Abstract
Germ-line stem cells (GSCs) serve as the source for gametogenesis in diverse organisms. We cloned and characterized the Drosophila piwi gene and showed that it is required for the asymmetric division of GSCs to produce and maintain a daughter GSC but is not essential for the further differentiation of the committed daughter cell. Genetic mosaic and RNA in situ analyses suggest that piwi expression in adjacent somatic cells regulates GSC division. piwi encodes a highly basic novel protein, well conserved during evolution. We isolated piwi homologs in Caenorhabditis elegans and humans and also identified Arabidopsis piwi-like genes known to be required for meristem cell maintenance. Decreasing C. elegans piwi expression reduces the proliferation of GSC-equivalent cells. Thus, piwi represents a novel class of genes required for GSC division in diverse organisms.
Keywords: Stem cell, germ line, piwi, cell fate, asymmetric division
The ability of stem cells to self-renew and to produce a large number of differentiated progeny is critical for the development and maintenance of a wide variety of tissues in organisms ranging from insects to mammals (for review, see Lin 1997; Lin and Schagat 1997; Morrison et al. 1997; Potten 1997). This self-renewing ability is controlled both by extrinsic signaling and by cell-autonomous mechanisms (for review, see Lin and Schagat 1997; Morrison et al. 1997). Cell-autonomous mechanisms have been elucidated in a few stem cell models such as neuroblasts and germ-line stem cells (GSC) in Drosophila (Deng and Lin 1997; Lin and Schagat 1997), whereas the role of extrinsic signaling in controlling stem cell division has been implicated in several systems (Morrison et al. 1997). For example, the proliferation and differentiation of mammalian stem cells in the hematopoietic, epidermal, and nervous systems depend on extrinsic signals that act on specific receptors on the stem cell surface (Morrison et al. 1997). In diverse organisms ranging from invertebrates to mammals, the proliferation of germ cells, some of which possess stem cell properties, has been postulated, and, in some cases, shown to be regulated by neighboring nonmitotic somatic cells (Lin 1997). Particularly, in Caenorhabditis elegans, cell–cell interactions between the somatic distal tip cell (DTC) at the end of each gonadal arm and the underlying mitotic germ-line nuclei via the lag-2/glp-1 signaling pathway provides a paradigm for soma-germ-line interaction (for review, see Kimble and Simpson 1997). The glp-1 pathway is required to maintain a population of mitotically active nuclei in the germ line. However, few molecules and/or mechanisms identified in a particular type of stem cells have been shown to be applicable to other stem cell systems. For example, the glp-1 equivalent pathway in Drosophila does not appear to play an obvious role in regulating GSC division and maintenance (Ruohola et al. 1991; Xu et al. 1992).
Drosophila has been an effective model for studying mechanisms that are conserved among diverse developmental systems. We show here that this is also the case for the study of stem cells. In Drosophila, stem cells exist in the germ line at the apical tip of the ovariole, the functional unit of the ovary. In the ovariole, GSCs are located in a specialized structure called the germarium, as first proposed by Brown and King (1962, 1964). The existence of GSCs was strongly supported by germ-line clonal analysis (Schüpbach et al. 1978; Wieschaus and Szabad 1979) and directly verified by laser ablation (Lin and Spradling 1993). In each germarium, two to three GSCs contact the somatic basal terminal filament cells. GSCs undergo oriented asymmetric divisions to produce a daughter stem cell that remains associated with the terminal filament and a differentiated daughter, the cystoblast, that becomes displaced one cell away from the terminal filament (Deng and Lin 1997; Lin and Spradling 1997). The germ-line cyst then becomes enveloped by follicle cells produced by somatic stem cells (Margolis and Spradling 1995) to form an egg chamber, which buds off the germarium, joins pre-existing egg chambers in a linear array to form an ovariole, and eventually develops into a mature egg (for review, see King 1970; Spradling 1993). This assembly line organization, with each egg chamber representing a differentiated stem cell product whose position along the ovariole corresponds to its birth order, provides a unique opportunity to study stem cell division.
The self-renewing asymmetric division of GSCs in the Drosophila ovary is known to be controlled both by an intracellular mechanism (Deng and Lin 1997) and by cell–cell interactions (Lin and Spradling 1993). The intracellular mechanism involves a cytoplasmic organelle termed the spectrosome that controls the orientation of GSC division (Lin et al. 1994; Deng and Lin 1997). The cell–cell interaction mechanism involves terminal filament cells, as shown by laser ablation studies (Lin and Spradling 1993). Recently, dpp has been shown as a key signaling molecule required for GSC division and maintenance (Xie and Spradling 1998). It is possible that the dpp signal emanates from somatic cells. Alternatively, the dpp signal may originate from the germ line or even within GSCs, like its mammalian homologs (Zhao et al. 1996). Thus, it remains to be determined what genes are expressed in somatic signaling cells to regulate GSC division and maintenance.
To explore further the stem cell mechanism in the germ line, three additional genes, piwi, pumilio (pum), and fs(1)Yb (Yb) have been recently identified to be essential for GSC maintenance in Drosophila (Lin and Spradling 1997; Forbes and Lehmann 1998; F.J. King and H. Lin, unpubl.). Among these genes, piwi is defined by recessive mutations that cause failure in GSC maintenance in both females and males (Lin and Spradling 1997). piwi mutant gonads contain a normal number of GSCs at the onset of gametogenesis. However, mutant adult gonads are devoid of GSCs and contain only a very small number of gametes that is approximately equal to or less than the wild-type number of GSCs. These observations reveal that piwi is essential for GSC maintenance in both males and females.
To investigate the nature of the piwi-mediated mechanism, we report here that piwi mediates a somatic signaling mechanism essential for the division and maintenance of GSCs in Drosophila. Moreover, we show that piwi encodes a novel basic protein well conserved in C. elegans and humans, and also conserved in Arabidopsis, in which two piwi-like genes are known to be required for meristem cell maintenance (Bohmert et al. 1998; Moussian et al. 1998). We demonstrate that the C. elegans piwi homolog is also required for germ-line proliferation and maintenance. Thus, piwi represents an essential stem cell gene existing in diverse organisms.
Results
piwi mutations eliminate the self-renewing division of germ-line stem cells
Previous studies showed that piwi1 mutant ovaries contain a normal number of mispositioned GSCs at the onset of oogenesis at the late third instar larval stage, which, however, leads to an equal or somewhat smaller number of gametes in the adult gonads that no longer contain GSCs (Lin and Spradling 1997). This failure of germ-line maintenance could be due to the following: (1) the differentiation of GSCs without self-renewing divisions; (2) a defect in the asymmetry of GSC division, producing aberrant germ cells that eventually degenerate; and/or (3) a secondary defect influenced by abnormal ovary differentiation.
To examine whether the failure of germ-line maintenance is a secondary defect due to abnormal ovary development, we examined the ovarian morphology of piwi2 and piwi3 mutants that also fail to maintain GSCs. The examination relied on Nomarski optics as well as markers that specifically identify germ cells, somatic cells, and spectrosomes/fusomes, germ-line specific organelles that indicate individual stages of germ cell development (Lin et al. 1994; Lin and Spradling 1995; see Materials and Methods). These mutant ovaries show normal morphology at the third instar larval stage (Fig. 1, cf. E and A). Their germ-line cells are normal in number and are correctly positioned along the medial plane of the ovary. Moreover, the expected number of terminal filaments are forming, so that at the pupal stage, the ovary differentiates normally, partitioning GSCs and their daughter cells correctly into individual germaria and ovarioles (Fig. 1, cf. F and C). GSCs are able to divide several times to provide a normal complement of germ cells to the germarium. Yet, GSCs subsequently fail to continue self-renewing divisions, and the existing germ-line cysts often degenerate during the late pupal stage (Fig. 1G) so that the adult ovarioles contain germaria lacking germ lines and fewer egg chambers than expected (Fig. 1H). These observations suggest that the failure of GSC maintenance in piwi mutants is not a secondary defect due to abnormal ovary development.
Figure 1.
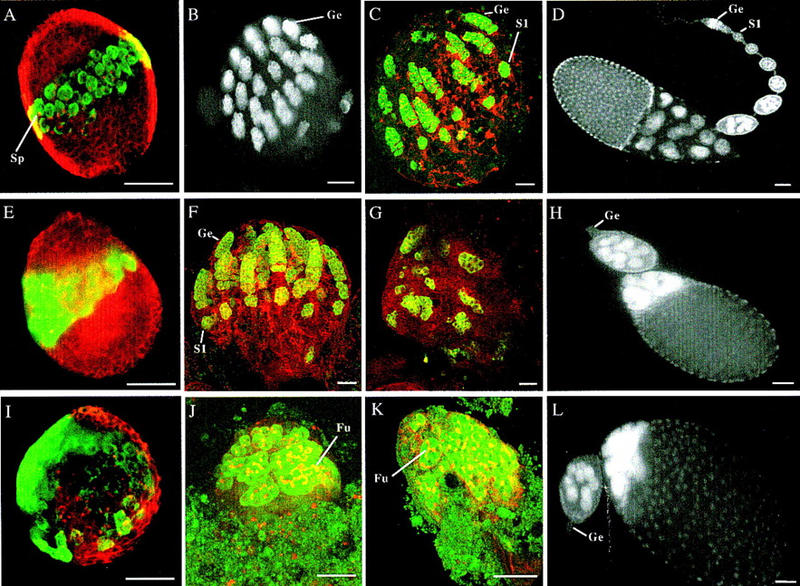
piwi is required for the self-renewing division of GSCs during oogenesis. (A,E,I) Confocal images showing third instar larval ovaries from wild-type, piwi2, and piwi1 mutant flies, respectively, stained for a germ-line-specific marker VASA in green and spectrin in red to mark somatic cells and spectrosomes/fusomes. GSCs are positioned medially along the anterioposterior axis in the wild-type (A) and piwi2 ovary (E); however, GSCs are mispositioned in the piwi1 mutant ovary (I). Spectrosomes (Sp) appear as red dots in germ cells. (B,J) Confocal images of 24-hr pupal ovaries from wild-type and piwi1 mutant flies, respectively, with B stained solely for VASA and J is stained for both VASA in green and spectrin in red. At this stage, the wild-type ovary has partitioned into individual germaria (Ge), which contain GSCs and early germ-line cysts. In piwi1 mutant ovaries, germ cells still appear to have been partitioned, although the partition is distorted by the highly differentiated large germ-line cysts containing elaborate fusomes (Fu). The number of cysts does not exceed the number of GSCs. (C,F,K) Confocal images of 48-hr pupal ovaries from wild-type, piwi2 and piwi1 mutant flies, respectively, stained with VASA in green and spectrin in red. By this stage, in both wild-type (C) and piwi2 mutant ovaries (F), germaria have developed fully and stage 1 egg chambers (S1) have budded off the germarium in synchrony because of continued GSC division. However, in piwi1 mutant ovaries (K), the number of germ-line cysts remains unchanged despite their continued differentiation. (G) Confocal image of a 72-hr pupal ovary from piwi2 mutant flies stained with VASA in green and spectrin stained in red. At this stage, GSCs fail to self-renew and some existing germ-line cysts begin to degenerate. (D,H,L), DAPI images of 0-to-1-day old adult ovarioles from wild-type, piwi2, and piwi1 mutant flies, respectively. Both piwi1 (L) and piwi2 (H) mutant ovarioles typically contain two normal or abnormal egg chambers connected to a germaria lacking germ lines (Ge). In contrast, wild-type ovarioles contain a long string of developing egg chambers produced continuously by the germarium (D). piwi3 phenotype is indistinguishable from that of piwi2 (data not shown). Bars in all panels, 50 μm.
To examine whether the failure of GSC maintenance is due to differentiation without self-renewing divisions or due to aberrant divisions followed by degeneration, we examined more closely the ovarian development of the piwi1 mutant. As shown previously (Lin and Spradling 1997), the piwi1 third instar larval ovaries contain a normal number of 50–70 presumed GSCs that are mispositioned (Fig. 1I). However, these GSCs still seem to, and must have, partitioned during subsequent pupal development (Fig. 1J,K), because the number of ovarioles formed in most adult ovaries is within the normal range (13 ± 3 in mutant versus 15 ± 3 in wild type). Hence, the effect of piwi1 on GSC mispositioning is not essential for oogenesis.
The main oogenic defect of the piwi1 mutant is the differentiation of GSCs without self-renewing divisions immediately following the initiation of oogenesis. At this stage in wild-type ovaries, GSCs in 16–23 newly formed germaria have initiated asymmetric divisions to generate multiple developing germ-line cysts (Fig. 1B). However, in mutant ovaries, GSCs and their immediate differentiated daughters, cystoblasts, are undetectable, as indicated by the absence of spectrosome-containing germ cells (Fig. 1J). Instead, most ovaries contain differentiated germ-line cysts whose number approximately equals that of GSCs. These cysts are much larger in size and contain two- to multicell stage fusomes, indicating their differentiating state (Fig. 1J,K). By the adult stage, most ovarioles contain only two normal or abnormal egg chambers derived from these cysts, but no other germ-line cells (Fig. 1L). This defect contrasts the development of wild-type ovaries, in which ovarioles contain a fully developed germarium and a stage 1 egg chamber by the 48-hr pupal stage (Fig. 1C) and have produced multiple egg chambers by the adult stage (Fig. 1D). This observation indicates that the mutant GSCs have differentiated into germ-line cysts without self-renewing divisions (Fig. 1J, K).
The abnormal fusome morphology often seen in cysts suggest that they contain more or less than the normal number of 16 cells (Fig. 1J), which we saw in adult egg chambers (Fig. 1L). This is consistent with previous observations of the piwi1 phenotype and the conclusion that piwi may have an additional function in subsequent cyst formation (Lin and Spradling 1997).
Molecular cloning of Drosophila piwi
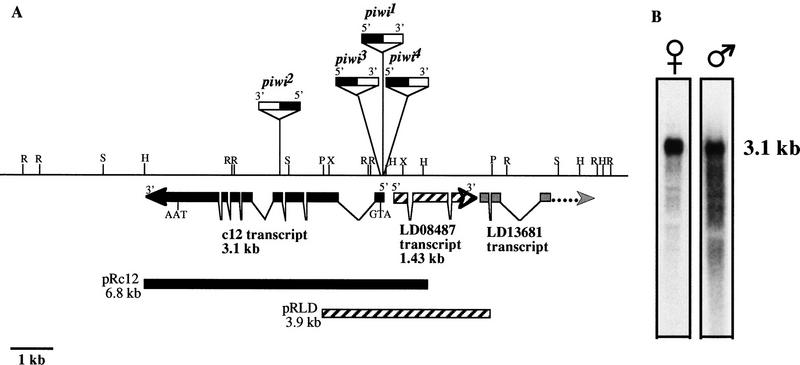
To study the molecular nature of the piwi-mediated mechanism, a 15-kb genomic region flanking the piwi insertions was cloned (see Materials and Methods). Candidate transcripts in the region were identified, and a 6.8-kb genomic fragment (pRc12) containing a 3.1-kb transcript interrupted by piwi mutations (transcript c12) was able to rescue the piwi phenotype completely (Fig. 2A,B, Table 1). An overlapping 3.9-kb genomic fragment (pRLD) containing a 1.43-kb transcript immediately adjacent to the piwi mutations fails to rescue the piwi phenotype (Fig. 2A; Table 1). Therefore, we conclude that the c12 transcript is the piwi mRNA.
Figure 2.
Molecular map of the piwi locus and piwi Northern analyses. (A) Molecular map of the piwi region. Restriction enzyme map for 15 kb of genomic DNA surrounding the piwi mutations is shown. Three transcripts are represented by thick arrows indicating their relative positions, sizes, splice sites, and direction of transcription. The relative insertion sites of the P-element alleles are indicated above the restriction map with orientation of insertion indicated by black (5′) and white (3′) boxes. The P elements are not shown according to scale, with piwi2 insertion partially mapped. (R) EcoRI; (H) HindIII; (P) PstI; (S) SalI; (X) XhoI. (B) piwi RNA is present in piwi1/CyO adult males and females. The female and male lanes contain ∼10 and 1.5 μg of poly(A)+ RNA, respectively. They are exposed for 3.5 and 12 hr, respectively.
Table 1.
piwi1rescue results by pRc12 and pRLD
|
|
piwi1/ piwi1
|
piwi1/ CyO
|
pRc12 (piwi1/piwi1)
|
pRLD (piwi1/piwi1)
|
|---|---|---|---|---|
| Females | 0 (136) | 100 (45) | 100 (39) | 0 (81) |
| Males | 0 (119) | 97.8 (47) | 88.4 (43) | 0 (64) |
The percentage of fertile flies for each genotype and sex are indicated. The number of individuals tested for fertility (n) is in parentheses. Data are pooled from six and four independent insertion lines on the X and third chromosomes for pRLD and pRc 12, respectively. Not included are two other pRc12 rescue lines that showed sex-specific restoration of fertility, presumably due to position effects stemming from the insertion site.
Drosophila PIWI represents a novel class of conserved proteins
To study the structure of the piwi gene, we isolated and sequenced two overlapping cDNA clones that together represent the 3.1-kb full-length c12 mRNA (see Materials and Methods). The c12 cDNA sequence contains a large ORF starting at +84 bp and encoding a protein of 843 amino acid residues with a predicted molecular mass of 97.2 kD (Fig. 3A). Multiple stop codons in all three frames are present upstream of the putative start codon (data not shown). Downstream of the ORF is a 432-bp 3′ untranslated region (UTR) followed by a 55-bp poly(A) tail not encoded in the genomic sequence.
Figure 3.


Alignment of predicted PIWI protein sequence with its homologs. (A) Sequence alignment of predicted PIWI proteins from Drosophila (PIWI), C. elegans (PRG-1 and PRG-2) and human (HIWI). The full-length amino acid sequence of PRG-1 and PRG-2 was deduced from the nucleotide sequence of two independent cDNA clones. Partial amino acid sequence of HIWI was deduced from the nucleotide sequence of a partial cDNA clone from a human testes library. The alignments were produced with CLUSTALW software (DNAStar). Identical residues to a consensus are highlighted. (B) The piwi box domain in 22 different proteins. The alignments were generated by Block Maker analysis (Henikoff et al. 1995) against amino acids 740–782 of the predicted piwi sequence. The amino acid sequences were deduced from genomic DNA sequences for C. elegans (Ce G) or from cDNA (EST) sequences (E) for Rattus sp. (Rs) and Oryza sativa (Os). GenBank accession numbers and cosmid designations (Ce) are indicated for each sequence. Residues identical to the consensus are highlighted. Shown below the alignments: Residues that are absolutely conserved are shown as uppercase letters and residues that are 80% or more conserved are shown as lowercase letters; dashes indicate spacing.
The PIWI protein is a highly basic (pI 9.6) novel protein with no obvious similarity to other known proteins or functional motifs in the databases. It is characterized by alternating basic and acidic regions and is particularly basic over the carboxy-terminal 100 amino acid residues. Hydropathy analysis indicates that the PIWI protein contains no significant local hydrophobic regions that could be potential signal peptide or transmembrane domains (data not shown). PSORT analysis (Nakai and Kanehisa 1992) predicts nuclear localization of the PIWI protein. The protein has 21 conserved protein kinase C phosphorylation sites, 14 casein kinase 2 phosphorylation sites, and 4 tyrosine phosphorylation sites, indicating its potential as a phosphorylation target.
To determine whether the PIWI protein is conserved during evolution, we searched for its homologous sequences at the protein level (see Materials and Methods) and identified two ORFs of unknown function from C. elegans and an expressed sequence tag (EST, GenBank accession no. AA430311) from a human testis cDNA library. We obtained and sequenced cDNAs for the two C. elegans genes, herein named prg-1 and prg-2 (piwi-related gene) to verify their homology to Drosophila piwi. The prg-1 and prg-2 genes share 40.1% and 38.5% amino acid identity to piwi, respectively, over their entire length. In the carboxy-terminal 104 amino acid region, the homology increases to 55.8% and 56.7%, respectively. Moreover, prg-1 and prg-2 are 90% identical to each other over their full length and 98% identical at the carboxyl terminus. This high degree of homology suggests that prg-1 and prg-2 may represent a gene duplication event. The two clones differ primarily in that prg-1 is 60 amino acids longer at the amino terminus than prg-2. Using ACeDB (Thierry-Mieg and Durbin 1992), we mapped prg-1 to chromosome I between unc-15 and gld-1 in cosmid D2030 and prg-2 to chromosome IV, between unc-44 and smg-7 on cosmid CO1G5. To isolate human piwi homologs, the human EST (0.9 kb) clone was sequenced and used to screen a human testis cDNA library. A resulting 2.3-kb partial cDNA, herein named hiwi (for human piwi), shows 47.1% identical amino acid sequence to the Drosophila piwi over its full length, with 58.7% identity at the carboxyl terminus (Fig. 3A). Interestingly, we did not find any piwi-related sequences from bacteria or yeast genomes whose entire sequences are known. This is consistent with the stem cell-related function of piwi and potentially of piwi-like genes specific for multicellular organisms.
In addition to the above piwi homologs, we identified a large number of putative and known proteins from various animals and plants that share significant homology with piwi solely at their respective carboxyl termini. This indicates that piwi is a member of a large novel gene family. Within this family are 13 additional putative C. elegans proteins and three Arabidopsis thaliana proteins, two of which are required for meristem cell divisions (Bohmert et al. 1998; Moussian et al. 1998). Particularly, between piwi and the three Arabidopsis genes, ZWILLE (ZLL), argonaute (ago), and argonaute-like (Bohmert et al. 1998; Moussian et al. 1998), an overall homology of ∼20% amino acid identity was observed. The homology increases to 32%–52% identity in four regions, 30–100 amino acid residues each, located throughout the length of the PIWI protein, including the highly conserved carboxy-terminal region. Given the role of ZLL and ago in meristem cell division (Bohmert et al. 1998; Moussian et al. 1998), this homology may have important implications for a conserved stem cell mechanism (see Discussion). PIWI, PRG-1, PRG-2, and HIWI differ from ZLL and AGO proteins, and especially from the 13 additional putative C. elegans proteins, predominantly at the amino terminus, suggesting that this region may be involved in piwi-specific function. The carboxy-terminal conservation suggests that this region of PIWI may contain a novel functional domain that plays an important role for the general activity of these proteins in diverse biochemical processes, with the amino terminus rendering the specificity of the activity.
To examine the carboxy-terminal region of homology more closely, we aligned these sequences using Block Maker, which reveals characteristic regions of protein families (Henikoff et al. 1995). Block Maker analysis identified a 43 amino acid domain conserved among all 22 proteins, within which 5 residues are absolutely conserved with defined spacing. Eight more residues are also conserved with defined spacing among all known genes across the phyla except for several C. elegans ORFs with unknown function (Fig. 3B). We refer to this region as the PIWI box and suggest that this protein signature represents a novel conserved functional motif. PIWI thus represents a novel class of evolutionarily conserved proteins with potentially conserved functions.
Drosophila piwi mRNA is present in both germ line and soma during oogenesis
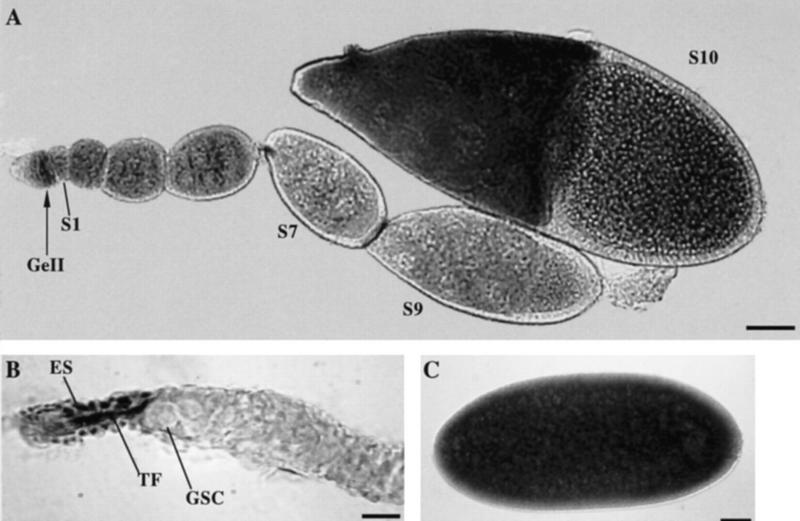
To investigate in which cells piwi is expressed to regulate GSC division, we examined the expression pattern of piwi during oogenesis by in situ hybridization of whole mount Drosophila ovaries with DIG-labeled DNA probes prepared from the piwi cDNA clone (see Materials and Methods). piwi mRNA is detected in the somatic terminal filament cells apical to GSCs in the germarium and anterior sheath cells (Fig. 4B) as well as in the germ line. In the germ line, it is first abundantly expressed in region 2 of the germarium in which 16-cell germ-line cysts are formed, persists at a lower level through stages 1–6 of oogenesis, is at its lowest level between stage 7–9, becomes strongly expressed again at stage 10, and eventually accumulates in early embryos (Fig. 4A,C; for staging, see King 1970). Given that removing sheath cells does not affect oogenesis, whereas the terminal filament cells play a role in regulating GSC division (Lin and Spradling 1993), it is likely that the somatic expression in the terminal filament is responsible for piwi function in regulating GSC division.
Figure 4.
piwi mRNA expression in Drosophila ovaries. (A) A complete Ore-R ovariole showing germ-line piwi expression in region II of the germarium (GeII), and in early stage egg chambers up to stage 10 (S10). (B) In the germarium, the piwi RNA is also detected in the terminal filament cells (TF) and epithelial sheath cells (ES) apical to GSCs. (C) The piwi RNA is uniformly present in early embryos. Bars, 50, 10, 50 μm for A, B, and C, respectively.
piwi expression in apical somatic cells regulates GSC division while its expression in the germ line provides a maternal component for embryogenesis
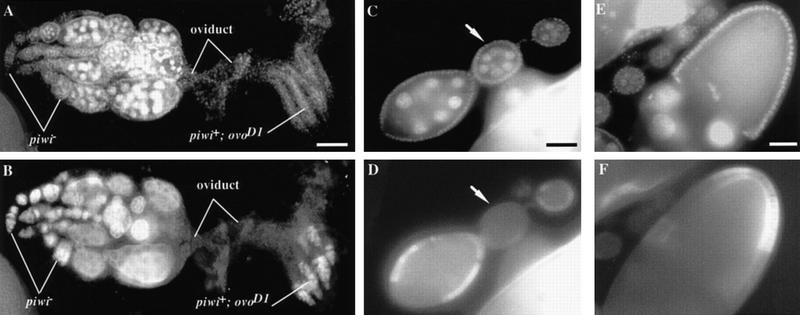
To examine the roles of somatic and germ-line piwi expression, we conducted genetic clonal analyses on piwi1 and piwi2 mutations. We first tested the germ-line requirement of piwi by germ-line clonal analysis using the FLP–DFS technique (Chou and Perrimon 1996; see Materials and Methods). The ovoD1 dominant mutation blocks oogenesis at the very beginning stage in a cell-autonomous manner, so that piwi+ ovoD1 females contain germ cells that never differentiate beyond the germarium (Fig. 5A,B). In this ovoD1 background, we generated germ-line cells homozygous for either the piwi1 or piwi2 mutation by applying a heat-shock treatment at the second and early third instar larval stage. The resulting adult females developed some completely normal ovarioles in which germ-line cells at all stages of oogenesis were observed (Fig. 5A,B) and mature eggs were continuously being produced (see Table 2). This result demonstrates that removing the piwi+ ovoD1 chromosome from the germ line allows oogenesis, including GSC division, to occur normally. Thus, the requirement of piwi for GSC division does not reside in the germ line but in somatic cells.
Figure 5.
piwi function is dispensable in the germ-line and differentiated follicle cells for GSC division. (A,B) piwi is dispensable in the germ line. A pair of ovaries containing ovarioles with homozygous piwi− germ-line clones and ovarioles with piwi+ ovoD1 germ line stained for DAPI to mark DNA (A) and for VASA to mark germ cells (B). The piwi+ ovoD1 germ-line cells are arrested at the beginning of oogenesis in the germarium because of the ovoD1 mutation. In contrast, in ovarioles in the left ovary containing piwi− germ-line clones, GSCs have divided normally, giving rise to a progression of morphologically wild-type egg chambers that eventually develop into mature eggs. This indicates that piwi is not required in the germ line for GSC division and subsequent steps of oogenesis. Bar in A, 100 μm. (C–F) piwi− clones in differentiated follicle cells do not effect normal GSC division and egg chamber development. Egg chambers were stained with DAPI (C,E) and a monoclonal anti-MYC antibody (D,F). Absence of Myc staining is indicative of homozygous piwi− clones. Induction of somatic clones around an entire early stage egg chamber such as the ones in C and D (see arrow) had no effect on GSC division and oogenesis. Similarly, removal of piwi function from some follicle cells of later stage egg chambers also had no effect on oogenesis (E,F). Therefore, piwi function is dispensable in differentiated follicle cells for oogenesis including GSC division. Bars in C and E, 50 μm.
Table 2.
piwi function in GSC division is soma-dependent while function in embryogenesis is germ line dependent
| FLP/FRT ovoD1-induced germ-line clones
|
Heat shock
|
N
|
Avg. no. of eggs laid/female
|
Embryos hatched (%)
|
|---|---|---|---|---|
| piwi1FRT/ovoD1, FRT | ||||
| × Ore-R males | + | 37 | 50.4 | 0 |
| piwi2FRT/ovoD1, FRT | ||||
| × Ore-R males | + | 26 | 66.7 | 0 |
| piwi1FRT/ovoD1, FRT × Ore-R males | − | 44 | 0 | N.A. |
| piwi2FRT/ovoD1, FRT × Ore-R males | − | 26 | 0 | N.A. |
| ovoD1, FRT/CyO × Ore-R males | + | 20 | 0 | N.A. |
| ovoD1, FRT/CyO × Ore-R males | − | 10 | 0 | N.A. |
In all tester and control crosses, a number (N) of newly eclosed females were allowed to lay eggs at 25°C for 2 days. In both piwi1and piwi2testers, germ line develops beyond that of ovoD1 mutant ovarioles only following heat shock, giving rise to numerous eggs. This indicates the generation of piwi− germ-line clones and the dispensability of piwi function in the germ line for normal GSC division. However, none of the eggs develop beyond embryogenesis, indicating that germ line piwi expression is required as a maternal component for embryogenesis. The ovoD1, FRT control females lay no eggs either with or without heat shock. (N.A.) Not applicable.
Eggs produced from homozygous piwi1 or piwi2 GSC clones are arrested in embryogenesis, not rescuable by the paternal piwi+ gene (see Table 2). Approximately 30% of the arrested embryos show severe mitotic defects during cleavage stage. The remaining embryos show various morphological defects during late embryogenesis, including a high frequency of severe deformation of the head region (data not shown). These defects demonstrate that piwi expression in the germ line provides an essential maternal contribution for embryogenesis.
To determine whether piwi is required in somatic cells outside the ovary for GSC maintenance, we transplanted wild-type germaria into the abdominal cavity of homozygous piwi2 females (see Materials and Methods), which produced 17 ± 0 egg chambers (n = 13) after 7 days of incubation in the host. Parallel transplantation into wild-type host yielded 16 ± 2 egg chambers (n = 5), indicating that the wild-type germaria continued oogenesis at a normal rate in the piwi2 females. Hence, piwi is not required in extra-ovarian cells for GSC maintenance.
To determine in which group of ovarian somatic cells piwi is required for GSC maintenance, we used a similar FRT-mediated technique for somatic clonal analysis of piwi1 and piwi2, marking the piwi+ chromosome with a transgenic Myc tag so that homozygous piwi− somatic cells are identified by their lack of Myc epitope expression (Xu and Rubin 1993; Fig. 5C–F; see Materials and Methods). By inducing mitotic recombination at the second and third instar larval stages, we generated piwi− somatic clones throughout ovarioles, with some egg chambers completely covered by piwi mutant follicle cells, (Fig. 5C,D). These egg chambers developed normally, indicating that piwi function is not required in follicle cells for egg chamber development. Because follicle cells are derived from their precursor cells in region II of the germarium (Margolis and Spradling 1995), this suggests that piwi is not required in somatic cells from germarial region II on for GSC division and ovarian development. These results, together with the piwi expression pattern in situ, suggest that piwi is required in somatic cells in the anterior-most tip of the germarium to regulate GSC division.
piwi-like genes in C. elegans are also required for germ-line self-renewal
The high degree of sequence homology between piwi and its homologs in other organisms suggests a potential functional conservation. We first tested this hypothesis in C. elegans. Guo and Kemphues (1995) have shown that the injection of specific antisense RNA into the germ-line syncytium of C. elegans eliminates maternal and zygotic gene activity, producing a gene-specific loss-of-function effect that may persist through several generations. This technique, as refined by Fire et al. (1998) and termed RNA-mediated interference (RNAi), was used to assess the function of prg-1 and prg-2. Given the extremely high homology between prg-1 and prg-2, we used an anti-prg-1 RNA for injection to interfere with the function of both genes (see Materials and Methods). Herein, we designate the F1 progeny of the injected worms as prg-RNAi worms for simplicity.
In wild-type C. elegans, two germ-line precursor cells, Z2 and Z3, give rise to ∼ 2000 germ cells in the adult hermaphrodite. Germ-line proliferation occurs throughout most of larval development (L1–L4) and continues in the adult. This proliferation and maintenance of the germ line requires signals from the DTC at the tip of each gonadal arm (see introductory section). During gonadal development, DTC migration results in the formation of two U-shaped gonadal arms by the L4 stage. Germ-line proliferation is limited to the distal end of each arm, forming a mitotic proliferation zone (MPZ), which serves as the GSC equivalent in C. elegans (Fig. 6B). Moving proximally, near the U turn of the gonad, germ cells enter meiotic prophase and then further differentiate into gametes at the proximal half of each arm, producing sperm in L4 and then oocytes in young adults.
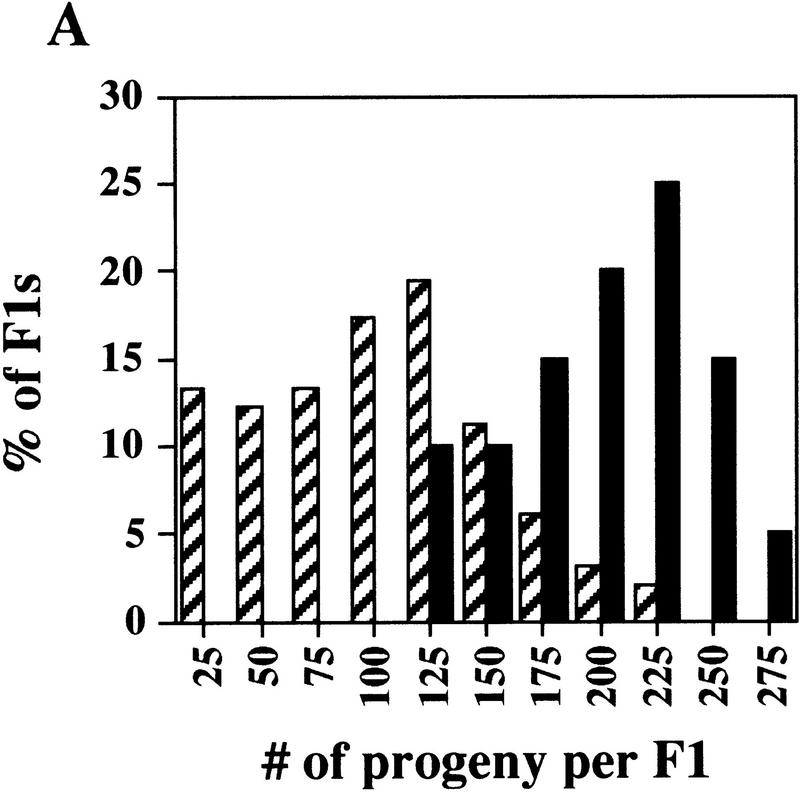
Figure 6.
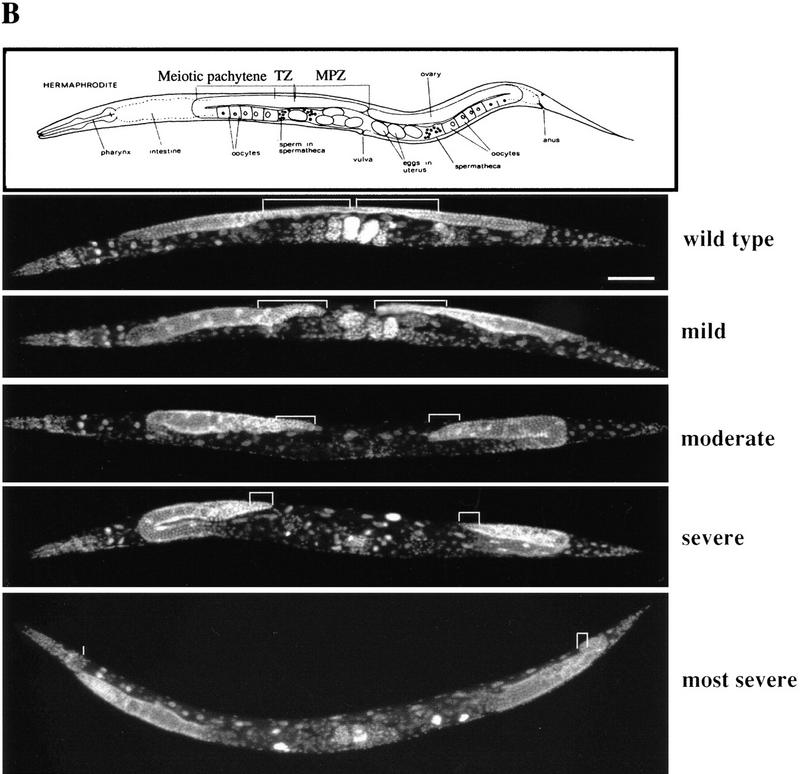
prg–RNAi significantly reduces germ-line proliferation in C. elegans. (A) Reducing prg activity affects fertility. The graph indicates the number of progeny produced by a single F1 worm from an RNAi prg-1 or pBST-injected mother vs. the percentage of F1 animals that produced that given number of progeny. Comparison of prg–RNAi (hatched bars) with pBST RNAi (solid bars) animals reveals two distinctive distributions. (B) Reducing prg activity results in reductions in both the MPZ and the mitotic index. Variable defects in both the size of the MPZ and the mitotic index were observed by DAPI staining. The MPZ in each micrograph is denoted in white brackets. In moderate to severely defective animals, a 50% reduction in the number of mitotic nuclei and a concomitant fivefold reduction in the mitotic index were typically observed. Bar, 100 μm for all panels.
We examined the phenotype of prg-RNAi worms both by quantitating their fertility and by assessing their gonadal and germ-line development via DAPI staining and Nomarski optics. As controls for RNA injection, worms were injected with BlueScript plasmid RNA sequences (see Materials and Methods).
The prg–RNAi worms show three major aspects of germ-line defects. First, as indicated by the distribution of the number of progeny per F1 animal (Fig. 6A), the fertility difference between the prg–RNAi and control worms is highly significant (t value = 7.93, P < 0.0001). On average, prg–RNAi worms produced 92 ± 53 progeny (n = 99), with 75.5% of prg–RNAi worms giving rise to <125 progeny. In contrast, control RNAi animals on average produced 191 ± 40 progeny (n = 20), with 90% of these animals giving rise to >125 progeny. The significant reduction of the fertility, as well as other defects described below, may still only reflect a partial loss of prg-1 and prg-2 function, because the RNAi technique is known to phenocopy partial rather than complete loss-of-function mutants (Kuwabara 1996).
Second, DAPI staining reveals a dramatic shortening of the MPZ in prg–RNAi worms (Fig. 6B). On average, adult prg–RNAi worms exhibited a 50% reduction in the number of mitotic nuclei as compared with control animals. Associated with the reduction of the mitotic zone is a gonadal shortening (Fig. 6B). Fifty-seven percent of prg–RNAi worms (n = 287) exhibited a moderate to severe shortening. In the most severe case, the gonadal arm never made the U turn. The number of sperm produced in these worms is greatly reduced as well (data not shown). In contrast, only 7% of the BlueScript–RNA-injected control animals (n = 548) exhibited a mild gonadal shortening. The above observed gonadal defects in prg–RNAi worms suggest that prg-1 and prg-2 are essential for germ-line proliferation and maintenance. Because the gonadal shortening may be due to a defect in DTC migration, it is also possible that prg-1 and prg-2 may play a role in proper gonadogenesis.
Third, in addition to the MPZ shortening, the mitotic index in the remaining mitotic zone is further reduced by 5.5-fold from 7.1 ± 2.2% (n = 1,339 nuclei, ns = 10 worms) in the control RNAi-injected worms to 1.3 ± 0.8% (n = 1,409 nuclei, ns = 10 worms) in prg–RNAi worms with mildly to moderately shortened gonads (see Materials and Methods). This indicates the important role of prg-1 and prg-2 in maintaining the mitotic ability of the germ-line nuclei.
Discussion
The ability of stem cells to self renew remains a mystery in modern biology. In this paper, we have presented the genetic and molecular analysis of piwi, a gene essential for stem cell self-renewal and maintenance in the Drosophila germ line. We have extended our analyses to its homologs in C. elegans, isolated a human homolog, and identified two piwi-like genes in Arabidopsis that are known to maintain meristem cells. This work reveals a new class of genes that may be essential for stem cell division in multicellular organisms ranging from invertebrates to humans and plants.
Drosophila piwi is required for the self-renewing divisions of stem cells in the germ line
The piwi gene has been shown previously to be required for both female and male GSC maintenance (Lin and Spradling 1997). In this study, we provide evidence that piwi achieves this predominantly by promoting the self-renewing division of GSCs. In a germarium, it normally takes at least 10 hr for a GSC division to occur and 5 days for a cystoblast to develop into a stage-one egg chamber (King 1970; Lin and Spradling 1993). Necrosis and apoptosis during oogenesis are also easily detectable. Thus, our examination of the piwi mutant phenotype at 24-hr intervals immediately prior to the onset of oogenesis is sufficient to monitor every major event in germ-line proliferation, differentiation, or cell death. Under these conditions, GSC divisions would have been detected as an increased number of germ-line cysts, whereas cell death would have been easily detected by looking for pyknotic nuclei and necrotic bodies via DAPI staining and Nomarski optics (Lin and Spradling 1993; Forbes et al. 1996a,b; Lin and Spradling 1997). Numerous cases of GSC divisions and cell death in the germ line during Drosophila oogenesis have been detected by use of these criteria (Lin and Spradling 1993; Forbes et al. 1996a,b; Lin and Spradling 1997). However, the current study, even though more systematic, did not detect either GSC division or cell death in piwi mutants (see Results). Instead, immediately following the onset of oogenesis, only differentiating cysts were detected in piwi1 mutant ovaries, with the cyst number generally corresponding to the number of GSCs. Although this does not rule out the possibility that infrequent GSC divisions or cell death might have been missed, which might account for the smaller-than-expected number of gametes sometimes observed in adult piwi mutants, these factors do not weaken the conclusion that the predominant defect of piwi mutants is the differentiation of GSCs without accompanying self-renewing division.
A number of elegant studies have identified genes that regulate germ-line proliferation. In mammalian systems, mechanisms mediated by paracrine factors such as the Steel factor (the ligand of the c-Kit receptor tyrosine kinase), leukemia inhibitory factor (LIF), and basic fibroblast growth factor (bFGF), control the proliferation and survival of primordial germ cells, that is, the precursors of GSCs (for review, see Lin 1997). In C. elegans, the LAG-2/GLP-1 pathway is essential for the self-renewal of a stem-cell equivalent population of germ-line nuclei in the gonad (for review, see Kimble and Simpson 1997). In the Drosophila ovary and testis, the BAM-mediated mechanism is involved in the further development of the differentiated GSC daughter (McKearin and Ohlstein 1995); the dpp pathway plays a similar role in testis (Matunis et al. 1997). More recently, this pathway has been shown to be essential for GSC division and maintenance in the ovary (Xie and Spradling 1998). Among these discoveries, piwi is the first gene known to mediate a somatic signaling mechanism to regulate the division and maintenance of GSCs in Drosophila (see introductory section). It therefore provides an important entry point for studying the somatic control of stem cell division in the germ line.
At present, we do not know if piwi is required for stem cell division in somatic tissues, because existing piwi mutations are strong, yet may not be null. Even if piwi is specific for GSCs, other piwi-like genes, however, may function in other stem cell systems (see below).
The piwi gene family and the conservation of a stem cell self-renewing mechanism during evolution
Our identification of prg-1 and prg-2 in C. elegans and hiwi in humans suggest that piwi represents a novel class of evolutionarily conserved genes with potentially important functions in GSC self-renewal. Among this class of genes, the significantly higher homology between piwi and hiwi as compared with that between piwi and prg-1/prg-2 suggests that hiwi function is closer to piwi. Consistent with this, GSC division and gametogenesis in humans are much more similar to that in Drosophila than that in C. elegans whose gonads contain syncytial mitotic germ-line nuclei that divide symmetrically and are capable of self-renewing only as a population. Our RNAi experiments, which presumably interfered with the activity of both prg-1 and prg-2 because of their extremely high homology, caused germ-line depletion similar to that in piwi mutants. This suggests that the piwi-mediated mechanism in germ-line self-renewal is conserved even in this evolutionarily distant organism without stereotypic GSCs.
The conservation of the piwi-mediated mechanism appears to extend to the plant kingdom as well. The overall homology between PIWI and ZLL and AGO (see Results) is worth noting. Intriguingly, ZLL is essential for maintaining stem cells of the shoot meristem in an undifferentiated state during the transition from embryo-specific development to repetitive organ formation through the self-perpetuating shoot meristem divisions (Moussian et al. 1998). AGO also plays an important role in maintaining normal apical shoot meristem function (Bohmert et al. 1998). Thus, the homology between piwi and ZLL and ago further suggests the existence of a family of novel genes essential for stem cell division in diverse organisms.
Given the functional conservation of the piwi family genes between distant species such as Drosophila, C. elegans, and Arabidopsis, it is tempting to speculate that the piwi-mediated mechanism is also conserved in mammals and humans less distant from Drosophila. Given that C. elegans and Drosophila are separated by >1000 million years of evolution (Vanfleteren et al. 1994), and human and Arabidopsis are even further apart, this functional conservation could reflect the existence of a very ancient mechanism for stem cell maintenance and proliferation in a multicellular ancestor.
The conserved piwi mechanism may mediate cell–cell interactions. In Drosophila, genetic mosaic and piwi expression analyses together suggest that piwi function is required in the apical nonmitotic somatic cells to control GSC division. Similarly, in A. thaliana, it is thought that ZLL is required to maintain the undifferentiated state of shoot meristem stem cells by relaying positional information, possibly by mediating cell–cell interactions within the center of the shoot meristem (Moussian et al. 1998). Further analysis of PIWI should shed light on this evolutionarily conserved cell–cell interaction mechanism.
Stem cells are characterized by two common properties that extend across diverse species: the capacity for self-renewal and the ability to give rise to numerous progeny that are fated for further differentiation (for review, see Lin 1997; Morrison et al. 1997). Although significant progress has been made in identifying genes important for stem cell function, no common molecular mechanism shared by diverse stem cell types in diverse organisms has been characterized with respect to those two basic stem cell properties. The piwi gene family may represent the first class of such genes. The analysis of the piwi gene family, therefore, provides an important first step towards the elucidation of molecular mechanisms underlying stem cell divisions.
Materials and methods
Drosophila strains and culture
All strains were grown at 25°C on yeast-containing molasses/agar medium. The piwi1, piwi3 (Lin and Spradling 1997), and piwi4 mutations are single PZ insertional mutations, whereas the piwi2 mutation is a single P-ry11 transposable insertion. Oregon R (Ore-R) serves as the wild-type strain for all experiments.
Phenotypic analysis of piwi mutants
piwi mutations were balanced over CyO–y+ and introduced into the y/y genetic background. The homozygous and heterozygous larvae were separated at the late third instar stage by scoring mouth hooks and denticle belts for y. They were then dissected to obtain the ovary immediately or were aged at 25°C to desired pupal stages (see Results) before dissection. Dissected ovaries were analyzed by immunofluorescence microscopy for defects in ovary differentiation and GSC division.
Immunocytochemistry and immunofluorescence microscopy
Wild-type and mutant ovaries from larval, pupal, or adult stages were dissected, fixed, and stained as described in Lin et al. (1994). For immunofluorescence staining, anti-VASA antibodies (Hay et al. 1990) were used to specifically mark germ cells at 1:2000 dilution. Anti-α-spectrin antibodies (Byers et al. 1987) were used to outline somatic cells and to mark spectrosomes and fusomes (Lin and Spradling 1995) at 1:200 dilution. The monoclonal anti-MYC epitope antibody 1-9E10.2 was described by Evan et al. (1985) and was used at 1:2 dilution. All the fluorescence-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratory and were used at 1:200 dilution. Immunofluorescently labeled samples were also counterstained with the DNA-specific dye DAPI as described in Lin and Spradling (1993). The immunologically labeled samples were examined by Nomarski and epifluorescence microscopy under a Zeiss Axioplan microscope equipped with a Star-1 cooled CCD camera (Photomatrics). Selected samples were further analyzed by confocal microscopy with a Zeiss LSM410 confocal microscope mounted on an Axiovert 100. Images from the Zeiss Axioplan were collected by IP Lab software and confocal images were processed by use of the LSM410 software. All images were processed by the Adobe Photoshop program.
Cloning of piwi and mapping of its mutations
piwi maps to the left arm of the second chromosome within polytene band 32C, as defined by insertions of single transposable elements (Lin and Spradling 1997). DNA fragments flanking the piwi1, piwi3, and piwi4 insertions were recovered by a plasmid rescue method (Wilson et al. 1989). Genomic DNA in the rescued plasmid was used to isolate additional genomic clones from a λ genomic DNA library (Maniatis et al. 1978) to constitute a 36-kb region of overlapping walk clones. The insertion sites for piwi1, piwi3, and piwi4 were first mapped by Southern blots of EcoRI-digested walk clones, and then precisely determined by DNA sequencing. The site of the piwi2 insertion was determined by sequencing its flanking DNA fragments isolated by inverse PCR of piwi2 genomic DNA with primers specific to the P-element termini (Ochman et al. 1990).
Genomic DNA fragments derived from the piwi locus were used to probe Northern blots of poly(A)+ RNA from wild-type male and female flies. Three nonoverlapping transcripts, α (1.1 kb), β (0.64 kb), and c12 (3.1 kb) were identified in the region and only c12 was interrupted by piwi mutations (Fig. 2A; α and β not shown). Genomic DNA (9.36 kb) that encompasses the c12 transcription unit was sequenced either with a Sequenase kit (U.S. Biochemical) or an ABI 377 Prism DNA Sequencer (Perkin-Elmer, Applied Biosystems). All sequence data was analyzed by the DNASTAR software package (DNAStar, Madison, WI). Two additional transcripts, LD08487 (1.43 kb) and LD13681, were identified on the right side of the P insertions by BLAST analysis (Altschul et al. 1990) of genomic sequence.
Two overlapping cDNA clones, which together represent the 3.1-kb full-length c12 mRNA and the LD08487 cDNA were sequenced. Comparison between the genomic and the cDNA sequences precisely determined the intron–exon organization of these two transcription units. The LD13681 transcription units were also mapped by partial sequencing.
Standard molecular biology techniques were carried out according to Sambrook et al. (1989).
Transgenic rescue
The 6.8-kb HindIII–EcoRV (pRc12; piwi) and 3.9-kb PstI (pRLD; LD08487) genomic fragments were subcloned into the P-element transformation vector pCaSpeR4 (Pirrotta 1988). Transgenic flies were produced according to Spradling and Rubin (1982), using w; Δ2-3 Sb e/TM6 e (Robertson et al. 1988) embryos as recipients. Transgenes were then separated from the Δ2-3 transposase and introduced into the homozygous piwi1 background for fertility tests by genetic crosses. Six independent pRc12 inserts and eight independent pRLD inserts were recovered. Rescue crosses were carried out at 25°C.
Northern blot analysis and piwi cDNA isolation
Poly(A)+ mRNA isolation, Northern blot preparation, and 32P-labeled probe preparation by random-hexamer extension were according to Sambrook et al. (1989). For Figure 2B, the probe was made from a 5.4-kb HindIII genomic fragment encompassing the piwi transcription unit. The same probe was used to isolate a 2.1-kb partial cDNA from the pNB40 third instar library of Nick Brown (Brown and Kafatos 1989). By use of the sites within the polylinker (HindIII at the 5′ end and NotI at the 3′ end), the 2.1-kb cDNA was subcloned into pBlueScript KS(+) (Stratagene), resulting in a construct denoted pDC1. A primer (pDC1 5′REV; 5′-ACGATAAGTTCTGTTAT-3′) was designed downstream of the 5′ end of pDC1 and was used in combination with library-specific primers to screen the Tulle Hazelrigg ovarian cDNA library (Friedman et al. 1990). A partially overlapping 500-bp PCR product was isolated, cloned into the pGEM-T vector (Promega), and used to probe the same library. An overlapping 1-kb cDNA clone was isolated and subcloned into the SalI site of pDC1 to produce a full-length 3.1-kb piwi cDNA construct denoted pDC2.
Characterization of piwi homologs from C. elegans and Homo sapiens
The BLAST algorithm (Altschul et al. 1990) was used to search the nonredundant databases at the National Center for Biotechnology Information (NCBI) at the National Library of Medicine. The Prosite Protein Motif Database (DNAStar) was also searched to identify domains shared between piwi and other known proteins. The PIWI ORF was used to query the SWISS-Prot database by BLASTp for homologous proteins. Significantly, the search identified two C. elegans-predicted proteins of unknown function. ESTs corresponding to these two gene products were then identified as D2030.6 (prg-1; GenBank accession no. Z73906) and C01G5.2 (prg-2; GenBank accession no. U50068) from GenBank. Dr. Yuji Kohara (National Institute of Genetics, Japan) kindly provided λ ZAPII phagemid clones corresponding to prg-1 and prg-2.
The piwi cDNA was further used to query the NCBI dbEST database and identified an EST, zw68h01.r1, isolated from a human testes cDNA library of Bento Soares (GenBank accession no. AA430311). This 0.9-kb EST clone was sequenced and used to screen an oligo-(dT)-primed human testis cDNA library (Clontech), which allowed a 2.3-kb cDNA clone corresponding to hiwi to be isolated and sequenced.
Whole-mount tissue in situ hybridization
Ovarian whole-mount tissue in situ hybridization was performed as described in Yue and Spradling (1992) with DIG-labeled (Genius Kit, Boerhinger Mannheim) full-length piwi cDNA from pDC2 as a probe. For detecting RNA in surface cells, ovaries were digested 5 min with 50 μg/ml proteinase K. Under this condition, only terminal filament cells and anterior sheath cells, but not any other surface cells, were stained with the piwi cDNA probe. The pBST control probe did not show any significant signal. For detecting RNA in inner germarial cells, the digestion was 7–10 min. Under this condition, the piwi probe revealed a hybridization pattern as shown in Results, the pBST control probe did not show any significant signal, whereas the positive control oskar probe showed specific staining in presumptive oocytes (Ephrussi et al. 1991; data not shown). Staging of egg chambers is according to King (1970).
Genetic clonal analyses and germarial transplantation
The piwi1 and piwi2 germline clones were generated with the FLP–DFS technique as described in Chou and Perrimon (1996). y w P[hsFLP]12; P[ovoD1]2L, FRT40A/CyO males were crossed to w; piwi1 FRT40A/CyO virgin females to produce y w P[hsFLP]12; P[ovoD1]2L, FRT40A/piwi1, FRT40A progeny. Identical crosses were performed with w; piwi2, FRT40A/CyO virgin females. The genotype of the flies were verified both by Southern analysis for the presence of piwi P insertion and the FRT as well as by sterility tests by backcrossing to the original piwi allele. Crosses were carried out for 2 days to produce transheterozygous progeny. After 2 days, adults were transferred to fresh vials. Larvae were heat shocked on days 3 and 4 for 1 hr in a 37°C water bath to induce mitotic recombination. The heat-shocked females with germ-line clones were crossed to Ore-R males for fertility tests and their ovarian phenotype examined by immunofluorescence microscopy.
To generate somatic clones of piwi1 and piwi2 the above described piwi1 FRT40A/CyO and piwi2 FRT40A/CyO strains were crossed to w P[hsFLP]9F; P[w+; hs–NM] FRT40A/CyO males as described in Xu and Rubin (1993). Females of the genotype w P[hsFLP]9F; P[w+; hs–NM], FRT40A/piwi, FRT40A were crossed to Ore-R males for fertility tests. Clonal induction was carried out as described above except that females of the appropriate genotype were subjected to an additional 20-min heat shock at 37°C 90 min prior to dissection to induce the expression of the Myc tag. Germarial transplantation was carried out as described by Lin and Spradling (1993).
RNAi experiments
Templates for in vitro transcription were cloned into pBluescript KS(+) and transcribed by either T7 or T3 RNA polymerases according to the manufacturer’s protocol (Megascript T7 and T3 kits, Ambion). prg-1 cDNA served as the experimental RNA for injection, whereas pBluescript KS(+) RNA was used as a negative control. RNA integrity was determined by gel electrophoresis; concentration was determined by a combination of UV spectrophotometry and ethidium bromide staining. Injections used an equimolar mixture of uncapped sense and antisense RNA at a concentration of 10 mg/ml. Gonadal injections of wild-type N2 Bristol hermaphrodites was done as described in Mello and Fire (1995). After recovery and transfer to standard solid medium, injected animals were transferred to fresh culture plates at 24-hr intervals to facilitate the identification of phenotypic differences. Germ lines of progeny produced between 6 and 48 hr after injection were examined for gamete differentiation either by DAPI staining or fertility. For DAPI analysis, adult hermaphrodites displaying RNAi-induced phenotypes were fixed overnight in Carnoy’s solution (60% ethanol; 30% acetic acid; 10% chloroform; Sulston and Hodgkin 1988). The mitotic index is defined as the ratio between the number of prophase, metaphase, anaphase, and telophase nuclei and the total number of nuclei in the MPZ.
Acknowledgments
We are grateful to Dr. Allan Spradling in whose laboratory H.L. initiated piwi cloning. We are indebted to Dr. Margaret Titus for her enthusiasm and help on the prg project, for generously supporting J.B. through her National Science Foundation grant (MCB-9507376), and for critical reading of the manuscript. We thank Ms. Lin-Hua Song for performing Drosophila germ-line transformation injections, Ms. Wei Deng for sequencing part of hiwi, Dr. Jonathan Friedman for use of his C. elegans injection facility, Dr. Yuji Kohara for providing prg-1 and prg-2 cDNA clones, Drs. Nick Brown and Tulle Hazelrigg for the Drosophila cDNA libraries, Dr. Dan Kiehart for anti-α-spectrin antibodies, Dr. Yuh-Nung Jan for anti-VASA antibodies, and Dr. Rick Fehon for anti-MYC antibodies. D.N.C. was a National Institutes of Health (NIH) predoctoral fellow during part of this work. This work was supported by a NIH grant (HD33760) to H.L., who is a recipient of the David and Lucille Packard Fellowship, an American Cancer Society Junior Faculty Research Award (JFRA-608) and a Basil O’Connor Award (5-FY95-1111) from the March of Dimes Birth Defects Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
The genomic and cDNA sequences of piwi, as well as the cDNA sequence of hiwi, have been submitted to GenBank under accession nos. AF104355, AF104354, and AF104260, respectively.
Footnotes
E-MAIL h.lin@cellbio.duke.edu; FAX (919) 684-5481.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EH, King RC. Oogonial and spermatogonial differentiation within a mosaic gonad of Drosophila melanogaster. Growth. 1962;26:53–70. [PubMed] [Google Scholar]
- ————— Studies on the events resulting in the formation of an egg chamber in Drosophila melanogaster. Growth. 1964;28:41–81. [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Byers TJ, Dubreuil RR, Branton D, Kiehart DP, Goldstein LSB. Drosophila spectrin II. Conserved features of the alpha-subunit are revealed by analysis of cDNA clones and fusion proteins. J Cell Biol. 1987;105:2103–2110. doi: 10.1083/jcb.105.5.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. Spectrosomes and fusomes are essential for anchoring mitotic spindles during asymmetric germ cell divisions and for the microtubule-based RNA transport during oocyte specification in Drosophila. Dev Biol. 1997;189:79–94. doi: 10.1006/dbio.1997.8669. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Forbes AJ, Lin H, Ingham PW, Spradling AC. hedgehog is required for the proliferation and specification of ovarian somatic cells prior to egg chamber formation in Drosophila. Development. 1996a;122:1125–1135. doi: 10.1242/dev.122.4.1125. [DOI] [PubMed] [Google Scholar]
- Forbes AJ, Spradling AC, Ingham PW, Lin H. The role of segment polarity genes during early oogenesis in Drosophila. Development. 1996b;122:3283–3294. doi: 10.1242/dev.122.10.3283. [DOI] [PubMed] [Google Scholar]
- Friedman KD, Rosen NL, Newman PS, Montgomery RR. Screening lambda gt11 libraries. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: A guide to methods and applications. San Diego, CA: Academic Press; 1990. pp. 253–260. [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development. 1990;109:425–433. doi: 10.1242/dev.109.2.425. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:17–26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- Kimble JE, Simpson P. The LIN-12/Notch signaling pathway and its regulation. Annu Rev Cell Dev Biol. 1997;13:333–361. doi: 10.1146/annurev.cellbio.13.1.333. [DOI] [PubMed] [Google Scholar]
- King RC. Ovarian development in Drosophila melanogaster. New York, NY: Academic Press; 1970. [Google Scholar]
- Kuwabara PE. Interspecies comparison reveals evolution of control regions in the nematode sex-determining gene tra-2. Genetics. 1996;144:597–607. doi: 10.1093/genetics/144.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. The tao of stem cells in the germline. Annu Rev Genet. 1997;31:455–491. doi: 10.1146/annurev.genet.31.1.455. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev Biol. 1993;159:140–152. doi: 10.1006/dbio.1993.1228. [DOI] [PubMed] [Google Scholar]
- ————— Fusome asymmetry and oocyte determination. Dev Genetics. 1995;16:6–12. doi: 10.1002/dvg.1020160104. [DOI] [PubMed] [Google Scholar]
- ————— A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Lin H, Schagat T. Neuroblasts: A model for asymmetric division of stem cells. Trends Genet. 1997;13:33–39. doi: 10.1016/s0168-9525(96)10050-0. [DOI] [PubMed] [Google Scholar]
- Lin H, Yue L, Spradling AC. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development. 1994;120:947–956. doi: 10.1242/dev.120.4.947. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Hardison RC, Lacy E, Lauer J, O’Connell C, Quon D, Sim GK, Efstradiatis A. The isolation of structural genes from libraries of eukaryotic DNA. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Margolis J, Spradling AC. Identification and behavior of epithelial stem cells in the Drosophila ovary. Development. 1995;121:3797–3807. doi: 10.1242/dev.121.11.3797. [DOI] [PubMed] [Google Scholar]
- Matunis E, Tran J, Gonczy P, Caldwell K, DiNardo S. punt and schnurri regulate a somatically derived signal that restricts proliferation of committed progenitors in the germline. Development. 1997;124:4383–4391. doi: 10.1242/dev.124.21.4383. [DOI] [PubMed] [Google Scholar]
- McKearin D, Ohlstein B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development. 1995;121:2937–2947. doi: 10.1242/dev.121.9.2937. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Morrison SJ, Shah NM, Anderson DJ. Regulatory mechanisms in stem cell biology. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Medhorn MM, Garza D, Hartl DL. Amplification of flanking sequences by Inverse PCR. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic Press; 1990. pp. 219–227. [Google Scholar]
- Pirrotta V. Vectors for P-element transformation in Drosophila. In: Rodriguez RL, Denhardt DT, editors. Vectors. A survey of molecular cloning vectors and their uses. London, UK: Butterworths; 1988. pp. 437–456. [Google Scholar]
- Potten CS. Stem cells. London, UK: Academic Press; 1997. [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engles WR. A stable source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruohola H, Bremer KA, Baker D, Swedlow JR, Jan LY, Jan YN. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991;66:433–449. doi: 10.1016/0092-8674(81)90008-8. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schüpbach T, Wieschaus E, Nöthiger R. A study of the female germ line in mosaics of Drosophila. Wilhelm Roux’s Arch Dev Biol. 1978;184:41–56. doi: 10.1007/BF00848668. [DOI] [PubMed] [Google Scholar]
- Spradling A. Developmental genetics of oogenesis. In: Bate M, Martinez Arias A, editors. Drosophila Development. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germline chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The nematode Caenorhabditis elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. pp. 598–600. [Google Scholar]
- Thierry-Mieg J, Durbin R. ACeDB, a C. elegans database. Cahiers IMABIO. 1992;5:15–24. [Google Scholar]
- Vanfleteren JR, Van de Peer Y, Blaxter ML, Tweedie SAR, Trotman C, Lu L, Van Hauwaert ML, Moens L. Molecular genealogy of some nematode taxa as based on cytochrome c and globin amino acid sequences. Mol Phylogenet Evol. 1994;3:92–101. doi: 10.1006/mpev.1994.1012. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Szabad J. The development and function of the female germline in Drosophila melanogaster, a cell lineage study. Dev Biol. 1979;68:29–46. doi: 10.1016/0012-1606(79)90241-0. [DOI] [PubMed] [Google Scholar]
- Wilson C, Pearson RK, Bellen HJ, O’Kane CJ, Grossniklaus U, Gehring WJ. P-element-mediated enhancer detection: Isolation and characterization of developmentally regulated genes in Drosophila. Genes & Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- Xie T, Spradling AC. decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94:251–260. doi: 10.1016/s0092-8674(00)81424-5. [DOI] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Xu T, Caron LA, Fehon RG, Artavanis-Tsakonas S. The involvement of the Notch locus in Drosophila oogenesis. Development. 1992;115:913–922. doi: 10.1242/dev.115.4.913. [DOI] [PubMed] [Google Scholar]
- Yue L, Spradling A. hu-li tai shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes & Dev. 1992;6:2443–2454. doi: 10.1101/gad.6.12b.2443. [DOI] [PubMed] [Google Scholar]
- Zhao G-Q, Deng K, Labosky PA, Liaw L, Hogan BLM. The gene encoding bone morphogenetic protein 8B is required for the initiation and maintenance of spermatogenesis in the mouse. Genes & Dev. 1996;10:1657–1669. doi: 10.1101/gad.10.13.1657. [DOI] [PubMed] [Google Scholar]