Abstract
Response to the gaseous plant hormone ethylene in Arabidopsis requires the EIN3/EIL family of nuclear proteins. The biochemical function(s) of EIN3/EIL proteins, however, has remained unknown. In this study, we show that EIN3 and EILs comprise a family of novel sequence-specific DNA-binding proteins that regulate gene expression by binding directly to a primary ethylene response element (PERE) related to the tomato E4-element. Moreover, we identified an immediate target of EIN3, ETHYLENE-RESPONSE-FACTOR1 (ERF1), which contains this element in its promoter. EIN3 is necessary and sufficient for ERF1 expression, and, like EIN3-overexpression in transgenic plants, constitutive expression of ERF1 results in the activation of a variety of ethylene response genes and phenotypes. Evidence is also provided that ERF1 acts downstream of EIN3 and all other components of the ethylene signaling pathway. The results demonstrate that the nuclear proteins EIN3 and ERF1 act sequentially in a cascade of transcriptional regulation initiated by ethylene gas.
Keywords: Plants, Arabidopsis, EIN3, ERF1, transcription
The plant hormone ethylene regulates a variety of stress responses and developmental adaptations in plants. This gaseous molecule is well known for its participation in physiological processes as diverse as fruit ripening, senescence, abscission, germination, cell elongation, sex determination, pathogen defense response, wounding, nodulation, and determination of cell fate (Abeles et al. 1992; Tanimoto et al. 1995; O’Donnell et al. 1996; Penninckx et al. 1996; Penmetsa and Cook 1997). Control of these processes by ethylene involves complex regulation of both ethylene biosynthesis and the ability of cells to perceive and respond to the hormone in an appropriate manner. Understanding the molecular events that lead to this diversity of plant responses is essential to elucidate how this gas modulates such functions.
The morphological changes evoked by continuous exposure of Arabidopsis seedlings to ethylene, the triple response, have allowed the identification of a number of components of the ethylene response pathway. Several classes of mutants impaired in their response to the hormone have been identified. Mutants that display a constitutive triple response phenotype may result either from ethylene overproduction (eto1, eto2, and eto3), or constitutive activation of the pathway (ctr1). Insensitive mutants are defective in their ability to perceive or respond to ethylene and include etr1, etr2, ein2, ein3, ein4, ein5/ain1, ein6, and ein7 mutants (Ecker 1995; McGrath and Ecker 1998; Sakai et al. 1998; Solano and Ecker 1998). On the basis of epistasis analysis, a genetic framework for the action of these genes has been established (Roman et al. 1995; Sakai et al. 1998). ETR1, ETR2, and EIN4 genes act upstream of CTR1, whereas EIN2, EIN3, EIN5/AIN1, EIN6, and EIN7 genes act downstream of CTR1.
Several of the early-acting ethylene signaling pathway genes have been cloned and characterized. ETR1, ETR2, and EIN4 encode members of a family of membrane proteins with significant similarity to two-component histidine kinase receptors found in bacteria and fungi (Chang et al. 1993; Hua et al. 1998; Sakai et al. 1998). Expression of ETR1 in yeast cells allows them to bind ethylene, consistent with its suggested role as an ethylene receptor (Schaller and Bleecker 1995; Chang 1996). The ETR1 family also includes two members (ERS1 and ERS2) that were cloned by sequence homology. When mutations that confer dominant ethylene insensitivity to ETR1 are introduced into these two genes, the resulting transgenic plants are ethylene insensitive, suggesting that ERS1 and ERS2 may also be ethylene receptors (Hua et al. 1995,1998). The CTR1 gene encodes a protein with similarity to the Raf-family of protein kinases, implicating a MAP-kinase cascade in the ethylene response pathway (Kieber et al. 1993). Coupling of bacterial-type receptor and Raf-like protein kinases in the osmosensing pathway in yeast is provided by phosphorelay proteins (Posas et al. 1996). While several proteins with both structural and functional similarity to response regulators have been identified in Arabidopsis (Imamura et al. 1998), the ethylene receptors ETR1 and ERS1 can interact physically with CTR1 (Clark et al. 1998); thus by-passing an absolute requirement for such intermediates.
Less is understood about the downstream components of the ethylene signaling pathway. Cloning and characterization of the EIN3 gene revealed that it encodes a nuclear-localized protein (Chao et al. 1997). Although sequence analysis failed to uncover homology to previously described proteins, EIN3 shares amino acid sequence similarity, conserved structural features, and genetic function with three EIN3-LIKE (EIL) proteins. Genetic studies revealed that EIL1 and EIL2 are able to functionally complement the ein3 mutation, suggesting their participation in the ethylene signaling pathway. High-level expression of EIN3 or EIL1 in transgenic wild-type or ein2 mutant plants conferred constitutive ethylene response phenotypes in all stages of development, indicating their sufficiency for activation of the pathway in the absence of ethylene. However, the function(s) of the EIN3/EIL family of proteins remains unknown. Further analysis of their biochemical activities has been hampered by the absence of candidate target genes.
Among the different classes of ethylene-responsive genes, the most extensively studied are those whose expression is activated by ethylene in response to pathogen attack. This class includes basic-chitinases, β-1,3-glucanases, defensins, and other pathogenesis-related (PR) proteins (Boller et al. 1983; Felix and Meins 1986; Broglie et al. 1989; Ohme-Takagi and Shinshi 1990; Samac et al. 1990; Eyal et al. 1993; Penninckx et al. 1996). Analysis of the promoters of several of these genes revealed a common cis-acting ethylene response element called the GCC box. This element was shown to be necessary and sufficient for ethylene regulation in a variety of plant species (Eyal et al. 1993; Hart et al. 1993; Meller et al. 1993; Ohme-Takagi and Shinshi 1995; Sessa et al. 1995; Shinshi et al. 1995; Sato et al. 1996). Efforts to isolate trans-acting factors in tobacco that bind the GCC box identified a family of proteins termed Ethylene-Responsive-Element-Binding-Proteins (EREBPs) (Ohme-Takagi and Shinshi 1995). These novel DNA-binding proteins interact in vitro with the GCC box through a domain homologous to that previously observed in the floral homeotic protein APETALA2 (Ecker 1995; Weigel 1995). In Arabidopsis, >30 genes belonging to this family have been identified by several groups (Wilson et al. 1996; Buttner and Singh 1997; Okamuro et al. 1997) and as a result of the Arabidopsis Genome Initiative (Bevan et al. 1997; Ecker 1998). On the basis of the available genomic sequence data, 125 members of this family have been estimated to exist in the Arabidopsis genome (Riechmann and Meyerowitz 1998). The expression of several members of this family has been reported to be regulated by ethephon, an ethylene-releasing compound (Ohme-Takagi and Shinshi 1995; Buttner and Singh 1997), raising the tantalizing possibility that one or more EREBPs may constitute an intermediate step(s) between EIN3/EILs and ethylene-inducible target genes such as basic-chitinase. However, evidence for the direct involvement of EREBPs in the ethylene signaling pathway in Arabidopsis is still lacking.
Here we report the cloning and characterization of ETHYLENE-RESPONSE-FACTOR1 (ERF1), an early ethylene responsive gene that encodes a GCC-box-binding protein. EIN3 expression is both necessary and sufficient for ERF1 transcription and, like EIN3 overexpression in transgenic plants, constitutive expression of ERF1 results in activation of a variety of ethylene response genes and phenotypes. Moreover, we demonstrate that EIN3 and EILs are novel sequence-specific DNA-binding proteins that bind a primary ethylene response element in the promoter of ERF1. Consistent with the biochemical studies, genetic analysis revealed that ERF1 acts downstream of EIN3 and all previously identified components in the ethylene gas signaling pathway.
Results
Cloning and characterization of ERF1
To identify targets of the EIN3/EILs proteins and to examine the role of EREBPs in the ethylene signaling pathway, a PCR-based approach was used to isolate members of the EREBP family in Arabidopsis. By use of oligonucleotides complementary to the tobacco EREBP1 sequence (Ohme-Takagi and Shinshi 1995), a 597-bp fragment was amplified from tobacco genomic DNA, and this fragment was used to screen an Arabidopsis cDNA library under low stringency. Among the positive clones, two classes of cDNAs showed high homology to the tobacco EREBP1/2 and EREBP3/4 genes. One of these genes, ERF1 (Fig. 1A) showed rapid induction in response to ethylene (Fig. 1B). More importantly, ERF1 mRNA began to accumulate after 15 min of hormone treatment of plants. Induction of ERF1 mRNA was also dependent on the presence of functional EIN3, as no expression was detected in ein3-1 mutants (Fig. 1B). To compare the kinetics of ERF1 induction with that of another known ethylene-inducible gene, the same blot was hybridized with PDF1.2, a member of the defensin gene family (Penninckx et al. 1996). Consistent with the hypothesis that ERF1 may be a regulator of these genes, maximal ERF1 expression occurred earlier than that of PDF1.2 (Fig. 1B). Consequently, we focused our studies on this gene as it fits the criteria for a candidate target of EIN3.
Figure 1.
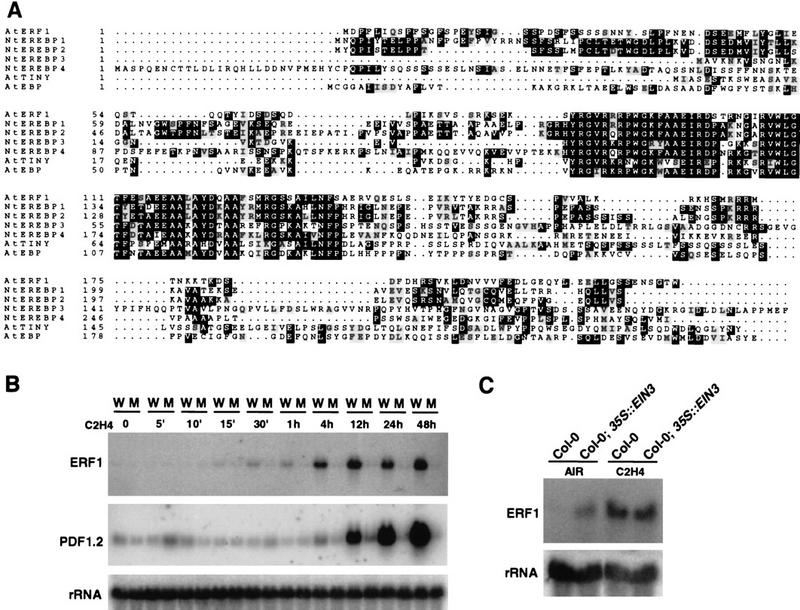
Cloning and ethylene inducibility of ERF1. (A) Sequence alignment of ERF1 and EREBP proteins from tobacco (EREBP1, EREBP2, EREBP3, and EREPB4; Ohme-Takagi and Shinshi 1995) and Arabidopsis [AtEBP (Buttner and Singh 1997) and AtTINY (Wilson et al. 1996)]. (B) RNA blot analysis of the induction of ERF1 mRNA expression by ethylene gas and comparison with the expression of PDF1.2. Total RNA was isolated from 4-week-old wild-type Col-0 (W) or ein3-1 (M) plants grown in air and exposed to ethylene gas for different times (0–48 hr). (C) RNA blot analysis of the expression of ERF1 in EIN3-overexpressing plants. Thirty micrograms of total RNA were loaded per lane in B and 60 μg in C.
It has been demonstrated previously that overexpression of the ethylene pathway gene EIN3 causes activation of all known ethylene response phenotypes (Chao et al. 1997). Although the level was somewhat lower than that achieved by exogenous ethylene treatment, ERF1 mRNA showed constitutive high-level expression in 35S::EIN3-expressing transgenic plants (Fig. 1C), indicating that EIN3 is sufficient for ERF1 expression. Taken together, the results indicate that EIN3 is both necessary and sufficient for expression of the early ethylene response gene ERF1.
ERF1 was mapped to chromosome III, in the ABI3 contig, by PCR amplification of YAC pools with ERF1-specific primers. Other than EIN3, none of the known ethylene signaling mutants map to this region.
Sequence-specific binding of EIN3 in the promoter of ERF1
To test whether the nuclear protein EIN3 is capable of DNA binding, we performed electrophoretic mobility shift assays (EMSAs) with in vitro-translated EIN3 protein and the 5′ promoter region of the ERF1 gene. A 6-kb fragment containing the ERF1 promoter was isolated from genomic sequences (BAC F11F14) and subcloned into pBluescript. Five overlapping fragments that covered ∼1.4-kb upstream of the ERF1 translation initiation site were amplified and radioactively labeled by PCR. As shown in Figure 2A, a slower migrating band was observed when one of the fragments (−1238 to −950) was incubated with reticulocyte lysates containing EIN3. The binding of EIN3 to the −1238 to −950 fragment was not competed by a 500-fold excess of poly[d(IC)] or poly[d(AT)], demonstrating the specificity of the EIN3–DNA interaction. To delimit further the EIN3 target site, this fragment was subdivided, and each subfragment was subjected to binding experiments. Only one of the subfragments (−1213 to −1179) was recognized specifically by EIN3, confirming further its sequence specificity (Fig. 2B).
Figure 2.
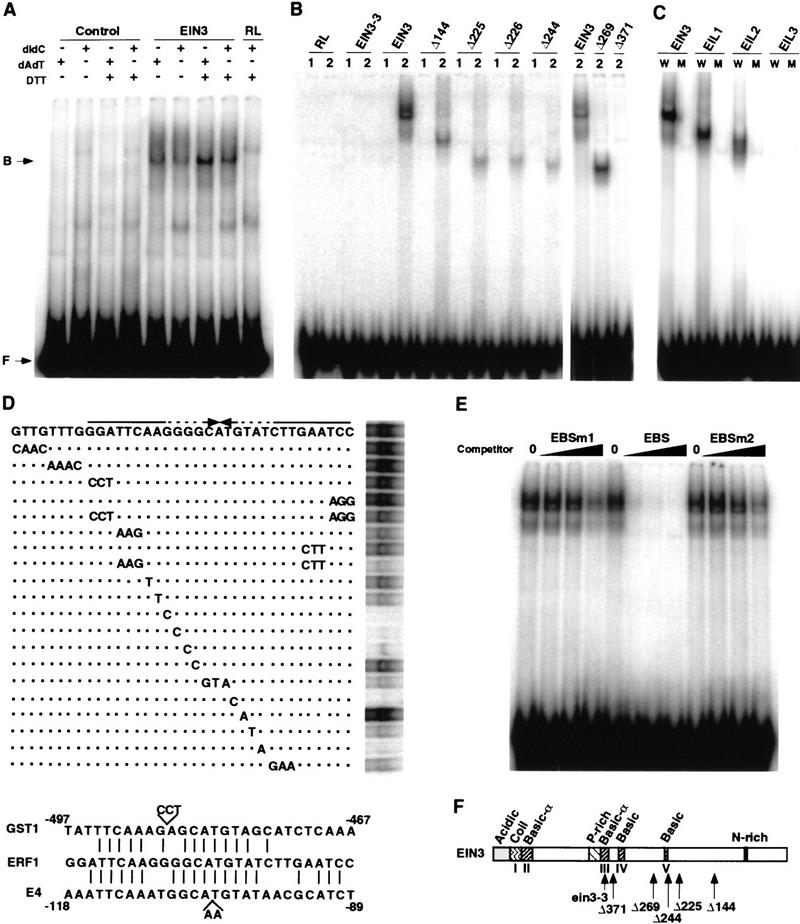
EIN3 is a sequence-specific DNA-binding protein. (A) EMSA of in vitro-translated EIN3 protein binding to the −1238 to −950 fragment of the ERF1 promoter. A control protein (control) or mock-translated reticulocyte lysates (RL) were used in the indicated lanes. (B) EMSA of EIN3-3 mutant protein, EIN3, and several carboxy-terminal deletion derivatives bound to fragments −1238 to −1204 (1) and −1213 to −1178 (2) of the ERF1 promoter. (C) Binding of EIL proteins to the EIN3 target site in the ERF1 promoter. EMSA was performed with in vitro-translated EIN3, EIL1, EIL2, and EIL3 proteins and the wild-type EIN3-binding site (W) or a mutant version (M) corresponding to the mutant G17 to C in Fig. 2D. (D; top) Scan mutagenesis of the EBS. Wild-type EBS is shown with the palindromic repeats indicated by arrows. Base changes in the mutants tested are indicated on the lines below. Dashed lines indicate mismatches. Dots indicate similar bases as in the wild-type EBS. (D; bottom) Sequence alignment of the EBS and a fragment of the promoters of the E4 and GST1 genes (including the ERE). (E) Competition of the EIN3–EBS complex formation by addition of an excess of unlabeled EBS or two mutant versions, EBSm1 and EBSm2 (see Materials and Methods). No competitor was added in the lanes labeled as 0. Black wedges represent increasing amounts of competitor (20, 60, and 200 ng). One nanogram of labeled EBS was used per lane. (F) Summary of EIN3 structural features and mutants used in EMSA experiments (adapted from Chao et al. 1997).
To demonstrate that EIN3 was in fact the protein present in the mobility-shifted band, a series of carboxy-terminal truncated EIN3 derivatives was generated and subjected to binding and EMSA with the 36-bp EIN3-binding fragment described above. A mobility shift that correlated with the molecular weight of each of the truncated proteins was observed, confirming the presence of EIN3 in the protein–DNA complex (Fig. 2B). The smallest protein that retained DNA-binding capacity was EIN3Δ269, delimiting the EIN3 DNA-binding domain to amino acids 1–359. In addition, a mutant version of EIN3 that contained the amino acid substitution encoded by the ein3-3 allele (Lys245 to Asn), was generated by in vitro translation of the corresponding mRNA. The amino acid substitution in the ein3-3 mutant lies within the basic domain III of the EIN3 protein (Chao et al. 1997). Interestingly, the mutant EIN3-3 protein was unable to recognize the 36-bp target site (Fig. 2B).
Further experiments demonstrated that the source of the EIN3 protein does not affect its ability to bind DNA. As with the in vitro-translated protein, baculovirus-expressed EIN3 was also able to recognize the 36-bp fragment containing the target sequence (data not shown).
To examine whether other members of the EIN3/EIL family are also able to bind DNA, we performed EMSA experiments with in vitro-translated EIL1, EIL2, and EIL3 proteins, and a DNA fragment containing the EIN3-binding site or a mutant version. Consistent with the ability of EIL1 and EIL2, but not EIL3, to complement the ein3-1 mutation in transgenic plants (Chao et al. 1997), both EIL1 and EIL2, but not EIL3, were able to recognize specifically the EIN3-binding site (Fig. 2C).
To define more precisely the sequence requirements of EIN3 binding, scanning mutagenesis of the 36-bp fragment was performed. As shown in Figure 2D, all mutations affecting the affinity of EIN3 for its target site resided within a 28-bp sequence with a palindromic structure. Single mutations within the core of this (imperfect) palindrome completely abolished binding, and mutations that affected both distal ends of the palindromic repeats greatly reduced the interaction with EIN3. Interestingly, the EIN3-binding site shows significant similarity to sequences present in the promoter regions required for ethylene responsiveness in the tomato E4 (Montgomery et al. 1993) and LEACO1 genes (Blume and Grierson 1997) and in the carnation GST1 gene (Itzhaki et al. 1994; Fig. 2D, bottom). In GST1, a 197-bp promoter fragment containing this sequence was also sufficient to confer ethylene responsiveness to a minimal CaMV 35S promoter in transient assays (Itzhaki et al. 1994).
To examine further the specificity of the binding to its target site, competition experiments were performed with an excess of unlabeled EIN3-binding site (EBS), or two mutated versions (EBSm1 and EBSm2) not recognized by EIN3 (see Materials and Methods). As shown in Figure 2E, the formation of the EIN3–EBS complex was more efficiently competed by an excess of unlabeled EBS than by any of the EBS mutant versions, further supporting the sequence specificity of the EIN3–EBS interaction.
EIN3 recognizes its target as a homodimer
The presence of palindromic repeats in the EBS suggested that EIN3 interacts with its target as a dimer. To address this question, full-length EIN3 and several carboxy-terminal deletion derivatives were translated in vitro, alone or in pair-wise combinations. The resulting translation or cotranslation products were tested for DNA binding to the EBS. As shown in Figure 3A, in addition to the bands corresponding to the full-size EIN3 and deletion derivatives bound to DNA, a band of intermediate mobility appeared when the cotranslation products were used. The intermediate band corresponds to the mobility shift for a heterodimer, indicating that these proteins bind to the EBS as dimers.
Figure 3.
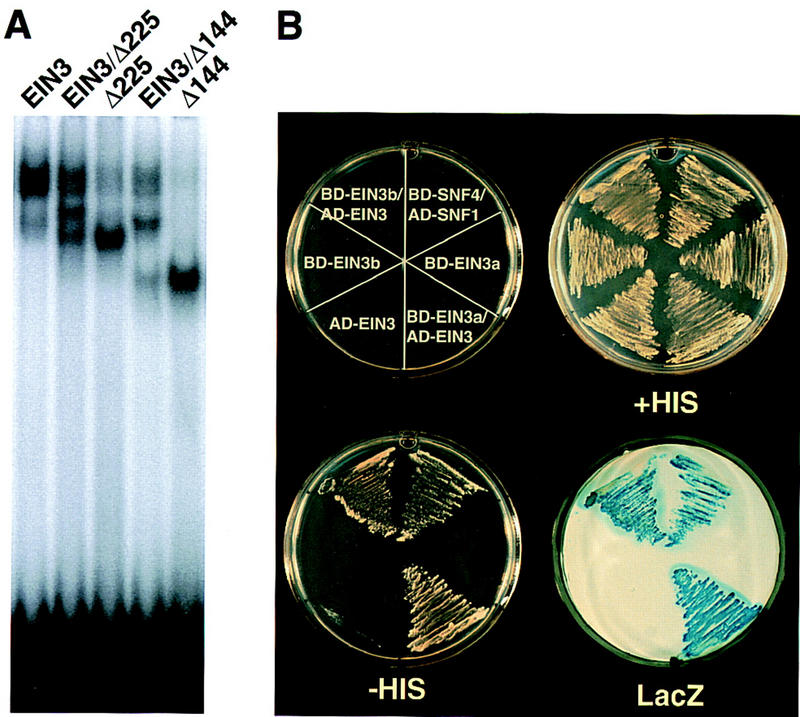
EIN3 homodimerization. (A) EMSA of full-size EIN3 and deletion derivatives binding to the EBS. Proteins were translated in vitro alone or in pair-wise combinations. (See Fig. 2F for description of EIN3 deletion derivatives.) (B) EIN3–EIN3 interactions assayed by the yeast two-hybrid system. Yeast cells transformed with the indicated constructs in the top left plate, were grown on synthetic complete (SC) medium (+HIS) or in SC medium without histidine (−HIS) and with 50 mm 3-aminotriazole (3-AT, Sigma) to repress basal activity of the his3 reporter gene. β-Galactosidase activity of the colonies grown in −HIS medium (LacZ) was determined by the filter-lift assay. SNF4/SNF1 were used as a positive control. Colonies from two independent transformation experiments are shown (BD-EIN3a and BD-EIN3b).
Additional evidence that EIN3 has the capacity to form dimers came from screening for EIN3-interacting proteins by use of the yeast two-hybrid system (Fields and Song 1990; Durfee et al. 1993). Consistent with EIN3 being a transcriptional activator, fusion of the full-size protein with the GAL4 DNA-binding domain (BD) activated transcription of the lacZ reporter gene (not shown), indicating that EIN3 possesses activation domains that are functional in yeast. To avoid this activation of the reporter gene, an EIN3 derivative containing amino acids 53–257, fused to the GAL4-BD, was used as a bait. As a prey, the GAL4 activation domain was fused to an Arabidopsis cDNA library constructed with mRNA from etiolated seedlings (Kim et al. 1997). Yeast strain Y190 transformed with the bait construct was subsequently transformed with the prey and 4 × 106 independent transformants were screened for positive interactions, as described by Kim et al. (1997). Twenty-six independent positive clones were obtained, the plasmids were recovered, and the cDNAs were sequenced. Among them, six different clones corresponded to EIN3. All positives were retested by direct transformation of yeast with both the original bait and the recovered prey. Figure 3B shows an example of the interaction with one of these positives that included amino acids 113–628 of EIN3. Because the bait contains EIN3 residues 53–257, the dimerization domain most likely resides between amino acids 113 and 257. These results also indicate that interaction of the EIN3 DNA-binding domain with DNA is not required for protein dimerization.
To assess whether EIN3 and EILs are capable of forming heterodimers, DNA-binding experiments with all combinations of cotranslated EIN3/EILs proteins were performed. While mobility-shifted bands corresponding in position to homodimeric forms of EIN3, EIL1, and EIL2 were observed, DNA–protein complexes with intermediate mobility were not seen, suggesting that these proteins do not form heterodimers (data not shown).
ERF1 is a GCC-box-binding protein
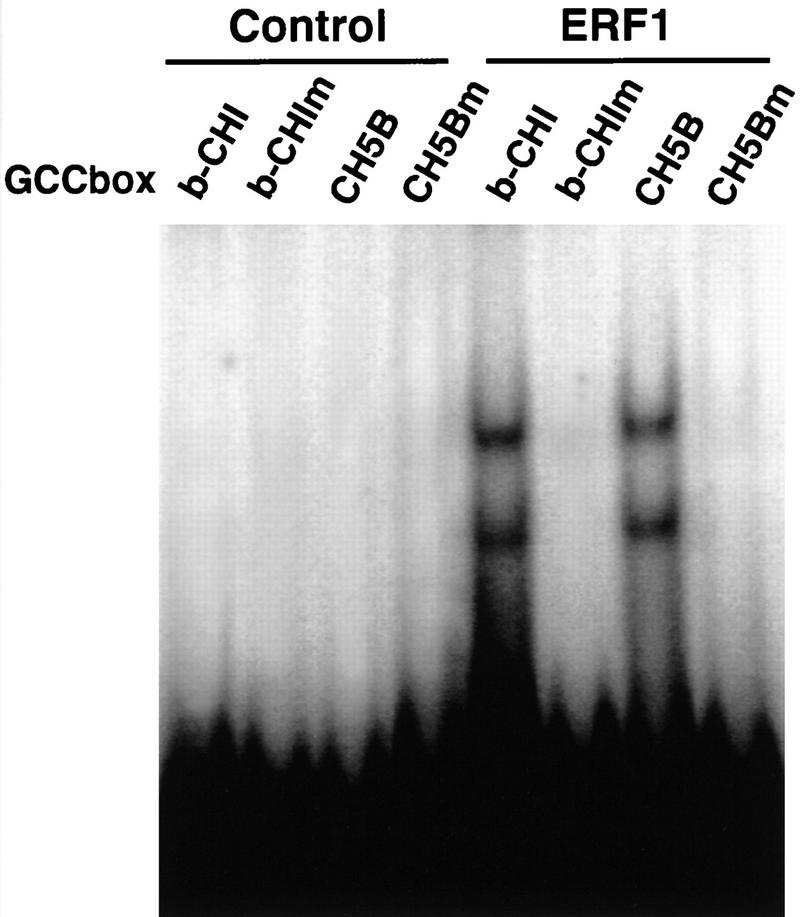
The results described above are consistent with EIN3 being a transcriptional activator that is both necessary and sufficient for ERF1 expression. ERF1, in turn, may be expected to direct the expression of target genes containing the GCC element. However, at least one EREBP, involved in the regulation of cold and drought response, is known to bind a DNA sequence unrelated to the GCC element (i.e., C-box/DRE element; Stockinger et al. 1997). To determine whether ERF1 contains a functional DNA-binding domain able to interact with the GCC box in a sequence-specific manner, we performed DNA-binding experiments with in vitro-translated ERF1 protein. Radiolabeled promoter fragments of the ethylene-regulated Arabidopsis basic-chitinase (Samac et al. 1990) and bean chitinase5B genes (Broglie et al. 1989) were incubated with ERF1 and analyzed by EMSA. To examine the specificity of the interaction, we also used a mutated version of the promoter fragments in which the cytosines of the GCC box were substituted with thymines (see Materials and Methods). As shown in Figure 4, ERF1 was able to bind specifically the promoter fragments containing the GCC element, whereas no binding was observed to the mutant sequences. The lower band in each lane containing ERF1 and the wild-type element likely corresponds to a truncated form of ERF1 as two major bands were obtained as products of the in vitro translation of ERF1 mRNA (not shown).
Figure 4.
ERF1 is a GCC box DNA-binding protein. EMSAs were performed using in vitro-translated ERF1 protein and promoter fragments from the Arabidopsis basic-chitinase (b-CHI) and bean chitinase5B (CH5B) genes. DNA fragments containing the GCC box or mutated versions (b-CHIm and CH5Bm) of these same elements were incubated with mock translated rabbit reticulocyte lysates (control) or those containing ERF1 protein.
Downstream activation of ethylene responses by ERF1
To evaluate further the role of ERF1 in the ethylene signaling pathway, transgenic plants constitutively expressing ERF1 mRNA under the control of the CaMV 35S promoter were constructed. T2 segregants of these transgenic lines were examined for ethylene response phenotypes. Of a total of 26 independent lines, plants from 9 lines displayed phenotypes similar to those observed in the constitutive ethylene response mutant ctr1, in EIN3- or EIL1-overexpressing plants, or in wild-type plants exposed to ethylene (Kieber et al. 1993; Chao et al. 1997). Etiolated 35S::ERF1 seedlings grown in hydrocarbon-free air showed inhibition of root and hypocotyl elongation, typical of treatment with ethylene (Fig. 5). However, the apical hook did not display exaggerated curvature typical of an ethylene response. The cotyledons of ERF1-expressing seedlings were still apressed, and many were still encapsulated in the seed coat. Consistent with this phenotype, HOOKLESS1, an ethylene response gene required for apical hook curvature (Lehman et al. 1996), was not expressed in ERF1-overexpressing plants (data not shown). Expression of only a partial seedling triple response phenotype in these lines is consistent with a role for ERF1 in mediating a subset of the ethylene responses. ERF1 may act along with other proteins (EREBPs and others) to fully mediate the various seedling responses to ethylene.
Figure 5.
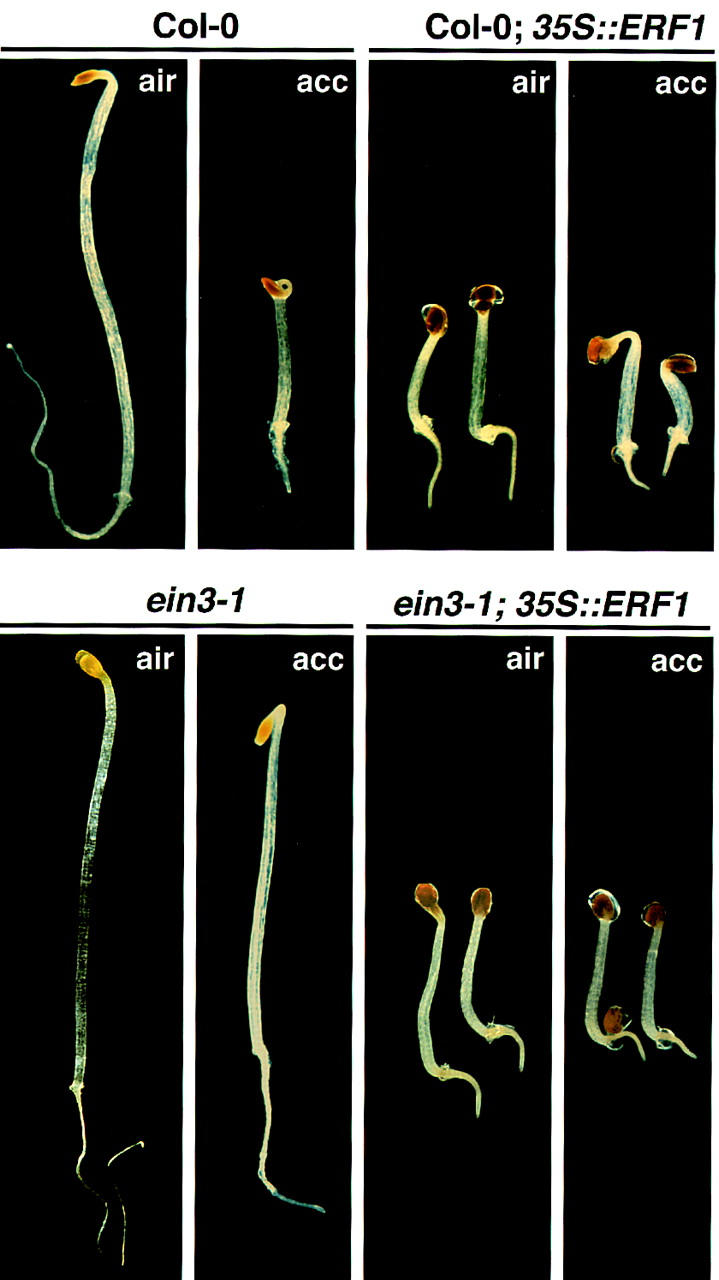
Constitutive activation of ethylene response phenotypes in 35S::ERF1-expressing seedlings. Three-day-old etiolated seedlings overexpressing ERF1 in wild-type (Col-0) and ein3 mutant backgrounds grown in agar plates with or without 10 μm acc (1-aminocyclopropane-d-carboxylic acid). Untransformed wild-type and ein3 mutant plants are also shown for comparison.
As adults, 35S::ERF1 transgenic plants showed an extreme dwarf phenotype similar to that of the constitutive ethylene response mutant ctr1 and EIN3/EIL1-overexpressing transgenic plants (Fig. 6). As in the case of the quadruple ethylene receptor knockout mutant (Hua and Meyerowitz 1998), plants from several ERF1-expressing lines showed extreme inhibition of cell enlargement and ultimately the plants wilted and died before bolting (data not shown).
Figure 6.
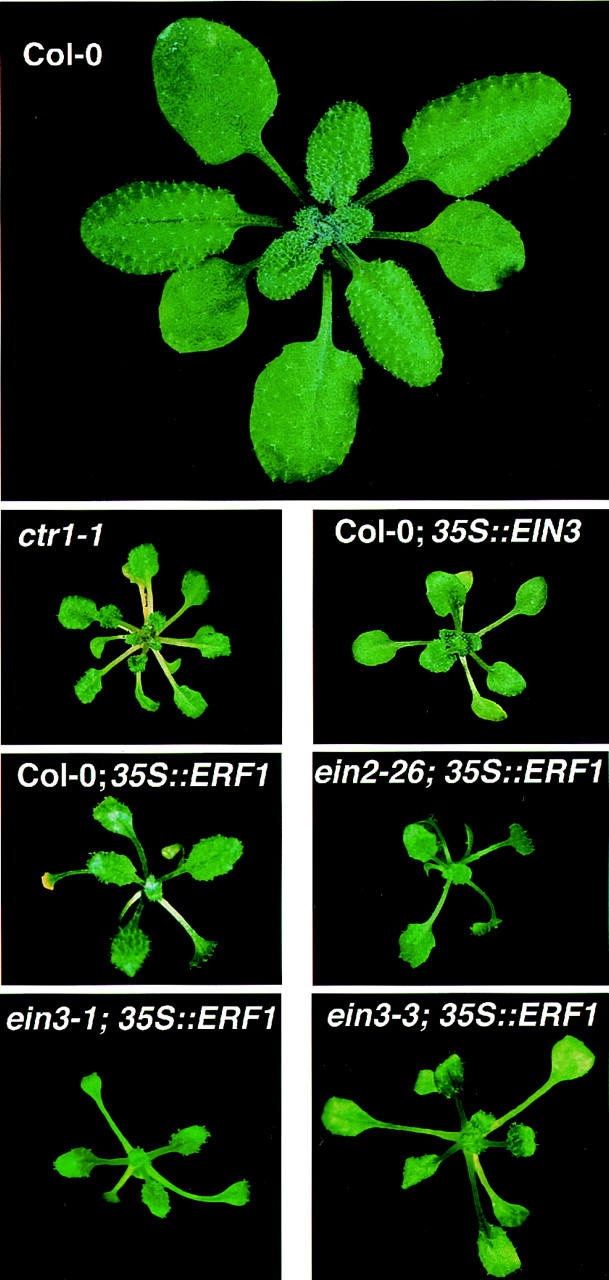
ERF1 acts downstream of EIN2 and EIN3 in the ethylene signaling pathway. Transgenic plants overexpressing ERF1 in wild-type (Col-0), ein2, ein3-1, and ein3-3 mutant backgrounds grown in continuous flowthrough cambers with hydrocarbon-free air for 5 weeks. Untransformed wild-type, ctr1-1, and EIN3-overexpressing plants are shown for comparison.
To determine whether the ethylene morphology displayed by these plants was the consequence of ethylene overproduction, or due to constitutive activation of the signaling pathway, the 35S::ERF1 gene was also introduced into several mutant backgrounds (ein2-5, ein2-17, ein2-26, ein3-1, ein3-3, and ein5-1) that suppress phenotypes resulting from ethylene overproduction (Roman et al. 1995). In all cases, the transgenic plants displayed a morphology indistinguishable from 35S::ERF1-expressing wild-type plants (Figs. 5 and 6; data not shown). These results indicate that the observed morphology evoked by expression of ERF1 was not a consequence of ethylene production but rather, like CaMV 35S::EIN3 expression (Chao et al. 1997), results from constitutive activation of the response pathway. Moreover, as there was an absence of the requirement for functional EIN2, EIN3, or EIN5 proteins for the constitutive activation phenotype, these results provide strong evidence for the downstream location of ERF1.
To confirm whether the observed morphology in the 35S::ERF1 lines was due to activation of ethylene responses, the expression of several ethylene-regulated genes was examined. As expected, no accumulation of two ethylene-inducible messages, basic-chitinase and PDF1.2, was detected in wild-type plants in the absence of the hormone, or in ethylene-insensitive mutants (ein2, ein3, or ein5). In contrast, high-level constitutive expression of both transcripts was observed in 35S::ERF1 transgenic plants regardless of the background (Fig. 7A). In the case of ein5-1, the lower expression of ERF1 in one of the two transgenic lines correlated with the lower expression of PDF1.2 and basic-chitinase and with the weaker constitutive ethylene response phenotype.
Figure 7.
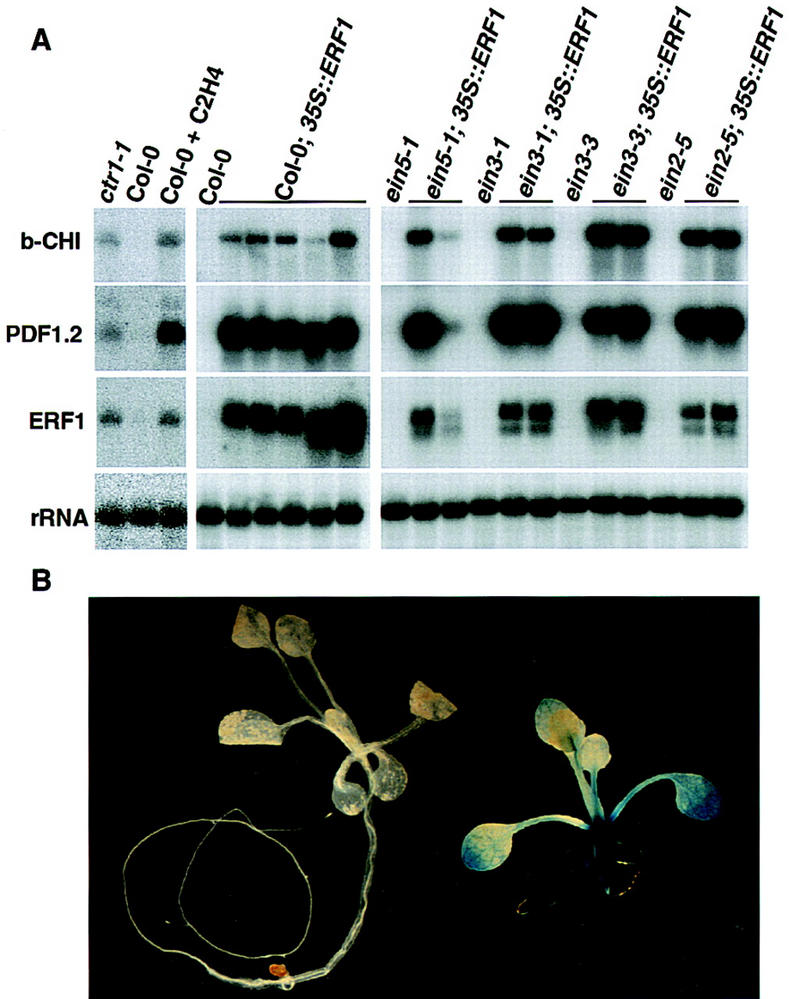
Transcriptional activation of ethylene-responsive genes by ERF1 (A) RNA blot analysis of the expression of ethylene-inducible genes in transgenic lines overexpressing ERF1 in wild-type (Col-0) and ethylene-insensitive mutant backgrounds. Five independent transgenic lines in Col-0 and two independent lines in each of the mutants are shown. Five micrograms of total RNA from 5-week-old plants grown in air were loaded per lane in the middle and right panels, and 50 μg in the left panel. The same blot was probed with ERF1, a loading control probe (rDNA), and ethylene-inducible genes PDF1.2 and basic-chitinase. (B) Constitutive activation of the ethylene-inducible CH5B–GUS reporter gene in ERF1-overexpressing transgenic plants. Three-week-old plants grown on agar plates were assayed for GUS activity.
The effect of 35S::ERF1 expression on a chitinase promoter–reporter fusion, CH5B::GUS (Broglie et al. 1989), was also examined. This well-characterized ethylene-responsive reporter gene has been shown to be a reliable marker for ethylene-evoked transcription in bean (Broglie et al. 1989) and Arabidopsis (Chen and Bleecker 1995). F1 plants derived from crosses between plants carrying the CH5B::GUS reporter gene and ERF1-overexpressing lines or wild-type plants were stained for GUS activity. As revealed by the intense staining in seedlings, high-level GUS activity was observed in the presence of 35S::ERF1, whereas no staining was detected in the control plants (absence of 35S::ERF1; Fig. 7B). Moreover, introduction of the 35S::ERF1 construct into an ethylene-repressible enhancer-trap reporter line also resulted in inhibition of GUS expression in the absence of ethylene (R. Solano, R. McGrath, and J.R. Ecker, unpubl.). These results confirm that ERF1 expression is sufficient to promote (or repress) transcription of ethylene-regulated target genes in a variety of tissues.
Discussion
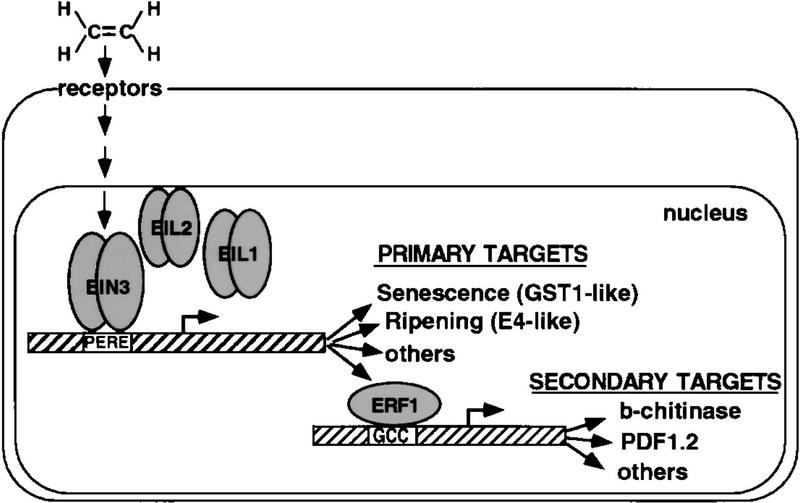
Cloning of EIN3 and related EIL genes identified a new family of proteins that are necessary and sufficient for signaling events initiated by ethylene (Chao et al. 1997). In this study, we show that EIN3 and EILs proteins comprise a novel family of sequence-specific DNA-binding proteins and define a primary ethylene response element (PERE) present in the promoters of several ethylene-regulated genes that is similar to the tomato E4 ethylene response element. EIN3 was both necessary and sufficient for the activation of ethylene responsive target genes and in particular for ERF1, a novel gene encoding an AP2/EREBP-type DNA-binding protein. Evidence is provided that sequences in the promoter of ERF1 serve as an immediate target of EIN3 binding and that constitutive expression of ERF1 activates the transcription of downstream effector genes, such as basic-chitinase and PDF1.2, to achieve the ethylene response (Fig. 8).
Figure 8.
Nuclear events in the ethylene gas signaling pathway. Model depicts the transcriptional regulatory cascade that mediates ethylene responses. Binding of ethylene (C2H4) to membrane receptors activates EIN3, and most likely EIL1 and EIL2, through a signaling cascade described elsewhere (Chao et al. 1997). EIN3 directs the expression of ERF1 and other primary target genes by binding directly, as a dimer to the E4-like PERE present in their promoters. ERF1 and probably other EREBPs bind to the GCC box (SERE) and activate the expression of secondary ethylene response genes such as basic-chitinase and defensin (PDF1.2). Although we favor this simple model, the results presented in this work do not exclude other more complicated models that may involve the existence of an intermediate between EIN3 and ERF1 or the existence of EIN3-interacting proteins that modulate EIN3 activity.
ERF1 is a downstream component in the ethylene signaling pathway
Previous efforts to understand the hormonal regulation of ethylene-regulated genes in several plant systems led to the identification of two kinds of ethylene response elements (EREs). One type of ERE was found to be responsible for ethylene-regulated expression of genes induced during ripening and senescence (Montgomery et al. 1993; Itzhaki et al. 1994). A second element, the GCC box, was identified as being necessary for ethylene inducibility in response to pathogen attack (for review, see Deikman 1997). On the basis of their ability to bind to the GCC element, a family of DNA-binding proteins (EREBPs) was identified in tobacco (Ohme-Takagi and Shinshi 1995). The fact that these genes were themselves transcriptionally activated by treatment with ethylene suggested that they might represent an intermediate step between the EIN3/EILs proteins and downstream effector genes such as basic-chitinase.
To identify targets of EIN3, members of the Arabidopsis EREBP family were cloned and characterized. ERF1 was rapidly induced in response to ethylene gas, with mRNA levels increasing within 15 min of ethylene treatment. ERF1 was constitutively expressed in the ethylene pathway mutant ctr1, and ethylene induction of ERF1 was completely dependent on a functional EIN3 protein, as no expression was detected in the ein3-1 mutant. Moreover, transgenic plants overexpressing EIN3 showed high-level expression of ERF1 mRNA. These results indicate that EIN3 is both necessary and sufficient for ERF1 expression and are consistent with ERF1 being a direct target of EIN3. The level of ERF1 mRNA expression in EIN3-overexpressing plants was somewhat lower than in ctr1 mutants or in ethylene treated wild-type plants. This indicates that although EIN3 is sufficient for ERF1 expression, other factors may be required for full ethylene-dependent ERF1 induction. In this regard, we have identified a DNA-binding protein that interacts with EIN3, using EIN3 as a bait in the two-hybrid screen (R. Solano, Q. Chao, and J. Ecker, unpubl.). This protein also binds to the ERF1 promoter in a sequence specific-manner, suggesting that it may be a partner of EIN3 and could be needed for full ERF1 expression in response to ethylene.
Thus far, loss-of-function mutations have not been reported for any member of the EREBP family. This finding, together with the large number of these genes present in Arabidopsis (125 estimated; Riechmann and Meyerowitz 1998), suggests a functional redundancy among members of this family. In the case of functionally redundant genes, loss-of-function alleles may not show a phenotype. A clear example of this is provided by studies of the ethylene receptors in Arabidopsis (Hua and Meyerowitz 1998). Implication in the ethylene signaling pathway of each of the five ETR1-related genes was made through the identification (or creation by site-specific mutagenesis) of dominant mutations (Chang et al. 1993; Hua et al. 1995, 1998; Sakai et al. 1998). While single loss-of-function mutations in these genes do not exhibit defects in ethylene response, triple and quadruple mutants display constitutive ethylene response phenotypes, revealing that ethylene responses are negatively regulated by the receptors (Hua and Meyerowitz 1998).
For this reason, we have used a gain-of-function strategy to address the in vivo function of ERF1. Gain-of-function mutations obtained by insertional mutagenesis of T-DNA or transposon elements carrying a CaMV 35S promoter (enhancer-trap/gene-trap) have proven to be a powerful tool for assessing the in vivo function of a gene. Constitutive ERF1 expression resulted in seedling and adult phenotypes very similar to those displayed by loss-of-function ctr1 mutants, plants overexpressing EIN3 or EIL1, or plants grown in ethylene (Kieber et al. 1993; Chao et al. 1997). Some significant differences, however, can be observed between EIN3- and ERF1-overexpressing plants. While ERF1 overexpression causes inhibition of hypocotyl and root cell elongation, seedlings lack an exaggerated apical hook. Consistent with this observation, HOOKLESS1, which contains a GCC element in its promoter, is not induced in ERF1 transgenic plants, suggesting that ERF1 is responsible for the activation of a subset of the ethylene-responsive GCC-box-containing target genes. Perhaps other EREBP family members may be responsible for activation of these target genes. Indeed, a transposon-induced gain-of-function mutant (tiny) that constitutively expresses an EREBP displays seedling phenotypes reminiscent of a partial ethylene response (Wilson et al. 1996), suggesting that TINY may be a partner of ERF1 in ethylene signaling. Alternatively, ERF1 may act in concert with other transcription factors in the activation of some promoters. In fact, interactions between EREBPs and bZIP transcription factors, as well as synergistic effects of their DNA target sites, the GCC box and G box, have been described previously (Hart et al. 1993; Sessa et al. 1995; Buttner and Singh 1997).
Another member of the Arabidopsis AP2/EREBP family (AtEBP) has been reported to be regulated by ethylene (Buttner and Singh 1997). Like ERF1, AtEBP is constitutively expressed in ctr1 mutants. However, unlike ERF1, AtEBP is probably not a direct target of EIN3 as its expression in response to ethylene can be blocked by cycloheximide (A. Stepanova and J.R. Ecker, unpubl.). This observation may suggest a regulatory cascade among members of the EREBP family in which AtEBP acts downstream of ERF1. Consistent with this idea, several of the EREBP genes contain GCC box elements in their promoters, suggesting that their expression might be autoregulated or controlled by other EREBPs. The existence of a transcriptional regulatory cascade is not constrained to EREBPs involved in ethylene signaling as it has also been inferred in the case of RAP genes (AP2/EREBP family members), which are not obviously involved in the response to ethylene (Okamuro et al. 1997).
EIN3 is a novel type of DNA-binding protein that regulates the expression of ERF1
Although EIN3 and EILs proteins do not share similarity with any known proteins, their nuclear location suggested their putative role as transcription factors (Chao et al. 1997). DNA-binding assays with in vitro-translated and baculovirus-expressed EIN3 protein demonstrated that EIN3 is able to bind to specific sequences in the ERF1 promoter. EMSA experiments with truncated forms of the protein confirmed the presence of EIN3 in the DNA–protein complex. A mutant protein corresponding to the ein3-3 allele of EIN3 was unable to recognize the target sequence. This mutation consists of a Lys to Asn substitution in the basic domain III, which may form part of the DNA-binding motif.
Two additional proteins that belong to the EIN3/EIL family, EIL1 and EIL2, were also able to recognize specifically the EIN3 target in the promoter of ERF1. Consistent with this result, EIN3 can be replaced functionally by EIL1 or EIL2 as overexpression of either of these genes in transgenic plants can complement the ein3-1 mutation (Chao et al. 1997). Deletion analysis of EIN3 has allowed us to define its DNA-binding domain within the amino-terminal half of the protein. This region is the most conserved among all four members of the family and does not contain any previously known DNA-binding motif. Defining of the EIN3/EILs DNA-binding domain will require further structural analysis of these proteins. Nevertheless, these four proteins, along with a fifth more recently identified homolog (EIL4; R. Solano and J.R. Ecker, unpubl.), possess several predicted α-helices, two of them rich in basic amino acids (Chao et al. 1997), that are good candidates to form a DNA-interaction surface.
Scanning mutagenesis of the DNA fragment containing the target site allowed us to determine the sequence requirements for the interaction. The defined target site includes two inverted repeats and is recognized by the protein as a dimer. Interestingly, the EIN3-binding site shares significant identity with sequences within the promoter region of the carnation GST1 gene that has been defined as necessary and sufficient for ethylene responsiveness. The conserved sequences are also present in the promoter regions required for ethylene responsiveness in the tomato E4 and LEACO1 genes (Montgomery et al. 1993; Itzhaki et al. 1994; Blume and Grierson 1997). This observation suggests that the EIN3 target site represents a PERE conserved in different species in which there are also orthologs of EIN3 (H. Klee; D. Cook, pers. comm.). Consistent with this hypothesis, one of the genes that contains this element (E4) has been previously proposed to be a primary ethylene response gene (Lincoln et al. 1987). The GCC element may be a secondary ethylene response element (SERE) present only in a subset of the ethylene-regulated genes (e.g., pathogenesis-related genes, HOOKLESS1 and some EREBPs) that may be regulated by a subgroup of the EREBP family of proteins. Experiments to test the functionality of the EIN3 target site as an autonomous ethylene response element are underway.
Cascades of transcription factors are a common theme in gene regulation, present in virtually all organisms from bacteria to humans, and involved in the regulation of processes as diverse as nitrogen fixation, embryogenesis, cell differentiation, response to extracellular signals or circadian rhythmicity (Kranz and Foster-Hartnett 1990; Jackle and Sauer 1993; Brun et al. 1996; Hwang et al. 1997; Allada et al. 1998; Darlington et al. 1998; Gekakis et al. 1998; Rutila et al. 1998). Rapid EIN3-dependent induction of ERF1 expression in response to ethylene, binding of EIN3 to the E4-like element in the ERF1 promoter and constitutive expression of ERF1 in EIN3-overexpressing plants provide three pieces of evidence supporting the hypothesis that ERF1 is an immediate target of EIN3. Binding of ERF1 to the GCC element in the promoters of ethylene-regulated genes, and constitutive activation of ethylene response genes and phenotypes in both etiolated seedling and adult plants in the ERF1 gain-of-function experiments provide additional evidence that ERF1 is a downstream ethylene signaling pathway gene.
The sequential action of EIN3 and ERF1 DNA-binding proteins adds a new level of complexity in the regulatory hierarchy of the ethylene-signaling pathway. Moreover, the strong similarity of the target site of EIN3 to an ERE previously identified in the promoters of ethylene response genes from a variety of species suggests that there is broad commonality among the downstream ethylene signaling events in plants. The existence of this hierarchy of transcription factors in the signaling pathway for ethylene may serve to amplify the input signal and/or provide a means to finely regulate the complex plant response to this gaseous plant growth regulator/stress signal.
Materials and methods
Strains and growth conditions
The Arabidopsis ecotype Columbia (Col-0) was the parent strain of all mutants and transgenic plants used in this work. Triple response screens were performed as described previously (Guzman and Ecker 1990). Plant growth in air and ethylene was carried out as described previously (Kieber et al. 1993).
Nucleic acid analysis
Total RNA extractions and Northern analysis were performed as described (Reuber and Ausubel 1996; Chao et al. 1997). β-Glucuronidase activity was assayed by incubation of the plants with the substrate of the enzyme (X-Gluc, 1 mg/ml) in sodium phosphate buffer for 18 hr. cDNA clones corresponding to ERF1 were isolated by hybridization of a size-selected cDNA library in λZAPII (Kieber et al. 1993). The probe, corresponding to a fragment of the tobacco EREBP1 gene, was obtained by PCR amplification with the following primers: EREBP1f, 5′-CACGCCATAGACATAATAC-3′, and EREBP1r, 5′-GCTACGATTCCTGTTCCTTCAG-3′.
ERF1 genomic sequences were isolated by hybridization of two BAC genomic libraries (TAMU and IGF; Choi et al. 1995; R. Wing, unpubl.; T. Altmann, unpubl.). The map position of ERF1 was obtained by PCR amplification of YAC pools with specific primers. PCR highlighted two YAC clones (CIC12H5 and CIC12H6) both located in the ABI3 contig.
Synthesis of proteins, DNA-binding reactions, and EMSA
Full-length ERF1, EIN3, and the EIN3 deletion derivatives were generated by in vitro translation (or cotranslation in the dimerization experiments) by use of the flexi-rabbit reticulocyte system (Promega) as described (Solano et al. 1997). PCR and Klenow labeling of promoter fragments and oligonucleotides, DNA-binding reactions and EMSAs were performed as described (Solano et al. 1995). In the competition experiments, the appropriate amount (see Fig. 2, legend) of the competitor (cold EBS, EBSm1, or EBSm2) was included in the reaction before the protein was added. Once the protein was added to the reaction, the mixture was incubated for 30 min on ice and then loaded onto the acrylamide gel.
Sequences of the mutant versions of EBS used in competition experiments were: EBSm1, 5′-GTTGTTTGGGATTCTTCGGGCATGTATCTTGAATCC-3′; EBSm2, 5′-GTTGTTTGGGATTCAAGCCCCATGTATCTTGAATCC-3′.
The promoter fragments of the Arabidopsis and bean basic-chitinase genes containing the GCC box were obtained by Klenow filling of the following overlapping primers:
b-CHIforward, 5′-GTTGATCACGAACCCGCCGCCTCATATTCATAATTA-3′; b-CHImutant, 5′-GTTGATCACGAACCCGTTGTTTCATATTCATAATTA-3′; b-CHIreverse, 5′TTTAACTTTAATTATGAATATGA-3′; CH5Bforward, 5′-CTTCACGCTTGGGAAGCCGCCGGGGTGGGCCCGCAG-3′; CH5Bmutant, 5′-CTTCACGCTTGGGAAGTTGTTGGGGTGGGCCCGCAG-3′; and CH5Breverse, 5′-AAACCTTTCTGCGGGCCCACCCC-3′.
Plant transformation
A 0.8-kb BamHI–KpnI fragment of ERF1 cDNA was cloned into BamHI–KpnI-digested pROK2 (Baulcombe et al. 1989). The C58 strain of Agrobacterium tumefaciens containing the above construct was used to transform Arabidopsis ecotype Col-0 and the ethylene insensitive mutants ein2-5, ein2-17, ein2-26, ein3-1, ein3-3, and ein5-1 by in planta vacuum infiltration (Bechtold et al. 1993). Kanamycin-resistant T1 plants were selected by plating seeds on Murashige and Skoog medium supplemented with 100 μg/ml kanamycin and transferring kanR seedlings to soil.
Yeast transformation and two-hybrid screening
Yeast strain Y190 was transformed by the PEG/lithium acetate method as described previously (Gietz et al. 1992). Growth conditions, screening procedures, and filter-lift assay for β-galactosidase activity were performed as described previously (Kim et al. 1997).
Acknowledgments
We thank A. Cashmore, J. Paz-Ares, S. Poethig, A. Theologis, and the members of the Ecker laboratory for critical reading of the manuscript and stimulating discussions. Plasmids and yeast strain for the two-hybrid screen were kindly provided by S. Elledge (Baylor University), and the Arabidopsis cDNA library in λ-ACT by A. Theologis (PGEC/UC. Berkeley). R.S. was a recipient of postdoctoral fellowships from the Human Frontiers of Science Program Organization and the Spanish Ministerio de Educacion y Ciencia, A.S. was a recipient of a predoctoral fellowship from the Department of Energy (ER20162-A003), and Q.C. was a recipient of a postdoctoral fellowship from the National Institutes of Health (1F32GM18003-01A1). This work was supported by a grant from the National Science Foundation (MCB-95-07166) to J.R.E.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note
GenBank accession numbers for the ERF1 cDNA and genomic sequences reported in this paper are AF076277 and AF076278, respectively.
Footnotes
E-MAIL jecker@atgenome.bio.upenn.edu; FAX 215-898-8780.
References
- Abeles F, Morgan P, Saltveit M. Ethylene in plant biology. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Allada R, White NE, Venus So W, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Baulcombe DC, Saunders GR, Bevan MW, Mayo MA, Harrison BD. Expression of biologically active viral satellite RNA from the nuclear genome of transformed plants. Nature. 1989;321:446–449. [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In Planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis plants. CR Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]
- Bevan M, Ecker J, Theologis A, Federspiel N, Davis R, McCombie D, Martienssen R, Chen E, Waterston B, Wilson R, Rounsley S, Venter C, Tabata S, Salanoubat M, Quetier F, Cherry JM, Meinke D. Objective: The complete sequence of a plant genome. Plant Cell. 1997;9:476–478. doi: 10.1105/tpc.9.4.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume B, Grierson D. Expression of ACC oxidase promoter-GUS fusions in tomato and Nicotiana plumbaginifolia regulated by developmental and environmental estimuli. Plant J. 1997;12:731–746. doi: 10.1046/j.1365-313x.1997.12040731.x. [DOI] [PubMed] [Google Scholar]
- Boller T, Gehri A, Mauch F, Vogeli U. Chitinase in bean leaves: Induction by ethylene, purification, properties, and possible function. Planta. 1983;157:22–31. doi: 10.1007/BF00394536. [DOI] [PubMed] [Google Scholar]
- Broglie KE, Biddle P, Cressman R, Broglie R. Functional analysis of DNA sequences responsible for ethylene regulation of a bean chitinase gene in transgenic tobacco. Plant Cell. 1989;1:599–607. doi: 10.1105/tpc.1.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun RP, Kim JB, Hu E, Altiok S, Spiegelman BM. Adipocyte differentiation: A transcriptional regulatory cascade. Curr Opin Cell Biol. 1996;8:826–832. doi: 10.1016/s0955-0674(96)80084-6. [DOI] [PubMed] [Google Scholar]
- Buttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. The ethylene signal transduction pathway in Arabidopsis: An emerging paradigm? Trends Biochem Sci. 1996;21:129–133. [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Chen Q, Bleecker A. Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol. 1995;108:597–607. doi: 10.1104/pp.108.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SD, Creelman R, Mullet J, Wing RA. Construction and characterization of a bacterial chromosome library from Arabidopsis thaliana. Weeds World. 1995;2:17–20. [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Deikman J. Molecular mechanisms of ethylene regulation of gene transcription. Physiologia Plantarum. 1997;100:561–566. [Google Scholar]
- Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn AE, Lee W-H, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- ————— Genes blossom from a weed. Nature. 1998;391:438–439. doi: 10.1038/35030. [DOI] [PubMed] [Google Scholar]
- Eyal Y, Meller Y, Levy S, Fluhr R. A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission zone-specific expression. Plant J. 1993;4:225–234. doi: 10.1046/j.1365-313x.1993.04020225.x. [DOI] [PubMed] [Google Scholar]
- Felix G, Meins FJ. Developmental and hormonal regulation of beta-1,3-glucanase in tobacco. Planta. 1986;167:206–211. doi: 10.1007/BF00391416. [DOI] [PubMed] [Google Scholar]
- Fields S, Song OK. A novel genetic system to detect protein protein interactions. Nature. 1990;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gietz D, Stjean A, Woods RA, Schiestl RH. Improved method for high-efficiency transformation of yeast-cells. Nucleic Acid Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart CM, Nagy F, Meins FJ. A 61 bp enhancer element of the tobacco beta-1,3-glucanase b gene interacts with one or more regulated nuclear proteins. Plant Mol Biol. 1993;21:121–131. doi: 10.1007/BF00039623. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz E. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz E. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Loftus TM, Mandrup S, Lane MD. Adipocyte differentiation and leptin expression. Annu Rev Cell Dev Biol. 1997;13:231–259. doi: 10.1146/annurev.cellbio.13.1.231. [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc Natl Acad Sci. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki H, Maxson JM, Woodson WR. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc Nat Acad Sci. 1994;91:8925–8929. doi: 10.1073/pnas.91.19.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackle H, Sauer F. Transcriptional cascades in Drosophila. Curr Opin Cell Biol. 1993;5:505–512. doi: 10.1016/0955-0674(93)90017-k. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz RG, Foster-Hartnett D. Transcriptional regulatory cascade of nitrogen-fixation genes in anoxygenic photosynthetic bacteria: Oxygen- and nitrogen-responsive factors. Mol Microbiol. 1990;4:1793–1800. doi: 10.1111/j.1365-2958.1990.tb02027.x. [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Cordes S, Read E, Fischer RL. Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proc Natl Acad Sci. 1987;84:2793–2797. doi: 10.1073/pnas.84.9.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath RB, Ecker JR. Ethylene signaling in Arabidopsis: Events from the membrane to the nucleus. Plant Physiol Biochem. 1998;36:103–113. [Google Scholar]
- Meller Y, Sessa G, Eyal Y, Fluhr R. DNA-protein interactions on a cis-DNA element essential for ethylene regulation. Plant Mol Biol. 1993;23:453–463. doi: 10.1007/BF00019294. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Goldman S, Deikman J, Margossian L, Fischer RL. Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proc Nat Acad Sci. 1993;90:5939–5943. doi: 10.1073/pnas.90.13.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell PJ, Calvert C, Atzorn R, Wasternack C, Leyser HMO, Bowles DJ. Ethylene as a signal mediating the wound response of tomato plants. Science. 1996;274:1914–1917. doi: 10.1126/science.274.5294.1914. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. Structure and expression of a tobacco beta-1,3-glucanase gene. Plant Mol Biol. 1990;15:941–946. doi: 10.1007/BF00039434. [DOI] [PubMed] [Google Scholar]
- ————— Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa RV, Cook DR. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Wurgler-Murphy S, Maeda T, Witten E, Thai T, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 ‘two-component’ osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Reuber TL, Ausubel FM. Isolation of arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell. 1996;8:241–249. doi: 10.1105/tpc.8.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, Venus So W, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samac DA, Hironaka CM, Yallaly PE, Shah DM. Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol. 1990;93:907–914. doi: 10.1104/pp.93.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F, Kitajima S, Koyama T, Yamada Y. Ethylene-induced gene expression of Osmotin-like protein, a neutral isoform of tobacco PR-5, is mediated by the AGCCGCC cis-sequence. Plant Cell Physiol. 1996;37:249–255. doi: 10.1093/oxfordjournals.pcp.a028939. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Sessa G, Meller Y, Fluhr R. A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol Biol. 1995;28:145–153. doi: 10.1007/BF00042046. [DOI] [PubMed] [Google Scholar]
- Shinshi H, Usami S, Ohme-Takagi M. Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol Biol. 1995;27:923–932. doi: 10.1007/BF00037020. [DOI] [PubMed] [Google Scholar]
- Solano R, Ecker JR. Ethylene gas: Perception, signaling and response. Curr Opin Plant Biol. 1998;1:393–398. doi: 10.1016/s1369-5266(98)80262-8. [DOI] [PubMed] [Google Scholar]
- Solano R, Fuertes A, Sanchez-Pulido L, Valencia A, Paz-Ares J. A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J Biol Chem. 1997;272:2889–2895. doi: 10.1074/jbc.272.5.2889. [DOI] [PubMed] [Google Scholar]
- Solano R, Nieto C, Avila J, Canas L, Diaz I, Paz-Ares J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 1995;14:1773–1784. doi: 10.1002/j.1460-2075.1995.tb07166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Nat Acad Sci. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto M, Roberts K, Dolan L. Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 1995;8:943–948. doi: 10.1046/j.1365-313x.1995.8060943.x. [DOI] [PubMed] [Google Scholar]
- Weigel D. The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell. 1995;8:659–671. doi: 10.1105/tpc.7.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G. A dissociation insertion causes a semidominant mutant that increases expression of TINY, an arabidopsis gene related to APETALA2. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]