Abstract
Development of yeast meiotic chromosome cores into full-length synaptonemal complexes requires the MEK1 gene product, a meiosis-specific protein kinase homolog. The Mek1 protein associates with meiotic chromosomes and colocalizes with the Red1 protein, which is a component of meiotic chromosome cores. Mek1 and Red1 interact physically in meiotic cells, as demonstrated by coimmunoprecipitation and the two-hybrid protein system. Hop1, another protein associated with meiotic chromosome cores, also interacts with Mek1 but only in the presence of Red1. Red1 displays Mek1-dependent phosphorylation, both in vitro and in vivo, and Mek1 kinase activity is necessary for Mek1 function in vivo. Fluorescent in situ hybridization analysis indicates that Mek1-mediated phosphorylation of Red1 is required for meiotic sister-chromatid cohesion, raising the possibility that cohesion is regulated by protein phosphorylation.
Keywords: Chromosome cores, lateral elements, meiosis, phosphorylation, sister-chromatid cohesion, synaptonemal complex
Meiosis generates haploid gametes from diploid parental cells through two rounds of chromosome segregation following a single round of DNA replication. Specialized interactions between homologous chromosomes—pairing, synapsis, and recombination—occur during meiotic prophase and are essential for the reductional segregation of chromosomes at meiosis I. Each homolog consists of two sister chromatids that remain associated throughout the first meiotic division.
During meiotic prophase, homologous chromosomes become physically associated along their lengths through a proteinaceous structure called the synaptonemal complex (SC; for review, see Roeder 1997). Early in SC assembly, each pair of sister chromatids develops a common protein core, referred to as an axial element. As chromosomes synapse, proteins assemble between the axial elements to form the central region of the SC. Axial elements are called lateral elements in the context of mature complex. The SC extends along the full lengths of chromosomes at pachytene and then disassembles in diplotene.
In Saccharomyces cerevisiae, several proteins associated with the SC have been characterized. The Zip1 protein localizes along the lengths of synapsed chromosomes and is a component of the SC central region (Sym et al. 1993). The Hop1 and Red1 proteins are components of axial and lateral elements (Hollingsworth et al. 1990; Smith and Roeder 1997). red1 mutants fail to form axial elements or SC (Rockmill and Roeder 1990); hop1 mutants assemble axial elements, but these do not synapse (Hollingsworth and Byers 1989; Loidl et al. 1994). The RED1 and HOP1 genes interact in genetic assays (Hollingsworth and Johnson 1993; Friedman et al. 1994; Hollingsworth and Ponte 1997), and the Red1 and Hop1 proteins colocalize on meiotic chromosomes (Smith and Roeder 1997), suggesting that these gene products participate in the same pathway or process.
Characterization of meiotic mutants in budding yeast and other organisms has provided information about the functions of the SC. The results suggest that the primary function of the SC central region is to regulate the distribution of meiotic crossovers along and among chromosomes (Sym and Roeder 1994). The functions of axial and lateral elements are less well defined, but there is some indication that these structures play important roles in meiotic chromosome segregation. Proper reductional segregation at meiosis I depends on the formation of chiasmata, which are chromatin bridges between homologous chromosomes that correspond in position to the sites of genetic crossovers. Chiasma function is presumed to depend on sister-chromatid cohesion; homologs are held together at crossover sites because of cohesion between sister chromatids in regions distal to chiasmata (Miyazaki and Orr-Weaver 1994). The yeast red1 mutant is apparently defective in sisterchromatid cohesion and/or chiasma function because chromosomes that have undergone crossing-over nevertheless fail to segregate correctly (Rockmill and Roeder 1990). The hamster Cor1 protein is another candidate for a lateral element protein that is involved in meiotic sister-chromatid cohesion, as Cor1 localizes to chromosome axes through metaphase I (Moens and Spyropoulos 1995). In fission yeast, the Rec8 protein is necessary both for formation of linear elements (lateral element-like structures) and for meiotic sister-chromatid cohesion (Molnar et al. 1995). Rec8 is homologous to the budding yeast Mcd1/Scc1 protein, which participates in sister-chromatid cohesion during mitosis (Guacci et al. 1997; Michaelis et al. 1997). Thus, sister-chromatid cohesion in mitosis and meiosis may be mechanistically related.
Despite progress in defining the role of SC proteins in sister-chromatid cohesion and meiotic chromosome development, little is known about how these functions are regulated. There is evidence, however, that protein phosphorylation is important. The rat SCP3 protein, a component of lateral elements, undergoes a change in phosphorylation during meiotic prophase (Lammers et al. 1994). The yeast Red1 protein interacts with a phosphatase in the two-hybrid-protein system (Tu et al. 1996). Additionally, the mouse Atm and Atr protein kinases, and mammalian homologs of the fission yeast Chk1 kinase, have been localized to meiotic chromosomes (Keegan et al. 1996; Flaggs et al. 1997).
The S. cerevisiae MEK1/MRE4 gene encodes a protein kinase homolog that is expressed specifically in meiosis. In a mek1 null mutant, meiotic recombination is reduced to ∼20% of the wild-type level (Rockmill and Roeder 1991; Leem and Ogawa 1992). The mek1 mutant forms multiple, short stretches of SC rather than full-length complexes (Rockmill and Roeder 1991). Genetic analysis indicates that MEK1 is in the same epistasis group as RED1 and HOP1 (Rockmill and Roeder 1991), and overproduction studies suggest that Mek1, Red1, and Hop1 interact with a defined stoichiometry (Hollingsworth and Ponte 1997; J.M. Bailis and G.S. Roeder, unpubl.). A Mek1–β-galactosidase fusion protein localizes to the nucleus (Burns et al. 1994), raising the possibility that Mek1 associates with meiotic chromosomes. Although it has been inferred from genetic data that Mek1 interacts with Red1 and Hop1 (Hollingsworth and Ponte 1997), the nature and function of these interactions have not been elucidated. Here, we demonstrate that the wild-type Mek1 protein functions as a kinase that phosphorylates Red1 and associates with both Red1 and Hop1. We propose that interaction of Mek1 with axial and lateral element components promotes SC morphogenesis and establishes meiotic sister-chromatid cohesion.
Results
Mek1 associates with meiotic chromosomes
To gain insight into the function of Mek1, anti-Mek1 antibodies were raised and used to localize the protein within meiotic cells. Meiotic chromosomes that are surface spread and stained with anti-Mek1 antibodies display numerous foci, indicating that Mek1 associates with chromosomes (Fig. 1). This staining pattern is specific for Mek1, as no antibody staining is observed in spread nuclei from a mek1 null mutant (JM82; data not shown).
Figure 1.
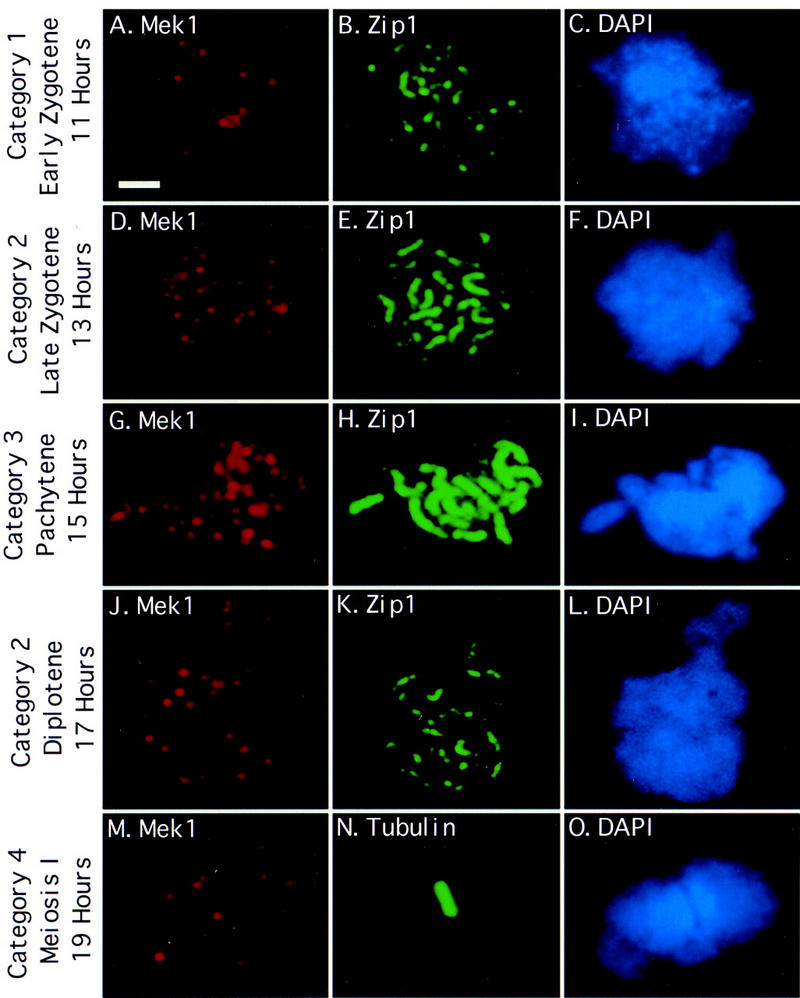
Mek1 localizes to chromosomes during and after synapsis. Spread meiotic chromosomes from wild type (BR2495) were stained with antibodies to Mek1 (A,D,G,J,M), antibodies to Zip1 (B,E,H,K) or tubulin (N), and DAPI (C,F,I,L,O). Categories of Mek1 staining are indicated at left (see text for explanation). Scale bar, 2 μm.
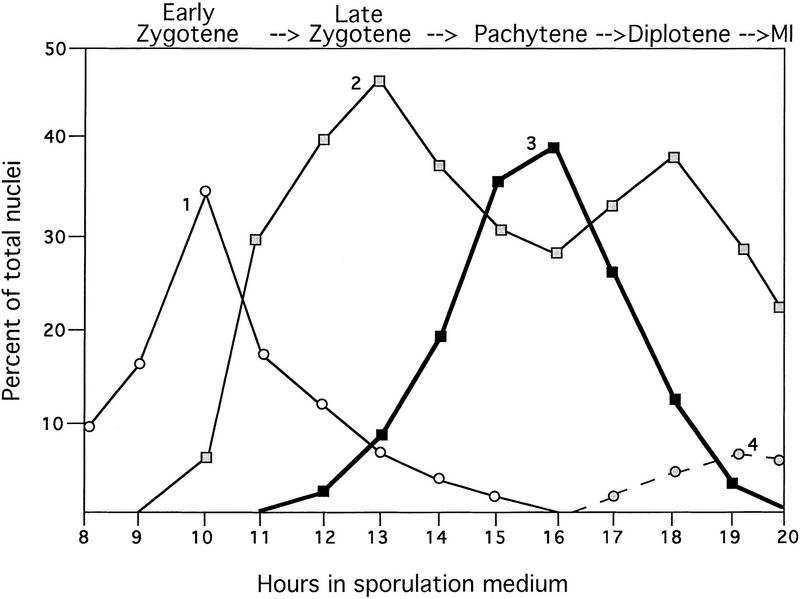
The timing and pattern of Mek1 localization was assessed by staining meiotic chromosomes from wild-type cells (BR2495) with antibodies to both Mek1 and Zip1. Chromosomal DNA was stained with 4′-6-diamino-2-phenolindole (DAPI). Zip1 localization indicates regions where chromosomes are synapsed (Sym et al. 1993; Smith and Roeder 1997). In early zygotene, spread chromosomes display numerous Zip1-staining foci, corresponding to sites of synapsis initiation. Later in zygotene, when chromosomes are partially synapsed, both Zip1 dots and linear stretches of Zip1 staining are apparent. At pachytene, chromosomes are fully condensed and Zip1 localizes continuously along the length of each pair of homologous chromosomes.
A comparison of the Mek1 and Zip1 localization patterns reveals that Mek1 foci first appear on chromosomes in early zygotene, reach a maximum number at pachytene, and disappear from chromosomes sometime after pachytene (Fig. 2). In early zygotene, ∼5%–10% of spread nuclei that display Zip1 foci fail to exhibit Mek1 staining. However, all early zygotene nuclei that display Mek1 foci also exhibit Zip1 foci (category 1, 24 ± 8 Mek1 foci per nucleus; Fig. 1A–C). Nuclei that display Zip1-staining dots and linear stretches contain an average of 30 (±8) Mek1 foci per nucleus (category 2; Fig. 1D–F,J–L). This category peaks at two points (Fig. 2), one before pachytene and the other after pachytene. The early peak is likely to represent nuclei in which chromosomes are partially synapsed (late zygotene); the later peak probably represents nuclei undergoing SC disassembly (diplotene). At pachytene (category 3), there are 40 (±4) Mek1 foci per nucleus (Fig. 1G–I). Surprisingly, Mek1 chromosomal localization persists in spread nuclei from late time points in which little or no Zip1 is detected (category 4, 17 ± 5 Mek1 foci per nucleus; Fig. 1M–O; data not shown).
Figure 2.
Time course analysis of Mek1 and Zip1 localization. Spread nuclei from wild type (BR2495) were prepared hourly during meiosis and double stained with antibodies to Mek1 and Zip1. The frequencies of the four categories of Mek1 localization are indicated; at least 100 nuclei were examined per time point. (Category 1) Early zygotene, punctate Zip1 staining; (category 2) Zip1 dots and linear stretches; first peak, late zygotene; second peak, diplotene; (category 3) pachytene, continuous Zip1 staining; (category 4) meiosis I (MI). The average number of Mek1 foci per nucleus in categories 1–4 is 24, 30, 40 and 17, respectively.
To determine whether Mek1 remains associated with chromosomes during the meiosis I division, spread chromosomes prepared at late time points in meiosis were double labeled with antibodies to Mek1 and tubulin. Mek1 foci are observed in spread nuclei that contain a short meiosis I spindle (Fig. 1M–O) but not in nuclei that have completed meiosis I or at meiosis II (data not shown). These results indicate that Mek1 gradually dissociates from chromosomes during the period from diplotene through metaphase I.
To determine whether Mek1 kinase activity is important for its localization, a conserved residue in the Mek1 kinase domain was changed to create the mek1–D290A mutation. The conserved aspartate residue (D290) is presumed to be the kinase catalytic site, and mutation of this residue in other protein kinases abolishes or significantly decreases protein kinase activity (Zhou and Elledge 1993; Sun et al. 1996). The spore viability of the mek1–D290A mutant is equivalent to that of the mek1::LYS2 null mutant (18.6% vs. 19.2%, 100 tetrads analyzed per strain). The mutant Mek1–D290A protein does, however, localize to chromosomes (Fig. 3M–P); Mek1 therefore localizes to chromosomes independently of its presumed kinase activity. Furthermore, the pattern and timing of localization of the mutant protein are similar to those of the wild-type Mek1 protein.
Figure 3.
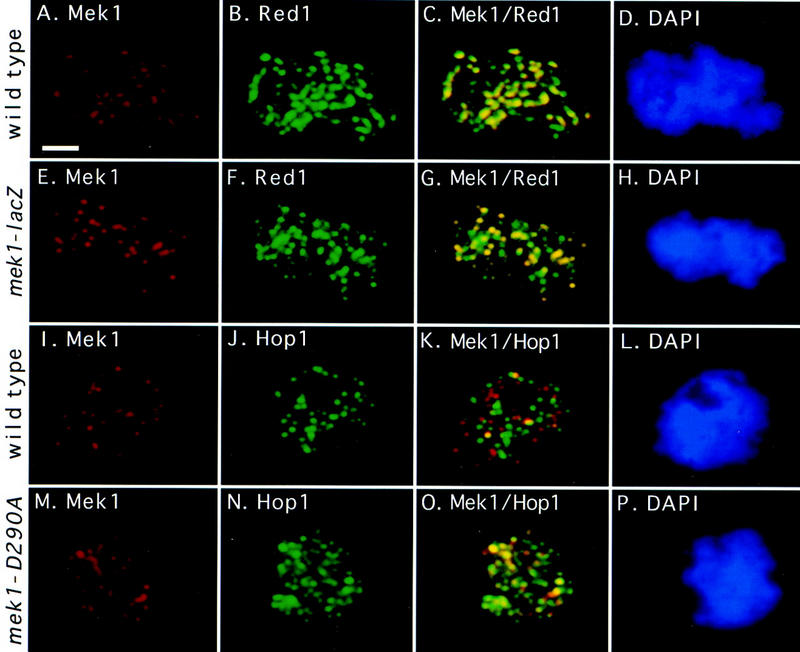
Mek1 localization pattern compared to Red1 and Hop1. A wild-type spread nucleus (BR2495) was stained with antibodies to Mek1 (A) and Red1 (B) and with DAPI (D); (C) fusion of A and B. A spread nucleus from the mek1–lacZ mutant (JM198) was stained with antibodies to β-galactosidase (E) and Red1 (F) and with DAPI (H); (G) fusion of E and F. A wild-type spread nucleus (BR2495) was stained with antibodies to Mek1 (I) and Hop1 (J) and with DAPI (L); (K) fusion of I and J. A spread nucleus from the mek1–D290A mutant (JM191) was stained with antibodies to Mek1 (M) and Hop1 (N) and with DAPI (P); (O) fusion of M and N. All nuclei are from a 13-hr time point. Areas of yellow in the merged images indicate overlap. Scale bar, 2 μm.
The Mek1–β-galactosidase fusion protein (Mek1–β-gal) shown previously to be nuclear (Burns et al. 1994) also localizes to chromosomes with a staining pattern that is indistinguishable from that of the authentic Mek1 protein (Fig. 3E–H). This fusion protein contains only the first 63 amino acids of Mek1 and lacks the entire Mek1 kinase domain, indicating that this region of Mek1 is sufficient for proper localization.
Mek1 colocalizes with Red1 and transiently with Hop1
To test whether Mek1 colocalizes with Red1 and/or Hop1, spread nuclei prepared from several time points in meiosis were double labeled with antibodies to Mek1 and Red1 or Mek1 and Hop1. The Red1 staining pattern is punctate in early prophase and then becomes somewhat continuous by pachytene (Smith and Roeder 1997). Hop1 foci reach a maximum number in early prophase; by late pachytene, Hop1 is largely dissociated from chromosomes (Smith and Roeder 1997). Red1 and Hop1 display extensive colocalization early in prophase, but the number of overlapping foci decreases as cells progress through prophase, and Hop1 dissociates from chromosomes while Red1 accumulates (Smith and Roeder 1997). In the BR2495 strain background, the dissociation of Hop1 from chromosomes correlates with the association of Zip1 with chromosomes and therefore with the formation of mature SC (Smith and Roeder 1997).
At least 95% of Mek1 foci in spread nuclei that are in early zygotene through pachytene colocalize with areas of Red1 staining (Fig. 3A–D). However, there are usually regions of Red1 staining that do not contain Mek1. After pachytene, Mek1 foci are present even though Red1 has mostly dissociated from chromosomes (Smith and Roeder 1997; data not shown). The Mek1–β-gal protein also colocalizes with Red1 (Fig. 3E–H), reinforcing the conclusion that the localization pattern of the Mek1–β-gal protein is similar to that of wild-type Mek1.
In contrast, very few Mek1 foci colocalize with Hop1 foci on spread chromosomes from wild type (Fig. 3I–L). At 11 hr (early zygotene), an average of 8% (range, 5%–27%) of Mek1 foci overlap with Hop1 foci. At 13 hr (late zygotene), an average of 36% (range, 23%–52%) of Mek1 foci overlap with Hop1, whereas an average of 18% overlap (range, 10%–25%) is observed at 14 hr (early pachytene). Fortuitous overlap of Mek1 with Hop1 is estimated at ∼6% or less (Gasior et al. 1998). The amount of overlap between Mek1 and Hop1 varies over time, as Hop1 is already abundant on chromosomes when Mek1 is first detected, and Hop1 is largely dissociated from chromosomes when Mek1 foci are at a maximum. This could be explained if either Mek1 and Hop1 localize to the same foci, but at different times, or if Hop1 associates with some of the Red1-staining foci, while Mek1 associates with other sites of Red1 localization.
Genetic assays support the view that Mek1 and Hop1 interact (Hollingsworth and Ponte 1997; J.M. Bailis and G.S. Roeder, unpubl.), suggesting either that Mek1 and Hop1 interact transiently or Mek1 replaces Hop1 as Hop1 dissociates from chromosomes. To test whether Mek1 kinase activity affects Hop1 dissociation from chromosomes, spread chromosomes from the mek1–D290A mutant were double labeled with antibodies to Mek1 and Hop1 (Fig. 3M–P). Hop1 associates with chromosomes on time in the mutant, but Hop1 dissociation from chromosomes appears to be delayed compared to wild type. In early zygotene (11 hr), similar numbers of Hop1 foci are observed in wild type and mutant (average of 61 foci per nucleus both in wild type and mutant). However, by early pachytene (14 hr), the average number of Hop1 foci per nucleus is 35 in wild type but somewhat higher (44 foci per nucleus) in the mutant. The delay in the dissociation of Hop1 from chromosomes is associated with an increased colocalization of Mek1 with Hop1. In early pachytene, 80% of Mek1 foci overlap with Hop1 in mek1–D290A, whereas just 18% of Mek1 foci on wild-type spread chromosomes overlap with Hop1 foci. This increased colocalization is not due to a delay in meiosis or to an increase in the number of Mek1 foci (data not shown). These results argue that Mek1 and Hop1 localize transiently to the same chromosomal foci and suggest that Mek1 kinase function promotes Hop1 dissociation from meiotic chromosomes.
Mek1 chromosomal localization depends on Red1 and Hop1
To determine whether Red1 and/or Hop1 are required for Mek1 localization to chromosomes, anti-Mek1 antibodies were used to stain spread chromosomes prepared from red1 and hop1 mutants. Both Mek1 and the Mek1–β-gal protein fail to localize to chromosomes in a red1 null mutant and localize to only ∼5% of spread nuclei from a hop1 null mutant (strains MY231, JM93, BR2498, and JM88; data not shown). When whole meiotic cells from the red1 or hop1 mutant are stained with antibodies to Mek1, Mek1 staining is detected in the cytoplasm rather than in the nucleus (data not shown).
The inability to detect Mek1 chromosomal localization in red1 and hop1 strains is not due to instability of the Mek1 protein. Anti-Mek1 immunoblots of whole meiotic cell extracts prepared from red1 or hop1 mutants detect the Mek1 protein in amounts similar to wild type (data not shown). In red1 or hop1 mutants containing the Mek1–β-gal protein, the level of β-galactosidase activity is similar to that of an otherwise wild-type (i.e., RED1 HOP1) strain [72.8 units of β-galactosidase in wild type (JM198), 76.2 units in red1 (JM93), and 74.6 units in hop1 (JM88)].
These data indicate that Red1 and Hop1 are required for Mek1 localization to the nucleus and to chromosomes. Taken together with the requirement of Red1 for Hop1 localization (Smith and Roeder 1997), these observations imply a specific order of protein association: Red1, then Hop1, and finally Mek1.
Red1 and Hop1 coimmunoprecipitate with Mek1
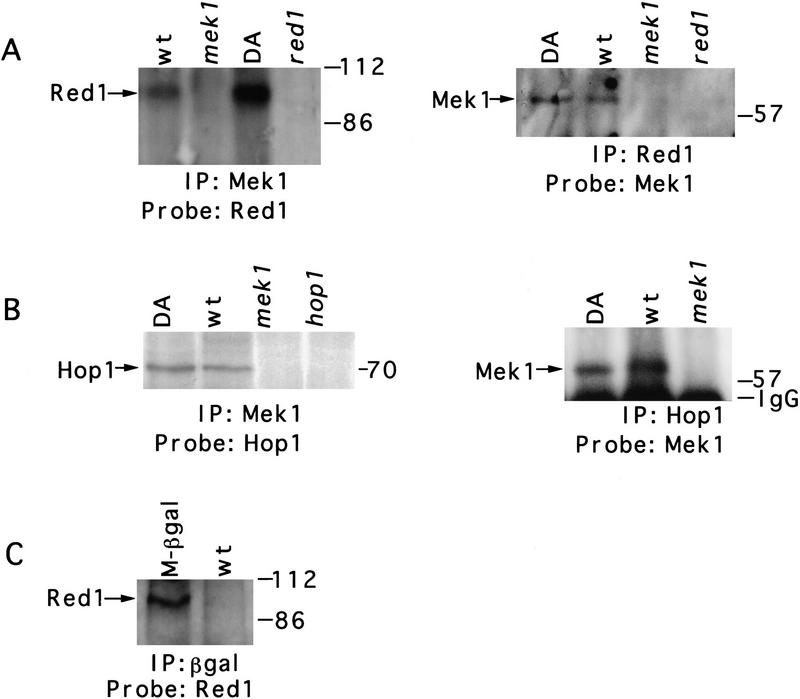
Immunoprecipitation experiments were carried out to determine whether Mek1 physically interacts with Red1 and Hop1. Extracts of sporulating cells were prepared (see Materials and Methods), and Mek1, Red1, or Hop1 was immunoprecipitated with appropriate antibodies and analyzed by SDS-PAGE and immunoblotting. Mek1 and Red1 coprecipitate from wild type (BR2495) and from mek1–D290A (JM191) (Fig. 4A). Mek1 and Hop1 also coimmunoprecipitate from these strains (Fig. 4B). Other proteins coimmunoprecipitate with Mek1 as well, as determined by silver staining of protein gels (data not shown). None of these proteins, including Red1 and Hop1, are detected in immunoprecipitates from a mek1 null mutant or from a wild-type extract immunoprecipitated using preimmune serum (data not shown).
Figure 4.
Association of Mek1 with Red1 and Hop1 in meiotic cells. (A) Anti-Mek1 antibodies coprecipitate Red1, and anti-Red1 antibodies coprecipitate Mek1. (B) Coimmunoprecipitation of Mek1 and Hop1. (C) Red1 coprecipitates with the Mek1–β-gal protein, which contains just the amino-terminal region of Mek1. Proteins were immunoprecipitated from 1 liter of cells that had been sporulated for 13 hr and then analyzed by SDS-PAGE and immunoblotting. Antibodies used for immunoprecipitation (IP) and for immunoblotting (Probe) are indicated on the bottom of each blot. When antibodies to Hop1 are used to coimmunoprecipitate Mek1, as in B, Mek1 is not detected in the hop1 mutant (data not shown). Molecular mass markers are shown at the right of each blot. (IgG) Immunoglobulin G. Strains used are wild type (BR2495), mek1–D290A (JM191), mek1::LYS2 (JM82), red1::URA3 (MY231), hop1::TRP1 (BR2498), and mek1–lacZ (JM198). (wt) Wild type; (DA) mek1–D290A; (M-βgal) Mek1–β-galactosidase.
Colocalization of the Mek1–β-gal protein with Red1 raised the possibility that the amino-terminal region of Mek1 interacts with Red1. To test this possibility, antibodies to β-galactosidase were used to immunoprecipitate Mek1 and associated proteins from meiotic cell extracts prepared from a strain in which the Mek1–β-gal protein is the only source of Mek1 (JM198). Red1 coprecipitates with Mek1–β-gal (Fig. 4C), as does Hop1 (data not shown).
Mek1 interacts with Hop1 in the presence of Red1
The two-hybrid protein system was used to test for a direct interaction of Mek1 with Red1 or Hop1. Mek1 and Red1 do interact in the two-hybrid system, and the first 63 amino acids of Mek1 are sufficient to establish this interaction (Table 1). However, an interaction between Mek1 and Hop1 is not detected in vegetative cells.
Table 1.
Two- and three-hybrid protein interactions
| Strains
|
DNA-binding domaina
|
Activation domain
|
Overproduced
|
β-Galactosidase activity
|
|
|---|---|---|---|---|---|
| vegetative
|
meiotic
|
||||
| Wild type (JM239) | Mek1 aa 1–497 (pJ54) | Red1 (pJ63) | 30.0 | 123.0 | |
| Wild type (JM239) | Mek1 aa 1–100 (pJ61) | Red1 (pJ63) | 26.8 | 91.6 | |
| Wild type (JM239) | Mek1 aa 1–63 (pJ71) | Red1 (pJ63) | 30.5 | 91.5 | |
| Wild type (JM239) | Mek1 aa 1–497 (pJ54) | Hop1 (pJ64) | 1.3 | 93.6 | |
| red1::URA3 (JM240) | Mek1 aa 1–497 (pJ54) | Hop1 (pJ64) | N.D. | 0.2 | |
| Wild type (JM239) | Mek1 aa 1–497 (pJ54) | Hop1 (pJ64) | Red1 (pJ89) | 42.0 | 93.1 |
| Wild type (JM239) | Mek1 aa 1–497 (pJ54) | vector (pACTII) | 1.0 | 0 | |
| Wild type (JM239) | Mek1 aa 1–497 (pJ54) | vector (pACTII) | Red1 (pJ89) | 1.1 | 0 |
| Wild type (JM239) | vector (pBG4D-1) | Red1 (pJ63) | 1.2 | 0 | |
| Wild type (JM239) | vector (pBG4D-1) | Hop1 (pJ64) | 1.2 | 0 | |
Numbers shown are averages obtained from triplicate cultures that were assayed for vegetative and meiotic β-galactosidase activity, measured as moles o-nitrophenyl-β-d-galactoside hydrolyzed per minute per milligram of protein (Durfee et al. 1993). Names of yeast strains and plasmids are indicated in parentheses. (N.D.) Not determined.
(aa) Amino acids.
If the interaction between Mek1 and Hop1 requires Red1, then a Mek1–Hop1 interaction should be detected in meiotic cells (where Red1 is expressed) or in vegetative cells producing Red1. This is indeed the case. A strong interaction between Mek1 and Hop1 is detected in meiotic cells from wild type, but not from a red1 mutant (Table 1). An interaction between Mek1 and Hop1 is also observed in vegetative cells overproducing Red1 (Table1). The greater intensity of interactions observed in meiotic cells compared to vegetative cells may be due to meiosis-specific protein modification.
Mek1 has protein kinase activity
To test whether the Mek1 homology to serine/threonine protein kinases is significant for its function, Mek1 protein kinase activity was examined. Mek1 and associated proteins were immunoprecipitated from meiotic cell extracts, and Mek1 phosphorylation capability was then assayed in vitro.
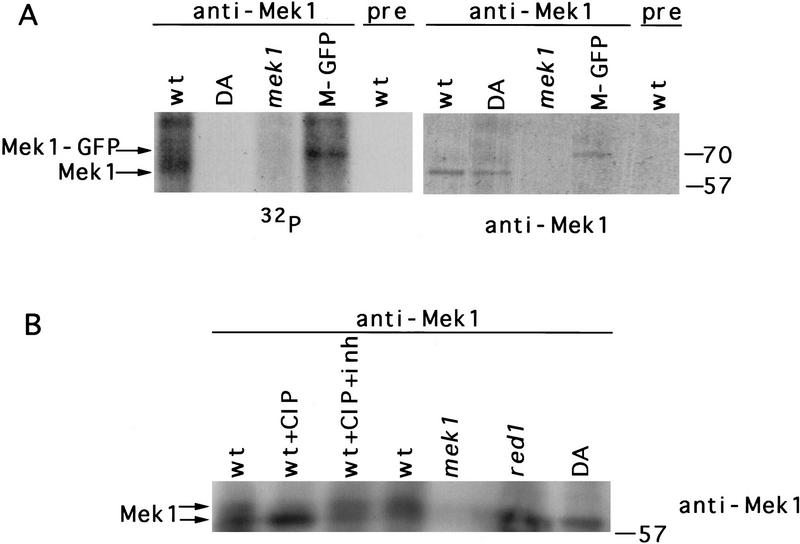
When anti-Mek1 immunoprecipitates are incubated in vitro with [γ-32P]ATP, a phosphorylated band of the approximate molecular mass predicted for Mek1 (∼60 kD) is observed; this band comigrates with the Mek1 protein as determined by immunoblotting (Fig. 5A). Evidence that the phosphorylated band is indeed Mek1 was obtained using a Mek1–green fluorescent protein (Mek1–GFP) fusion as the only source of Mek1. The 75-kD Mek1–GFP also undergoes phosphorylation in vitro (Fig. 5A). Other phosphorylated bands in the autoradiograph are likely to represent other proteins that coprecipitate with Mek1. In contrast to the wild-type Mek1 protein, the mutant Mek1–D290A protein displays little or no phosphorylation in vitro, although immunoblotting indicates that the amount of protein present is similar to that in wild type (Fig. 5A).
Figure 5.
Mek1 is a phosphoprotein that displays kinase activity. (A) Mek1 kinase activity in vitro. Anti-Mek1 immunoprecipitates were incubated with [γ-32P]ATP, separated by an 8% SDS–polyacrylamide gel, and analyzed by autoradiography and immunoblotting with anti-Mek1 antibodies. (B) Gel mobility shift analysis of Mek1. Anti-Mek1 immunoprecipitates were untreated, treated with CIP, or treated with CIP plus phosphatase inhibitor (CIP + inh). Conditions of electrophoresis (20 V for 16 hr) were slower than those used in A. The Mek1 protein was detected by immunoblotting. Molecular mass markers (in kD) are at right. Strains used are wild type (BR2495), mek1–D290A (JM191), mek1:: LYS2 (JM82), MEK1::GFP (JM98), and red1 (MY231). (wt) Wild type; (DA) mek1–D290A; (M-GFP) Mek1–GFP; (pre) preimmune serum.
A gel mobility shift assay was used to examine whether the Mek1 protein is itself phosphorylated in vivo. Mek1 that is immunoprecipitated from wild-type meiotic cell extracts is present in two mobility forms (Fig. 5B). The slower migrating form is due to protein phosphorylation, as treatment with calf intestinal alkaline phosphatase (CIP) converts most Mek1 to the faster migrating form (Fig. 5B). Only the faster migrating form of Mek1 is present in the mek1–D290A mutant, suggesting that Mek1 kinase activity is required for Mek1 phosphorylation in vivo.
Red1 displays Mek1-dependent phosphorylation
Because Mek1 exhibits protein kinase activity and Mek1 interacts with Red1 and Hop1, Mek1-dependent phosphorylation of Red1 and Hop1 was tested. When Red1 and Hop1 are precipitated from wild-type meiotic cell extracts with antibodies to Red1 or to Hop1, respectively, these proteins undergo phosphorylation in vitro (Fig. 6A,B). This phosphorylation depends on Mek1, as little or no phosphorylation of Red1 or Hop1 is observed in immunoprecipitates from mek1 mutants. Mek1-dependent phosphorylation of Red1 and Hop1 in vitro is also observed if anti-Mek1 antibodies are used to immunoprecipitate Red1 and Hop1 (data not shown).
Figure 6.
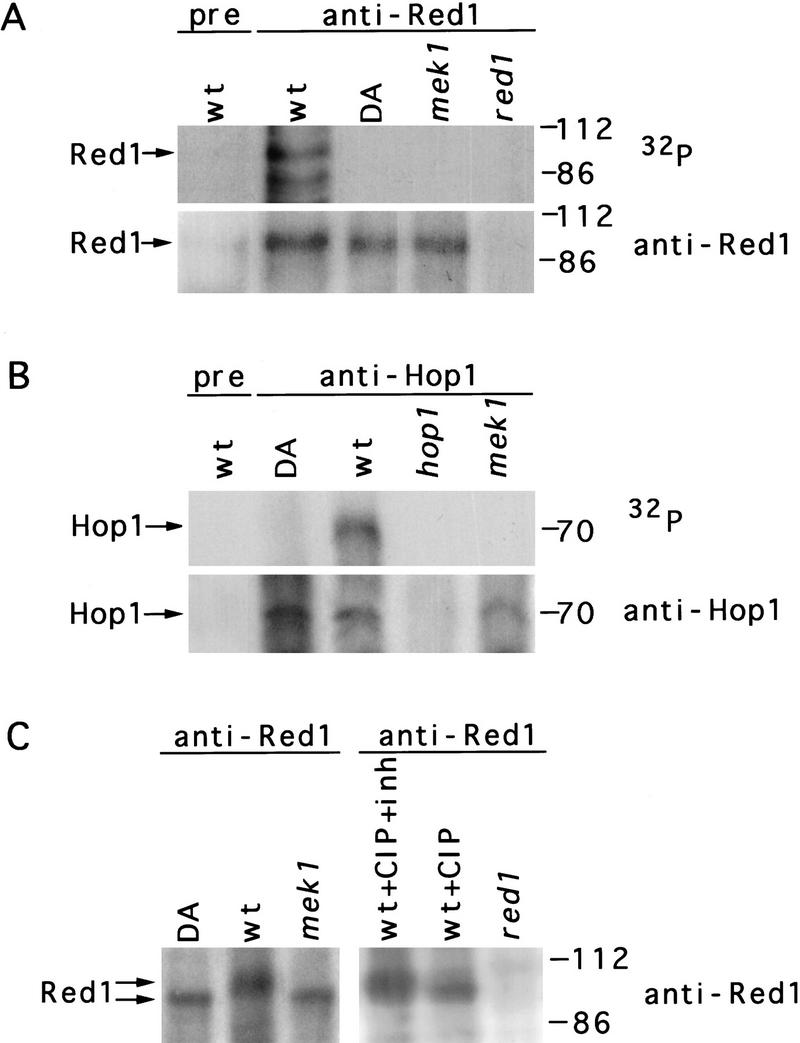
Mek1-dependent phosphorylation of Red1. (A) In vitro phosphorylation of Red1 protein immunoprecipitated with anti-Red1 antibodies. (B) In vitro phosphorylation of Hop1 immunoprecipitated with antibodies to Hop1. Immunoprecipitates were incubated with [γ-32P]ATP, then analyzed by SDS-PAGE, autoradiography, and immunoblotting with antibodies to either Red1 (A) or Hop1 (B). Additional phosphorylated bands may represent other proteins in the immune complex. (C) Gel mobility shift analysis of Red1. Anti-Red1 immunoprecipitates were untreated, treated with CIP, or treated with CIP plus inhibitor (CIP + inh). Immunoprecipitates were analyzed by slower conditions of electrophoresis (20 V for 16 hr) than used in A. The Red1 protein was detected by immunoblotting. Molecular mass markers (in kD) are at right. Strains used are wild type (BR2495), mek1–D290A (JM191), mek1::LYS2 (JM82), and red1::URA3 (MY231). (wt) Wild type; (DA) mek1–D290A; (pre) preimmune serum.
To confirm that the proteins phosphorylated in vitro are indeed Red1 and Hop1, these proteins were tagged with a hemagglutinin (HA) epitope to generate Red1–HA and Hop1–HA fusion proteins. When Red1 is immunoprecipitated from a strain heterozygous for the RED1–HA fusion gene, both the band presumed to represent Red1 and an additional band of slightly greater molecular mass are observed on the autoradiograph and on the immunoblot. This extra band is recognized by both anti-HA antibodies and anti-Red1 antibodies, indicating that it is the Red1–HA protein (data not shown). Similar results were obtained with Hop1–HA (data not shown).
Gel mobility shift assays were employed to assess Red1 phosphorylation in vivo. When Red1 protein is immunoprecipitated from meiotic cell extracts using anti-Red1 antibodies, two mobility forms of Red1 are observed (Fig. 6C). The slower migrating form is due to phosphorylation, as treatment with CIP converts much of the Red1 protein to the faster migrating form (Fig. 6C). In mek1 mutants, only the faster migrating form of Red1 is observed (Fig. 6C). Thus, Red1 is a phosphoprotein that displays Mek1-dependent phosphorylation in vivo.
Although Mek1-dependent phosphorylation of Hop1 is observed in vitro, no obvious gel mobility shift of Hop1 was detected in vivo (data not shown). It is possible that Hop1 is nonspecifically phosphorylated in vitro but this phosphorylation is not biologically relevant.
Mek1 kinase activity is required for sister-chromatid cohesion
One function proposed for Red1 is meiotic sister-chromatid cohesion (Rockmill and Roeder 1990; Smith and Roeder 1997). To determine whether this is the case and whether Mek1 contributes to sister-chromatid cohesion, precocious separation of sister chromatids was analyzed on spread pachytene chromosomes by fluorescent in situ hybridization (FISH) (Table 2).
Table 2.
FISH analysis of meiotic sister-chromatid cohesion
| Strain
|
Class A 1 •
|
Class B doublet ••
|
Class C 2 • •
|
Class D doublet + 1 •• •
|
Class E 3 • • •
|
Percent PSSC
|
|---|---|---|---|---|---|---|
| Wild type (BR2495) | 74.0 | 13.0 | 13.0 | 0 | 0 | 0 |
| mek1Δ (JM82) | 48.0 | 14.0 | 19.0 | 13.0 | 6.0 | 19.0 |
| mek1–D290A (JM191) | 49.0 | 17.0 | 17.5 | 15.0 | 2.0 | 17.0 |
| red1Δ (MY231) | 38.5 | 8.5 | 31.5 | 11.0 | 10.5 | 21.5 |
| hop1Δ (BR2498) | 49.0 | 12.0 | 36.0 | 2.0 | 1.0 | 3.0 |
| mek1Δ red1Δ (JM235) | 56.5 | 9.0 | 16.0 | 11.0 | 7.5 | 18.5 |
Spread meiotic nuclei were prepared after 15 hr in sporulation medium and then hybridized with a probe for chromosome III. Two hundred nuclei were scored for the number and position of condensed FISH signals; the percentage observed for each class is shown (see text for explanation). The percentage of precocious separation of sister chromatids (PSSC) was calculated as the sum of class D and class E nuclei divided by the total number of nuclei scored.
In a wild-type pachytene nucleus, chromosomes are homologously paired and sister chromatids are tightly associated, resulting in a single FISH signal (class A). The presence of a doublet (class B) indicates either that homologs are paired, but not yet fused, or that homologs have started to separate during meiosis I. During and after the meiosis I division, but before meiosis II, two separate signals (class C) are observed. Class C may also represent prophase nuclei in which homologs have failed to pair. Mutants that display defects in homolog pairing, such as red1 and hop1 (Nag et al. 1995), are expected to show an increase in class C nuclei, even if sister chromatids remain tightly associated throughout meiosis I.
Two novel classes of FISH signals are observed in red1 and mek1 mutants and rarely in hop1. Class D nuclei exhibit a doublet plus an additional signal, whereas class E nuclei contain three separate signals. In both classes, one pair of sister chromatids has separated precociously. The increase in class D and E nuclei in mek1 and red1 indicates that these mutants suffer defects in sister-chromatid cohesion (Table 2). The mek1 red1 double mutant displays a defect in cohesion similar in magnitude to each of the single mutants, indicating that Mek1 and Red1 affect sister-chromatid cohesion in the same way. These observations raise the possibility that Mek1-dependent phosphorylation of Red1 establishes meiotic sister-chromatid cohesion.
Discussion
Mek1 interacts with components of meiotic chromosomes
We have demonstrated that the Mek1 kinase localizes to meiotic chromosomes and specifically promotes morphogenesis of the SC and meiotic sister-chromatid cohesion. The colocalization of Mek1 with Red1 and the dependency of Mek1 localization on both Red1 and Hop1 imply that Mek1 associates with axial elements and SC lateral elements. Mek1 chromosomal localization, like that of Red1 and Hop1, does not depend on the initiation of meiotic recombination or the formation of mature SC (J.M. Bailis and G.S. Roeder, unpubl.).
Mek1 and Red1 can associate in the absence of Hop1 in the two-hybrid system, and Mek1 and Red1 both localize to the nucleolus, where Hop1 is not present (Smith and Roeder 1997; unpubl.). Also, the hop1 mutant displays only a minor defect in sister-chromatid cohesion, as expected if Mek1-dependent phosphorylation of Red1 is important for cohesion, and Mek1 and Red1 can interact in the absence of Hop1. Two observations, however, suggest that Hop1 normally stabilizes the association of Mek1 with Red1. First, Mek1 localization to chromosomes is detected in only a small fraction of nuclei from the hop1 mutant. Second, anti-Mek1 antibodies appear to precipitate less Red1 protein from a hop1 mutant than from wild type (J.M. Bailis and G.S. Roeder, unpubl.).
Interaction between Mek1 and Hop1 is detected only if Red1 is also present. Hop1, like Mek1, is dependent on Red1 for its localization to chromosomes (Smith and Roeder 1997). The localization requirements of Hop1 and Mek1 support a specific order of protein assembly: Red1, then Hop1, and finally Mek1. These proteins may associate with each other on chromosomes, or they may first assemble into a complex that then localizes to chromosomes. The order of protein assembly is consistent with the severity of the SC defects observed in the mutants. red1 fails to form any SC or axial elements (Rockmill and Roeder 1990), whereas hop1 forms axial elements that do not synapse (Hollingsworth and Byers 1989; Loidl et al. 1994). mek1 undergoes extensive SC formation (Rockmill and Roeder 1991).
Experiments presented here indicate that the first 63 amino acids of Mek1 are sufficient to interact with Red1. The Mek1 amino terminus contains a forkhead-associated homology domain (FHA) (amino acids 47–102; Hofmann and Bucher 1995). FHA domains are also present in the yeast mitotic protein kinases Rad53 and Dun1 (Zhou and Elledge 1993; Sun et al. 1996, 1998); the interaction of the FHA2 domain of Rad53 with Rad9 (Sun et al. 1998) suggests that this motif serves as a protein-binding domain. However, the first 63 amino acids of Mek1 do not contain the whole FHA domain; only one of three conserved sequence motifs within the FHA domain is present in this region of Mek1.
Much as Rad53 and Dun1 are involved in sensing DNA damage and transducing signals during vegetative growth (Zhou and Elledge 1993; Sun et al. 1996), the interaction between Mek1 and meiotic chromosomal proteins may function to sense and/or transduce signals about the progress of synapsis and/or meiotic recombination (Xu et al. 1997). Consistent with this hypothesis, a red1 or a mek1 mutation restores sporulation to mutants that arrest at pachytene due to a checkpoint triggered by defects in recombination and/or synapsis (Xu et al. 1997). Mek1 kinase function is required to effect this pachytene checkpoint, as the mek1–D290A mutation also restores sporulation to zip1 (J.M. Bailis and G.S. Roeder, unpubl.). Xu et al. (1997) have suggested that the chromosomal context of Mek1 and Red1 is important for monitoring meiotic recombination; this context may be established through Mek1-dependent phosphorylation of Red1.
Hollingsworth and Ponte (1997) have proposed that phosphorylation of Hop1 or Red1 by Mek1 promotes SC assembly. In a specific non-null mek1 mutant (mek1-974), overproduction of Red1 decreases recombination and viability, whereas overproduction of Hop1 improves recombination and viability. These observations led to the speculation that there is an excess of Red1 relative to Hop1 on chromosomes in the mek1 mutant compared to wild type. This hypothesis is inconsistent with our observation that Hop1 appears to assemble onto chromosomes normally in the absence of Mek1 but does not dissociate efficiently. However, our observations can be reconciled with those of Hollingsworth and Ponte (1997). We have found that Red1 overproduction decreases recombination in wild type and in a zip1 mutant and also decreases spore viability in zip1 (J.M. Bailis, A.V. Smith, and G.S. Roeder, in prep.), which strongly suggests that the effect of Red1 overproduction observed by Hollingsworth and Ponte (1997) is not specific to the mek1-974 mutant. If the mek1-974 mutant exhibits decreased Mek1 kinase activity, then overproduction of Hop1 might create more opportunities for Mek1 to phosphorylate Red1 and/or Hop1 by increasing the number of Red1–Hop1 complexes available as substrates. Alternatively, the Mek1-974 mutant protein might be defective in interacting with Red1 and/or Hop1, and overproduction of Hop1 might promote Mek1 binding by increasing the number of Red1–Hop1 complexes available.
Mek1 function depends on its kinase activity
Mek1 protein kinase activity is necessary for its meiotic function, as the mek1–D290A mutant displays a reduction in spore viability equivalent to that of the mek1 null mutant. The simplest explanation of these results is that Mek1 directly phosphorylates Red1; however, an indirect interaction cannot be ruled out. It is possible that other protein kinases coprecipitate with Mek1 and that Mek1, like its mitotic counterparts, functions as part of a protein kinase cascade. Also, it is not yet clear what signal serves to trigger Mek1 phosphorylation/activation, or whether this signaling requires other protein kinases. In many cases, kinases require phosphorylation or autophosphorylation to translocate to the nucleus (for review, see Jans 1995). However, Mek1 localizes to chromosomes in both the mek1–lacZ and mek1–D290A mutants (which lack kinase activity), suggesting that phosphorylation of Mek1 (and Red1) need not occur until after Mek1 becomes associated with chromosomes. The fact that Mek1 is not phosphorylated in a red1 mutant (J.M. Bailis and G.S. Roeder, unpubl.) is consistent with this hypothesis.
Mek1 may promote Hop1 dissociation from chromosomes
Hop1 remains associated with chromosomes longer in mek1 mutants than in wild type, suggesting that Mek1 kinase activity aids in Hop1 dissociation from meiotic chromosomes, an event that is correlated with the production of mature SC (Smith and Roeder 1997). Because Hop1 is required to promote interhomolog recombination (Hollingsworth and Byers 1989; Schwacha and Kleckner 1994), and Hop1 demonstrates DNA-binding capability (Hollingsworth et al. 1990; Kironmai et al. 1998), it is possible that Hop1 directly associates with DNA undergoing recombination. The interaction of Mek1 with Red1 and/or Hop1 might release Hop1 from the DNA, allowing recombination events to proceed and synapsis to be completed. This interpretation is consistent with evidence that Mek1 monitors meiotic recombination (Xu et al. 1997) and that Hop1 dissociation from DNA is slow in an vitro assay from which Mek1 is absent (Kironmai et al. 1998). However, Hop1 does eventually dissociate from chromosomes in the absence of Mek1, indicating that Mek1 kinase activity is not a prerequisite for Hop1 release from chromosomes.
Mek1-dependent phosphorylation of Red1 may establish meiotic sister-chromatid cohesion
FISH analyses of precocious separation of sister chromatids provide evidence that Red1 plays a role in meiotic sister-chromatid cohesion and suggests that phosphorylation of Red1 by Mek1 regulates this function. Meiotic sister-chromatid cohesion may involve the phosphorylation of SC components in organisms other than yeast. The hamster Cor1 protein is a lateral element component that is presumed to play a role in sister-chromatid cohesion (Moens and Spyropoulos 1995), and the rat homolog of Cor1, SCP3, has been shown to be a phosphoprotein (Lammers et al. 1994). Additionally, two forms of the yeast Mcd1/Scc1 protein are detected by Western blots, suggesting that regulation of mitotic cohesion may also involve protein phosphorylation (Guacci et al. 1997).
It is interesting to note that a defect in sister-chromatid cohesion is associated with a decrease in meiotic recombination, suggesting a functional relationship between cohesion and recombination. In the red1 and mek1 mutants, recombination is reduced to ∼20% of the wild-type level (Rockmill and Roeder 1990, 1991; Leem and Ogawa 1992). Drosophila ord mutants and Schizosaccharomyces pombe rec8 mutants also display defects in both recombination and meiotic sister-chromatid cohesion (Miyazaki and Orr-Weaver 1994; Molnar et al. 1995). Recombination might be reduced if cells fail to discriminate sisters from nonsisters and therefore fail to favor interhomolog (i.e., nonsister) interactions (Smith and Roeder 1997). Alternatively, failure to establish sister-chromatid cohesion may result in chromosomal axes that are developed improperly and therefore serve as less suitable substrates for the initiation of interhomolog recombination (Schwacha and Kleckner 1997; Xu et al. 1997). On the other hand, it is possible that recombination contributes to cohesion. Consistent with this possibility, insertion of a recombination hot spot into a yeast artificial chromosome enhances sister-chromatid cohesion (Sears et al. 1994). Also, a defect in sister-chromatid cohesion is observed in the dmc1 mutant (Rockmill and Roeder 1994), which lacks a meiosis-specific homolog of the RecA strand exchange enzyme (Bishop et al. 1992).
Our proposal that Mek1 functions in meiotic sister-chromatid cohesion may also explain why the mek1 mutant forms short stretches of SC instead of full-length complexes (Rockmill and Roeder 1991). In the spo76 mutant of Sordaria (Moreau et al. 1985), extensive SC forms, but there are regions where axial elements are unsynapsed. These axial elements are often split into two, thinner elements; however, lateral elements within SC never appear to be subdivided. These observations suggest that Spo76 functions in sister-chromatid cohesion and that synapsis cannot occur in regions where sisters are not properly associated (Moreau et al. 1985). Thus, the interruptions in SC observed in the mek1 mutant may correspond to regions where meiotic sister-chromatid cohesion has failed.
Materials and methods
Plasmids
Standard methods were used in plasmid constructions (Sambrook et al. 1989). pB121 carries the null mek1::LYS2 mutation (Rockmill and Roeder 1991). pJ68, constructed by K. Nandabalan (Yale University, New Haven, CT), contains the SpeI–PvuII fragment of MEK1 inserted at the XbaI site of pGEX-KG (Guan and Dixon 1991), such that MEK1 is downstream of glutathione S-transferase and fused in-frame (GST–Mek1). pR1284 carries an in-frame fusion of the lacZ gene inserted after 189 nucleotides of the MEK1 coding region (Burns et al. 1994).
The mek1–D290A mutation was constructed by PCR using the method of Zhou and Elledge (1993). The SpeI–HpaI region of MEK1 containing this mutation was substituted into the SpeI–HpaI sites of pJ77, which contains the EcoRI–PvuII fragment of MEK1 inserted into the EcoRI–EcoRV sites of YIp5 (Rothstein 1991). The resulting plasmid, pJ30, contains the mek1–D290A mutation as confirmed by sequencing.
An in-frame fusion of MEK1 with GFP was constructed as follows. PCR was used to generate a NotI site immediately preceding the MEK1 stop codon and a SalI site immediately following the stop codon. An HpaI–SalI fragment of MEK1 (containing the NotI site) was cloned into pB124, which contains the MEK1 EcoRI–PvuII fragment inserted at the EcoRI–EcoRV sites of pBR322 (Rockmill and Roeder 1991). Then, the NotI (filled in)–XbaI fragment of MEK1 was inserted into the NheI (filled in)–SalI sites of a multicopy plasmid containing the GFP gene (pCB431; Chua and Roeder 1998), creating pJ23.
For plasmids used in the two-hybrid assay, the RED1-coding region was amplified such that a BamHI site was introduced immediately preceding the start codon, and a SalI site was introduced directly before the stop codon. PCR was used to generate a BglII site in HOP1 just before the start codon and a SalI site immediately before the stop codon. pJ63 contains the RED1 PCR product (cut with BamHI and SalI) cloned into the BamHI–XhoI sites of pACTII, which contains the GAL4 activation domain (Bai and Elledge 1997). The HOP1 PCR product was cut with BglII and SalI and inserted into the BamHI–SalI sites of pACTII to generate pJ64. The MEK1-coding region was amplified such that a BamHI site was introduced before the start codon and a SalI site was generated before the stop codon; this PCR product was cut with BamHI and SalI and ligated into the BamHI–SalI sites of pBG4D-1, which contains the GAL4 DNA-binding domain (Durfee et al. 1993), to produce pJ54. pJ61 encodes the first 100 codons of Mek1 (on a BamHI–SpeI fragment of pJ54) in the BamHI–XbaI sites of pBG4D-1. A MEK1 fragment encoding the first 63 amino acids of Mek1 was generated by PCR, cut with BamHI and SalI (the SalI site was introduced by PCR), and cloned into the BamHI–SalI sites of pBG4D-1 to generate pJ71. The NaeI–AatII fragment containing HIS3 from pRS423 (Sikorski and Hieter 1989) was cloned into the PmlI–AatII sites of pYADE4 (Brunelli and Pall 1993), producing pJ88. Then, to generate pJ89, which contains RED1 downstream of the ADH1 promoter, the BamHI–SalI RED1 PCR product (above) was inserted into pJ88 at the BamHI–SalI sites.
Yeast strains
Yeast strains were constructed and maintained with standard procedures and media (Sherman et al. 1986). Transformation of yeast was carried out by the lithium acetate procedure of Gietz et al. (1995). Gene disruptions were confirmed by Southern blot analysis.
All strains used for cytology and for immunoprecipitations are isogenic with BR2495 (Rockmill and Roeder 1990). Homozygous mutant red1::URA3 and hop1::TRP1 strains are MY231 and BR2498, respectively (Smith and Roeder 1997). Homozygous mutant mek1::LYS2 (JM82) and mek1–lacZ (JM198) strains were constructed using pB121 and pR1284, respectively. JM191 (homozygous for mek1–D290A) was generated by two-step transplacement (Rothstein 1991) using pJ30. The homozygous mek1::LYS2 red1::URA3 double mutant (JM235) was constructed using pB121, and then pV180 (Smith and Roeder 1997). The haploid parents of JM198 were transformed with either pNH32-1 (hop1::TRP1; Hollingsworth and Byers 1989) or pV180, and then mated to produce the homozygous mutant mek1-lacZ red1::URA3 (JM93) and mek1–lacZ hop1::TRP1 (JM88) diploids. Strain JM98 (MEK1::GFP) is JM82 carrying pJ23.
Strains containing the Red1–HA and Hop1–HA fusion proteins were constructed by PCR as described (Schneider et al. 1995). The PCR products were transformed into yeast, and correct integration was confirmed by PCR and immunoblotting. Strains JM241 and JM142 are BR2495 that is heterozygous for Red1–HA or Hop1–HA, respectively.
The yeast strain used for the two-hybrid protein system is JM239, a diploid constructed by inducing mating type switching of PJ69-4A (James et al. 1996) and mating MATa and MATα haploid strains. The red1::URA3 mutant version of JM239 (JM240) was constructed using pV180.
Mek1 antibodies
GST–Mek1 protein was purified from Escherichia coli containing pJ68 by the methods of Guan and Dixon (1991). GST–Mek1 was then used to raise antibodies in rabbits and mice at the Pocono Rabbit Farm and Laboratory (Canadensis, PA).
Cytology
Meiotic chromosomes were spread and stained with antibodies as described (Chua and Roeder 1998). Anti-Mek1 antibodies were affinity purified from serum (Chua and Roeder 1998) and used at a 1:40 dilution. Anti-rabbit antibodies conjugated to biotin, or anti-mouse antibodies conjugated to biotin (Zymed Laboratories), were used at a 1:200 dilution as secondary antibodies. Antibodies labeled with streptavidin conjugated to Texas red (Jackson ImmunoResearch Laboratories) were used at a 1:200 dilution to detect primary and secondary antibodies (Page and Snyder 1992).
Anti-Red1 and anti-Hop1 antibodies (Smith and Roeder 1997), as well as anti-Zip1 antibodies (Sym et al. 1993), were used at 1:100 dilution. Anti-β-galactosidase antibodies (ICN Biochemicals) were used at 1:40 dilution. Antibodies other than anti-Mek1 antibodies were detected using rabbit or mouse antibodies conjugated to Oregon green (Molecular Probes). Chromosomal DNA was stained with 1 μg/ml DAPI. Images were recorded and analyzed using a Leitz DMRB microscope and a Photometrics Imagepoint CCD camera.
To determine the amount of overlap between Mek1, Red1, and Hop1, the total number of Mek1 foci per spread nucleus per time point were compared with the number of Mek1 foci that also contain Red1 or Hop1. Mek1 overlap with Red1 was examined for at least 200 nuclei. Mek1 overlap with Hop1 was assessed for 200 nuclei both in wild type and in the mek1–D290A mutant. Fortuitous colocalization was assessed by rotating an overlay corresponding to one of the two signals by 90°C or 180°C and determining the amount of overlap between the misoriented images, as described by Gasior et al. (1998).
Immunoprecipitations and Western blots
To prepare meiotic cell extracts, cells were grown to saturation in one liter of YPD (Sherman et al. 1986) supplemented with 60 μg/ml uracil and 40 μg/ml adenine; cells were then pelleted and resuspended in 3 liters of sporulation medium (2% potassium acetate). After 13–14 hr, cells were collected by centrifugation and resuspended in 3 ml of lysis buffer (50 mm Tris, 250 mm NaCl, 5 mm EDTA, 50 mm NaF, 0.1% NP-40 at pH 7.8) to which protease inhibitors (100 μg/ml PMSF, 20 μg/ml aprotinin, 10 μg/ml antipain, 10 μg/ml leupeptin, 10 μg/ml pepstatin A; Sigma) and 1.5 mm dithiothreitol (DTT) were added. Glass beads (425–600 μm) (Biospec Products; Sigma) were added to three-fourths of the original volume. Cell lysis was accomplished by vortexing at 4°C for 15 intervals of 1 min each, with tubes incubated on ice for 3–5 min between periods of vortexing. After centrifugation at 15,000 rpm in a SS-34 Sorvall rotor for 20 min at 4°C, supernatants (containing at least 50 mg of total protein) were recovered and used for immunoprecipitations.
Immunoprecipitations were carried out at 4°C on a nutator (Adams/ALA Scientific Instruments). Anti-Mek1 serum (1:200), anti-Red1 serum (1:500), anti-Hop1 serum (1:500), anti-β-galactosidase antibody (1:500), or preimmune serum (1:500) was added to each supernatant. After 2 hr, 40 μl of Protein A–Sepharose (Pierce) was added, and incubations were allowed to proceed for an additional hour. At the start of immunoprecipitations, protease inhibitors were added to final concentrations twice that originally included for cell lysis. Immune complexes were collected by centrifugation, washed twice in IP buffer (20 mm Tris, 100 mm NaCl, 2 mm EDTA, 0.05% Tween 20 at pH 7.8), boiled in 6× sample buffer (70% Tris-Cl at pH 6.8, 30% glycerol, 10% SDS, 0.09 % DTT, 0.01% bromophenol blue), and fractionated by an 8% SDS–polyacrylamide gel at 100 V for 3–4 hr (Figs. 4, 5A, and 6A,B) or at 20 V for 16 hr (Fig. 5B and 6C).
Gels were blotted onto nitrocellulose (Schleicher & Schuell) using the Mini-Protean II system (Bio-Rad). For Western blot analysis (Towbin et al. 1979), filters were incubated in blocking buffer (5% dry milk and 0.1% Tween 20 in Tris-buffered saline; Sambrook et al. 1989). After 1 hour, primary antibody was added to a final dilution of 1:200–1:500. Primary antibody was detected using anti-rabbit antibody conjugated to alkaline phosphatase (Jackson ImmunoResearch Laboratories) at a 1:200 dilution, followed by CDP-Star (Boehringer Mannheim). Blots were exposed to Biomax film (Kodak). The Red1 protein migrates at ∼100 kD, as predicted based on sequence; Hop1 migrates as a 70-kD protein, as described (Hollingsworth et al. 1990).
Kinase assays
For in vitro kinase assays, immune complexes were resuspended in 75 μl kinase buffer (50 mm Tris, 10 mm MgCl2 at pH 7.0) after two washes in IP buffer. Nonradioactive ATP was added to 1 μm; 20 μCi of [γ-32P]ATP (sp. act. 3000 Ci/mmole, Amersham) was added and reactions were allowed to proceed for 20 min at room temperature. SDS-PAGE and blotting to nitrocellulose were carried out as described above. Filters were exposed to film overnight to 2 days to assess [γ-32P]ATP incorporation.
For gel mobility shift analysis, immune complexes were resuspended in 75 μl phosphatase buffer (50 mm Tris, 20 mm MgCl2, 40 mm KCl at pH 8.0) containing 100 μg/ml PMSF. Sixty units of CIP (Boehringer Mannheim) and 14 μl of 10× phosphatase buffer (Boehringer Mannheim) were added to two samples from wild-type immunoprecipitates; 5 mm β-glycerophosphate, a phosphatase inhibitor (Sigma), was also added to one of these samples. Samples were incubated at 30°C for 10 min, and proteins were then analyzed as described above.
Two-hybrid protein assays
JM239 derivatives carrying the various combinations of MEK1 and RED1 or MEK1 and HOP1 plasmids (see Table 1) were grown to saturation in 2× synthetic complete medium lacking leucine and tryptophan. Strain JM239 carrying MEK1-, HOP1-, and RED1-containing plasmids was grown in 2× synthetic complete medium (Sherman et al. 1986) lacking leucine, tryptophan, and histidine. One-half of each culture was diluted 1:100, then grown for an additional 12 hr and collected by centrifugation. The other half of each culture was diluted 1:1 in YPAD and grown for 10 hr, then transferred to sporulation medium and collected 15 hr later (approximately pachytene). β-Galactosidase assays were performed on cells in log phase of vegetative growth (Durfee et al. 1993) and on meiotic cells (Chua and Roeder 1998).
FISH
FISH was carried out as described by Chua and Roeder (1998). A 25-kb fragment of Chromosome III was labeled with digoxigenin and then hybridized to spread chromosomes. Antibodies against digoxigenin conjugated to rhodamine were used to detect the chromosome III probe. Chromosomal DNA was stained with DAPI.
Acknowledgments
We thank Jennifer Fung, Janet Novak, Beth Rockmill, and Albert Smith for critical comments on the manuscript. We are grateful to Mike Snyder and David Stern for advice on immunoprecipitations. The Howard Hughes Biopolymer/Keck Foundation Biotechnology Resource Laboratory at Yale University provided oligonucleotides and performed sequence analysis. This work was supported by American Cancer Society grant VM-7G to G.S.R. and by the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL shirleen.roeder@yale.edu; FAX (203) 432-3263.
References
- Bai C, Elledge SJ. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- Bishop D, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Brunelli JP, Pall ML. A series of yeast shuttle vectors for the expression of cDNAs and other DNA sequences. Yeast. 1993;9:1299–1308. doi: 10.1002/yea.320091203. [DOI] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi E-Y, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization and gene disruption in Saccharomyces cerevisiae. Genes & Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Flaggs G, Plug AW, Dunks KM, Mundt KE, Ford JC, Quiggle MR, Taylor EM, Westphal CH, Ashley T, Hoekstra M F, Carr AM. Atm-dependent interactions of a mammalian Chk1 homolog with meiotic chromosomes. Curr Biol. 1997;7:977–986. doi: 10.1016/s0960-9822(06)00417-9. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Hollingsworth NM, Byers B. Insertional mutations in the yeast HOP1 gene: Evidence for multimeric assembly in meiosis. Genetics. 1994;136:449–464. doi: 10.1093/genetics/136.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wong AK, Kora Y, Shinohara A, Bishop DK. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes & Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The FHA domain: A putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N, Byers B. HOP1: A yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Johnson AD. A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by overexpression of two other meiosis-specific genes: RED1 and REC104. Genetics. 1993;133:785–797. doi: 10.1093/genetics/133.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L. Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;147:33–42. doi: 10.1093/genetics/147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Goetsch L, Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans D. The regulation of protein transport to the nucleus. Biochem J. 1995;311:705–716. doi: 10.1042/bj3110705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan KS, Holtzman DA, Plug AW, Christenson ER, Brainerd EE, Flaggs G, Bentley NJ, Taylor EM, Meyn MS, Moss SB, Carr AM, Ashley T, Hoekstra MF. The Atr and Atm protein kinases associate with different sites along meiotically pairing chromosomes. Genes & Dev. 1996;10:2423–2437. doi: 10.1101/gad.10.19.2423. [DOI] [PubMed] [Google Scholar]
- Kironmai KM, Muniyappa K, Friedman DB, Hollingsworth NM, Byers B. DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1424–1435. doi: 10.1128/mcb.18.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers JHM, van Aalderen M, Peters AHFM, van Pelt AAM, de Rooij DG, de Boer P, Offenberg HH, Dietrich AJJ, Heyting C. A change in the phosphorylation pattern of the 30000-33000 Mr synaptonemal complex proteins of the rat between early and mid-pachytene. Chromosoma. 1994;104:154–163. doi: 10.1007/BF00352179. [DOI] [PubMed] [Google Scholar]
- Leem S-H, Ogawa H. The MRE4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:449–457. doi: 10.1093/nar/20.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Moens PB, Spyropoulos B. Immunocytology of chiasmata and chromosomal disjunction at mouse meiosis. Chromosoma. 1995;104:175–182. doi: 10.1007/BF00352182. [DOI] [PubMed] [Google Scholar]
- Molnar M, Bahler J, Sipiczki M, Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing, and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau PJF, Zickler D, Leblon G. One class of mutants with disturbed centromere cleavage and chromosome pairing in Sordaria macrospora. Mol & Gen Genet. 1985;198:189–197. [Google Scholar]
- Nag DK, Scherthan H, Rockmill B, Bhargava J, Roeder GS. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995;141:75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Snyder M. CIK1: A developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes & Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- ————— The yeast med1 mutant undergoes both meiotic homolog nondisjunction and precocious separation of sister chromatids. Genetics. 1994;136:65–74. doi: 10.1093/genetics/136.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: It takes two to tango. Genes & Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. In: Molecular cloning: A laboratory manual. Nolan C, Ford N, Irwin N, Ferguson M, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher B. Use of polymerase chain reaction epitope tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;90:1123–1135. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- ————— Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Sears DD, Hieter P, Simchen G. An implanted recombination hot spot stimulates recombination and enhances sister chromatid cohesion of heterologous YACs during yeast meiosis. Genetics. 1994;138:1055–1065. doi: 10.1093/genetics/138.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Roeder G S. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes & Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in response to the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht J, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J, Song W, Carlson M. Protein phosphatase type 1 interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4199–4206. doi: 10.1128/mcb.16.8.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes & Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]