Abstract
Cancer cells display a reprogramming of metabolism that facilitates growth but addicts them to key enzyme activities. Two studies in Nature and Nature Genetics find that the gene encoding the serine biosynthetic enzyme phosphoglycerate dehydrogenase (PHGDH) is amplified in a subset of cancers and contributes to tumor cell growth.
Cancer cells possess metabolic properties that distinguish them from nonmalignant cells. Of enduring interest is their propensity to take up glucose in large amounts and convert it to lactic acid – the so-called Warburg effect (Koppenol et al., 2011). The causes and benefits of this phenomenon have been the focus of study and speculation for many years (Vander Heiden et al., 2009). Since the mid-1990s, which brought a resurgence of interest in cancer metabolism, a large body of work has demonstrated that enhanced glycolysis is a common consequence of the perturbed signal transduction that accompanies transforming mutations: that is, mutations in tumor suppressors and oncogenes lead, and metabolism follows. These mechanistic links with well-established driver mutations support the consensus that enhanced glycolysis is both a hallmark of malignant transformation and a potential therapeutic target (Hanahan and Weinberg, 2011).
But recent evidence implies that metabolic reprogramming can also occur as the result of genomic modifications of metabolic enzymes, and that these alterations independently contribute to tumorigenesis. Two new studies published in Nature Genetics and Nature (Locasale et al., 2011; Possemato et al., 2011) report that the gene encoding phosphoglycerate dehydrogenase (PHGDH) is amplified in a significant subset of human tumors. The two studies converged on PHGDH from different starting points. Possemato et al. identified it from a loss-of-function RNA-interference screen for metabolic genes required for tumorigenesis in an orthotopic model of breast cancer. Locasale et al. studied the fate of glucose-derived carbon in cancer cell lines and observed significant flux into metabolites downstream of PHGDH. Both studies then mined databases of copy number alterations in cancer to determine that the PHGDH gene on chromosome 1p12 is amplified in ~6% of breast cancers and 40% of melanomas. Subsequent experiments showed that a much larger fraction of tumors had elevated PHGDH protein levels, including 70% of estrogen receptor-negative breast tumors. High PHGDH expression with or without genomic amplification was associated with dependence on the enzyme for cell growth, suggesting that subsets of tumor cells are addicted to flux through this metabolic pathway.
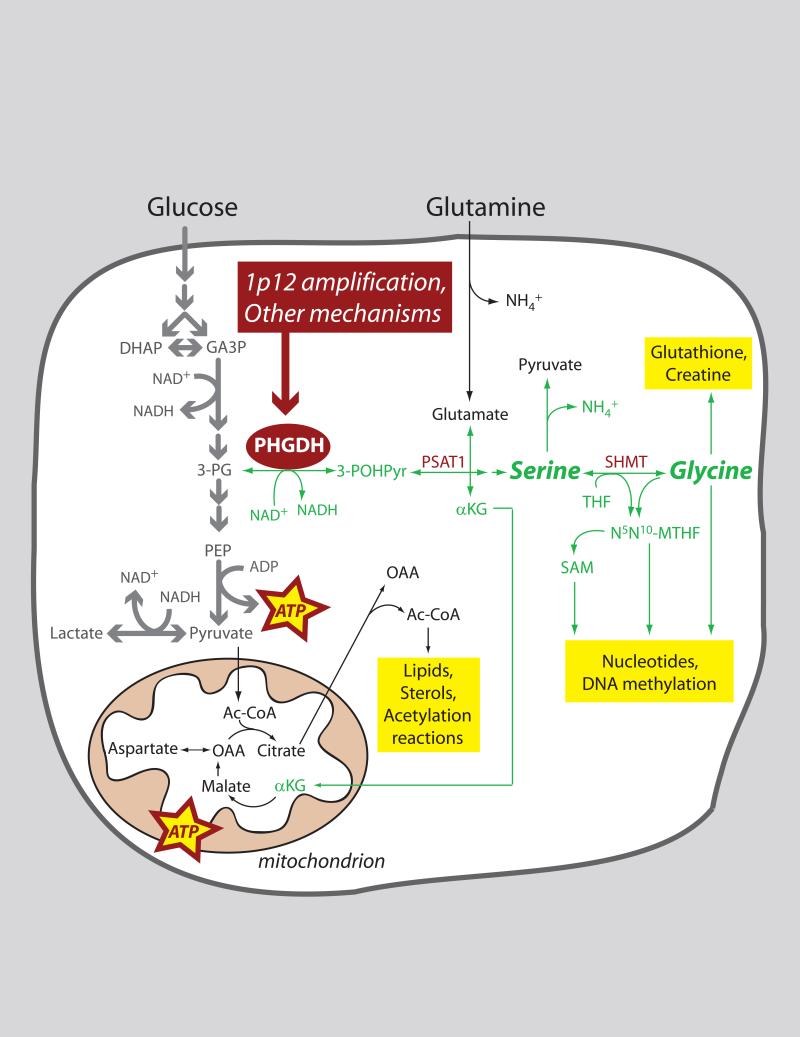
PHGDH is a fascinating and at first glance surprising metabolic target in cancer. It catalyzes entry into what amounts to a metabolic side-street, diverting flux away from the “superhighway” of tumor cell glycolysis (Figure 1). Thus it would appear to reduce energy formation from glucose. But the PHGDH pathway also provides many advantages for growing cells. First, de novo synthesis of serine and glycine, precursors for a variety of biosynthetic pathways, requires the removal of 3-phosphoglycerate from glycolysis via PHGDH. Second, conversion of serine to glycine by serine hydroxymethyltransferase (SHMT) is a major source of methyl groups for the one-carbon pools required for biosynthesis and DNA methylation. Furthermore, there were a few previous hints that serine/glycine biosynthesis might be important in tumorigenesis. In breast tumors, expression of several enzymes in the pathway, including PHGDH itself, correlated with metastasis in mice and poor clinical outcomes in humans (Pollari et al., 2011). Also, two isoforms of SHMT are transcriptional targets of the oncogene c-Myc, and over-expressing this enzyme stimulated proliferation in c-Myc deficient cells (Nikiforov et al., 2002).
Figure 1. Role of PHGDH in tumor cell metabolism.
Many tumors have enhanced expression of the metabolic enzyme phosphoglycerate (PHGDH), in some cases because the gene encoding this enzyme is amplified on chromosome 1p12. PHGDH diverts flux away from glycolysis by oxidizing 3-phosphoglycerate (3-PG) to 3-phospho-hydroxypyruvate (3-POHPyr). Subsequent metabolism of this 3-carbon intermediate feeds into the serine and glycine pools, providing numerous precursor molecules and other substrates required for cell growth and proliferation. Abbreviations: Ac-CoA, acetyl-CoA; αKG, α-ketoglutarate; DHAP, dihydroxyacetone phosphate; GA3P, glyceraldehyde-3-phosphate; NH4+, ammonium ion; N5N10-MTHF, N5,N10-methylene tetrahydrofolate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PSAT1, phosphoserine aminotransferase-1; SAM, S-adenosylmethionine; SHMT, serine hydroxymethyltransferase; THF, tetrahydrofolate.
Yet both serine and glycine are abundant in the plasma, so it was not immediately clear what could be gained by up-regulating their de novo synthesis at the expense of glycolysis. Possemato et al. made the counterintuitive observation that silencing PHGDH expression did not deplete intracellular serine, suggesting that some other output of the pathway must contribute to cell growth. Another metabolite generated during serine biosynthesis is α-ketoglutarate (αKG), produced at the transamination step that converts 3-phosphohydroxypyruvate to 3-phosphoserine (Figure 1). αKG is also the entry point through which glutamine supplies carbon to the tricarboxylic acid (TCA) cycle during cell growth, enabling the production of many essential biosynthetic precursors. Surprisingly, as much as half of all glutamine-derived αKG in PHGDH-overexpressing cells was generated via the PHGDH pathway. The compelling implication is that PHGDH could serve as a metabolic gatekeeper both for macromolecular biosynthesis downstream of glutamine metabolism (i.e. cell growth) and serine-dependent DNA synthesis (i.e. cell proliferation).
Is PHGDH a feasible therapeutic target in breast cancer, melanoma, or other diseases? In the Possemato study, silencing PHGDH in established breast tumors reduced their growth in mice, a promising finding. However, as is often the case for core metabolic enzymes, global PHGDH deficiency is associated with severe neurologic dysfunction in children (Jaeken et al., 1996). But perhaps using agents with poor access to the central nervous system would be adequately tolerated in cancer patients, or perhaps the exquisite dependence of selected tumors on PHGDH could provide a safe therapeutic window.
PHGDH joins a growing list of putative metabolic oncogenes and tumor suppressors whose alteration in the genome re-orchestrates the metabolic roadmap and may influence the development of malignant properties (Frezza et al., 2011). In a few cases, the relationship between enzyme mutation and oncogenesis is well established. Some familial cancer syndromes are caused by mutations in fumarate hydratase (FH) or in the succinate dehydrogenase (SDH) complex. In affected individuals, a loss-of-function mutation is inherited on one allele, and the other allele is deleted or otherwise mutated in the tumor, establishing these genes as tumor suppressors. More recently, mutations in two isoforms of isocitrate dehydrogenase (IDH1 and IDH2) were identified through genome-sequencing in gliomas and acute myeloid leukemia. Somatic acquisition of specific active-site mutations in one allele of either enzyme conveys a gain-of-function enzymatic activity, producing a metabolite, 2-hydroxyglutaric acid, with suspected effects on cell signaling, epigenetics and transformation (Figueroa et al., 2010). Thus mutant forms of IDH1 and IDH2 may function as bona fide oncogenes. Can the same be said of PHGDH? Locasale et al. found that PHGDH overexpression stimulates anchorage independence and disturbs polarity in mammary epithelial cells, features strongly correlated with malignancy. Precisely how mutations in any of these enzymes contribute to transformation and tumorigenesis is an extremely active area of research. Surely these genetically-determined “metabolic outliers” hold important clues about the biological basis of malignancy and the specific metabolic features selected during tumorigenesis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Pollard PJ, Gottlieb E. Inborn and acquired metabolic defects in cancer. J Mol Med. 2011;89:213–220. doi: 10.1007/s00109-011-0728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Jaeken J, Detheux M, Van Maldergem L, Foulon M, Carchon H, Van Schaftingen E. 3-Phosphoglycerate dehydrogenase deficiency: an inborn error of serine biosynthesis. Arch Dis Child. 1996;74:542–545. doi: 10.1136/adc.74.6.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011 doi: 10.1038/ng.890. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollari S, Kakonen SM, Edgren H, Wolf M, Kohonen P, Sara H, Guise T, Nees M, Kallioniemi O. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat. 2011;125:421–430. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. doi: 10.1038/nature10350. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]