. 1998; Shi et al. 2000, 2002). By comparison, the number of mutants affected in the level of salt tolerance in species other than Arabidopsis which are already available for the scientific community is rather scarce. Occasional spontaneous mutants or, alternatively, those generated by chemical (e.g. EMS) or physical (e.g. X- and γ-rays or fast neutrons) methods could provide the basis for advancing in the knowledge of physiological processes related to salinity and drought tolerance. For example, by analysing tomato salt-hypersensitive (tss) mutants, Borsani et al. (2001) were able to identify two loci: TSS1, which is essential for potassium nutrition and salt tolerance, and TSS2, which plays an important role in the interactions between salt tolerance and abscisic acid signalling. Certainly, after the detection of a spontaneous or induced mutant there is still a long way to cover before the cloning and functional characterization of the target gene. As a matter of fact, in the absence of obvious candidate genes, the isolation of the altered gene in the mutant genome through a positional cloning strategy still requires a huge and drawn out effort. Fortunately, alternative approaches, such as insertional mutagenesis, allow this problem to be overcome, as the tagging of the endogenous gene in an insertional mutant expedites its further isolation. Insertional mutagenesis by T-DNA was first implemented in Arabidopsis due to its ability to admit in planta transformation methods (Clough and Bent 1998). Nowadays, this approach has also been implemented for functional genomic studies in other species, including crops such as rice (Jeong et al. 2002; Chen et al. 2003; Peng et al. 2005; Wang et al. 2005) despite the fact that not a long time ago this important species was considered as recalcitrant for regeneration and transformation in in vitro culture.
Currently, we are performing a programme on insertional mutagenesis of the wild tomato species S. pennellii. The strategy for the identification of genes controlling agronomic interesting traits in tomato and those determining salinity and drought tolerance in the tomato wild-related species is based on the use of gene traps because of the theoretical advantages provided by this system. In this regard, insertional mutagenesis with T-DNA constitutes a powerful genomic tool for the identification of genes and the analysis of their function. The integration of T-DNA within the structural sequence or the regulatory elements of a given gene leads to its disruption and the consequent loss-of-function or, depending on the characteristics of the T-DNA-insert, gain-of-function or alteration of its level of expression (Krysan et al. 1999; Kuromori et al. 2009). Upon detection of the mutant phenotype in T0 (in the case of dominant, semidominant or additive effects) or T1 (recessive) the cloning of the mutated gene is very much facilitated as it is tagged by the T-DNA. In comparison with classical insertional mutagenesis, the trapping systems (Springer 2000; Ramachandran and Sundaresan 2001) can be particularly useful for the identification of genes involved in tolerance to salinity, drought and other abiotic stresses. The advantage of using enhancer, promoter or gene traps resides in their dual nature. Like any other T-DNA, these traps act as insertional mutagens, but also, when the T-DNA is integrated in the appropriate orientation, the reporter gene becomes under the control of the regulatory elements of the tagged endogenous gene. In this way, by analysing the reporter gene expression it is possible to get a precise picture of the spatial and temporal expression pattern of the endogenous gene tagged by the trap. Furthermore, with this strategy there are two alternative ways for identifying genes related to abiotic stress, through the phenotypic effect promoted by the T-DNA insert in stress conditions (e.g. detection of hypersensitive mutants) or through the specific expression of the reporter gene in some plant organs, in both control and stress conditions.
This paper describes the first results of the insertional mutagenesis programme we are performing with S. pennellii in order to identify genes responsible for the high level of tolerance to salinity and drought exhibited by some accessions of this tomato wild-related species. With the aim of collecting a sufficient number of insertional mutants, we first developed an efficient protocol for transformation of this tomato wild-related species which allows a transformation rate close to that attained in model species. Here, we report this high-throughput method as well as the results of the screening of about 2200 T-DNA insertion lines generated with an enhancer trap in salinity and drought conditions.
Materials and methods
Generation of S. pennellii T-DNA enhancer-trap lines
Two accessions of S. pennellii have been used to generate the collection of T-DNA enhancer-trap lines: ‘20164’ (kindly provided by Dr Maria Jose Diez, COMAV, Valencia, Spain) and ‘PE47’, which display a high salt-tolerance level (Bolarin et al. 1991; Alarcon et al. 1993; Santa-Cruz et al. 1999).
The enhancer trap vector used for transformation was pD991 (kindly provided by Dr. Thomas Jack; Department of Biological Sciences, Dartmouth College, USA). This vector contains a cassette consisting of a minimal promoter fused to the bacterial gene uidA that encodes the enzyme β-glucuronidase (GUS). The minimal promoter is a truncated version of the 35S promoter of cauliflower mosaic virus (35SCaMV), which contains a TATA box and transcription start site and that is unable to drive reporter gene expression alone but can be activated by neighbouring enhancer elements (Springer 2000). After integration, enhancer or promoter proximal elements in genomic DNA adjacent to the T-DNA insertion cause an increase in transcription from the −60 promoter, resulting in an increased level of transcription of the uidA gene, often in cell-, tissue-, or organ-specific expression patterns as directed by the adjacent genomic enhancer. These patterns can be detected by staining for GUS activity using the chromogenic substrate 5-bromo-4-chloro-3-indole-β-d-glucuronide (X-gluc) (Campisi et al. 1999).
A method for the acquisition of S. pennellii transgenic plants was previously developed in our laboratory (Gisbert et al. 1999). Several modifications were introduced in order to increase the transformation efficiency and generate a great number of T-DNA lines. First, transgenic buds were obtained in a different organogenic culture medium (SPO) based on a modified MS medium (Murashige and Skoog 1962), containing 4.3 g/l MS salts (Duchefa, Haarlem, Netherlands), 30 g/l sucrose, 1 mg/l thiamine, 100 mg/l myo-inositol, 4 mg/l indole-3-acetic acid (IAA), 4 mg/l kinetin, 1 mg/l zeatin riboside, Shahin vitamins (Shahin 1985), 8 g/l bacteriologic TC agar (Duchefa, Haarlem, Netherlands), pH 5.7 and supplemented with kanamycin (100 mg/l) and timentin (300 mg/l). Leaves from axenic seedlings were used as source of plant explants as described by Gisbert et al. (1999), although the preculture step was removed. Leaf explants were inoculated with the LBA4404 strain of Agrobacterium tumefaciens for 10 min. After infection, leaf explants were co-cultured for 2 days at 23 ± 1°C in the dark. Explants were washed in medium containing cefotaxime (500 mg/l). Explants were maintained in the organogenic culture medium at 23 ± 1°C under a 16 h photoperiod.
Three to four weeks after organogenic response induction, leaf explants formed buds, which were separated as putative individual events and transferred to the same culture medium. After subculture of the buds, the elongated shoots were transferred to rooting culture medium consisting of MS basal medium supplemented with 0.1 mg/l IAA, 100 mg/l kanamycin and 300 mg/l cefotaxime. All plants regenerated from a single poked area of an inoculated leaf explant were considered to be derived from a single independent transformation event. Thus, only one regenerated plant from a single poke was counted as an independent transgenic event to ensure that each regenerate represented an independent transgenic event. The molecular verification of the putative transgenic plants was performed by PCR and the number of T-DNA inserts in selected insertional mutants was determined by Southern blot using the methods described in Gimenez-Caminero et al. (2010). Transformation frequency was estimated as the number of independent transgenic events divided by the total number of inoculated leaf explants, then multiplied by 100.
Ploidy level evaluation and maintenance of diploid S. pennellii T-DNA enhancer-trap lines
In order to select the diploid plants from the T-DNA insertion lines, the ploidy level in transgenic plants was evaluated by DNA quantification in a flow cytometer. Ploidy was determined in young leaf fresh tissue sliced with a razor blade into thin strips, smaller than 0.5 mm, in a glass Petri dish containing 0.4 ml of nuclei isolation buffer (high-resolution DNA kit, solution A: nuclei isolation, Partec, Münster, Germany). The nuclei extract was mixed with 1 ml of staining buffer (high resolution DNA kit, solution B: DAPI staining; Partec, Münster, Germany) and filtered through 50-μm nylon mesh (Nyblot). The filtrates (more than 5,000 nuclei per extract) were analysed using a Partec PA-II flow cytometer. From every diploid transformed plant (T0), several clonal replicates were obtained by culturing axillary buds in rooting medium in order to maintain the in vitro collection as well as acclimatize a sufficient number of replicates in greenhouse to identify dominant insertion mutants and obtain T1 seeds by hand-pollination.
Phenotypic characterization, histochemical GUS assays, and physiological trait analysis in non-stress and abiotic stress conditions
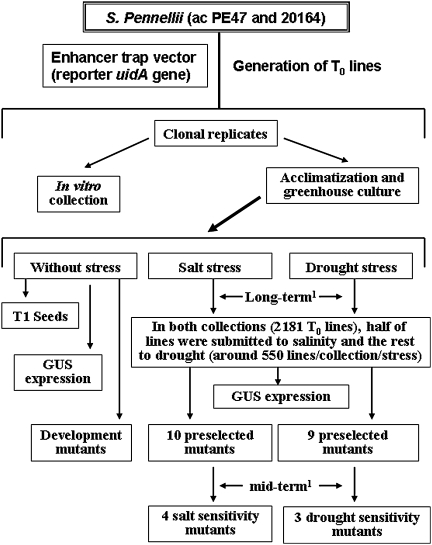
The identification of insertional mutants affected in the tolerance to salinity and drought has been carried out in T0 where mutants with dominant, semi-dominant or additive effects are identified. A summary of the screening strategy of the collections of S. pennellii enhancer trap lines is shown in Fig. 1. Acclimatized plants of each of the T-DNA lines were grown in a greenhouse of 1,000 m2 surface area, equipped with a cooling system to hold the inner temperature down to 24°C. Plants were grown in pots (35 l) filled with coconut fibre, using a drip-irrigation system, with a programmed flow of 3 l/h. The culture practices have been previously described (Estañ et al. 2005).
Fig. 1.
Summary of the screening process of the collections of S. pennellii enhancer trap lines carried out in non-stress and abiotic stress conditions. 1Long-term and mid-term experiments are described in “Materials and methods”
All lines were grown without stress (control conditions) in order to collect T1 seeds, identify mutants affected in vegetative and reproductive development, as well as to select lines with specific expression of the reporter uidA gene in plant organs. Samples of different tissues were collected to carry out histochemical GUS activity assays. The samples were transferred to multiple well plates containing GUS staining solution [100 mM sodium phosphate at pH 7.0, 10 mM ethylenediamine-tetraacetic acid (EDTA), 0.1% Triton X-100, 0.5 mg/ml X-Gluc, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide and 20% methanol] and incubated at 37°C for 20–24 h. The staining solution was replaced with 70% ethanol to remove the chlorophyll. After chlorophyll removal, the tissues were stored in 70% ethanol and examined under a zoom stereomicroscope (MZFLIII; Leica).
Both collections (2181 T0 lines) were also evaluated under salinity or drought conditions. The screening strategy has been to grow in the same greenhouse culture the lines without and with stress, which allowed us to know whether T-DNA lines affected by abiotic stress were or not affected in developmental traits. According to the culture area, it has been necessary to evaluate the 2181 T0 lines in four successive cultures. In each culture, around 550 lines (the half of one collection) were grown to long-term in control and either saline or drought conditions. In this manner, the half of lines of each collection was submitted to salinity and the rest to drought. At least four replicates per treatment were used. The evaluation under salt stress has been carried out at long-term (90 days), and 100 mM NaCl was added to the irrigation solution. Drought sensitivity was also evaluated at long-term (90 days), and the drought stress was imposed by watering plants with 30% of nutrient solution, compared with the volume applied to unstressed controls. In the selected T-DNA lines, plant material was frozen in liquid N2 and stored at 80°C for molecular and physiological characterization, according to previously described methods (Estañ et al. 2005; Gimenez-Caminero et al. 2010). In the preliminary screening, 10 and 9 lines were selected by their salt and drought sensitivity, respectively. The preselected mutants were newly evaluated at mid-term under controlled conditions, as previously described (Estañ et al. 2005). The mutants affected by salt stress were grown at 200 mM NaCl between 15 and 30 days. In the mutants affected by drought, the treatment was applied by withholding irrigation, so water stress intensity increased with the time, and two successive dehydration–rehydration cycles were applied. The salt sensitivity of salt and drought stress was corroborated in four and three mutants, respectively.
Measurements of the photosynthetic rate were made using a portable analyser (Model LI-6400, Li-Cor Inc., Lincoln, NE). Measurements of the chlorophyll were also made using a portable analyser. The water content and the Na+ and K+ concentrations in the leaf tissue were determined as described previously (Estañ et al. 2005). Briefly, a part of this material was weighed and dried further to determine its water content, which was calculated on a dry weight basis. Another part of the plant material was immediately frozen with liquid nitrogen. When the material was thawed, leaf sap was obtained by centrifugation and used for ion analysis.
Results
Development of a high-throughput transformation method for accessions of S. pennellii and generation of T-DNA lines
Previous studies had shown that the accession ‘PE47’ of S. pennellii displayed a high level of salt-tolerance both in vivo (i.e. plants grown in greenhouse conditions under saline stress) and in vitro (e.g. primary and secondary calli as well as excised shoot apices incubated in culture media supplemented with high concentrations of NaCl) (Bolarin et al. 1991; Alarcon et al. 1993; Cano et al. 1996, 1998; Santa-Cruz et al. 1999). A new mid-term trial with different accessions of this tomato wild-related species was performed in order to evaluate their levels of tolerance to salinity conditions. After 30 days of treatment with 150 mM NaCl in the irrigation solution, the accession ‘20164’ was selected as the most tolerant (data not shown). Further experiments indicated that plants from this accession were able to grow and thrive after long periods (up to 4 months) of irrigation with high levels of salinity (200 mM NaCl) (Fig. 2). Interestingly, subsequent trials in greenhouse revealed that plants from this accession also showed a high level of tolerance to water-deficit stress applied by either withholding irrigation or watering volume reduction (data not shown). As a result, the accessions ‘PE47’ and ‘20164’ were chosen for the insertional mutagenesis programme.
Fig. 2.
Salt tolerance in the accession ‘20164’ of S. pennellii. a, b No difference was observed between plants grown in control (a) and stress conditions (b) after irrigation with 200 mM NaCl for 4 months. c No symptoms of wilting, senescence or necrotic spots were seen in adult leaves of plants grown under salt stress (right) as compared with those grown in control conditions (left)
Agrobacterium-mediated transformation with leaf explants from S. pennellii plants grown in axenic conditions allowed the acquisition of transgenic plants of this tomato wild-related species (Gisbert et al. 1999). However, the transformation efficiency with the method previously developed (about 6.1%) was insufficient to tackle a large-scale program on insertional mutagenesis. Thus, several experiments were conducted in order to increase the number of shoot-buds regenerated from leaf explants, shorten or avoid the step of shoot elongation and improve the process of explant inoculation (data not shown). The finding of a more balanced combination of growth regulators in the organogenic culture medium (4 mg/l indole-3-acetic acid, 4 mg/l kinetin, 1 mg/l zeatin riboside) allowed us to establish a more efficient regeneration method (explants from both accessions producing multiple shoot-buds per explant; Fig. 3) with respect to the previous one (average of 4.7 shoot-buds per explant). Moreover, using this procedure the step for shoot elongation did not require the formulation of a new culture medium, whereas it was necessary in the previous one (Gisbert et al. 1999), and the whole regeneration process (including shoot rooting) was greatly shortened as it takes no longer than 80–90 days.
Fig. 3.
A high-throughput transformation method for the generation of S. pennellii T-DNA lines. a Leaf explants developing multiple shoot-buds on kanamycin-free organogenic culture medium. b Absence of growth and organogenesis in non-inoculated explants sown in kanamycin containing culture medium. c–e Inoculated explants developing shoot-buds in selective conditions. Despite the great number of transformation events per inoculated explant only those sufficiently far away to one another were selected to avoid the possibility to recover plants coming from the same transformation event. f Shoot elongation process on kanamycin-containing culture medium. g Collection of T-DNA lines in the growth chamber. h, i Acclimatization and cultivation of T-DNA lines in the greenhouse
The improvement in the regeneration process along with the use of timentin instead of cefotaxime in the organogenic culture medium (Mamidala and Nanna 2009) and some other changes related to the previous stage of leaf explant inoculation (see “Materials and methods”) allowed, after co-cultivation with Agrobacterium tumefaciens, the great majority of explants to develop several shoot-buds per explant on kanamycin-containing medium (Fig. 3c–e). Despite the great number of transformation events per inoculated explant (>5 in 58% of explants), only those sufficiently distant from one another were selected to avoid the possibility of recovering plants coming from the same transformation event (Fig. 3f). For this reason the average of transgenic plants per explant was voluntarily limited to 1.7 and 1.2 for ‘20164’ and ‘PE47’ accessions, respectively. After determining the ploidy level of regenerated plants the actual transformation efficiencies (number of diploid transgenic plants per inoculated explants) proved to be 135 and 102% for ‘20164’ and ‘PE47’ accessions, respectively (Table 1). Interestingly, only 4.2% of escapes were detected in the regenerated plants (21.4% with the previous transformation method) as shoot apices of the great majority of putative transgenics were able to grow and root in selective conditions, while those regenerated in control experiments (i.e. from non-inoculated explants) and those coming from wild-type plants (i.e. plants derived from the original accessions) did not. Several clonal replicates were obtained by culturing axillary buds in rooting medium in order to maintain the in vitro collection (Fig. 3g) as well as acclimatize a sufficient number of replicates and grow in greenhouse (Fig. 3h, i) to identify dominant insertion mutants and obtain T1 seeds by hand-pollination. Moreover, the presence of the nptII marker gene was confirmed by PCR and the number of T-DNA inserts in selected insertional mutants was determined by Southern blot hybridization.
Table 1.
Transformation efficiency in two accessions of Solanum pennellii
| S. pennellii accession | Number of inoculated explants | Total number of transgenic plants (2n + 4n) | Number (%) of 2n transgenic plants | Transformation efficiencya (%) |
|---|---|---|---|---|
| ‘20164’ | 816 | 1,363 | 1,105 (81%) | 135 |
| ‘PE47’ | 1,056 | 1,223 | 1,076 (88%) | 102 |
| Total | 1,872 | 2,586 | 2,181 | 117 |
aThe transformation efficiency is calculated on the basis of the number of diploid plants regenerated per inoculated explant
Screening of the collection of S. pennellii enhancer trap lines in abiotic stress conditions and preliminary identification of hypersensitive mutants to salinity and drought
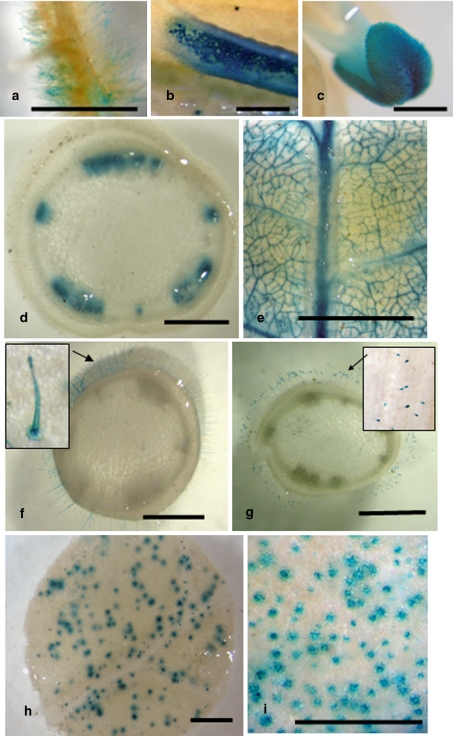
The screening of T-DNA lines was performed either in salinity conditions or in drought stress (see “Materials and methods”). Apart from the search for hypersensitive mutants to salinity or water-deficit stress, we screened the collection of T-DNA lines looking for expression of the reporter uidA gene in specific plant organs. The enhancer trap used to generate the T-DNA lines from S. pennellii plants contains the structural region of the reporter uidA gene with a minimal promoter that does not allow a detectable level of transcription adjacent to the T-DNA right border (Campisi et al. 1999). Thus, when the T-DNA is integrated in the appropriate orientation inside or close to the 3′ end of an endogenous gene, the reporter comes under the control of the regulatory elements of the tagged endogenous gene. Expression of the reporter gene was detected in different vegetative and reproductive organs of plants grown in control conditions of S. pennellii T-DNA lines (Fig. 4a–c) which could be interesting for the study of developmental-related traits. Under abiotic stress, reporter gene expression in transport elements of different organs has been observed in several S. pennellii T-DNA lines (Fig. 4d, e), as well as in trichomes and stomata, as such structures may have relevant roles in maintaining ionic and osmotic homeostasis. Regarding trichomes, ten T-DNA lines with reporter expression in those structures have been selected for further studies (Fig. 4f, g). Three of them were previously selected on the basis of phenotypic analysis due to their visible high density of trichomes. Moreover, in one of them the reporter expression increased when the mutant was submitted to salt treatment (data not shown). Finally, we have also looked for reporter expression in stomata of S. pennellii T-DNA lines, and have identified 8 out of 1,105 trap lines analysed of the S. pennellii acc. ‘20164’ (Fig. 4h, i). Although it is to be expected that the identification of genes specifically expressed in transport elements, trichomes and stomata of S. pennellii T-DNA lines will be helpful in studies related to salinity and/or drought stress, the best way to infer the relevance of a given gene in some of the mechanisms underlying a high level of tolerance in abiotic stress situations is to find that the single alteration of that gene promotes a dramatic drop in the level of tolerance (e.g. a hypersensitive mutant).
Fig. 4.
Reporter gene expression in specific plant organs of different S. pennellii T-DNA lines. a Root hairs. b Pollen grains and anther tissue. c Stigma. Vascular bundles of stem (d) and leaf (e) under stress conditions. f, g Trichomes. Close-ups show reporter expression in all cells of these specialized structures (f) or just in the terminal cell (g). Stomata of leaves (h) and pedicel (i) Bar 1 mm
The phenotypic analysis of the T-DNA lines, either in salinity conditions or drought stress applied to long-term (see “Materials and methods”), enabled the selection of several hypersensitive mutants in either salinity condition (named S olanum p ennellii-s alt-s ensitive mutants, SPss-) or water-deficit stress (called d rought-s ensitive mutants, SPds-). Moreover, in order to know whether the stress sensitivity showed by the preselected mutants was also observed using different stress regimes, new experiments were performed to mid-term. Thus, from the 19 presumptive hypersensitive mutants which were detected in the preliminary screening, 7 (four hypersensitive to salinity and three to water-deficit stress) had cross-validated their sensitivity in both stress regimes, which have been firstly selected for further characterization. One of the mutants selected by its salt sensitivity among the S. pennellii T0 lines proceeding from the accession PE47 is the SPss-2 mutant (Fig. 5a, b), which shows necrosis in the great majority of leaves, from the bottom to the upper part of the plant, after 15 days of 200 mM NaCl treatment. At this time, no visible symptoms of toxicity induced by salinity are observed in WT plants (see close-ups Fig. 5b). At present, we are studying the salt response of the segregant population T1, and the preliminary results suggest that there is a segregation 3:1 of mutant plants (salt sensitive) and plants with WT phenotype, which corresponds to a dominant mutation as observed in T0 plants. Moreover, Southern blot hybridization confirmed that mutant had one T-DNA insert. Regarding sensitive mutants to drought stress, the sensitivity in the SPds-1 mutant is manifested by senescence of leaves from the basal to the upper part of the plant (Fig. 5c, d), ending with the total collapse unless the water supply is re-established, a feature that was not observed in wild-type plants grown in the same conditions (Fig. 5d). It is noteworthy that this mutant exhibited higher expression of the reporter gene in stem vascular bundles as well as in stomata of both leaflet and rachis from plants grown in drought conditions (Fig. 5e–h). As Southern blot hybridization indicated that SPds-1 mutant has four T-DNA inserts, it has been backcrossed with wild-type plants in order to carry out the co-segregation analysis and the identification of the T-DNA insert responsible for the phenotype of hypersensitivity.
Fig. 5.
S. pennellii mutants showing sensitivity to salinity and drought. a, b Plants grown in control conditions (a) and 200 mM NaCl for 15 days (b) of one selected line from ac. PE47, named SP salt sensitive-2; WT plants were unaffected by salt treatment at this time (close-ups), whereas mutant showed necrosis in leaves spreading from the bottom to the upper part of the plant. c, d Drought sensitivity of one selected line from ac. 20164, named SP drought sensitive-1; control plants were well-watered (c) and drought plants were watered with 30% of the nutrient solution used in the control plants for 60 days (d). The hypersensitivity symptoms were manifested by the wilting and senescence of basal leaves, which spread from the basal to the upper part of the plant, whereas WT plants only showed reduced leaf area but not symptoms of senescence (close-ups). Moreover, the reporter gene expression (GUS) was increased by stress; expression in stem vascular bundles and stomata of non-stressed plants (e, f) and drought stressed plants (g, h). Bar 1 mm
Identification of S. pennellii insertional mutants altered in developmental traits
T-DNA lines of S. pennellii were cloned on a regular basis in order to cultivate clonal replicates of each T-DNA line in both control and stress conditions. In this way it was possible to know if a certain phenotype of hypersensitivity was or was not related to a developmental alteration. Moreover, the cultivation of T-DNA lines in control conditions allowed us to look for mutants altered in the developmental traits which could be related to abiotic stress. In this respect, we focused on the search for mutants altered in photosynthetic rate and root development (Fig. 6b, c). In the first mutant, the photosynthetic rate was reduced from 12 (WT) to 4 μmol/m2/s (mutant). The change in photosynthetic rate may serve as a criterion for selection of mutants related to abiotic stress according to the results reported by Carrari et al. (2003) and Nunes-Nesi et al. (2005). Research using these mutants affected in their photosynthetic rate could shed light on the relationship between salt tolerance and ability for Na+ storage in chloroplasts. In this respect, studies by Barrero-Gil et al. (2007) support a major role for chloroplasts in cytoplasmic sodium clearance by means of its accumulation in those organelles, whose volume represents more than half of the total volume of cells in leaf tissue. Alternatively, S. pennellii mutants altered in root development can be envisaged as a complementary way of identifying new ways of adaptation to abiotic stress situations as it is generally assumed that there is a relationship between root mass and ability to thrive in arid soils (Romero-Aranda et al. 2001; Chaves et al. 2002). Other interesting mutants for the genetic dissection of developmental traits have also been identified, like those affected in growth habit, identity of flower whorls and inflorescence architecture (Fig. 6d–f). In this regard, S. pennellii mutants with larger size fruit can be particularly relevant as the identification of genes which are related to, or were selected in, the process of domestication is a relevant issue for both basic research and breeding programs. In the mutant of S. pennellii altered in fruit size, named Sp fruit size-2 and proceeding from ac. 20164, the fruit size is increased around three times (the mean size was 10.6 × 9.2 mm) with respect to the WT fruits (the mean size was 4.1 × 3.9 mm). Southern blot analysis indicated that SPfs-2 mutant has one T-DNA insert. Moreover, segregation 3:1 for fruit size has been observed in T1 progenies, which is associated with the expression level of the reporter gene (data not shown), and homozygous lines are being identified.
Fig. 6.
S. pennellii mutants altered in developmental traits. a Leaves from wild-type plants. b Mutant with chlorophyll-deficient leaves. c Comparison between root development in vitro of a mutant (right) with respect to wild-type plants (left). d Two clonal replicates of an mutant (right) altered in growth habit with respect to that of wild-type plants (left). e Inflorescence architecture of an mutant (right) and that of a wild-type plant (left). f Homeotic conversion of floral whorls in a T-DNA line (down) as compared to normal floral development in wild type plants (up). g, h Fruits of WT (g) and a mutant with larger size fruit (h). Crosses between WT and mutant were carried out using the mutant as donor parental. i On a WT plant, one fruit of WT (left) and one of the hybrid WT × mutant (right), with a fruit size around three times greater than that of the WT
We also detected unexpected phenotypes like that exhibited by the insertional mutant S . p ennellii s ucculent l eaves-1 (SPsl-1) proceeding from the accession PE47 (Fig. 7). The higher chlorophyll content and the succulence of the leaves were clearly observed both in vitro and in vivo (Fig. 7). Thus, the chlorophyll contents were 33.7 ± 2.0 and 54.9 ± 6.9 in the WT and mutant SPsl-1 plants, respectively, grown in vivo. The leaf water content was also lower in the WT than in the mutant SPsl-1 plants (8.9 ± 0.5 and 11.2 ± 0.6 g per g DW, respectively). Moreover, it is interesting to point out that SPsl-1 mutant, which is as tolerant to salinity as wild-type, it seems to use a mechanism of adaptation to salinity more similar to cultivated than wild tomato species, as its leaves accumulate less Na+ and have a lower Na+/K+ ratio than those of wild-type plants after 17 days of 200 mM NaCl treatment (Fig. 7). Thus, studies with this mutant can provide new insights on how to change complex mechanisms which are critical for adaption to unfavourable conditions through the alteration in key genes.
Fig. 7.
Phenotype of the S. pennellii succulent leaves-1 mutant (ac. PE47). The higher chlorophyll content and the succulence of the leaves were clearly observed both in vitro and in vivo. Under salinity (17 days of 200 mM NaCl treatment), the mutant shows lower Na+ accumulation and Na+/K+ ratio in its leaves than WT (M = SPsl-1 mutant)
Discussion
The first prerequisite to tackle a program on insertional mutagenesis by T-DNA is the development of a transformation method allowing the acquisition of a sufficient number of insertion lines. Dan et al. (2006) developed a high-throughput transformation system for Micro-Tom. On the basis of the high transformation efficiency (57%) and the time required (2–3 months), the authors estimated that it was possible to generate about 2,000 T-DNA lines/person/year, a sufficient number to undertake an insertional mutagenesis program with this tomato model genotype. With the method we have developed accessions for S. pennellii; the transformation efficiency is twofold higher (117%) and the process takes no longer than 3 months (Table 1). Therefore, the main limitation to implement a programme of insertional mutagenesis in the wild tomato species has been solved with the high-throughput transformation method. In the near future, this approach based on insertional mutagenesis from tolerant accessions may allow the identification and tagging of genes which may be crucial for the high level of tolerance exhibited by these accessions, for example, those involved in the ionic homeostasis. In this sense, it has been reported that A. thaliana might not be the best choice for understanding how ion transporters involved in long-distance transport work in salt tolerance in crop plants (Essah et al. 2003; Tester and Bacic 2005). Thus, when performing the functional analysis of SlSOS1, the tomato orthologue of AtSOS1 gene, Olias et al. (2009) indicated that one limitation of the A. thaliana in planta physiological characterization is its very small size. In fact, the role of SOS1 in xylem loading and/or unloading, Na+ export by roots and retention in stems was studied in tomato SlSOS1-silenced plants because the very short stem of Arabidopsis plants did not allow a precise dissection of the relative content of Na+ in stem versus leaf. Thus, the previous report that AtSOS1 could be putatively involved in long-distance Na+ transport (Shi et al. 2002) was actually confirmed using tomato for the functional analysis of its corresponding orthologue.
Actually the transformation method is so efficient that the limitations in our program on insertional mutagenesis do not arise because we could generate more than sufficient number of T-DNA lines, but as a consequence of the intrinsic difficulties related to the characterization of plant material. S. pennellii accessions have a growth habit similar to that of tomato cultivars, making the in vivo evaluation a time-consuming process. Despite this we decided to perform the identification of hypersensitive mutants in greenhouse in order to avoid results which could be mere artefacts of unnatural treatments (Munns and Tester 2008), and to examine plant responses under control and abiotic stress conditions in natural growth conditions (Hirayama and Shinozaki 2010). In this manner, we have been able to identify lines with alterations in plant development and lines affected in the tolerance to saline and drought stress, as well as to follow the mutagenesis insertional programme by collecting seeds of T1 progenies. Taking into account that S. pennellii is a facultative allogamous species, thus requiring hand-pollination, to get T1 progenies from T-DNA lines is a labour-consuming process. For this reason until now we have focused on the identification of dominant mutants in T0 plants although it is expected that the number of hypersensitive recessive mutants will be higher in the screening we are performing with T1 progenies. We are also trying to develop an efficient transformation system for the mass generation of Solanum cheesmaniae T-DNA lines as its autogamous nature will obviate hand-pollination. In any case, the very characteristics of wild accessions imply some constraints that are not found when using plant models. In return, their use is advantageous in matters relevant to the final goal of our studies.
The S. pennellii accessions used in this work are well adapted to thrive in unfavourable conditions, but the adaptive mechanisms they have developed with evolution to cope to abiotic stress situations may be very different. Extreme salt and drought tolerance of S. pennellii accessions has been attributed to evolutionary adaptations leading to better protection from reactive oxygen species (Frary et al. 2010 and references therein) as well to improved mechanisms of osmotic adjustment and ionic homeostasis (Bolarin et al. 1995; Santa-Cruz et al. 1999; Perez-Alfocea et al. 2000), although the genetic basis for those adaptations remains unknown. We foresee that the step-by-step in situ genetic dissection of these traits by means of an insertional mutagenesis program in these sources of variation will provide new insights in this field. In this respect, the results on the phenotypic characterization of some presumptive S. pennellii mutants affected in the level of tolerance to either salinity (SPss-2) or drought (SPds-1) suggested that the tagged-genes have a key role in the tolerance to either salinity or drought and that the hypersensitive phenotype is an exclusive consequence of abiotic stress. It is necessary to take into consideration that some identified mutants could not be insertional ones, or well have several inserts as it was observed in rice (Peng et al. 2005). However, independently of the fact that a sensitive mutant was of insertional nature or not, the identification of the altered gene responsible for this sensitivity would continue being interesting. The only difference would be that in the first case the gene is tagged enormously facilitating the cloning, whereas a positional cloning strategy should be envisaged if the T-DNA was not responsible of the mutation, which requires a huge and drawn out effort.
Phenotyping the S. pennellii T-DNA collection has also led us to the selection of different lines with expression of the reporter uidA gene in specific plant organs (Fig. 4). As regards abiotic stress, we mainly focused these assays on the search for reporter gene expression in vascular bundles (root, stem, pedicel and rachis), since they play a key role in processes related to ion transport, as well as in trichomes and stomata. The interest of the mutants with expression in trichomes is based on the assumption that these structures may allow (a) the accumulation of water when plants are faced with a dry environment, and (b) the maintenance of the ionic homeostasis by means of sequestration of toxic ions when plants are grown in a saline ambient (Agarie et al. 2007). More recently, studies on gene expression patterns suggested that trichomes play several roles in the defence against both biotic and abiotic stresses in tobacco (Harada et al. 2010). It is well known that trichomes of S. pennellii are involved in the resistance mechanisms against certain plagues (Simmons et al. 2003; Simmons and Gurr 2005). However, it remains to be determined if any of the different types of S. pennellii trichomes play some role in abiotic stress conditions. Hopefully, the identification and study of several T-DNA lines with specific reporter expression in these specialized structures derived from the epidermal cell layer will shed light on this important issue. On the other hand, water vapour escapes when stomata are open and plants must therefore control stomatal closing to prevent excess water loss. In this respect, the key role that the regulation of stomatal aperture plays in osmotic stress conditions (Chaerle et al. 2005; Roelfsema and Hedrich 2005), which is a common feature of both salinity and drought (Casson and Gray 2008) is well established. For this reason we have looked for reporter expression in stomata, and have identified eight T-DNA lines to date. In the drought sensitivity SPds-1 mutant, it was observed a higher GUS expression in stomata of the drought-treated plants (Fig. 5).
Finally, it is interesting to point out that mutants altered not only in the level of tolerance but also in the mechanism underlying the ability to grow in abiotic stress situations have also been found when lines were grown under non-stress conditions. For example, the S. pennellii mutant succulent leaves-1 (SPsl-1) shows its characteristic phenotype both in stress and in control conditions and (Fig. 7), as an additional difference with respect to the previous mutants, maintains the salt tolerance level displayed by the wild-type plants. Interestingly, preliminary results of the physiological characterization of SPsl-1 mutant suggest that its tolerance mechanism is different to that of the wild-type plants. The original accessions of S. pennellii have developed ionic homeostasis mechanisms, allowing the accumulation of Na+ ions without experiencing the harmful effects that these toxic ions would produce in non-tolerant plants (Cano et al. 1996; Rus et al. 2000). At the same time, the accumulation of Na+ is used for the osmotic adjustment, which is not only efficient but, in addition, is also a less energetic cost-requiring process than that associated with accumulation of osmolytes (Alarcon et al. 1993; Bolarin et al. 2001). Surprisingly, in the SPsl-1 mutant, the levels of Na+ and the ratio Na+/K+ in leaves are significantly lower than in wild-type plants, most probably because of its greater capacity to accumulate water in its succulent leaves (Fig. 7). It is difficult to foresee that this new adaptation to stress conditions could have any practical value (e.g. tomato breeding) because of the profound changes associated with vegetative and reproductive growth. It also needs to be demonstrated whether such dramatic changes are merely the consequence of the alteration in one (or a few) genes as a result of insertion mutations.
Other mutants of this tomato wild-relative detected in control conditions may also be interesting for different purposes. For example, insertional mutants with alterations in vegetative growth, flower and inflorescence architecture and, specially, those affected in fruit size could be relevant for developmental-related studies (Fig. 6). A spontaneous mutant of S. pennellii (Aco1) altered in fruit size has been previously characterized (Carrari et al. 2003). Moreover, two genes involved in tomato fruit size have been isolated using a positional cloning strategy, fw.2.2, the first case in which a quantitative trait locus (QTL) was resolved in the corresponding gene (Frary et al. 2000) and fasciated, which affects tomato fruit size through its effect on locule number (Cong et al. 2008). Unlike what occurs in tomato cultivars, which display an enormous variability in shape and fruit size, accessions of S. pennellii are characterized by the uniformity in both traits, as it occurs in other wild tomato relatives. Thus, the finding of S. pennellii altered in fruit size (e.g. Sp fruit size-2; Fig. 6g–i) may be particularly interesting for the identification of new genes involved in, or related to, those were selected in the process of tomato domestication. In conclusion, the probabilities for identifying key genes involved in different development processes as well as in salinity and drought tolerance are really high enough with the T-DNA collection generated from the wild tomato species S. pennellii, as the integrated knowledge on many processes involved not only in stress responses but also in energy regulation, metabolic processes and developmental processes, are required to produce crops tolerant to abiotic stress and providing high yields. In order to advance in this aim, not only the collection of T-DNA lines is being increased but also homozygous lines of the selected insertion-tagged mutants are being obtained for carrying out the phenotypic, molecular, physiological and genetic characterization of different mutants.
Acknowledgments
This work was supported by the Spanish Ministry of Science and Innovation through Grant No. AGL2009-13388-C03, and by the Council of Science and Technology from the Region of Murcia (Spain) (Fundación SENECA) through grant no. 04553/GERM/06. We thank Dr. Thomas Jack for providing the enhancer trap vector pD991 (Department of Biological Sciences, Dartmouth College, Hanover, NH 03755, USA). We thank Dr. María José Díaz (COMAV-UPV, CPI, Edif. 8E, E-46023, Valencia, Spain) for supplying the accession ‘20164’ of Solanum pennellii.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
A. Atarés and E. Moyano have contributed equally.
References
- Agarie S, Shimoda T, Shimizu Y, et al. Salt tolerance, salt accumulation, and ionic homeostasis in an epidermal bladder-cell-less mutant of the common ice plant Mesembryanthemum crystallinum. J Exp Bot. 2007;58:1957–1967. doi: 10.1093/jxb/erm057. [DOI] [PubMed] [Google Scholar]
- Alarcon JJ, Sanchez-Blanco MJ, Bolarin MC, Torrecillas A. Water relations and osmotic adjustment in Lycopersicon esculentum and L. pennellii during short-term salt exposure and recovery. Physiol Plant. 1993;89:441–447. doi: 10.1111/j.1399-3054.1993.tb05196.x. [DOI] [Google Scholar]
- An R, Chen QJ, Chai MF, Lu PL, Su Z, Qin ZX, Chen J, Wang XC. AtNHX8, a member of the monovalent cation: proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter. Plant J. 2007;49:718–728. doi: 10.1111/j.1365-313X.2006.02990.x. [DOI] [PubMed] [Google Scholar]
- Ashraf M, Harris PJC. Abiotic stresses: plant resistance through breeding and molecular approaches. New York: Haworth Press; 2005. [Google Scholar]
- Ashraf M, Athar HR, Harris RJC, Kwon TR. Some prospective strategies for improving crop salt tolerance. Adv Agron. 2008;97:45–110. doi: 10.1016/S0065-2113(07)00002-8. [DOI] [Google Scholar]
- Barrero-Gil J, Rodriguez-Navarro A, Benito B. Cloning of the PpNHAD1 transporter of Physcomitrella patens, a chloroplast transporter highly conserved in photosynthetic eukaryotic organisms. J Exp Bot. 2007;58:2839–2849. doi: 10.1093/jxb/erm094. [DOI] [PubMed] [Google Scholar]
- Bolarin MC, Fernandez FG, Cruz V, Cuartero J. Salinity tolerance in four wild tomato species using vegetative yield-salinity response curves. J Am Soc Hortic Sci. 1991;116:286–290. [Google Scholar]
- Bolarin MC, Santa-Cruz A, Cayuela E, et al. Short-term solutes change in leaves and roots of cultivated and wild tomato seedlings under salinity. J Plant Physiol. 1995;147:463–468. [Google Scholar]
- Bolarin MC, Estañ MT, Caro M, et al. Relation between tomato fruit growth and fruit osmotic potential under salinity. Plant Sci. 2001;160:1153–1159. doi: 10.1016/S0168-9452(01)00360-0. [DOI] [PubMed] [Google Scholar]
- Borsani O, Cuartero J, Fernandez JA, Valpuesta V, Botella MA. Identification of two loci in tomato reveals distinct mechanisms for salt tolerance. Plant Cell. 2001;13:873–887. doi: 10.1105/tpc.13.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bot AJ, Nachtergaele FO, Young A. Land resource potential and constraints at regional and country levels. World Soil Resources Reports 90. Rome: FAO; 2000. [Google Scholar]
- Campisi L, Yang Y, Yi Y, Heilig E, Herman B, Cassista AJ, Allen DW, Xiang H, Jack T. Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 1999;17:699–707. doi: 10.1046/j.1365-313X.1999.00409.x. [DOI] [PubMed] [Google Scholar]
- Cano EA, Perez-Alfocea F, Moreno V, Bolarin MC. Responses to NaCl stress of cultivated and wild tomato species and their hybrids in callus cultures. Plant Cell Rep. 1996;15:791–794. doi: 10.1007/BF00232231. [DOI] [PubMed] [Google Scholar]
- Cano EA, Perez-Alfocea F, Moreno V, Caro M, Bolarin MC. Evaluation of salt tolerance in cultivated and wild tomato species through in vitro shoot apex culture. Plant Cell Tissue Organ Cult. 1998;53:19–26. doi: 10.1023/A:1006017001146. [DOI] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR. Reduced expression of Aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol. 2003;133:1322–1335. doi: 10.1104/pp.103.026716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S, Gray JE. Influence of environmental factors on stomatal development. New Phytol. 2008;178:9–23. doi: 10.1111/j.1469-8137.2007.02351.x. [DOI] [PubMed] [Google Scholar]
- Chaerle L, Saibo N, van der Straeten D. Tuning the pores: towards engineering plants for improved water use efficiency. Trends Biotechnol. 2005;23:308–315. doi: 10.1016/j.tibtech.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Chaves MM, Pereira JS, Maroco J, et al. How plants cope with water stress in the field. Photosynthesis and growth. Ann Bot. 2002;89:907–916. doi: 10.1093/aob/mcf105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Jin WZ, Wang MY, Zhang F, Zhou J, Jia OJ, Wu YR, Liu FY, Wu P. Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 2003;36:105–113. doi: 10.1046/j.1365-313X.2003.01860.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cong B, Barrero LS, Tanksley SD. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat Genet. 2008;40:800–804. doi: 10.1038/ng.144. [DOI] [PubMed] [Google Scholar]
- Cuartero J, Bolarin MC, Asins MJ, Moreno V. Increasing salt tolerance in the tomato. J Exp Bot. 2006;57:1045–1058. doi: 10.1093/jxb/erj102. [DOI] [PubMed] [Google Scholar]
- Cuartero J, Bolarin MC, Moreno V, Pineda B. Molecular tools for enhancing salinity tolerance in plants. In: Jain SM, Brar DS, editors. Molecular techniques in crop improvement. Berlin: Springer; 2010. pp. 373–405. [Google Scholar]
- Dan YH, Yan H, Munyikwa T, Dong J, Zhang YL, Armstrong CL. MicroTom-a high-throughput model transformation system for functional genomics. Plant Cell Rep. 2006;25:432–441. doi: 10.1007/s00299-005-0084-3. [DOI] [PubMed] [Google Scholar]
- Essah PA, Davenport R, Tester M. Sodium influx and accumulation in Arabidopsis. Plant Physiol. 2003;133:307–318. doi: 10.1104/pp.103.022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estañ MT, Martinez-Rodriguez MM, Perez-Alfocea F, Flowers T, Bolarin MC. Grafting raises the salt tolerance of tomato through limiting the transport of sodium and chloride to the shoot. J Exp Bot. 2005;56:703–712. doi: 10.1093/jxb/eri027. [DOI] [PubMed] [Google Scholar]
- Flowers TJ. Improving crop salt tolerance. J Exp Bot. 2004;55:307–319. doi: 10.1093/jxb/erh003. [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD. Salinity tolerance in halophytes. New Phytol. 2008;179:945–963. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- Foolad MR (2007) Genome mapping and molecular breeding of tomato. Int J Plant Genomics. doi:10.1155/2007/64358 [DOI] [PMC free article] [PubMed]
- Frary A, Nesbitt TC, Frary A, et al. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- Frary A, Gol D, Keles D, et al. Salt tolerance in Solanum pennellii: antioxidant response and related QTL. BMC Plant Biol. 2010;10:58. doi: 10.1186/1471-2229-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati M, Simoni L, Pavesi G, et al. Gene trap lines identify Arabidopsis genes expressed in stomatal guard cells. Plant J. 2008;53:750–762. doi: 10.1111/j.1365-313X.2007.03371.x. [DOI] [PubMed] [Google Scholar]
- Gimenez-Caminero E, Pineda B, Capel J et al (2010) Functional analysis of the Arlequin mutant corroborates the essential role of the arlequin/tagl1 gene during reproductive development of tomato. PLoS ONE. doi:10.1371/journal.pone.0014427 [DOI] [PMC free article] [PubMed]
- Gisbert I, Arrillaga I, Roig LA, Moreno V. Acquisition of a collection of Lycopersicon pennellii (Corr. D’Arcy) transgenic plants with uidA and nptII marker genes. J Hortic Sci Biotechnol. 1999;74:105–109. [Google Scholar]
- Gong QQ, Li PH, Ma SS, Rupassara SI, Bohnert HJ. Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 2005;44:826–839. doi: 10.1111/j.1365-313X.2005.02587.x. [DOI] [PubMed] [Google Scholar]
- Harada E, Kim JA, Meyer AJ, et al. Expression profiling of tobacco leaf trichomes identifies genes for biotic and abiotic stresses. Plant Cell Physiol. 2010;51(10):1627–1637. doi: 10.1093/pcp/pcq118. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Indorf M, Cordero J, Neuhaus G, Rodriguez-Franco M. Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 2007;51:563–574. doi: 10.1111/j.1365-313X.2007.03162.x. [DOI] [PubMed] [Google Scholar]
- Jeong DH, An SY, Kang HG, Moon S, Han JJ, Park S, Lee HS, An KS, An GH. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002;130:1636–1644. doi: 10.1104/pp.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S. Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T-halophila. Plant Cell Environ. 2006;29:1220–1234. doi: 10.1111/j.1365-3040.2006.01502.x. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–2290. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Takahashi S, Kondou Y, Shinozaki K, Matsui M. Phenome analysis in plant species using loss-of-function and gain-of-function mutants. Plant Cell Physiol. 2009;50(7):1215–1231. doi: 10.1093/pcp/pcp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud JP, Aldon D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008;56:575–589. doi: 10.1111/j.1365-313X.2008.03622.x. [DOI] [PubMed] [Google Scholar]
- Mamidala P, Nanna RS. Influence of antibiotics on regeneration efficiency in tomato. Plant Omics. 2009;2:135–140. [Google Scholar]
- Medina J, Rodriguez-Franco M, Penalosa A, Carrascosa MJ, Neuhaus G, Salinas J. Arabidopsis mutants deregulated in RCI2A expression reveal new signaling pathways in abiotic stress responses. Plant J. 2005;42:586–597. doi: 10.1111/j.1365-313X.2005.02400.x. [DOI] [PubMed] [Google Scholar]
- Mowla SB, Cuypers A, Driscoll SP, Kiddle G, Thomson J, Foyer CH, Theodoulou FL. Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J. 2006;48:743–756. doi: 10.1111/j.1365-313X.2006.02911.x. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, et al. Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 2005;137:611–622. doi: 10.1104/pp.104.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olias R, Eljakaoui Z, Li J, De Morales PA, Marin-Manzano MC, Pardo JM, Belver A. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009;32:904–916. doi: 10.1111/j.1365-3040.2009.01971.x. [DOI] [PubMed] [Google Scholar]
- Peng H, Huang HM, Yang YZ, Zhai Y, Wu JX, Huang DF, Lu TG. Functional analysis of GUS expression patterns and T-DNA integration characteristics in rice enhancer trap lines. Plant Sci. 2005;168:1571–1579. doi: 10.1016/j.plantsci.2005.02.011. [DOI] [Google Scholar]
- Perez-Alfocea F, Balibrea E, Alarcon JJ, et al. Composition of xylem and phloem exudates in relation to the salt-tolerance of domestic and wild tomato species. J Plant Physiol. 2000;156:367–374. [Google Scholar]
- Ramachandran S, Sundaresan V. Transposons as tools for functional genomics. Plant Physiol Biochem. 2001;39:243–252. doi: 10.1016/S0981-9428(01)01243-8. [DOI] [Google Scholar]
- Roelfsema MR, Hedrich R. In the light of stomatal opening: new insights into ‘the Watergate’. New Phytol. 2005;167:665–691. doi: 10.1111/j.1469-8137.2005.01460.x. [DOI] [PubMed] [Google Scholar]
- Romero-Aranda R, Soria T, Cuartero J. Tomato plant-water uptake and plant-water relationships under saline growth conditions. Plant Sci. 2001;160:265–272. doi: 10.1016/S0168-9452(00)00388-5. [DOI] [PubMed] [Google Scholar]
- Rus AM, Rios S, Olmos E, et al. Long-term culture modifies the salt responses of callus lines of salt tolerant and salt-sensitive tomato species. J Plant Physiol. 2000;157:413–420. [Google Scholar]
- Sakamoto H, Matsuda O, Iba K. ITN1, a novel gene encoding an ankyrin-repeat protein that affects the ABA-mediated production of reactive oxygen species and is involved in salt-stress tolerance in Arabidopsis thaliana. Plant J. 2008;56:411–422. doi: 10.1111/j.1365-313X.2008.03614.x. [DOI] [PubMed] [Google Scholar]
- Santa-Cruz A, Acosta M, Rus A, Bolarin MC. Short-term salt tolerance mechanisms in differentially salt tolerant tomato species. Plant Physiol Biochem. 1999;37:65–71. doi: 10.1016/S0981-9428(99)80068-0. [DOI] [Google Scholar]
- Shahin EA. Totipotency of tomato protoplasts. Theor and Appl Genet. 1985;69:235–240. doi: 10.1007/BF00662431. [DOI] [PubMed] [Google Scholar]
- Shi HZ, Ishitani M, Kim CS, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi HZ, Xiong LM, Stevenson B, Lu TG, Zhu JK. The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell. 2002;14:575–588. doi: 10.1105/tpc.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons AT, Gurr GM. Trichomes of Lycopersicon species and their hybrids: effects on pests and natural enemies. Agric For Entomol. 2005;7:265–276. doi: 10.1111/j.1461-9555.2005.00271.x. [DOI] [Google Scholar]
- Simmons AT, Gurr GM, McGrath D, Nicol HI, Martin PM. Trichomes of Lycopersicon spp. and their effect on Myzus persicae (Sulzer) (Hemiptera: Aphididae) Aust J Entomol. 2003;42:373–378. doi: 10.1046/j.1440-6055.2003.00376.x. [DOI] [Google Scholar]
- Springer PS. Gene traps: tools for plant development and genomics. Plant Cell. 2000;12:1007–1020. doi: 10.1105/tpc.12.7.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Bacic A. Abiotic stress tolerance in grasses. From model plants to crop plants. Plant Physiol. 2005;137:791–793. doi: 10.1104/pp.104.900138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Xue YB, Li JY. Towards molecular breeding and improvement of rice in China. Trends Plant Sci. 2005;10:610–614. doi: 10.1016/j.tplants.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, et al. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 2006;140:1437–1450. doi: 10.1104/pp.105.070508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Ding L, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK, Liu JP, Xiong LM. Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]