Abstract
Mitochondrial biogenesis is essential for cell viability. Growth hormone receptor knockout (GHRKO), calorie restriction, and surgical visceral fat removal constitute experimental interventions to delay aging and increase life span. We examined the expression of known regulators of mitochondriogenesis: peroxisome proliferator–activated receptor γ co-activator 1α (PGC-1α), adenosine monophosphate (AMP)–activated protein kinase (AMPK), sirtuin-1 (SIRT-1) and sirtuin-3 (SIRT-3), endothelial nitric oxide synthase (eNOS), nuclear respiratory factor-1, mitochondrial transcription factor A (TFAM), and mitofusin-2 (MFN-2) in the skeletal muscles and hearts of control and calorie-restricted female GHRKO mice and in the kidneys of male GHRKOs after visceral fat removal or sham surgery. Expression of PGC-1α in skeletal muscles, AMPK, SIRT-1, SIRT-3, eNOS, and MFN-2 in the heart and PGC-1α, AMPK, SIRT-3, eNOS, and MFN-2 in kidneys was increased in GHRKO mice but was not affected by calorie restriction or visceral fat removal. GHRKO mice have increased expression of key regulators of mitochondriogenesis, which is not improved further by calorie restriction or visceral fat removal.
Keywords: GHRKO mice, Mitochondrial biogenesis, Gene expression, Calorie restriction, Visceral fat removal
ALTERATIONS in the somatotrophic and insulin signaling pathway are among the most important potential mechanisms of extended longevity. One of the genetic interventions that leads to prolonged longevity in mice is targeted disruption of the growth hormone receptor/growth hormone–binding protein gene (Ghr/bp gene) (1). Growth hormone (GH) receptor/–binding protein knockout mice (GHRKO; Ghr/bp−/−) are homozygous for this mutation (1). These long-lived animals are GH resistant and characterized by reduced weight and body size, not detectable GH receptor levels, high serum GH concentration, low or normal glucose, and greatly reduced plasma levels of insulin-like growth factor-1 and insulin (1–7) with enhanced insulin sensitivity (8). Moreover, GHRKO mice have improved resistance to oxidative stress, reduced oxidative damage (6,9,10) and lower incidence, and delayed onset of fatal neoplastic diseases (11).
Mitochondria are complex essential eukaryotic organelles, which play a crucial role, among others, in energy homeostasis and metabolism. Most importantly, they generate adenosine triphosphate—the cellular energy carrier (via oxidative phosphorylation; OxPhos). Moreover, mitochondria contain essential enzymes indispensable for multiple biochemical processes, including lipid, cholesterol, steroid, heme, and nucleotide synthesis. Mitochondria are also fundamental regulators of apoptosis. One of the elements of apoptosome, whose formation leads to caspase cascade activation, is cytochrome c, which is involved in mitochondrial complex IV of the electron transport chain.
Table 1.
p Values for Genes Expression in Skeletal Muscles of Normal (N) and Growth Hormone Receptor/–Binding Protein Knockout (GHRKO; KO) Mice Fed AL or Subjected to 40% CR
| Gene |
p Value |
|||||
| A vs B | C vs D | A vs C | B vs D | AB vs CD (N vs KO) | AC vs BD (AL vs CR) | |
| PGC-1α | NS | NS | NS | .049 | .034 | NS |
| AMPK | NS | NS | NS | NS | NS | NS |
| SIRT-1 | NS | NS | NS | NS | NS | NS |
| SIRT-3 | NS | NS | NS | NS | NS | NS |
| eNOS | NS | NS | NS | NS | NS | NS |
| NRF-1 | NS | NS | NS | NS | .011 | NS |
| TFAM | NS | NS | NS | NS | .021 | NS |
| MFN-2 | NS | NS | NS | NS | NS | NS |
| COXIV | NS | NS | NS | NS | NS | .048 |
Note: A represents N-AL mice, B represents N-CR mice, C represents KO-AL mice, and D represents KO-CR mice. AL = ad libitum; CR = calorie restriction; NS = not significant.
Table 2.
p Values for Genes Expression in Hearts of Normal (N) and Growth Hormone Receptor/–Binding Protein Knockout (GHRKO; KO) Mice Fed AL or Subjected to 40% CR
| Gene |
p Value |
|||||
| A vs B | C vs D | A vs C | B vs D | AB vs CD (N vs KO) | AC vs BD (AL vs CR) | |
| PGC-1α | NS | NS | NS | NS | NS | NS |
| AMPK | NS | NS | NS | <.001 | .001 | NS |
| SIRT-1 | NS | NS | NS | NS | .037 | NS |
| SIRT-3 | NS | NS | NS | .002 | .011 | NS |
| eNOS | NS | NS | NS | NS | .024 | NS |
| NRF-1 | NS | NS | NS | NS | NS | NS |
| TFAM | NS | NS | NS | NS | NS | NS |
| MFN-2 | NS | .029 | NS | .003 | .028 | NS |
| COXIV | NS | NS | .003 | NS | <.001 | NS |
Note: A represents N-AL mice, B represents N-CR mice, C represents KO-AL mice, and D represents KO-CR mice. AL = ad libitum; CR = calorie restriction; NS = not significant.
Table 3.
p Values for Genes Expression in Kidneys of Normal (N) and Growth Hormone Receptor/–Binding Protein Knockout (GHRKO; KO) Sham Operated (sham) or Subjected to VFR
| Gene |
p Value |
|||||
| A vs B | C vs D | A vs C | B vs D | AB vs CD (N vs KO) | AC vs BD (sham vs VFR) | |
| PGC-1α | NS | NS | .013 | .001 | <.001 | NS |
| AMPK | NS | NS | NS | .023 | .011 | NS |
| SIRT-1 | NS | NS | NS | NS | NS | NS |
| SIRT-3 | NS | NS | .006 | .009 | <.001 | NS |
| eNOS | NS | NS | .005 | <.001 | <.001 | NS |
| NRF-1 | NS | NS | NS | NS | NS | NS |
| TFAM | NS | NS | NS | .023 | .012 | NS |
| MFN-2 | NS | NS | .042 | .019 | <.001 | NS |
Note: A represents N-sham mice, B represents N-VFR mice, C represents KO-sham mice, and D represents KO-VFR mice. NS = not significant; VFR = visceral fat removal.
The process by which new mitochondria are formed, known as the biogenesis of mitochondria, is essential for proper cell viability (12). Dysfunction of mitochondrial biogenesis affects oxidative stress resistance, maintenance of energy production, and metabolism regulation, and it may result in the development of degenerative diseases that are often age associated.
Peroxisome proliferator–activated receptor γ (PPARγ) co-activator 1α (PGC-1α) is the key regulator (“master regulator”) of mitochondrial biogenesis. PGC-1α plays an important role in the activation and control of mitochondrial biogenesis through physiological signals integration (13) and coordination of multiple transcription factors activities (14).
Many other factors are also involved in the activation and regulation of mitochondrial biogenesis, including adenosine monophosphate (AMP)–activated protein kinase (AMPK), sirtuins (sirtuin-1 [SIRT-1] and sirtuin-3 [SIRT-3]), endothelial nitric oxide synthase (eNOS), nuclear respiratory factor-1 (NRF-1), and mitochondrial transcription factor A (TFAM). Mitochondriogenesis activity increases in response to calorie restriction (CR) (15–18) through stimulation of AMPK, eNOS, and SIRT-1 and, consequently, stimulation of PGC-1α activity.
AMP-activated protein kinase (AMPK) plays a role of a cellular energy sensor and is activated by an increase in intracellular AMP/adenosine triphosphate ratio (19), which can be induced by CR or endurance exercise. Activated AMPK phosphorylates PGC-1α and enhances PGC-1α activity (20).
Sirtuins (nicotinamide adenine dinucleotide [NAD+]–dependent deacetylases) are evolutionarily conserved enzymes found in all eukaryotic and in many prokaryotic organisms. The enzymes in question are involved in many biological processes, including DNA repair and the maintenance of chromosome stability. Among them, the key role is played by SIRT-1—a mammalian homolog of the yeast “silent information regulator 2” (Sir2), which mediates effects of CR on yeast life span. SIRT-1 deacetylates and activates PGC-1α, thus stimulating mitochondrial biogenesis. This factor is activated in states of nutrient deprivation, including fasting and CR (21). For this reason, SIRT-1 is sometimes referred to as a nutrient deprivation sensor. SIRT-3 is a member of the sirtuin family that use NAD+ as a co-substrate. It was shown that SIRT-3 can play an important role in the regulation of longevity in mice (22). Moreover, SIRT-3 may regulate mitochondrial metabolism, adaptive thermogenesis, energy homeostasis, and apoptosis (23,24).
Nitric oxide, generated by eNOS, activates mitochondrial biogenesis through the transcriptional activation of PGC-1α (25,26). Another proposed mechanism of nitric oxide–dependent activation of mitochondrial biogenesis is cyclic guanosine monophosphate (cGMP)-mediated upregulation of certain transcriptional factors (27). The increase of cGMP level also can lead to expression of active SIRT-1 (16), indirectly stimulating mitochondrial biogenesis.
Nuclear respiratory factor-1 (NRF-1) is a nuclear DNA-binding factor, which among other functions, regulates transcription of OxPhos genes. It binds to a regulatory site of the cytochrome c promoter (28). Moreover, NRF-1 regulates expression of mitochondrial transcription factor A (TFAM) and plays an important role in nervous system and muscle development (29).
Mitochondrial transcription factor A (TFAM) is a DNA-binding protein that is encoded by the nuclear genome. The main role of TFAM is the coordination of nuclear and mitochondrial genomes both of which contain genes necessary for mitochondrial biogenesis. Moreover, TFAM is known to be essential for the maintenance of mitochondrial DNA (30).
Another important process involved in the regulation of mitochondrial activity and biogenesis is mitochondrial fusion and maintenance of the mitochondrial network architecture (31,32). One of the factors playing an essential role in this process is mitofusin-2 (MFN-2) (31,32). Interestingly, decreased expression of MFN-2 may be linked to metabolic disturbances that result in obesity development (31).
In spite of its important role in the regulation of physiological processes, the biogenesis of mitochondria has not been analyzed in GHRKO mice. In this context, we decided to examine the expression of PGC-1α, AMPK, SIRT-1, SIRT-3, eNOS, NRF-1, TFAM, and MFN-2 genes in the skeletal muscles, hearts, and kidneys of GHRKO mice. Apart from the messenger RNA (mRNA) levels of key regulators of mitochondriogenesis, we also assessed the gene expression and protein level of subunit IV of one of mitochondrial activity markers—cytochrome c oxidase (COXIV). Additionally, we assessed the cytochrome c oxidase (COX) activity in normal and GHRKO mice. Furthermore, NAMPT (nicotinamide phosphoribosyltransferase; visfatin) protein level was analyzed. NAMPT is responsible for conversion of nicotinamide to NAD+ being a co-substrate for SIRT-1, which activates PGC-1α.
The analysis of all mentioned factors seems to be particularly important in long-lived GHRKO mice because it is known that decline in energy production in various species is strongly associated with aging.
CR is a well-known experimental intervention to delay aging and increase life span (33). The characteristics of normal mice subjected to CR resemble certain phenotypic features of GHRKO mice, including reduced plasma insulin-like growth factor-1 and insulin levels and enhanced sensitivity to insulin. Similarly, longevity of normal mice subjected to CR resembles that of GHRKO mice fed ad libitum (34). However, it is still unclear which of the physiological alterations induced by CR and by Ghr/bp gene disruption are responsible for extended longevity. Bonkowski and colleagues (34) have shown that CR extends longevity in normal but not in GHRKO mice. This suggests that mechanisms linking GH resistance and CR to aging must overlap. This is why we decided to analyze the effect of CR on the process of mitochondrial biogenesis in these long-lived mutants (Experiment 1).
Surgical visceral fat removal (VFR; removal of epididymal and perinephric fat depots) is another intervention that has been reported to improve insulin signaling in normal mice and rats and extend longevity in rats (35–37), thus mimicking the effects of CR. Unexpectedly, our preliminary results showed that VFR promoted insulin resistance in GHRKO mice, although it had the expected beneficial effect on insulin signaling in normal mice (38). In the context of these observations and similarities between CR and VFR in their effects on life span and the insulin signaling pathway, we also decided to examine the effect of VFR on the biogenesis of mitochondria in GHRKO mice (Experiment 2).
MATERIALS AND METHODS
Animals
The normal and GHRKO mice used in the present study were produced in our breeding colony, developed using animals kindly provided by Dr. J.J. Kopchick (Ohio University). Animals were produced by mating knockout (−/−) males with heterozygous (+/−) females. Normal (+/−) and GHRKO animals (−/−) were separated by phenotypic characteristics. All animal procedures were approved by the Laboratory Animal Care and Use Committee at the Southern Illinois University School of Medicine (Springfield, IL). The mice were housed under temperature- and light-controlled conditions (22 ± 2°C, 12-hour light/12-hour dark cycle) and fed Lab Diet 5001 chow (PMI Nutrition International, Richmond, IN) containing, among others, 4.5% fat and 23.4% protein.
Experiment 1
Calorie restriction.—
Starting at 2 months of age, 46 normal and GHRKO female mice were grouped according to average body weight within the phenotype and divided into four experimental groups: normal fed ad libitum (N-AL; 10 animals), normal CR (N-CR; 14 animals), GHRKO ad libitum (KO-AL; 10 animals), and GHRKO-CR (KO-CR; (12 animals). Food-restricted animals were subjected to gradually introduced 40% CR (which corresponded to 60% of the food consumed by their AL counterparts). Water was available at all times to all animals. At 8 months of age, the animals were fasted overnight, anesthetized using isoflurane, bled by cardiac puncture, and euthanized by decapitation. Hind-limb skeletal muscles and hearts were rapidly collected, quickly frozen on dry ice, and stored at –80°C until processed.
Experiment 2
Visceral fat removal.—
At the age of approximately 6 months, 43 normal and GHRKO male mice were grouped according to average body weight within the phenotype and divided into four experimental groups: normal sham operated (N-sham; 11 animals), normal subjected to VFR (N-VFR; 11 animals), GHRKO sham operated (KO-sham; 10 animals), and GHRKO subjected to VFR (KO-VFR; 11 animals). The animals were anesthetized with ketamine/xylazine, shaved, and prepared in the usual sterile fashion. Moreover, mice were supplied with ibuprofen in drinking water starting 2 days before and up to 3 days after the surgery. Tap water was available at all times to all animals. In the VFR group, the epididymal fat pads were removed using blunt dissection through a vertical midline incision, and perinephric fat pads were removed via flank incisions. We removed as much epididymal or perinephric fat as was possible without compromising blood supply to the testes and to the adrenals. For sham operations, the abdominal cavity and both sides of the back were incised and the visceral fat was mobilized but not removed. Two months after VFR or sham operations, the animals were fasted overnight, anesthetized using ketamine/xylazine, bled by cardiac puncture, and euthanized by decapitation. Kidneys were rapidly collected, quickly frozen on dry ice, and stored at −80°C until processed.
RNA Extraction and Complementary DNA Transcription
After pulverizing the skeletal muscle and heart tissues in liquid nitrogen and homogenizing the kidney samples, RNA was extracted using guanidinium thiocyanate–phenol–chloroform method based on Chomczynski–Sacchi procedure (39). RNA quantity and quality were analyzed on 1.5% agarose gel using electrophoresis. Potentially contaminating residual genomic DNA was eliminated using deoxyribonuclease I (Promega, Madison, WI). Reverse transcription was performed, and complementary DNA was synthesized using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instruction.
Real-Time Polymerase Chain Reaction
The real-time polymerase chain reaction was carried out using the Smart Cycler instrument (Cepheid, Sunnyvale, CA) with iQ SYBR Green Supermix (Bio-Rad Laboratories). The three steps of the PCR included denaturation at 94°C for 2 minutes, annealing at 62°C for 30 seconds with fluorescence reading, and extension at 72°C for 30 seconds. In addition, a melting curve was done for each reaction to evaluate the potential of nonspecific products. β2-microglobulin (B2M) was used as a housekeeping gene; it has been previously validated in our laboratory as the most appropriate gene for normalizing the data (40). The genes expression was assessed by measurement of steady state levels of mRNA. Relative expression from real-time polymerase chain reaction was calculated from the equation: 2A−B/2C−D (where A = cycle threshold [Ct] number for the gene of interest in the first control sample, B = Ct number for the gene of interest in the analyzed sample, C = Ct number for the housekeeping gene in the first control sample, and D = Ct number for housekeeping gene in the analyzed sample). The first control was expressed as 1.00 by this equation, and all other samples were calculated in relation to this value. Then, the results in the control group (N-AL or N-sham) were averaged, and all other outputs were divided by the mean value of the relative expression in the control group to yield the fold change of the expression of genes of interest compared with the control group.
For real-time polymerase chain reaction, the following primers were used (gene bank sequence number for each forward primer): B2M—forward (NM_009735): 5′-aagtatactcacgccaccca, backward: 5′-aagaccagtccttgctgaag; PGC-1α—forward (NM_008904): 5′-tacgcaggtcgaacgaaact, backward: 5′-acttgctcttggtggaagca; AMPK—forward (AF036535): 5′-cacttgtctgcatctctcca, backward: 5′-cttgaggaacttgaggatcc; SIRT-1—forward (AY377984): 5′-gtaatgtgaggagtcagcac, backward: 5′-ttggacattaccacgtctgc; SIRT-3—forward (NM_001127351): 5′-actacaggcccaatgtcact, backward: 5′-ttcaaccagctttgaggcag; eNOS—forward (NM_008713): 5′-cagctgtgtccaacatgct, backward: 5′-tcatactcatccatgcactg; NRF-1—forward (AF098077): 5′-gcattgagctactgacagac, backward: 5′-ctgtgtcctggatcttcctt; TFAM—forward (NM_009360): 5′-tggaacacagccacatgctt, backward: 5′-accatggtggcaaactgtct; MFN-2—forward (NM_133201): 5′-ccacaaagtgagtgaacgtc, backward: 5′-atccaccagaaagctggtgc; and COXIV—forward (NM_009941): 5′-acagcccttggcttgatgta, backward: 5′-tggcctgaaagcttccacta.
Protein Extraction and Western Blotting
Total proteins were obtained from tissue homogenates. Approximately 100 mg samples of skeletal muscles and kidney tissue were homogenized in 1 ml ice-cold T-PER Tissue Protein Extraction Reagent (Pierce Biotechnology, Rockford, IL), with Protease Inhibitor Cocktail Kit (Pierce Biotechnology), Phosphatase Inhibitor Cocktail 1 (Sigma-Aldrich, St. Louis, MO), and Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich). After mixing, homogenates were centrifuged at 16,000g for 30 minutes. Protein concentrations were assessed using Pierce BCA (bicinchoninic acid) Protein Assay Kit (Pierce Biotechnology) in accordance with manufacturer’s protocol.
Western blot procedure was performed using two primary antibodies: COXIV (Cell Signaling Technology, Beverly, MA) and Visfatin (BioVision, Inc., Mountain View, CA) and secondary goat antirabbit antibodies (Calbiochem, La Jolla, CA). Monoclonal anti-β-actin antibody (Sigma-Aldrich Corp.) was used, after stripping the membrane, as a control for protein loading.
For Western blotting, protein extracts were mixed with XT Sample Buffer (Bio-Rad Laboratories, Inc.) and heated in a thermocycler at 99°C for 5 minutes and then cooled to 4°C. Forty micrograms of the protein was separated electrophoretically using Criterion XT Precast Gel (26 wells; Bio-Rad Laboratories, Inc.) for 90 minutes at 150 V. Subsequently, proteins were wet transferred for 60 minutes at 100 V onto nitrocellulose membranes (Bio-Rad Laboratories, Inc.) at 4°C. After the transfer, membranes were rinsed briefly in Tris-buffered saline (TBS; pH 7.6) and blocked with 5% nonfat dry milk or 1% bovine serum albumin in TBS containing 0.05% Tween 20 (TBST) for 1 hour at room temperature. After blocking, membranes were washed with TBST three times for 15 minutes each time. Then, the membranes were incubated with the primary antibody specific for the protein of interest diluted in the appropriate blocking solution at 4°C overnight with shaking. After incubation, the blots were washed three times (15 minutes each) with TBST and incubated with an appropriate horseradish peroxidase–conjugated secondary antibody for 1 hour at room temperature. Horseradish peroxidase activity from secondary antibody was detected using the Amersham ECL Plus Western Blotting Detection Reagents (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire, UK). Photos of blots were taken with Image Reader LAS-4000 (FujiFilm, Tokyo, Japan) and quantified for statistical analysis using Multi Gauge version 3.0 software (FujiFilm Life Science, Japan). The protein level was expressed as the arbitrary unit per square millimeter. The arbitrary unit is a unit to measure the emission amount of chemiluminescence material read using LAS. It represents the relative density value accumulated as linear data by a charge-coupled device camera in the image surface.
Cytochrome c Oxidase (COX) Enzyme Activity
COX activity (regarded as a measure of mitochondrial content) was evaluated as described previously (41). Briefly, tissue homogenates were diluted in a buffer containing 0.1 M KH2PO4 and 2 mM EDTA (pH 7.2) and sonicated for 3 × 3 seconds on ice. The enzyme activity was determined by the maximal rate of oxidation of fully reduced cytochrome c, measured by the change in absorbance at 550 nm (Beckman DU-64 spectrophotometer).
Statistical Analysis
The data are expressed as mean ± standard error of the mean (SEM). Two-way analysis of variance was used to evaluate the effects of the genotype and intervention (CR or VFR, respectively). A t test was used to evaluate the effects of interventions within genotypes and genotypes within interventions. A value of p < .05 was considered significant. We used the Bonferroni test—post hoc test for analyzing of differences between group means. All statistical calculations were performed using SPSS version 17.0 (SPSS, Chicago, IL) with α = .05. All graphs were made using Prism 4.02 (GraphPad Software, San Diego, CA).
RESULTS
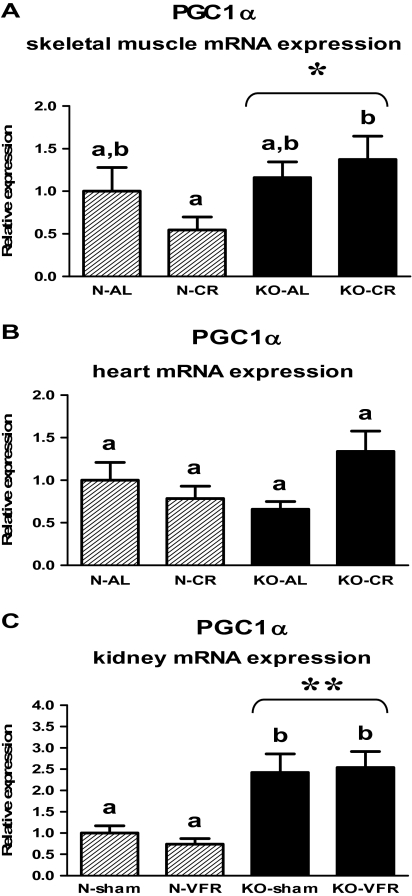
Expression of PGC-1α gene was increased in the skeletal muscles of GHRKO (KO) mice as compared with normal (N) animals, independent of CR (p = .034); additionally, the level of PGC-1α mRNA was upregulated in KOs subjected to CR (KO-CR) in comparison with N-CR animals (p = .049; Figure 1A, Table 1). The PGC-1α gene expression also was increased in the kidneys of KO as compared with N mice (p < .001; Figure 1C, Table 3). Moreover, there was upregulation of renal PGC-1α mRNA in KO-sham and KO-VFR mice as compared with the corresponding N mice (p = .013, p = .001, respectively; Figure 1C, Table 3). In the heart, there was no statistically significant difference between N and KO mice, and the expression of PGC-1α was not affected by CR (Figure 1B, Table 2).
Figure 1.
PGC-1α messenger RNA (mRNA) expression in skeletal muscle (A), heart (B), or kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A and B—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; C—Experiment 2). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .034 vs N mice (the significance for genotype) and **p < .001 vs N mice (the significance for genotype).
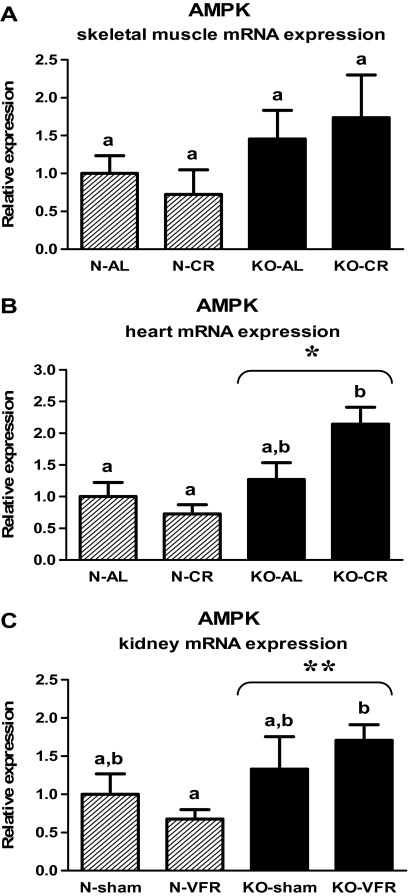
In contrast to the findings concerning PGC-1α, the AMPK expression was increased in the hearts of KO mice as compared with N animals (p = .001; Figure 2B, Table 2). Moreover, in heart tissue, KO-CR mice exhibited an increased level of AMPK mRNA in comparison with N-CR (p < .001; Figure 2B, Table 2). The elevated AMPK gene expression in KO as compared with N mice also was observed in the kidneys (p = .011), with the upregulated level of AMPK mRNA in KO-VFR versus N-VFR mice (p = .023; Figure 2C, Table 3). Expression of AMPK in skeletal muscles was affected neither by disruption of the growth hormone receptor (Ghr) gene nor by CR (Figure 2A, Table 1).
Figure 2.
AMPK messenger RNA (mRNA) expression in skeletal muscle (A), heart (B), or kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A and B—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; C—Experiment 2). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .001 vs N mice (the significance for genotype) and **p = .011 vs N mice (the significance for genotype).
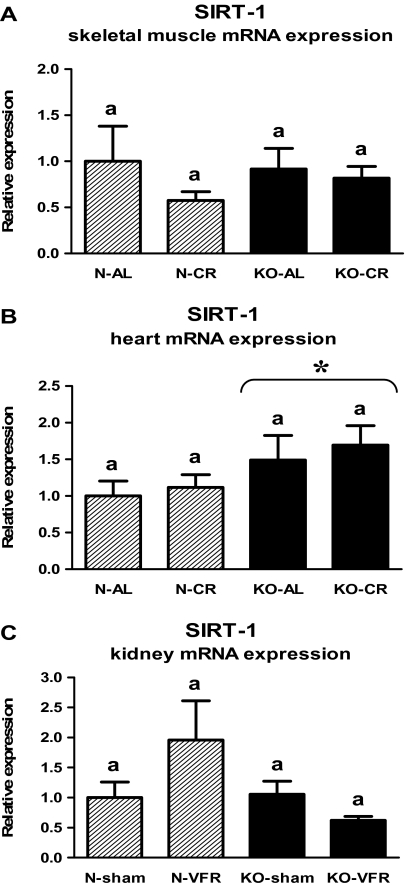
The level of SIRT-1 mRNA in the skeletal muscles (Figure 3A, Table 1) or in the kidneys (Figure 3C, Table 3) did not differ between KO and N mice. Moreover, the SIRT-1 gene expression was not altered by CR or VFR in either tissue (Figure 3A and C, Table 1 and 3). In contrast, SIRT-1 expression was increased in the heart of KO mice as compared with N animals (p = .037; Figure 3B, Table 2).
Figure 3.
SIRT-1 messenger RNA (mRNA) expression in skeletal muscle (A), heart (B), or kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A and B—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; C—Experiment 2). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a—values that share the same letter in the superscript are not statistically significant. *p =.037 vs N mice (the significance for genotype).
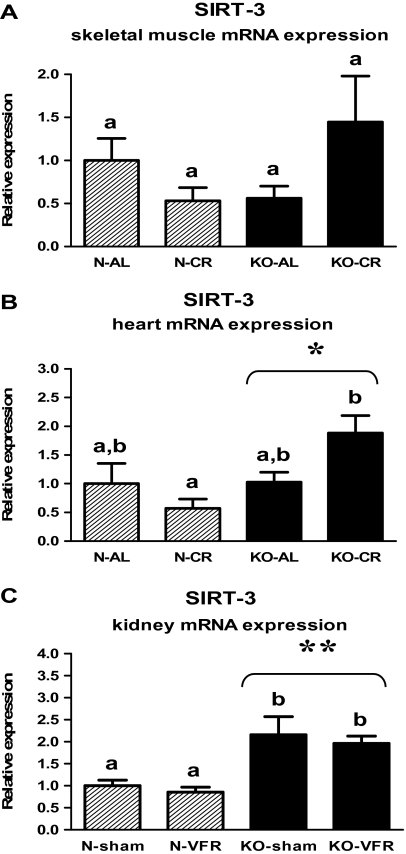
The mRNA levels of another member of the sirtuin family—SIRT-3—also was increased in the heart of KO mice in comparison with N animals (p = .011; Figure 4B, Table 2). Furthermore, there was the upregulation of SIRT-3 expression in KO-CR as compared with N-CR (p = .002; Figure 4B, Table 2). The increased level of SIRT-3 mRNA also was observed in the kidneys of knockouts versus N mice (p < .001; Figure 4C, Table 3). KO-sham and KO-VFR mice exhibited the upregulated SIRT-3 expression in the kidneys in comparison with the corresponding N animals (p = .006, p = .009, respectively; Figure 4C, Table 3). The level of SIRT-3 mRNA in skeletal muscles was not affected by genotype or CR (Figure 4A, Table 1). However, a divergent trend in SIRT-3 gene expression in this tissue could be observed (tendency toward a decrease of SIRT-3 expression in N-CR mice and an increase in KO-CR mice as compared with the corresponding AL animals; Figure 4A, Table 1).
Figure 4.
SIRT-3 messenger RNA (mRNA) expression in skeletal muscle (A), heart (B), or kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A and B—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; C—Experiment 2). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .011 vs N mice (the significance for genotype) and **p < .001 vs N mice (the significance for genotype).
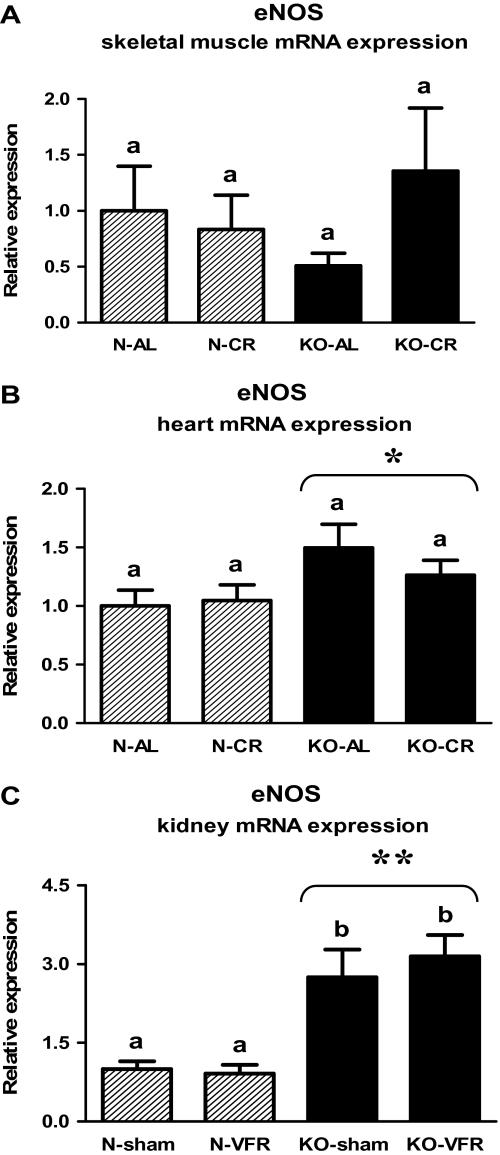
Similarly to AMPK and SIRT-3, the eNOS gene expression was increased in both the heart (Figure 5B, Table 2) and the kidney tissue (Figure 5C, Table 3) of knockouts versus N mice (p = .024, p < .001, respectively). The level of eNOS mRNA in the kidneys was increased in KO-sham or KO-VFR as compared with the corresponding N mice (p = .005, p < .001, respectively; Figure 5C, Table 3). The disruption of Ghr gene or CR did not alter the eNOS expression in the skeletal muscles (Figure 5A, Table 1).
Figure 5.
Endothelial nitric oxide synthase (eNOS) messenger RNA (mRNA) expression in skeletal muscle (A), heart (B), or kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A and B—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; C—Experiment 2). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .024 vs N mice (the significance for genotype) and **p < .001 vs N mice (the significance for genotype).
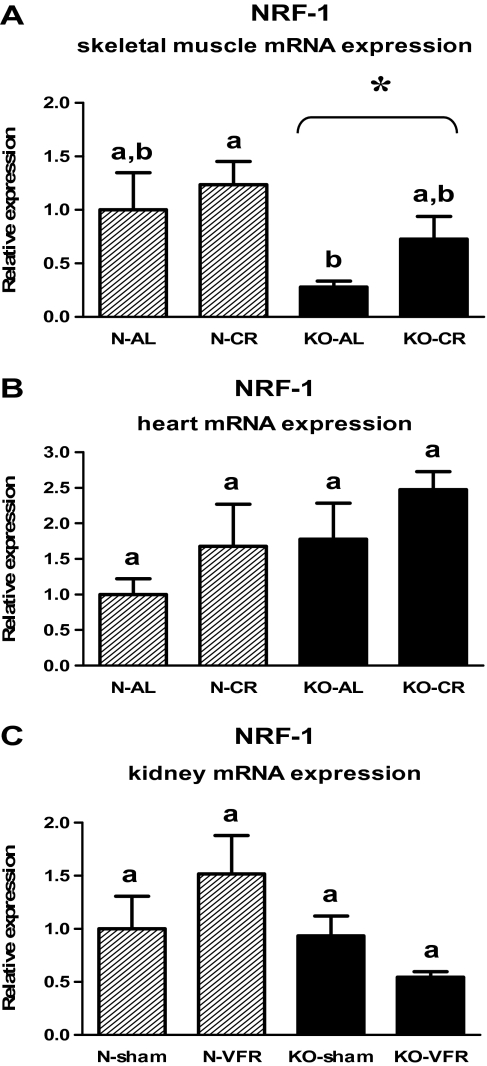
The level of NRF-1 mRNA in heart (Figure 6B, Table 2) or kidneys (Figure 6C, Table 3) was not affected by genotype and CR or VFR. KO mice exhibited the altered NRF-1 gene expression only in the skeletal muscles in which NRF-1 mRNA level was decreased as compared with N mice (p = .011; Figure 6A, Table 1).
Figure 6.
Nuclear respiratory factor-1 (NRF-1) messenger RNA (mRNA) expression in skeletal muscle (A), heart (B), or kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A and B—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; C—Experiment 2). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .011 vs N mice (the significance for genotype).
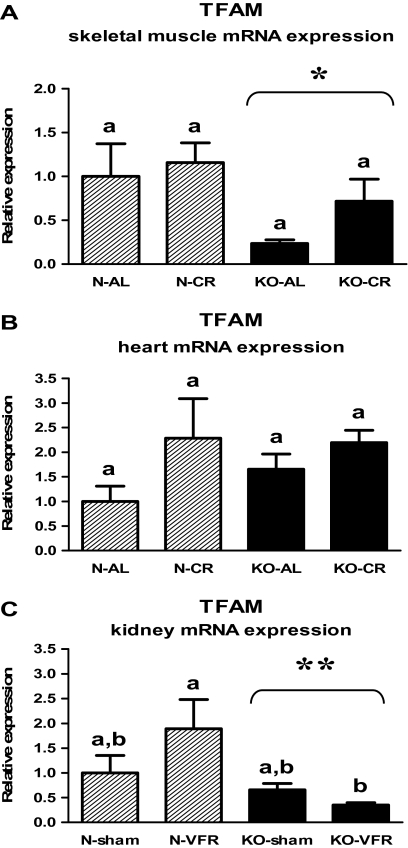
The decreased TFAM expression was observed in the skeletal muscles (Figure 7A, Table 1) and kidneys (Figure 7C, Table 3) of KO mice as compared with N animals (p = .021, p = .012, respectively). In the latter tissue, the level of TFAM mRNA was downregulated in KO-VFR mice versus N-VFR animals (p = .023; Figure 7C, Table 3). No statistical differences of TFAM gene expression, depending on genotype or CR, were observed in the heart (Figure 7B, Table 2).
Figure 7.
TFAM messenger RNA (mRNA) expression in skeletal muscle (A), heart (B), or kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A and B—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; C—Experiment 2). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .021 vs N mice (the significance for genotype) and **p = .012 vs N mice (the significance for genotype).
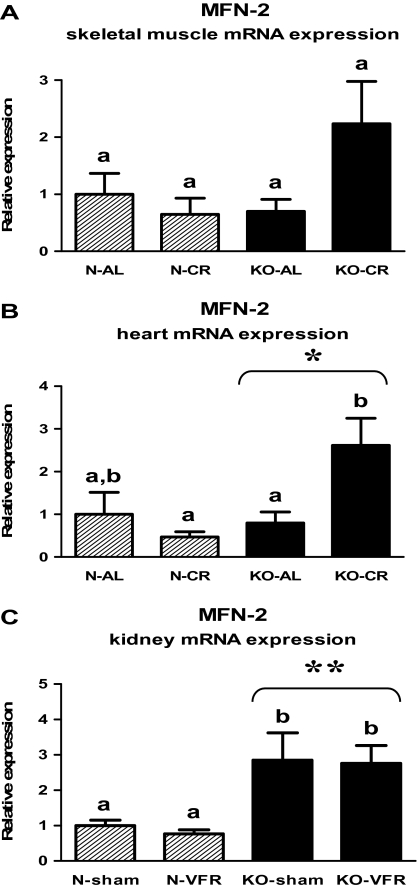
The knockout of the Ghr gene caused the increased MFN-2 gene expression in the heart (Figure 8B, Table 2) and kidneys (Figure 8C, Table 3) (p = .028, p < .001, respectively). KO-CR mice exhibited the elevated level of MFN-2 mRNA in the heart as compared with KO-AL or N-CR mice (p = .029, p = .003, respectively; Figure 8B, Table 2). In the kidneys, there was increased MFN-2 expression in KO-sham or KO-VFR mice in comparison with the corresponding N mice (p = .042, p = .019, respectively; Figure 8C, Table 3). The level of MFN-2 mRNA in skeletal muscles was not affected by genotype or CR (Figure 8A, Table 1).
Figure 8.
MFN-2 messenger RNA (mRNA) expression in skeletal muscle (A), heart (B), or kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A and B—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; C—Experiment 2). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .028 vs N mice (the significance for genotype) and **p < .001 vs N mice (the significance for genotype).
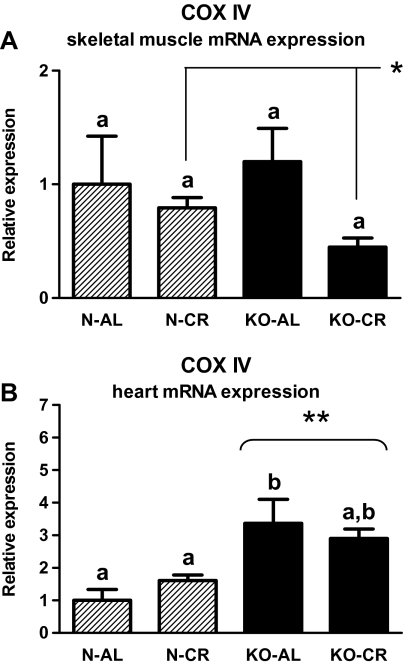
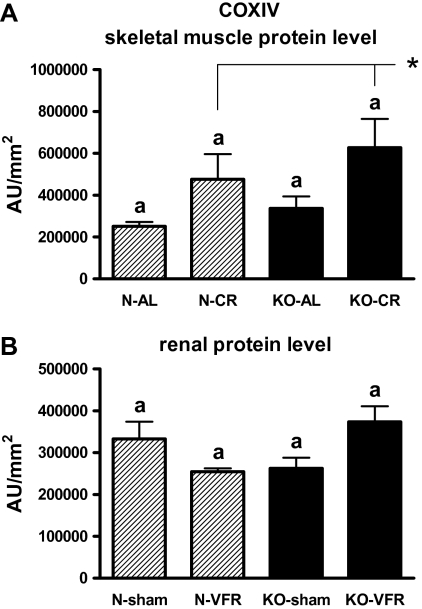
The increased skeletal muscle COXIV protein level was demonstrated in CR as compared with AL animals (p = .019; Figure 10A) even though COXIV gene expression was reduced in CR versus AL mice in this tissue (Figure 9A, Table 1; p = .048). COXIV gene expression was increased in heart of KOs versus N mice (p < .001; Figure 9B, Table 2). KO-AL mice demonstrated the elevated level of COXIV mRNA in the heart as compared with N-AL animals (p = .003; Figure 9B, Table 2).
Figure 9.
COXIV messenger RNA (mRNA) expression in skeletal muscle (A) and heart (B) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL) or subjected to 40% calorie restriction (CR; Experiment 1). The data from real-time polymerase chain reaction were normalized by the housekeeping gene β2-microglobulin (B2M) and expressed as the relative expression. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .048 vs AL mice (the significance for diet intervention) and **p < .001 vs N mice (the significance for genotype).
Figure 10.
COXIV protein level in skeletal muscle (A) and kidney (B) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; B—Experiment 2). The protein level was expressed as the arbitrary unit (AU) per square millimeter. Values are means ± SEM. a—values that share the same letter in the superscript are not statistically significant. *p = .019 vs AL mice (the significance for diet intervention).
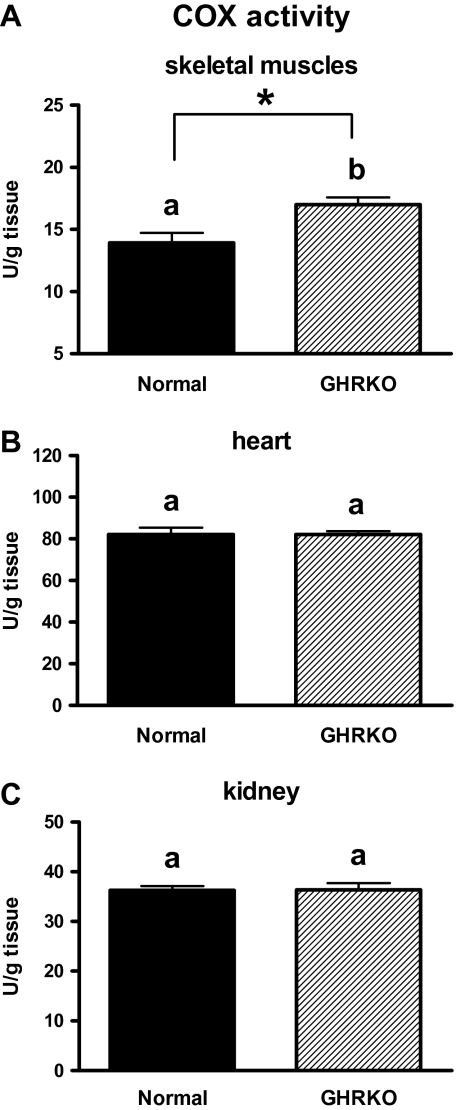
The COX activity was increased in skeletal muscles of GHRKO mice as compared with N animals (Figure 11A; p = .007). No differences in the heart (Figure 11B) and kidney (Figure 11C) COX activity between GHRKO and N mice were detected.
Figure 11.
COX activity in skeletal muscle (A), heart (B), and kidney (C) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO) mice. The COX activity was expressed as the units (U) per gram of tissue. Values are means ± SEM. a and b—values that do not share the same letter in the superscript are statistically significant (p < .05). *p = .007.
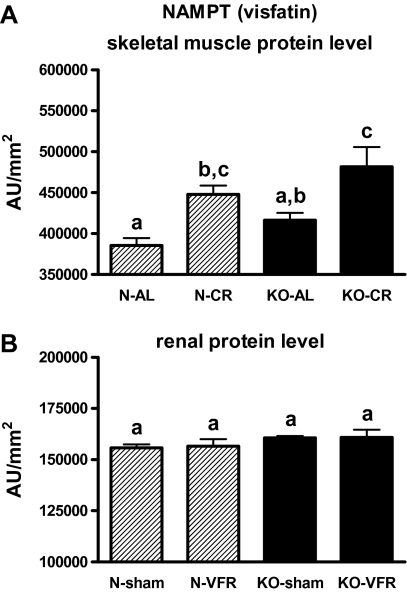
N-CR and KO-CR mice exhibited increased skeletal muscle level of NAMPT (visfatin) protein in comparison with the corresponding AL mice (p = .042, p = .03; Figure 12A). No significant changes in renal NAMPT protein levels were observed (Figure 12B).
Figure 12.
NAMPT protein level in skeletal muscle (A) and kidney (B) of normal (N) and growth hormone receptor/–binding protein knockout (GHRKO; KO) mice fed ad libitum (AL), subjected to 40% calorie restriction (CR; A—Experiment 1), sham operated (sham), or subjected to visceral fat removal (VFR; B—Experiment 2). The protein level was expressed as the arbitrary unit (AU) per square millimeter. Values are means ± SEM. a–c—values that do not share the same letter in the superscript are statistically significant (p < .05).
DISCUSSION
Long-lived GHRKO mice with targeted disruption of the growth hormone receptor (Ghr) gene (1) are a very important model system for studies of aging and control and regulation of life span. Various biochemical and physiological mechanisms, including enhanced insulin sensitivity and resistance to cancer or oxidative stress, were considered as potential reasons for the remarkably extended longevity of these animals (3,5–8,11,42). However, mitochondrial biogenesis—an essential process for proper cell viability—has not been analyzed in these mutants.
In the present study, the expression of PGC-1α—the “master regulator” of mitochondriogenesis and a coactivator of various transcription factors—was increased in skeletal muscles and kidneys of GHRKO (KO) mice. The upregulated PGC-1α expression presumably confers a major benefit on these mutants and corresponds to the results obtained by Al-Regaiey and colleagues (43) in livers of male GHRKO mice. Decreased PGC-1α expression may lead, through alterations of transcription of various genes, to lipid accumulation in skeletal muscles and finally to insulin resistance, obesity, and diabetes (44). However, Hancock and colleagues (45) recently showed that high-fat diet in Wistar rats caused insulin resistance despite the increase in muscle mitochondria and improved mitochondrial function. This high-fat diet led to fatty acids elevation, triggering proliferator-activated receptor δ (PPAR δ) and resulting in an increase of PPAR δ and PGC-1α protein levels (45). Interestingly, this PPAR δ overexpression did not cause an increase of PGC-1α mRNA levels (45). Therefore, it could be speculated that unaltered PGC-1α mRNA expression, as seen in the hearts of normal (N) versus KO mice, may not exclude an increase in PGC-1α protein level. Interestingly, insulin resistance may be one of the elements of the antioxidant defense mechanism, protecting cells from oxidative damage due to nutrient excess (46). In conclusion, the interactions between insulin resistance and mitochondrial physiology are still not clear (47) and require further studies.
The mice with targeted disruption of the Ghr gene exhibited increased levels of AMPK mRNA in the hearts and kidneys. The elevated AMPK expression may be beneficial for proper heart function in these mice. Activation of AMPK in the heart appears to be necessary for cardiomyocyte survival during ischemia (48), increases energy production, and inhibits apoptosis, thus protecting the heart during the ischemic stress (49). Moreover, Russell and colleagues (50) showed severe left ventricular contractile dysfunction with increased apoptosis and necrosis in AMPK-deficient hearts in mice. The increased cardiac AMPK expression may protect from cardiomyocytes hypertrophy induced by angiotensin II because it has been previously shown that this kind of hypertrophy is accompanied by decreased activation of AMPK (51). In the present study, no changes in AMPK mRNA level were observed in skeletal muscles of KO mice. This unchanged AMPK gene expression may be consistent with the results of the study performed by Gonzalez and colleagues (52), showing that AMPK activity in “m. gastrocnemius” of C57BL/6 mice was unaffected by age or hypoxemia.
The increased SIRT-1 mRNA level in the heart of KO mice, detected in the present study, can also be assumed to be beneficial for the knockouts. Hsu and colleagues (53) showed that transgenic mice with cardiac-specific overexpression of SIRT-1 demonstrated delayed aging and protection against oxidative stress in the heart. However, such effects of SIRT-1 were observed only in transgenic mice with low-to-moderate SIRT-1 overexpression (54). SIRT-1 plays a role in sustaining normal immune function and in delaying the onset of autoimmune disease (55). Moreover, deletion of SIRT-1 results in embryonic lethality, causes chromosome abnormalities, and leads to impaired DNA damage repair (56). Interestingly, SIRT-1 activators improved insulin sensitivity in obese mice as well as in fatty Zucker rats and even are considered as a potential medicament for treating type 2 diabetes (57). Additionally, SIRT-1 is said to be a novel target for atherosclerosis prevention and treatment (58).
In the present study, the level of another member of the sirtuin family—SIRT-3—also was increased in the heart of GHRKO mice in comparison with N controls, and this upregulation may benefit these mutants. Sundaresan and colleagues (59) showed that SIRT-3 protected the mouse heart by blocking the cardiac response to some hypertrophic stimuli. KO mice also exhibited an increased level of SIRT-3 mRNA in the kidneys as compared with normal mice. This result is consistent with the previous observations showing high SIRT-3 expression in numerous metabolically active tissues, including kidney, brown fat, liver, and brain (60). In contrast, SIRT-1 or SIRT-3 expression did not differ between KO and N mice in the skeletal muscles. One of possible explanations of these results can be based on the observation that AMPK enhances SIRT-1 activity (61), and thus, unchanged muscular AMPK expression would correspond to unaltered SIRT-1 (and potentially also SIRT-3) mRNA level in this tissue in KOs. Unaltered expression of some genes may not exclude changes in the corresponding protein levels (45,62).
The next alteration in GHRKO mice that is potentially protective and beneficial for extended longevity is the increased level of eNOS mRNA in the heart and the kidneys. Lee-Young and colleagues (63) confirmed that some metabolic disturbances, including insulin resistance, can be associated with reduced eNOS expression. In another study, the authors showed that eNOS knockout mice [eNOS(−/−)] had defects in arteriogenesis after ischemia (64). It is worth emphasizing that eNOS(−/−) mice are characterized by hypertension, increased heart rate variability, impaired angiogenesis, age-dependent left ventricle hypertrophy, cardiac valve abnormalities, and insulin resistance (65) with fasting hyperinsulinemia and hyperlipidemia (66). eNOS knockout mice also may have renal dysfunction or damage similar to what has been shown after ischemia/reperfusion injury (67). Moreover, lack of eNOS may accelerate glomerular and tubulointerstitial injury, being characterized by a loss of glomerular and peritubular capillaries (68). In contrast, the prevention of the ischemia-reperfusion injury in skeletal muscle by maintaining vascular integrity was observed in transgenic mice overexpressing eNOS in endothelial cells (69). Furthermore, eNOS overexpression prevented the development of renovascular hypertension in C57BL/6 mice (70).
It is known that PGC-1α induces transcription of NRF-1, leading to the increased expression of TFAM (71). Unexpectedly, the NRF-1 or TFAM gene expressions were decreased in the skeletal muscles of KO mice as compared with N animals, even though the PGC-1α expression was increased in this tissue in knockouts. Moreover, the level of TFAM mRNA also was decreased in the kidneys of KOs in comparison with N mice, although the expression of PGC-1α was elevated in this tissue. Therefore, it could be hypothesized that PGC-1α may potentially act in KO mice via other transcription factors. It was previously shown that mice with a disrupted TFAM gene develop dilated cardiomyopathy and atrioventricular heart conduction blocks and die at a very young age (72). In contrast, TFAM overexpression ameliorates the cardiac dysfunctions caused by myocardial infarction (73,74). Although consistent with reduced level of NRF-1, the unexpected decrease of TFAM gene expression in KOs may suggest a different role of this factor in long-lived GHRKO mice. Presumably, mechanisms of TFAM action and of the disruption of Ghr gene may overlap. It could be speculated that upregulated TFAM activity is not necessary for maintenance of proper cell viability in KO mice. Interestingly, Pesce and colleagues (75) demonstrated the increase of TFAM amount (and reduction in mitochondrial DNA content) in aged rat “soleus” skeletal muscle, and they suggest that this increase may be a cell compensatory response to decrease of mitochondrial DNA observed during aging. Similar observations were obtained in aged rat frontal cortex by Picca and colleagues (personal communication). Therefore, the decrease of TFAM expression, as seen in the present study, may also be regarded as beneficial for KO mice and potentially reflect unaltered (not decreased) mitochondrial DNA content.
Mice with the targeted disruption of Ghr gene exhibited increased MFN-2 gene expression in the heart and kidneys in comparison with normal animals. Fang and colleagues (76) showed the downregulated MFN-2 expression in hypertrophied cardiomyocytes. Moreover, the decreased MFN-2 expression was observed in patients with obesity and type 2 diabetes (31,77). Therefore, the changes observed in the present study can be, by analogy, regarded as beneficial for KO mice.
The level of COXIV protein was increased in skeletal muscles in CR versus AL animals, although no differences in this protein level between KO and N mice were detected. Importantly, the COX activity was significantly increased in KO mice as compared with N animals in this tissue. It could be hypothesized that an increase of the COX activity may be regarded as beneficial for GHRKO mice in the context of mitochondriogenesis regulation and potentially for life-span extension.
COXIV gene expression was increased in the heart of KOs versus N mice, even though the COX activity in this tissue of KO mice did not differ from N animals. Also, no differences in renal COXIV and NAMPT (visfatin) protein levels as well as in renal COX activity between KO and N mice were detected. Comparison of these data (showing no changes) with the increased gene expression of several key regulators of mitochondriogenesis in corresponding tissues of KO mice, as was noticed earlier, may create some difficulties for their unambiguous explanation. Nevertheless, some authors show in their studies different patterns of expression of mitochondriogenesis regulators. For example, Yin and colleagues (78) demonstrated increased COXIV expression and, at the same time, unchanged PGC-1 gene expression and protein level in rat model of hypoxic/ischemic brain injury. Therefore, by analogy, the results observed by us in kidneys and hearts (ie, the increase of gene expressions and, at the same time, unchanged COX activity) of GHRKO mice may not exclude an important role of these tissues in mitochondriogenesis regulation. However, taking into account the above-mentioned observations concerning COX, one could speculate that the KOs’ skeletal muscles may play an essential role in the regulation of mitochondrial biogenesis and other physiological processes. We have previously demonstrated the decreased expression level of several proapoptotic genes and proteins in the skeletal muscles of GHRKO mice what may be regarded as beneficial for these knockouts (79).
CR has been previously shown to affect numerous factors involved in the mitochondrial biogenesis (including AMPK, eNOS, sirtuins, and PGC-1α), leading to the increase of its activity (80). Recently, Someya and colleagues (81) have shown that SIRT-3 is required for the reduction of oxidative DNA damage in several tissues under CR conditions and prevents age-related hearing loss in wild-type mice. The authors suggest that a SIRT-3–dependent mechanism may be one of the most important mechanisms involved in aging retardation by CR in mammals (81). However, in the present study, expression of none of the examined genes was affected by CR. Nevertheless, these results are not unexpected. In contrast to extended life span and improved insulin sensitivity in normal mice subjected to CR, an identical intervention failed to enhance insulin sensitivity or increase average or mean life span of GHRKO mice (34). Additionally, CR failed to further modify the alterations in insulin signaling in the GHRKOs’ livers as compared with normal mice (82). The insulin signaling cascade was also unaffected by CR in the heart of GHRKO mice (83). Moreover, Gonzalez and colleagues (84) showed that CR did not alter AMPK activity in the heart, skeletal muscles, and liver in mice. In another study, 6-month CR in Fischer-344 rats did not change the total level of SIRT-1 (85). Furthermore, we have recently demonstrated that decreased expression of certain proapoptotic genes and proteins in GHRKO mice, potentially beneficial for extended longevity in these mutants, was not affected by CR (79). Importantly, Hancock and colleagues (86) demonstrated that 30% CR did not induce an increase in mitochondria in heart, brain, liver, adipose tissue, or skeletal muscle in male Wistar rats, as assessed by measurements of several key mitochondrial proteins, including cytochrome c, citrate synthase, and cytochrome c oxidase subunit IV (COXIV).
Another intervention that has been reported to improve insulin signaling in normal mice and rats and extend longevity in rats is surgical VFR (35–37). Similarly to CR, VFR did not affect the expression of the examined genes in GHRKO or normal mice. These results may suggest that mechanisms linking GH resistance, CR, and VFR to aging presumably overlap. Moreover, it should be emphasized that some tissues including the heart and kidney are already at an extremely high metabolic rate, and therefore, further adaptations may be less feasible.
In summary, GHRKO mice are characterized by increased expression of key regulators of mitochondrial biogenesis. This effect may be regarded as beneficial for proper cell viability and, as a probable result, for extended longevity in these long-lived mutants. However, this potentially beneficial characteristic did not improve further with CR or VFR, which attributes the primary role in mitochondriogenesis to GH resistance. Furthermore, the gender of mice used in our study was also of no consequence for the alterations of the expression of the examined genes. Further studies are needed to determine how the levels of expression of mitochondriogenesis-related genes in different tissues and organs can contribute to the control and regulation of longevity.
FUNDING
The present study was supported by National Institute on Aging (AG 19899, U19 AG023122, AG31736, and AG032290), the Ellison Medical Foundation, Southern Illinois University School of Medicine, Claude D. Pepper Older Americans Independence Center (1P30AG028740), and Polish Ministry of Science and Higher Education (N N401 042638).
Acknowledgments
The authors would like to thank Steve Sandstrom for helping with the editing of the manuscript.
References
- 1.Zhou Y, Xu BC, Maheshwari HG, et al. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse) Proc Natl Acad Sci USA. 1997;94:13215–13220. doi: 10.1073/pnas.94.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopchick JJ, Laron Z. Is the Laron mouse an accurate model of Laron syndrome? Mol Genet Metab. 1999;68:232–236. doi: 10.1006/mgme.1999.2890. [DOI] [PubMed] [Google Scholar]
- 3.Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene disrupted mice. Endocrinology. 2000;141:2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- 4.Bartke A, Chandrashekar V, Bailey B, Zaczek D, Turyn D. Consequences of growth hormone (GH) overexpression and GH resistance. Neuropeptides. 2002;36:201–208. doi: 10.1054/npep.2002.0889. [DOI] [PubMed] [Google Scholar]
- 5.Coschigano KT, Holland AN, Riders ME, List EO, Flyvbjerg A, Kopchick JJ. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased lifespan. Endocrinology. 2003;144:3799–3810. doi: 10.1210/en.2003-0374. [DOI] [PubMed] [Google Scholar]
- 6.Bartke A, Brown-Borg H. Life extension in the dwarf mouse. Curr Top Dev Biol. 2004;63:189–225. doi: 10.1016/S0070-2153(04)63006-7. [DOI] [PubMed] [Google Scholar]
- 7.Coschigano KT. Aging-related characteristics of growth hormone receptor/binding protein gene-disrupted mice. Age. 2006;28:191–200. doi: 10.1007/s11357-006-9004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu JL, Coschigano KT, Robertson K, et al. Disruption of growth hormone receptor gene causes diminished pancreatic islet size and increased insulin sensitivity in mice. Am J Physiol Endocrinol Metab. 2004;287:E405–E413. doi: 10.1152/ajpendo.00423.2003. [DOI] [PubMed] [Google Scholar]
- 9.Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289:E23–E29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- 10.Sun LY, Steinbaugh MJ, Masternak MM, Bartke A, Miller RA. Fibroblasts from long-lived mutant mice show diminished ERK1/2 phosphorylation but exaggerated induction of immediate early genes. Free Radic Biol Med. 2009;47:1753–1761. doi: 10.1016/j.freeradbiomed.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeno Y, Hubbard GB, Lee S, et al. Reduced incidence and delayed occurrence of fatal neoplastic diseases in growth hormone receptor/binding protein knockout mice. J Gerontol A Biol Sci Med Sci. 2009;64:522–529. doi: 10.1093/gerona/glp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 13.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 14.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 15.Lambert AJ, Wang B, Yardley J, Edwards J, Merry BJ. The effect of aging and caloric restriction on mitochondrial protein density and oxygen consumption. Exp Gerontol. 2004;39:289–295. doi: 10.1016/j.exger.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Nisoli E, Tonello C, Cardile A, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Lluch G, Hunt N, Jones B, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci USA. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Civitarese AE, Carling S, Heilbronn LK, et al. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reznick RM, Zong H, Li J, et al. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab. 2007;5:151–156. doi: 10.1016/j.cmet.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benigni A, Corna D, Zoja C, et al. Disruption of the Ang II type 1 receptor promotes longevity in mice. J Clin Invest. 2009;119:524–530. doi: 10.1172/JCI36703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 24.Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669–2677. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- 25.Leary SC, Shoubridge EA. Mitochondrial biogenesis: which part of “NO” do we understand? Bioessays. 2003;25:538–541. doi: 10.1002/bies.10298. [DOI] [PubMed] [Google Scholar]
- 26.Nisoli E, Clementi E, Paolucci C, et al. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 27.Brown GC. Nitric oxide and mitochondria. Front Biosci. 2007;12:1024–1033. doi: 10.2741/2122. [DOI] [PubMed] [Google Scholar]
- 28.Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 29.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 30.Larsson NG, Wang J, Wilhelmsson H, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 31.Bach D, Pich S, Soriano FX, et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem. 2003;278:17190–17197. doi: 10.1074/jbc.M212754200. [DOI] [PubMed] [Google Scholar]
- 32.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 33.Weindruch R, Sohal RS. Seminars in medicine of the Beth Israel Deaconess Medical Center. Caloric intake and aging. N Engl J Med. 1997;337:986–994. doi: 10.1056/NEJM199710023371407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonkowski MS, Rocha JS, Masternak MM, Al Regaiey KA, Bartke A. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci USA. 2006;103:7901–7905. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barzilai N, She L, Liu B-Q, Vuguin P, Cohen P, Wang J, et al. Surgical removal of visceral fat reverses hepatic insulin resistance. Diabetes. 1999;48:94–98. doi: 10.2337/diabetes.48.1.94. [DOI] [PubMed] [Google Scholar]
- 36.Shi H, Strader AD, Woods SC, Seeley RJ. The effect of fat removal on glucose tolerance is depot specific in male and female mice. Am J Physiol Endocrinol Metab. 2007;293:E1012–E1020. doi: 10.1152/ajpendo.00649.2006. [DOI] [PubMed] [Google Scholar]
- 37.Muzumdar R, Allison DB, Huffman DM, et al. Visceral adipose tissue modulates mammalian longevity. Aging Cell. 2008;7:438–440. doi: 10.1111/j.1474-9726.2008.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masternak MM, Bartke A, Wang F. The Role of Visceral Fat on Insulin Signaling in Long-Living Mice. Presented at: 39th Annual Meeting of the American Aging Association; June 47, 2010; Portland, OR. Abstract 22, page 40. [Google Scholar]
- 39.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 40.Masternak MM, Al-Regaiey KA, Del Rosario Lim MM, et al. Caloric restriction and growth hormone receptor knockout: effects on expression of genes involved in insulin action in the heart. Exp Gerontol. 2006;41:417–429. doi: 10.1016/j.exger.2006.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cogswell AM, Stevens RJ, Hood DA. Properties of skeletal muscle mitochondria isolated from subsarcolemmal and intermyofibrillar regions. Am J Physiol Cell Physiol. 1993;264:C383–C389. doi: 10.1152/ajpcell.1993.264.2.C383. [DOI] [PubMed] [Google Scholar]
- 42.Harper JM, Salmon AB, Chang Y, Bonkowski M, Bartke A, Miller RA. Stress resistance and aging: influence of genes and nutrition. Mech Ageing Dev. 2006;127:687–694. doi: 10.1016/j.mad.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Al-Regaiey KA, Masternak MM, Bonkowski M, Sun L, Bartke A. Long-lived growth hormone receptor knockout mice: interaction of reduced insulin-like growth factor I/insulin signaling and caloric restriction. Endocrinology. 2005;146:851–860. doi: 10.1210/en.2004-1120. [DOI] [PubMed] [Google Scholar]
- 44.Liang H, Ward WF. PGC-1α: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30:145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 45.Hancock CR, Han D-H, Chen M, et al. High-fat diets cause insulin resistance despite an increase in muscle mitochondria. Proc Natl Acad Sci USA. 2008;105:7815–7820. doi: 10.1073/pnas.0802057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoehn KL, Salmon AB, Hohnen-Behrens C, et al. Insulin resistance is a cellular antioxidant defense mechanism. Proc Natl Acad Sci USA. 2009;106:17787–17792. doi: 10.1073/pnas.0902380106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kraegen EW, Cooney GJ, Turner N. Muscle insulin resistance: a case of fat overconsumption, not mitochondrial dysfunction. Proc Natl Acad Sci USA. 2008;105:7627–7628. doi: 10.1073/pnas.0803901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolinsky VW, Dyck JRB. Role of AMP-activated protein kinase in healthy and diseased hearts. Am J Physiol Heart Circ Physiol. 2006;291:H2557–H2569. doi: 10.1152/ajpheart.00329.2006. [DOI] [PubMed] [Google Scholar]
- 49.Dyck JRB, Lopaschuk GD. AMPK alterations in cardiac physiology and pathology: enemy or ally? J Physiol. 2006;574:95–112. doi: 10.1113/jphysiol.2006.109389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russell RR, 3rd, Li J, Coven DL, et al. AMP-activated protein kinase mediates ischemic glucose uptake and prevents postischemic cardiac dysfunction, apoptosis, and injury. J Clin Invest. 2004;114:495–503. doi: 10.1172/JCI19297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stuck BJ, Lenski M, Böhm M, Laufs U. Metabolic switch and hypertrophy of cardiomyocytes following treatment with angiotensin II are prevented by AMP-activated protein kinase. J Biol Chem. 2008;283:32562–32569. doi: 10.1074/jbc.M801904200. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez AA, Kumar R, Mulligan JD, Davis AJ, Saupe KW. Effects of aging on cardiac and skeletal muscle AMPK activity: basal activity, allosteric activation, and response to in vivo hypoxemia in mice. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1270–R1275. doi: 10.1152/ajpregu.00409.2004. [DOI] [PubMed] [Google Scholar]
- 53.Hsu CP, Odewale I, Alcendor RR, Sadoshima J. Sirt1 protects the heart from aging and stress. Biol Chem. 2008;389:221–231. doi: 10.1515/BC.2008.032. [DOI] [PubMed] [Google Scholar]
- 54.Alcendor RR, Gao S, Zhai P, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 55.Sequeira J, Boily G, Bazinet S, et al. sirt1-null mice develop an autoimmune-like condition. Exp Cell Res. 2008;314:3069–3074. doi: 10.1016/j.yexcr.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 56.Wang R-H, Sengupta K, Li C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Milne JC, Lambert PD, Schenk S, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu W, Fu Y-C, Chen C-J, Wang X, Wang W. SIRT1: a novel target to prevent atherosclerosis. J Cell Biochem. 2009;108:10–13. doi: 10.1002/jcb.22240. [DOI] [PubMed] [Google Scholar]
- 59.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palacios OM, Carmona JJ, Michan S, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1α in skeletal muscle. Aging. 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Canto C, Gerhart-Hines Z, Feige JN, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hawley JA, Holloszy JO. Exercise: it’s the real thing. Nutr Rev. 2009;67:172–178. doi: 10.1111/j.1753-4887.2009.00185.x. [DOI] [PubMed] [Google Scholar]
- 63.Lee-Young RS, Ayala JE, Hunley CF, et al. Endothelial nitric oxide synthase is central to skeletal muscle metabolic regulation and enzymatic signaling during exercise in vivo. Am J Physiol Regul Integr Comp Physiol. 2010;298:R1399–R1408. doi: 10.1152/ajpregu.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu J, deMuinck ED, Zhuang Z, et al. Endothelial nitric oxide synthase is critical for ischemic remodeling, mural cell recruitment, and blood flow reserve. Proc Natl Acad Sci USA. 2005;102:10999–11004. doi: 10.1073/pnas.0501444102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mungrue IN, Bredt DS, Stewart DJ, Husain M. From molecules to mammals: what’s NOS got to do with it? Acta Physiol Scand. 2003;179:123–135. doi: 10.1046/j.1365-201X.2003.01182.x. [DOI] [PubMed] [Google Scholar]
- 66.Duplain H, Burcelin R, Sartori C, et al. Insulin resistance, hyperlipidemia, and hypertension in mice lacking endothelial nitric oxide synthase. Circulation. 2001;104:342–345. doi: 10.1161/01.cir.104.3.342. [DOI] [PubMed] [Google Scholar]
- 67.Milsom AB, Patel NSA, Mazzon E, et al. Role for endothelial nitric oxide synthase in nitrite-induced protection against renal ischemia-reperfusion injury in mice. Nitric Oxide. 2010;22:141–148. doi: 10.1016/j.niox.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 68.Nakayama T, Sato W, Kosugi T, et al. Endothelial injury due to eNOS deficiency accelerates the progression of chronic renal disease in the mouse. Am J Physiol Renal Physiol. 2009;296:F317–F327. doi: 10.1152/ajprenal.90450.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ozaki M, Kawashima S, Hirase T, et al. Overexpression of endothelial nitric oxide synthase in endothelial cells is protective against ischemia-reperfusion injury in mouse skeletal muscle. Am J Pathol. 2002;160:1335–1344. doi: 10.1016/s0002-9440(10)62560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gava AL, Peotta VA, Cabral AM, Vasquez EC, Meyrelles SS. Overexpression of eNOS prevents the development of renovascular hypertension in mice. Can J Physiol Pharmacol. 2008;86:458–464. doi: 10.1139/y08-044. [DOI] [PubMed] [Google Scholar]
- 71.Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 72.Wang J, Wilhelmsson H, Graff C, et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 73.Ikeuchi M, Matsusaka H, Kang D, et al. Overexpression of mitochondrial transcription factor A ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- 74.Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7:39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 75.Pesce V, Cormio A, Fracasso F, Lezza AM, Cantatore P, Gadaleta MN. Age-related changes of mitochondrial DNA content and mitochondrial genotypic and phenotypic alterations in rat hind-limb skeletal muscles. J Gerontol A Biol Sci Med Sci. 2005;60:715–723. doi: 10.1093/gerona/60.6.715. [DOI] [PubMed] [Google Scholar]
- 76.Fang L, Moore XL, Gao XM, Dart AM, Lim YL, Du XJ. Down-regulation of mitofusin-2 expression in cardiac hypertrophy in vitro and in vivo. Life Sci. 2007;80:2154–2160. doi: 10.1016/j.lfs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 77.Bach D, Naon D, Pich S, et al. Expression of Mfn2, the Charcot-Marie-Tooth neuropathy type 2A gene, in human skeletal muscle: effects of type 2 diabetes, obesity, weight loss, and the regulatory role of tumor necrosis factor alpha and interleukin-6. Diabetes. 2005;54:2685–2693. doi: 10.2337/diabetes.54.9.2685. [DOI] [PubMed] [Google Scholar]
- 78.Yin W, Signore AP, Iwai M, Cao G, Gao Y, Chen J. Rapidly increased neuronal mitochondrial biogenesis after hypoxic-ischemic brain injury. Stroke. 2008;39:3057–3063. doi: 10.1161/STROKEAHA.108.520114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gesing A, Masternak MM, Wang F, Lewinski A, Karbownik-Lewinska M, Bartke A. Decreased expression level of apoptosis-related genes and/or proteins in skeletal muscles, but not in hearts, of growth hormone receptor knockout mice. Exp Biol Med. 2011;236:156–168. doi: 10.1258/ebm.2010.010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lopez-Lluch G, Irusta PM, Navas P, de Cabo R. Mitochondrial biogenesis and healthy aging. Exp Gerontol. 2008;43:813–819. doi: 10.1016/j.exger.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Someya S, Yu W, Hallows WC, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bonkowski MS, Dominici FP, Arum O, et al. Disruption of growth hormone receptor prevents calorie restriction from improving insulin action and longevity. PLoS One. 2009;4:e4567. doi: 10.1371/journal.pone.0004567. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Giani JF, Bonkowski MS, Muñoz MC, et al. Insulin signaling cascade in the hearts of long-lived growth hormone receptor knockout mice: effects of calorie restriction. J Gerontol A Biol Sci Med Sci. 2008;63:788–797. doi: 10.1093/gerona/63.8.788. [DOI] [PubMed] [Google Scholar]
- 84.Gonzalez AA, Kumar R, Mulligan JD, Davis AJ, Weindruch R, Saupe KW. Metabolic adaptations to fasting and chronic caloric restriction in heart, muscle, and liver do not include changes in AMPK activity. Am J Physiol Endocrinol Metab. 2004;287:E1032–E1037. doi: 10.1152/ajpendo.00172.2004. [DOI] [PubMed] [Google Scholar]
- 85.Shinmura K, Tamaki K, Bolli R. Impact of 6-mo caloric restriction on myocardial ischemic tolerance: possible involvement of nitric oxide-dependent increase in nuclear Sirt1. Am J Physiol Heart Circ Physiol. 2008;295:H2348–H2355. doi: 10.1152/ajpheart.00602.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hancock CR, Han D-H, Higashida K, Kim SH, Holloszy JO. Does calorie restriction induce mitochondrial biogenesis? A reevaluation. FASEB J. 2011;25:785–791. doi: 10.1096/fj.10-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]