Abstract
Mass spectrometry-based studies of proteins that are post-translationally modified by O-linked β-N-acetylglucosamine (O-GlcNAc) are challenged in effectively identifying the sites of modification while simultaneously sequencing the peptides. Here we tested the hypothesis that a combination of high-energy C-trap dissociation (HCD) and electron transfer dissociation (ETD) could specifically target the O-GlcNAc modified peptides and elucidate the amino acid sequence while preserving the attached GlcNAc residue for accurate site assignment. By taking advantage of the recently characterized O-GlcNAc-specific IgG monoclonal antibodies and the combination of HCD and ETD fragmentation techniques, O-GlcNAc modified proteins were enriched from HEK293T cells and subsequently characterized using the LTQ Orbitrap Velos™ ETD (Thermo Fisher Scientific) mass spectrometer. In our dataset, 83 sites of O-GlcNAc modification are reported with high confidence confirming that the HCD/ETD combined approach is amenable to the detection and site assignment of O-GlcNAc modified peptides. Realizing HCD triggered ETD fragmentation on a linear ion trap/Orbitrap platform for more in-depth analysis and application of this technique to other post-translationally modified proteins are currently underway. Furthermore, this report illustrates that the O-GlcNAc transferase appears to demonstrate promiscuity with regards to the hydroxyl-containing amino acid modified in short stretches of primary sequence of the glycosylated polypeptides.
Keywords: O-GlcNAc, HCD, ETD, tandem mass spectrometry, site assignment, post-translational modification, glycosylation
Introduction
Glycosylation on serine and threonine by a single O-linked β-N-acetylglucosamine (O-GlcNAc) moiety is a widespread post-translational modification observed on cytosolic and nuclear proteins. O-GlcNAc modification is a nutrient/stress-sensing modification that regulates proteins involved in a wide array of biological processes, including transcription, signal transduction, and metabolism1–2. Cycling of O-GlcNAc is regulated by the concerted actions of O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA)3, and the fluctuation of O-GlcNAc levels has been associated with the etiology of type II diabetes, cardiovascular disease, and neurodegenerative disorders4–7. Elucidating the molecular structure of O-GlcNAc modified proteins not only is crucial in revealing their site-specific functional roles but also is necessary in facilitating further discovery of the involvement of O-GlcNAc in major biological networks.
In order to compensate for the substoichiometric occupancy of O-GlcNAc modification3, 8–9, numerous techniques have been developed for the detection and enrichment of O-GlcNAc modified proteins, such as immunoblotting10–12, lectin affinity chromatography13–14, and chemoenzymatic approach15–16. Facilitated by the advances in analytical technology, the identification of O-GlcNAc modified proteins following specific enrichment techniques are mostly accomplished by tandem mass spectrometry. However, since some of the enrichment techniques are performed at the protein level, O-GlcNAc modified peptides often remain underrepresented in a proteolyzed mixture during a “bottom-up” proteomic experiment. Furthermore, due to the susceptibility of β-O-glycosidic bond to gas-phase collisional fragmentation17–19, the GlcNAc residue is readily cleavable under collision-induced dissociation (CID) generating dominant neutral loss ions that suppress the production of peptide backbone fragments and renders the site of modification unknown. Especially when analyzing a peptide mixture that may contain peptides with multiple sites of O-GlcNAc modification, a neutral loss MS approach cannot accurately characterize a modified peptide without introducing unnecessary ambiguity in anticipating the number of modification sites. Therefore, a reliable characterization of O-GlcNAc modified proteins, especially in terms of generating high quality peptide fragmentation that includes the sites of modification, cannot be easily achieved in typical CID-oriented MS experiments. To date, proteomic analyses have identified more than 700 O-GlcNAc modified proteins in diverse functional classes5, however, only a small percentage (<12%) of those proteins were assigned with modification sites. Recent advances in mass spectrometry, such as the introduction of high-energy C-trap dissociation (HCD) coupled with the Orbitrap and electron transfer dissociation (ETD) have provided the capability to perform unambiguous characterization of peptides with various modifications. When applied to post-translationally modified peptides, HCD tend to generate characteristic immonium or oxonium ions at low m/z region, such as phosphotyrosine immonium ion20 and HexNAc oxonium ion18–19, 21, which can serve as diagnostic tools for certain types of modification, whereas ETD produces sufficient c- and z-ions for confident peptide sequencing while often preserving the modified residue for accurate site assignment.
In our study, we took advantage of the recently characterized O-GlcNAc-specific IgG monoclonal antibodies10 and the combination of HCD and ETD fragmentation techniques, and mapped a total of 83 O-GlcNAc modification sites following high-stringency filtering from the enriched HEK293T cell extracts. The HCD/ETD combined approach is easily amenable to the detection and site localization of O-GlcNAc modification and provides insight into O-GlcNAc transferase (OGT) site utilization suggesting that the enzyme displays a degree of promiscuity. Applicability of the HCD/ETD approach to other type of glycosylated peptides, such as O-Mannose and O-GalNAc modified peptides, were also investigated in our study.
Experimental Procedures
Monoclonal antibodies
The three monoclonal antibodies used in our current study, 18B10.C7(3), 9D1.E4(10) and 1F5.D6(14), were generated and characterized as described in our previous study10.
Standard and synthetic peptides
In our experiments, three O-GlcNAc modified standard peptides were used, which are CREB [TAPTs(GlcNAc)TIAPG], CKII [PGGSTPVs(GlcNAc)SANMM], and BPP [PSVPVs(GlcNAc)GSAPGR]; and three O-Mannose and O-GalNAc modified synthetic peptides were used, which are Ac-IRt(Man)t(Man)t(GalNAc)SGVPR-NH2, Ac-PTTt(GalNAc)PLK-NH2, and Ac-RIRTT t(Man)SGVPR-NH2.
Protein digestion
1 mg of bovine serum albumin (Fisher) was incubated with 10 mM DTT and 40 mM NH4CO3 at 56 °C for 1 hr. After cooling to room temperature, the suspension was incubated for 45 min in dark with 55 mM iodoacetamide dissolved in 40 mM NH4CO3. After denaturing and alkylation, the sample was digested overnight at 37 °C using sequencing grade modified trypsin (Promega). The reaction was quenched by 0.1% trifluoroacetic acid, and the resulting peptides were divided into 4 aliquots (250 μg each), desalted using Vydac C18 Silica spin columns (Nest Group) and dried in SpeedVac22.
Preparation of multiple-antibody-enriched HEK293T cell extract
The O-GlcNAc proteome enrichment from HEK293T cell pellets were prepared as previously described by Teo et al.10. Briefly, HEK293T cells were obtained from ATCC and maintained in Dulbecco’s modified Eagle’s medium (4.5 g/L glucose; Cellgro/Mediatech, Inc.) supplemented with 10% fetal bovine serum (GIBCO/Invitrogen) in 37 °C incubator humidified with 5% CO2. Two 15 cm plates of HEK293T cells were treated with 50 μM of O-GlcNAcase inhibitor, PUGNAc, for 24 h, washed off of the plates with ice-cold PBS and stored as a pellet at −80 °C until used. To prepare the nucleocytosolic fraction for immunoprecipitation, the PUGNAc-treated HEK293T cell pellets were resuspended in 4 volumes of hypotonic buffer (5 mM Tris-HCl, pH 7.5, Protease inhibitor cocktail) and transferred into a 2 ml homogenizer. After incubating on ice for 10 min, the cell suspension was subjected to dounce homogenization followed by 5 min incubation on ice. One volume of hypertonic buffer (0.1 M Tris-HCl, pH 7.5, 2 M NaCl, 5 mM EDTA, 5 mM DTT, Protease inhibitor cocktail) was then added to the lysate. The lysate was incubated on ice for 5 min followed by another round of dounce homogenization. The resulting lysates were transferred to microcentrifuge tubes containing PUGNAc (final concentration 10 μM) and centrifuged at 18000 g for 25 min at 4 °C. Protein concentration was determined using Bradford protein assay (Bio-Rad, Hercules, CA). Prior to immunoprecipitation, the lysates were supplemented with Igepal CA-630 (1%) and SDS (0.1%), and precleared with a mixture of normal mouse IgG AC and protein A/G PLUS agarose at 4 °C for 30 min. The precleared supernatant was incubated at 4 °C in the presence of antibodies of interest (conjugated MAbs 18B10.C7(3), 9D1.E4(10) and 1F5.D6(14)) for 4 h at 4 °C. After adding protein A/G PLUS agarose, the samples were incubated for another 2 h at 4 °C and extensively washed with IP wash buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Igepal CA-630, 0.1% SDS). To elute proteins from the agarose, glycine (0.1 M, pH 2.5) was added and the eluates were immediately neutralized with Tris-HCl (1 M, pH 8.0). The samples were then reduced and alkylated as described above and subjected to LysC (Roche) digestion at 37 °C for 24 h. After digestion, the samples were desalted and dried as described above.
LC-MS/MS analysis of the peptide mixtures
Two peptide mixtures were analyzed in the experiment. The first mixture was produced by mixing 2 nmol of each O-GlcNAc modified standard peptides (sequence as described above) with an equal molar concentration of bovine serum albumin digest. The second mixture was produced by mixing equal molar amounts of three synthetic O-Mannose and O-GalNAc modified peptides (sequence as described above). Each peptide mixture was resuspended in 1 μl of solvent B (0.1% formic acid/80% acetonenitrile) and 39 μl of solvent A (0.1% formic acid) and loaded on a 75 μm i.d. × 105 mm C18 reverse phase column (packed in house, YMC GEL ODS-ǺQ120 S-5, Waters) by nitrogen bomb. Peptides were eluted directly into the nanospray source of an LTQ Orbitrap XL™ (Thermo Fisher Scientific) with a 160-min linear gradient consisting of 5–100% solvent B over 100 min at a flow rate of approximately 250 nl/min. The spray voltage was set to 2.0 kV and the temperature of the heated capillary was set to 200 °C. Full MS scans were acquired from m/z 150 to 2000 at a resolution of 60000 (FWHM at m/z 400), with a maximum ion injection time of 100 ms, and an automatic gain control (AGC) setting of 700000 ions. AGC was set to 30000 ions for MS/MS analysis (CID and ETD modes) in the ion trap and to 500000 ions for the MS/MS analysis (HCD mode) in the Orbitrap. The HCD normalized collision energy was set to 35%, and fragment ions were detected in the Orbitrap at a resolution of 7500 (FWHM at m/z 400) using 1 microscan, with a maximum injection time of 100 ms. For ion trap CID MS/MS, isolation of 2 amu, 1 microscan with a maximum injection time of 100 ms were used; for ion trap ETD MS/MS, isolation of 2 amu, 1 microscan with a maximum injection time of 300 ms were used. ETD fragmentations were performed based on charge state with the anion AGC target set at 300000. A dynamic exclusion window was applied which prevents the same m/z value from being selected for 6 seconds after its acquisition. Data acquisition was conducted in the fashion of an Orbitrap MS followed by top 4 data-dependent Orbitrap HCD MS/MS, ion trap ETD MS/MS, and ion trap CID MS/MS triple-play using Xcalibur® (ver. 2.0.7, Thermo Fisher Scientific).
LC-MS/MS analysis of the enriched HEK293T cell extract
Two LC-MS/MS experiments were performed on an LTQ Orbitrap Velos™ ETD mass spectrometer (Thermo Fisher Scientific) with nano-ESI source that was coupled to a Surveyor™ MS Pump with a flow splitter. Peptides were separated on a 75 μm i.d. × 200 mm spraytip Magic C18 column (Michrom Bioresources) with a 240-min linear gradient consisting of elution by solvent B from 5–20% over 120 min and from 20–40% over 70 min at a flow rate of approximately 300 nL/min. The LTQ Orbitrap Velos™ mass spectrometer was operated at S-lenses setting of 50 %, the heated capillary temperature of 200 °C, resolution of 60000 (FWHM at m/z 400) in full MS, with a maximum ion injection time of 300 ms, and AGC setting of 1000000 ions. AGC was set to 10000 ions for MS/MS analysis in the ion trap and to 50000 ions for the MS/MS analysis in the Orbitrap. HCD normalized collision energy was set to 35% and fragment ions were detected in the Orbitrap at a resolution of 7500 (FWHM at m/z 400) using 1 microscan, with a maximum injection time of 200 ms. ETD reactions were performed based on charge state with the anion AGC target set at 200000. Three different LC-MS/MS acquisition methods were performed: (1) Orbitrap MS followed by top 5 data-dependent Orbitrap HCD MS/MS and ion trap ETD MS/MS double-play; (2) Orbitrap MS followed by top 5 data-dependent Orbitrap HCD MS/MS. Data were acquired using Xcalibur® (ver. 2.0.7, Thermo Fisher Scientific).
Data analysis
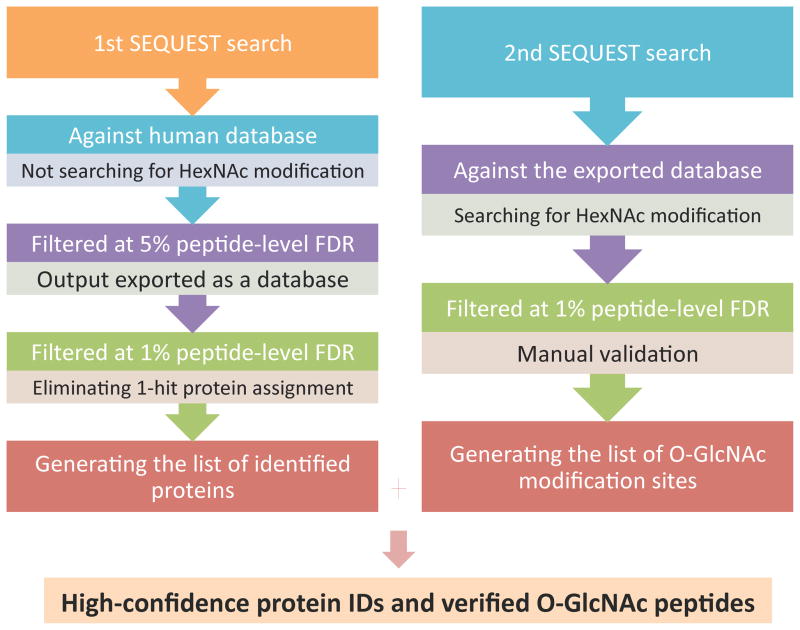
For the peptide mixtures, the raw spectra were interpreted manually. For the enriched HEK293T sample, database searches were performed using SEQUEST® (Proteome Discoverer ver. 1.2, Thermo Fisher Scientific), with the following search parameters: 20-ppm tolerance for monoisotopic precursor mass, and 1.2-Da or 0.01-Da tolerance for monoisotopic fragment masses of ion trap MS/MS or Orbitrap MS/MS spectra, respectively; LysC (fully digested) specified as the enzyme; a maximum of 3 missed cleavage sites, 3 differential amino acids per modification and 3 differential modifications per peptide allowed. Each raw spectral file was searched twice against different databases with corresponding settings of dynamic modifications (Figure 1): the first search was performed against Uniprot human database (70337 entries, updated at Dec. 21, 2009), allowing for methionine oxidation (+15.995 Da) and cysteine carbamidomethylation(+57.021 Da); the second search was performed against the output exported from the first search after being filtered at 5% peptide-level FDR, allowing for methionine oxidation (+15.995 Da), cysteine carbamidomethylation (+57.021 Da), and HexNAc modification on serine/threonine (203.079 Da). Random database searches were performed against the corresponding reversed databases using the same program. The output from the first search was filtered at 1% peptide-level FDR, and the single-hit protein assignments within the filtered result were eliminated to generate the list of identified proteins; the output from the second search was filtered at 1% peptide-level FDR, and all the assignments of O-GlcNAc modification sites were manually validated.
Figure 1. Database search strategy for the enriched HEK293T sample.
Each raw file was searched twice against respective databases using SEQUEST. The first search was against the entire human database without allowing for HexNAc modification, the output of which was initially filtered at 5% peptide-level FDR to generate a limited protein database and further filtered at 1% protein-level FDR for high-confidence protein identification. The final list of identified proteins was produced after eliminating the 1-hit protein assignments from the 1% FDR-filtered result. The second search was performed against the exported database from the first search allowing for HexNAc modification, the output of which was filtered at 1% peptide-level FDR to generate a list of O-GlcNAc peptides. All the assignments of O-GlcNAc modification on the list were manually validated to produce the final list of O-GlcNAc sites.
A multiple-engine database search strategy was applied to the same raw spectra files obtained from the HEK293T cell sample to increase the sensitivity of data analysis. In this strategy (Figure S1), the database searches were performed in parallel with three search engines: SEQUEST® (Proteome Discoverer ver. 1.2, Thermo Fisher Scientific), Mascot® (ver. 2.3, www.matrixscience.com), and ProteinProspector (v5.7.3, prospector.ucsf.edu). With each search engine, database searches comprised of two consecutive sections: the first search was performed against the same human database as described above, allowing for N-terminal and cysteine carbamidomethylation (+57.021 Da), methionine oxidation (+15.995 Da) and dethiomethylation (−48.003 Da) as dynamic modifications; the second search was against the exported database from the first search with 5% peptide-level FDR filter, and allowed for HexNAc modification on serine/threonine (+203.079 Da) and phosphorylation on serine/threonine/tyrosine (+79.966 Da) in addition to the 3 dynamic modifications specified in the first search. Other search parameters and random database searches were set up as described above. The list of identified proteins was generated only from the second database searches and was filtered at 1% peptide-level FDR. The list of O-GlcNAc modified peptides was the combination of multiple-engine database search results and each modification site was further verified manually.
Results and Discussion
The MS approach employed in this study for sequencing O-GlcNAc modified peptides and assigning the modification sites is a combination of HCD and ETD fragmentation. When analyzing O-GlcNAc modified peptides, the unique advantage of HCD fragmentation is the generation of distinct HexNAc oxonium ions (m/z 204.09)18–19, 21 and a series of HexNAc fragments (m/z 186, m/z 168, m/z 144, m/z 138 and m/z 126) which are significant to the diagnosis of O-GlcNAc modified peptides yet not necessarily observed in ion trap tandem mass spectra because of the dependence of ion trap scan range on precursor m/z values. Moreover, the product ions formed during HCD are detected in the Orbitrap analyzer, therefore not only do they overcome the 1/3 cut-off rule of an ion trap, they also exhibit higher mass accuracy and lower chemical noise. However, HCD spectra alone are not as informative as those acquired in the linear ion trap pertaining to peptide sequence-related ion products, which has been attributed to the increased collision energy leading to ion scattering and a lack of peptide backbone fragments23.
In order to provide more informative spectra for peptide sequencing as well as the site assignment of O-GlcNAc modification, ETD was employed in combination with HCD. By transferring an electron from a radical anion to a protonated peptide, ETD has been proven advantageous for analyzing relatively large, non-tryptic peptides compared to CID. In the particular case of post-translationally modified peptides, ETD has the capacity to preserve labile modifications attached to peptide backbones, such as phosphorylation and glycosylation, allowing for the detection of multiple modifications within the context of one another, as well as producing almost complete series of peptide backbone fragments for peptide sequencing at the same time24. Especially when facilitated by supplementary collisional activation converting the nondissociative electron transfer products into c- and z-type fragment ions, the effect of precursor ion charge states have on dissociation efficiency, which is one of the limitations imposed by the intrinsic charge-reducing process in electron-based fragmentation methods, has been remarkably reduced25.
By utilizing the HexNAc signature ions generated by HCD and the extensive peptide sequence information provided by ETD, we analyzed glycosylated standard peptides to investigate the applicability of HCD/ETD method in identification and site localization of O-GlcNAc modified peptides. Furthermore, we applied the same MS approach to a complex biological sample to prove the effectiveness and robustness of this method.
Characterization of O-GlcNAc modified standard peptides
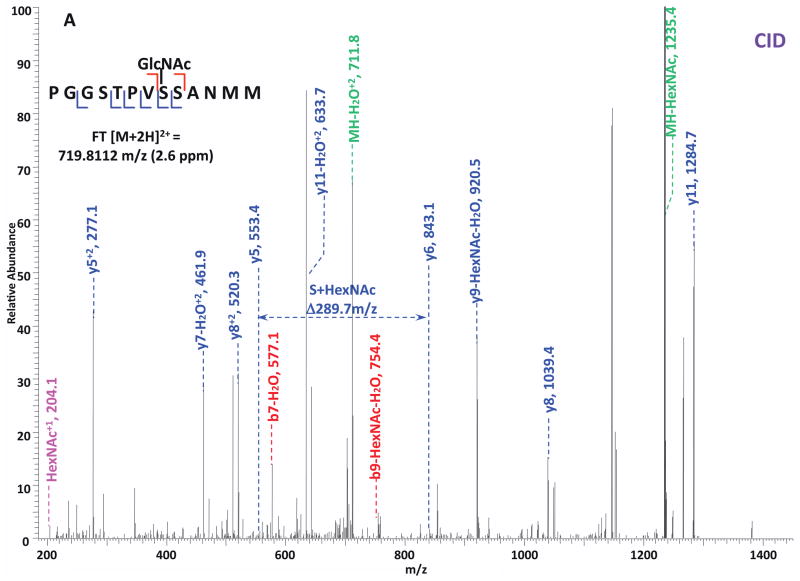
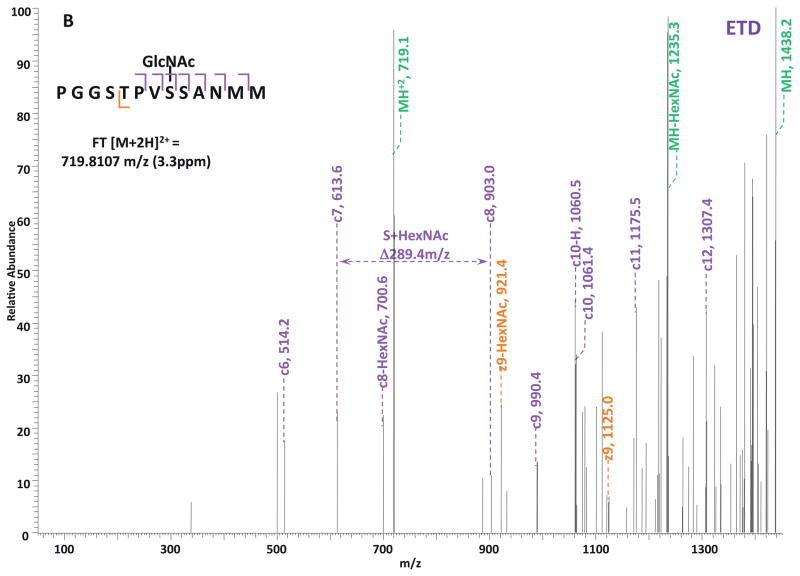
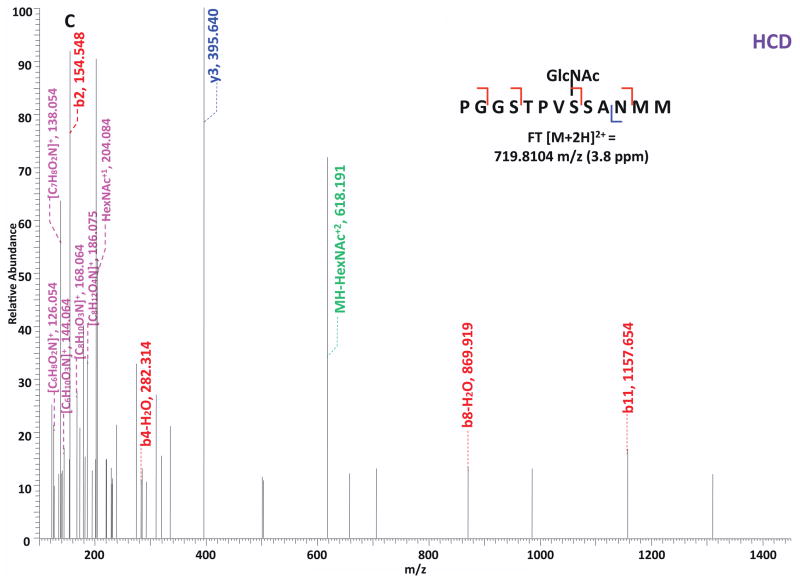
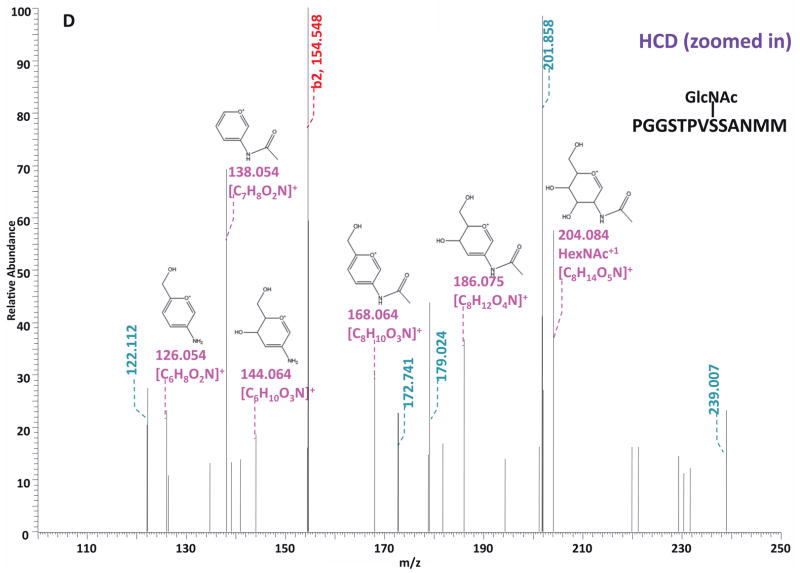
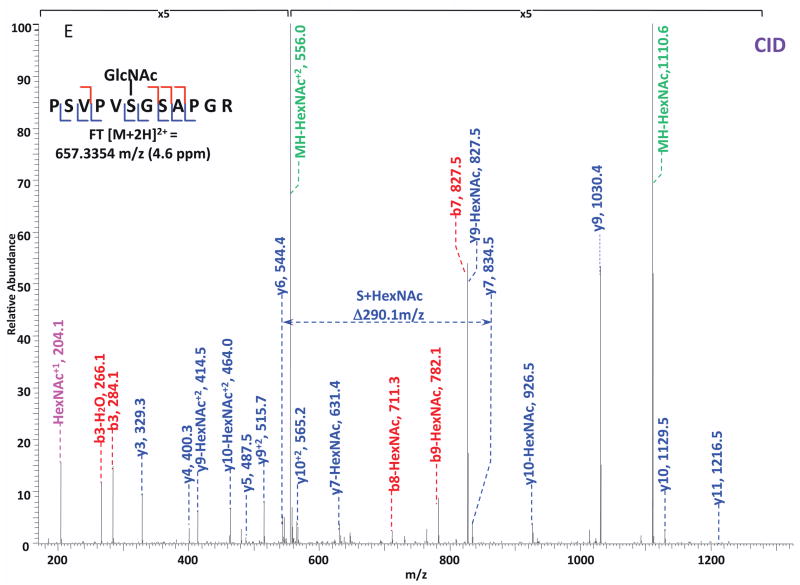
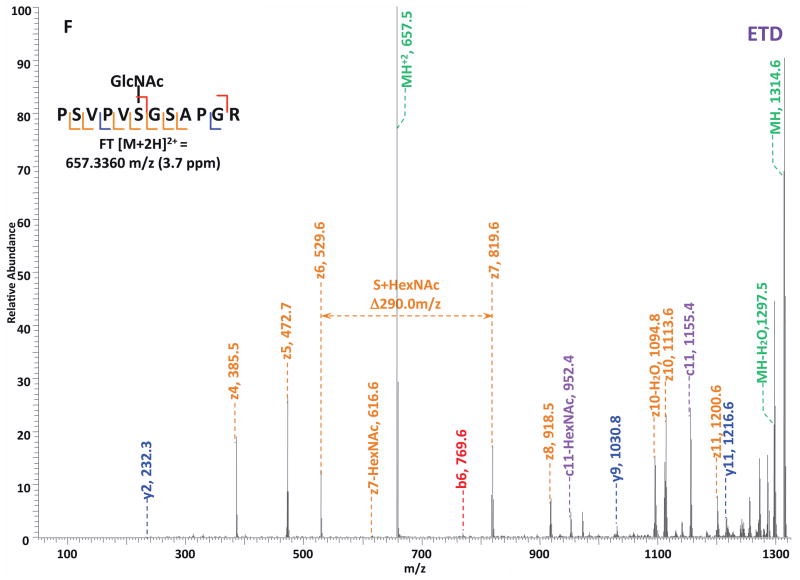
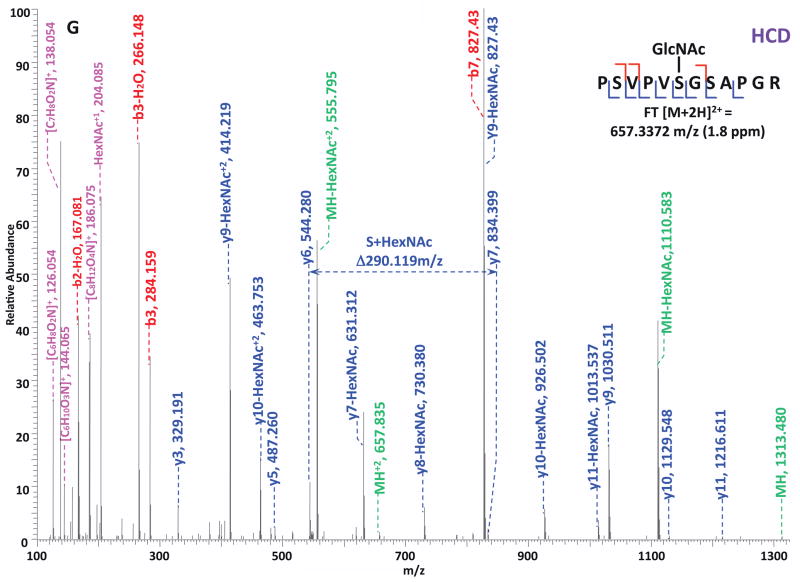
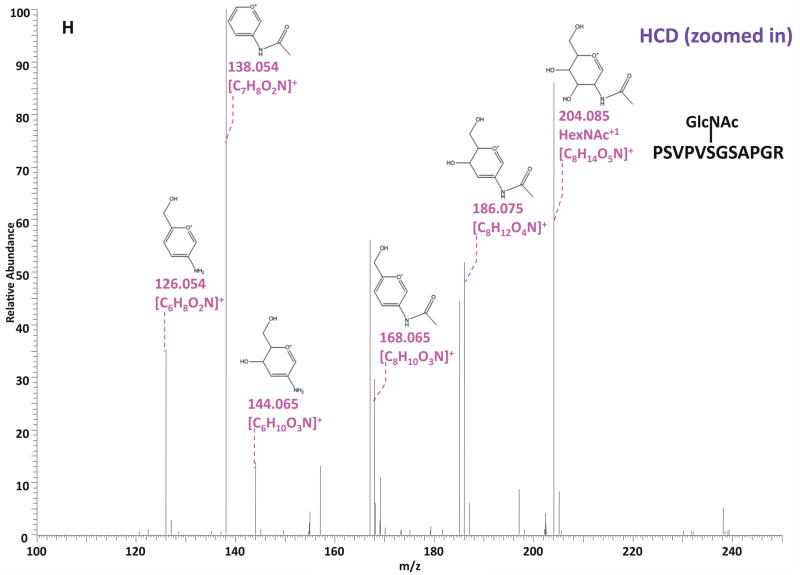
O-GlcNAc modified standard peptides in the mixture were analyzed by an alternating CID/ETD/HCD fragmentation method. As indicated in Figure 2, the most dominant peaks formed during CID fragmentation of glycopeptides CKII and BPP resulted from the loss of HexNAc residue from respective precursor ions (Figure 2A and 2E). As a result of the intensive HexNAc-loss ions, b- and y-type ions generated by backbone fragmentation that are required for peptide sequencing and site assignment are severely suppressed, making it difficult to accurately and confidently characterize O-GlcNAc peptides. In Figure 2B and 2F, almost a complete series of c- or z-type ions were observed in the ETD spectrum allowing for the elucidation of both respective peptide sequence and modifications sites. During the HCD fragmentation of CKII and BPP glycopeptides (Figure 2C and 2G), the high m/z fragment ions were either missing or at very low intensity. However, the HexNAc oxonium ion (m/z 204) and its fragments (m/z 186, m/z 168, m/z 144, m/z 138, and m/z 126) were produced at pronounced high intensity (Figure 2D and 2H, Table S1). This distinctive fragment pattern of HexNAc residue allows for unambiguous identification of O-GlcNAc modified peptides which are generally underrepresented in protein mixtures extracted from biological samples3, 8–9.
Figure 2. Respective CID, ETD, and HCD spectra of standard O-GlcNAc modified peptides CKII and BPP.
(A)–(B) CID and ETD spectra of O-GlcNAc modified CKII peptide, respectively; (C)–(D) HCD and zoomed in HCD spectra of O-GlcNAc modified CKII peptide; (E)–(F) CID and ETD spectra of O-GlcNAc modified BPP peptide, respectively; (G)–(H) HCD and zoomed in HCD spectra of O-GlcNAc modified CKII peptide. Note: “-HexNAc” or “-H2O” indicates the loss of HexNAc or H2O. Low m/z range HCD displays a distinctive pattern of HexNAc fragments (D and H).
When O-GlcNAc modified peptides are subjected to CID, it is probable, yet not necessary, that the GlcNAc residue is cleaved from the peptide backbone and forms an oxonium ion at m/z 204.09. When undergoing HCD, the GlcNAc residue will likely always be cleaved and generate the oxonium ion at m/z 204.09. This characteristic ion product can be used as a diagnostic tool to identify the O-GlcNAc modified peptides. Moreover, not only will the HexNAc oxonium ion be present in the HCD spectrum, a series of its fragment ions will also be produced that are indicative of HexNAc residue, such as m/z 186, m/z 168, m/z 144, m/z 138, and m/z 12626. The presence of HexNAc oxonium ion and the series of its fragment ions can be used to target O-GlcNAc modified peptides during an MSn experiment and selectively trigger an equal order CID or ETD fragmentation of the same precursor ion for more detailed peptide characterization. Compared to the dominant neutral loss ions created upon the loss of HexNAc residues during CID, which suppress the formation of peptide sequencing-related ions, ETD has the capability to preserve labile modifications and render more complete ion series for peptide sequencing and site assignment. Using the HexNAc oxonium ion as an indicator to target O-GlcNAc modified peptides with modification favorable ETD fragmentation, peptide sequencing and site localization can be achieved with improved selectivity and sensitivity.
Characterization of O-GlcNAc modified proteins enriched from HEK293T cell extract
186 proteins were identified at 1% pepFDR from the multiple-antibody enriched HEK293T cell extract (Table S2). In comparison with the recently published work from our group10 that used the same sample, 67% (124/186) of the identified proteins were consistent with the result from previous experiment, including heterogeneous nuclear ribonucleoproteins, BAT2 domain-containing protein 1, ribosomal proteins, heat shock proteins, and nuclear pore complex proteins. Of the 33% (62/186) of the dataset that was only observed in our current study, 17 proteins were supported in the literature2–6, 14, 27 as being O-GlcNAc modified and 45 were identified as novel O-GlcNAc proteins (Table 1). Furthermore, while the previous experiment did not discover any O-GlcNAc modification sites, our current study identified 83 O-GlcNAc sites on 172 glycopeptides from 13 proteins (Table S3). By comparison to the literature8, 13–15, 28–33, 13 of the 83 sites had been previously assigned leaving 70 novel O-GlcNAc sites (Table 2); and 11 of the 13 characterized proteins were also previously observed as modified but not site-mapped in the previously work from our group.
Table 1.
Novel O-GlcNAc Proteins Identified in HEK293T Cells
| UniProt Accession | Protein Name | Gene Name | Coverage by #AA (%) | # Peptides |
|---|---|---|---|---|
| Q9Y265-1 | Isoform 1 of RuvB-like 1 | RUVBL1 | 21.05 | 16 |
| P35268 | 60S ribosomal protein L22 | RPL22 | 21.88 | 12 |
| P22061-1 | Isoform 1 of Protein-L-isoaspartate(D-aspartate) O-methyltransferase | PCMT1 | 9.69 | 10 |
| P62081 | 40S ribosomal protein S7 | RPS7 | 21.65 | 10 |
| P62263 | 40S ribosomal protein S14 | RPS14 | 18.54 | 9 |
| Q07021 | Complement component 1 Q subcomponent-binding protein, mitochondrial | C1QBP | 15.6 | 9 |
| P26196 | Probable ATP-dependent RNA helicase DDX6 | DDX6 | 7.45 | 8 |
| P62750 | 60S ribosomal protein L23a | RPL23A | 17.95 | 8 |
| C9JU56 | Putative uncharacterized protein RPL31 | RPL31 | 35.65 | 7 |
| P25398 | 40S ribosomal protein S12 | RPS12 | 41.67 | 7 |
| P60866 | 40S ribosomal protein S20 | RPS20 | 19.33 | 7 |
| P62829 | 60S ribosomal protein L23 | RPL23 | 28.57 | 7 |
| C9JLW8 | HCG1818442, isoform CRA_c | hCG_1818442 | 11.34 | 6 |
| P30050-1 | Isoform 1 of 60S ribosomal protein L12 | RPL12 | 17.58 | 6 |
| P62913-2 | Isoform 2 of 60S ribosomal protein L11 | RPL11 | 23.73 | 6 |
| Q9NYF8-3 | Isoform 3 of Bcl-2-associated transcription factor 1 | BCLAF1 | 11.51 | 6 |
| C9J0W3 | Putative uncharacterized protein SMARCE1 | SMARCE1 | 17.2 | 5 |
| P24534 | Elongation factor 1-beta | EEF1B2 | 12 | 5 |
| Q6MZS5 | Putative uncharacterized protein DKFZp686A13234 | DKFZp686A13234 | 8.32 | 5 |
| Q92922 | SWI/SNF complex subunit SMARCC1 | SMARCC1 | 4.07 | 5 |
| Q9Y310 | UPF0027 protein C22orf28 | C22orf28 | 5.94 | 5 |
| O95881 | Thioredoxin domain-containing protein 12 | TXNDC12 | 16.86 | 4 |
| P62266 | 40S ribosomal protein S23 | RPS23 | 15.38 | 4 |
| Q9NPF5 | DNA methyltransferase 1-associated protein 1 | DMAP1 | 8.57 | 4 |
| P13929-3 | Isoform 3 of Beta-enolase | ENO3 | 10.49 | 3 |
| P49207 | 60S ribosomal protein L34 | RPL34 | 22.22 | 3 |
| Q5T4L4 | Ribosomal protein S27 | RPS27 | 16.67 | 3 |
| Q8IXM2 | Uncharacterized potential DNA-binding protein C17orf49 | C17orf49 | 26.16 | 3 |
| Q96K80 | Zinc finger CCCH domain-containing protein 10 | ZC3H10 | 8.99 | 3 |
| C9JMM0 | Putative uncharacterized protein CBX3 | CBX3 | 48.39 | 2 |
| P30040 | Endoplasmic reticulum protein ERp29 | ERP29 | 8.43 | 2 |
| P37108 | Signal recognition particle 14 kDa protein | SRP14 | 16.18 | 2 |
| P50991 | T-complex protein 1 subunit delta | CCT4 | 5.38 | 2 |
| Q8TAQ2-2 | Isoform 2 of SWI/SNF complex subunit SMARCC2 | SMARCC2 | 1.77 | 2 |
Table 2.
Novel O-GlcNAc Sites Identified in HEK293T Cells
| UniProt Accession | Protein Name | Novel Sites |
|---|---|---|
| O95487-2 | Isoform 2 of Protein transport protein Sec24B | T327; T341 |
| P35658-2 | Isoform 2 of Nuclear pore complex protein Nup214 | S1202; S1904; S1905; S1907; T1915; S1916 |
| P49790 | Nuclear pore complex protein Nup153 | T535; S893, S895; S1017; S1018; T1026; T1041 |
| P51610-1 | Isoform 1 of Host cell factor | T405; S620; S622; S623; T625; S628; S638; T640; T651; T652; T658 |
| P52594-2 | Isoform 2 of Arf-GAP domain and FG repeats-containing protein 1 | S291 |
| P52948-4 | Isoform 4 of Nuclear pore complex protein Nup98-Nup96 | S262; T264 |
| Q14157-4 | Isoform 4 of Ubiquitin-associated protein 2-like | S445 |
| Q2KHR3-1 | Isoform 1 of Glutamine and serine-rich protein 1 | T1271 |
| Q5T6F2 | Ubiquitin-associated protein 2 | T487; S494 |
| Q5T8P6-3 | Isoform 3 of RNA-binding protein 26 | S657; S667 |
| Q6MZP7-1 | Isoform 1 of Protein lin-54 homolog | T109 |
| Q7Z589 | Isoform 1 of Protein EMSY | S228; T264; T272 |
| Q8IWZ3-1 | Isoform 1 of Ankyrin repeat and KH domain-containing protein 1 | S1817 |
| Q9H4A3-2 | Isoform 2 of Serine/threonine-protein kinase WNK1 | S1849 |
| Q9Y520 | Isoform 7 of BAT2 domain-containing protein 1 | S2196 |
By comparing the results from the two respective LC-MS/MS experiments, 37 O-GlcNAc peptides were observed by the HCD-alone method, and 168 glycopeptides were found by the HCD/ETD method (Table 3). After manual validation, 7 O-GlcNAc sites were assigned in the HCD-alone analysis, and 83 sites were assigned in the HCD/ETD analysis.
Table 3.
Comparison of Two Methods for O-GlcNAc Site Mapping
| Method | O-GlcNAc Peptides | Manually Confirmed Sites | Presence of HexNAc Signature Ions |
|---|---|---|---|
| HCD/ETD | 172 | 83 | 100% |
| HCD-alone | 37 | 7 | 100% |
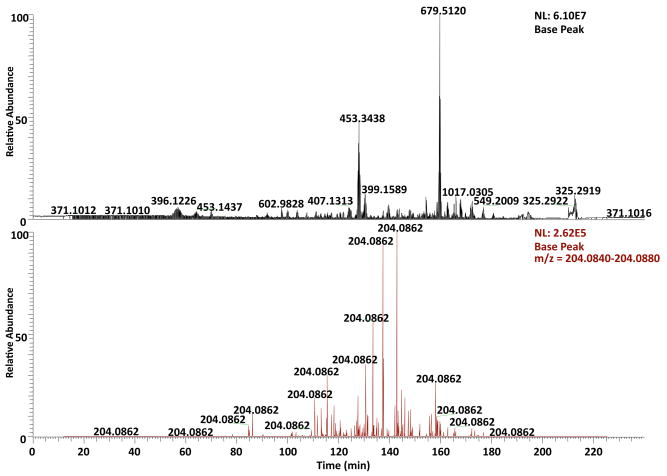
We also evaluated the abundance of ionized and fragmented O-GlcNAc peptides in the sample by analyzing the occurrence of HexNAc oxonium ions in the spectral data collected from HCD-alone and HCD/ETD experiments. Generally, the respective ion chromatograms at m/z 204.086 (bottom panel) of the HCD-alone (Figure S1) and HCD/ETD (Figure 3) experiments were extracted and compared to the total ion chromatograms (top panel). The extracted ion chromatograms illustrates the retention time profiles of the O-GlcNAc peptides analyzed by the mass spectrometer and indicates the acquisition of HCD spectra of those peptides. In both figures (Figure 3 and S1), the noticeable difference of more than two orders of magnitude in intensity between the extracted and total ion chromatograms demonstrated the low abundance of O-GlcNAc modified peptides in the ionized mixture, and further suggests the sensitivity of detection by HCD and the importance of selectively targeting O-GlcNAc peptides for fragmentation as a manner of enrichment.
Figure 3. Comparison between the extracted ion chromatogram at m/z 204.086 and the total ion chromatogram of the HCD/ETD LC-MS/MS experiment on the enriched HEK293T sample.
The extracted ion chromatogram at m/z 204.086 (bottom panel) delineates the retention time profile of O-GlcNAc modified peptides eluted over the entire gradient. In comparison with the total ion chromatogram, the normalized peak height of the extracted ion chromatogram indicates the substoichiometric nature of O-GlcNAc modification and therefore suggests the importance of selective fragmentation on O-GlcNAc peptides.
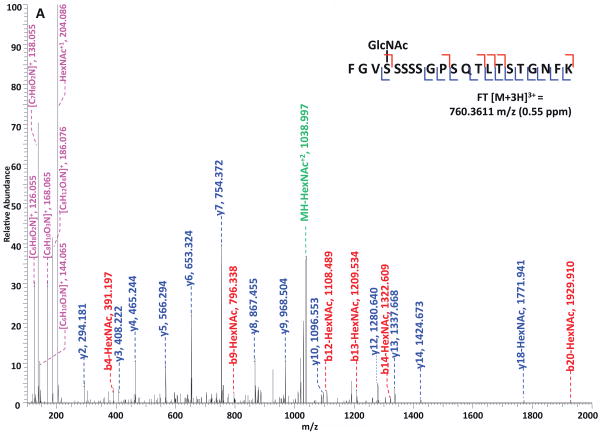
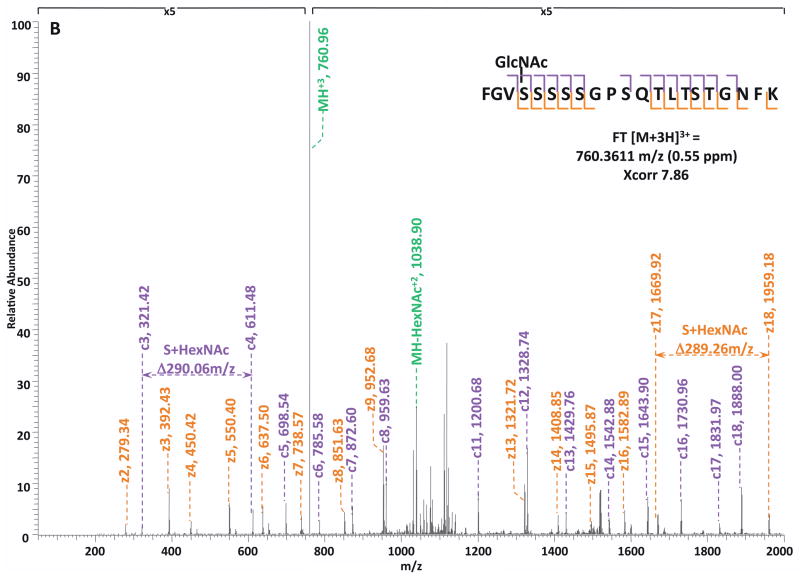
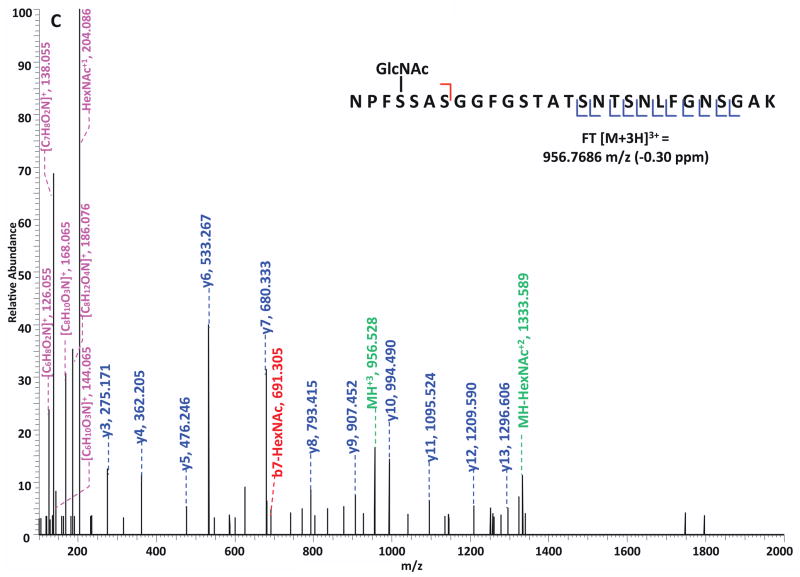
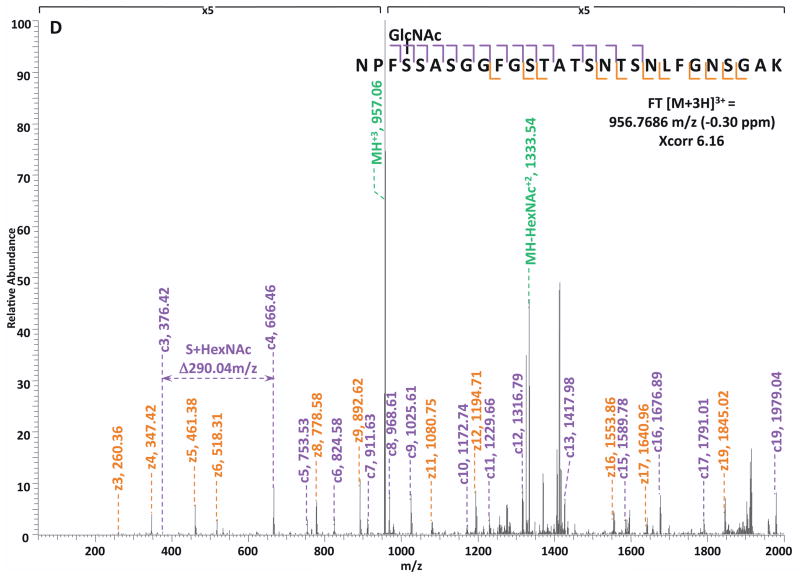
Specifically, in the HCD-alone experiment, each HCD spectrum of the eluted O-GlcNAc peptides showed the oxonium ions of HexNAc at m/z 204 and some other signature ions at m/z 186, m/z 168, etc, including the spectra of the 37 peptides assigned by the database search program that survived 1% FDR filtering. However, because of the lack of primary fragment ions caused by the high-energy during the fragmentation, most O-GlcNAc peptides, even when being indicated by the HexNAc signature ions in the spectra, were not sequenced by the database search algorithm due to the poor ion match ratios. The 37 O-GlcNAc peptides in our dataset that were sequenced still suffer from low ratio of ion match and therefore low scores (Table S4), especially because of the loss of GlcNAc during HCD, the majority of the glycopeptides don’t have indicative b- or y-ions in their spectra for the manual validation of O-GlcNAc site localization. This observation suggests that even though HCD is powerful in generating high quality spectral data to indicate the presence of O-GlcNAc peptides, it is not capable of providing sufficient information for accurately mapping the sites of modification. In the HCD/ETD experiment, where 172 O-GlcNAc peptides were identified and 83 sites were mapped on them, all of the O-GlcNAc peptides were characterized by the ETD spectra and each of those ETD spectra corresponds with an HCD spectrum observing a pattern of HexNAc signature ions. As an example, in Figure 4, panel A and panel B are the consecutively acquired HCD and ETD spectra of the same triple-charged precursor m/z 760.3611. As noted in Figure 4A, most b- and y- ions were generated without the attachment of GlcNAc (b-HexNAc or y-HexNAc) and were at relatively low intensity, therefore can not be used to assign the site of modification. At the low m/z range of the spectrum, a strong HexNAc oxonium ion peak was observed at m/z 204.086, and the other five signature ions of HexNAc were also found at high intensities, indicating the presence of O-GlcNAc modification on the peptide. In Figure 4B, an almost complete series of c- and z- ions were produced in the ETD spectrum of the same peptide and the site of O-GlcNAc modification was unambiguously indicated by corresponding c- and z- ion pairs.
Figure 4. Corresponding HCD and ETD spectra of O-GlcNAc modified peptides identified in the enriched HEK293T sample.
(A–B) HCD and corresponding ETD spectra of peptide FGVS(GlcNAc)SSSSGPSQTLTSTGNFK; (C–D) HCD and corresponding ETD spectra of peptide NPFS(GlcNAc)SASGGFGSTATSNTSNLFGNSGAK. Note: “-HexNAc” or “-H2O” indicates the loss of HexNAc or H2O. The HCD spectra of both peptides exhibit the signature ion pattern of HexNAc fragments at the low m/z range, and both ETD spectra sufficient c- and z- ions with HexNAc modification for site assignment.
We further investigated the prevalence of each HexNAc signature ion by summarizing the occurrence of them in the 172 O-GlcNAc peptides identified by the HCD/ETD analysis (Table 4). As presented in Table 4, the HexNAc oxonium ion at m/z 204 and its fragments at m/z 138 and m/z 126 were observed in the spectra of all 172 peptides; signature ions at m/z 186 and m/z 168 were also present in over 90% of the spectra, whereas the signature ion at m/z 144 was observed in less than 75% of the spectra. By examining the patterns of the HexNAc signature ions observed from the 172 O-GlcNAc peptides (Table 5), the most prevalent pattern is the presence of all 6 signature ions, which was observed in approximately 85% of the spectra; the second most prevalent pattern is the presence of 5 signature ions with m/z 144 missing, indicating that m/z 144 is an unfavorable product of HexNAc fragmentation. The patterns which have more than one signature ion missing were observed at low prevalence, suggesting that most fragments from HexNAc under HCD are stable and constant. Further comparison between the extracted ion chromatograms at respective signature ions of HexNAc (m/z 204, m/z 186, m/z 168, m/z 144, etc.) is illustrated in Figure S2, where the general difference in intensity between the chromatograms is consistent with the prevalence of each signature ion, for example, the intensity of the extracted ion chromatogram at m/z 204.086 is the highest among the 6 chromatograms whereas that at m/z 144.064 is the lowest.
Table 4.
HexNAc Signature Ion Occurrence in HCD/ETD Analysis
| 204 | 186 | 168 | 144 | 138 | 126 | |
|---|---|---|---|---|---|---|
| O-GlcNAc Peptides | 172 | 164 | 164 | 148 | 172 | 172 |
| Corresponding Sites | 83 | 76 | 77 | 70 | 83 | 83 |
| Prevalence | 100% | 95.35% | 93.02% | 74.42% | 100% | 100% |
Table 5.
HexNAc Signature Ion Patterns in HCD/ETD Analysis
| Signaure Ion Patterns | Prevalence | O-GlcNAc Peptides | Corresponding Sites | |||||
|---|---|---|---|---|---|---|---|---|
| 204 | 186 | 169 | 144 | 138 | 126 | |||
| √ | √ | √ | √ | √ | √ | 84.88% | 146 | 68 |
| √ | – | √ | √ | √ | √ | 1.16% | 2 | 4 |
| √ | √ | √ | – | √ | √ | 9.30% | 16 | 13 |
| √ | √ | – | – | √ | √ | 1.16% | 2 | 4 |
| √ | – | – | – | √ | √ | 3.49% | 6 | 6 |
Observed; - Not observed.
The consistent and reliable production of HexNAc signature ions by HCD suggests the possibility and plausibility of targeting O-GlcNAc modified peptides with selective fragmentation; the two examples presented in Figure 4B and 4D confirmed the capability of ETD in characterizing O-GlcNAc modified peptides. Based on the result of analysis on the occurrence of 6 HexNAc signature ions, we further propose that while it is capable to target O-GlcNAc peptides by only the HexNAc oxonium ion at m/z 204, it would increase the accuracy of selection and decrease the possibility of false positives by monitoring more than one signature ions, such as the combination of m/z 204, m/z 138 and m/z 126, which were observed in all 172 O-GlcNAc peptides detected in the HCD/ETD experiment.
We further analyzed the same raw spectra files obtained from the HEK293T sample using a multiple-engine database search approach to improve the sensitivity of data analysis (Figure S3). Basically, database search programs assign an MSn spectrum with the most probable peptide match, and the assigned spectra generally constitute only a fraction of all the spectra acquired in an LC-MSn analysis. Variations in database searching algorithms for assigning peptides to MSn spectra have been known to provide different identification results34. By combining search results from different search engines have been proven to lead to a larger number of protein identification with an increased rate of peptide assignments35–37. By processing the files in this manner, 200 proteins were identified at 1% pepFDR after combining and validating the output from three independent database searches. In comparison with the result from our recently published work10 that used the same sample, 66% (132/200) of the identified proteins were consistent with the previous experiment. Among the rest 34% (68/200) of the dataset which had only been observed in our current study, 18 proteins were supported in the literature as being O-GlcNAc modified and 50 were identified as novel O-GlcNAc proteins. Furthermore, the multiple-engine search strategy indentified 165 O-GlcNAc sites on 178 glycopeptides from 40 proteins (Table S5), including the 83 sites that were assigned by SEQUEST-only database search. By comparison to the literature8, 13–15, 28–33, 29 of the 165 sites have been previously assigned leaving 136 novel O-GlcNAc sites; and 22 of the 40 characterized proteins were also observed in the previously work from our group10, whereas 18 of the 40 proteins are novel. By taking advantage of the multiple-engine database search methodology, the numbers of identified proteins, peptides and modification sites have been greatly increased and the sensitivity of the acquired raw data has been dramatically improved.
Clustered O-GlcNAc sites on single peptide sequence suggests promiscuity of O-GlcNAc transferase (OGT)
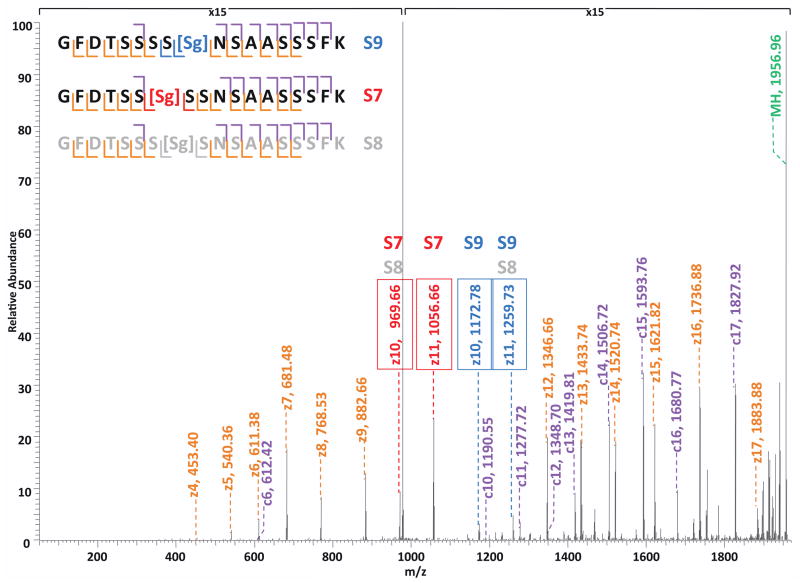
O-GlcNAc modification is analogous to serine/threonine phosphorylation in many respects 15, 32. However, unlike phosphorylation, which is catalyzed by almost 500 kinases encoded in the human genome 38, O-GlcNAc modification is catalyzed by the products of a single human gene 39–40. Studies have shown that OGT glycosylation is quite specific 41, and furthermore that the catalytic subunit of OGT achieves both high specificity and a remarkable diversity of substrates through forming a complex with a variety of targeting proteins via its tetratricopeptide repeat (TPR) protein-protein interaction domains42–44. In our dataset, we discovered several cases of clustered sites of O-GlcNAc modification exhibited by co-eluted peptides, such as proteins Nup214, Nup153, host cell factor and EMSY (Table S3). One such example, presented in Figure 5, shows multiple isobaric glycopeptides were eluted at the same time and had acquired the same charge state. In one ETD spectrum, there were possibly three mono-GlcNAc modified peptides being co-eluted, and the only difference between them is the site of the modified serine residue on the peptide (S7 in red, S8 in grey, and S9 in blue). By interpreting the spectrum, we found that the ions at m/z 969 and m/z 1056 correspond to the z10 and z11 ions of the S7-modified peptide, and the ions at m/z 1172 and m/z 1259 correspond to the z10 and z11 ions of the S9-modified peptide. In fact, all the ions observed in the spectrum can be explained as the fragments of S7-modified and S9-modified peptides, confirming the presence of both. However, since the ions at m/z 969 and m/z 1259 can also be interpreted as z10 and z11 ions of the S8-modified peptide and there is no unique ion to exclude its presence, the co-eluting mixture can have up to three O-GlcNAc modified peptides. Thus, there are three sequential sites on this single peptide that can all be recognized by OGT, and throughout our dataset, this peptide was not found to be modified by di- or tri-GlcNAc. This observation of the diversity in peptide substrates was observed in multiple cases and suggests the promiscuity of OGT (Table S3).
Figure 5. ETD spectrum of co-eluted mono-O-GlcNAc modified peptides GFDTSSSSs(GlcNAc)NSAASSSFK and GFDTSSs(GlcNAc)SSNSAASSSFK.
Spectrum illustrates heterogeneity of the singly O-GlcNAc modified peptide suggestive of O-GlcNAc transferase having promiscuity in terms of site of modification.
Signature ion patterns observed from other types of glycosylated peptides under HCD
In order to explore the applicability of HCD/ETD MS approach on other types of O-glycosylation, synthetic O-Mannose and O-GalNAc modified peptides were analyzed in the same fashion as O-GlcNAc modified peptides using the combined HCD/ETD fragmentation. Indicative ion patterns at low m/z range were observed, respectively, when O-Mannose and O-GalNAc peptides underwent HCD: the O-GalNAc modified peptide exhibited similar signature ions at m/z 204, m/z 186, m/z 168, m/z 144, m/z 138, and m/z 126 (Figure S4A and S4B); the O-Mannosylated peptide exhibited the Hexose oxonium ion at m/z 163, and a series of its fragments at m/z 145, m/z 127, m/z 115, and m/z 109 (Figure S4A and S4C, Table S1). The utility of HCD has been demonstrated for the characterization of protein tyrosine-phosphorylation20, protein N-glycosylation45, and the quantification of iTRAQ-labeled phosphopeptides46–47. Our observation of the distinctive ion patterns of O-GlcNAc, O-Mannose, and O-GalNAc peptides proved the applicability of HCD in targeting O-glycosylated peptides. Although, based on our data, it appears that O-GlcNAc and O-GalNAc cannot be distinguished from each other solely by their signature ion patterns. Furthermore, the signature ion-trigger strategy has the potential to be applied to the analysis of post-translational modifications besides glycosylation and phosphorylation, such as methylation, bromylation, hydroxylation and other modifications that have previously been shown to produce specific fragment ions48.
Conclusion
The signature ion patterns of HexNAc and Hexose revealed under HCD condition can be utilized to selectively target HexNAc and/or Hexose modified proteins, and when combined with ETD, which preserves labile post-translational modification on proteins, the reliability and accuracy in glycoprotein identification and site localization can be greatly improved. In our study, we investigated the applicability of the combined HCD/ETD MS approach in characterizing O-GlcNAc modified proteins from a complex biological sample and successfully identified a minimum of 83 modification sites. Additionally, with a multiple-engine database search method, we were able to increase the sensitivity of our discovery drastically to reach a total of 165 sites of O-GlcNAc modification.
By analyzing the data from two parallel experiments performed respectively by HCD-alone and HCD/ETD method, we were able to prove the consistency and stability of signature ions produced from O-GlcNAc peptides under HCD, and demonstrate the superior sensitivity and accuracy of the HCD/ETD method in sequencing and site-mapping of O-GlcNAc modified peptides. Considering that the neutral loss of HexNAc from O-GlcNAc peptides under HCD may also be used as a trigger for selective fragmentation, the signature ion-trigger mechanism has the advantage of producing the constant m/z values of signature ions regardless of the occupancy of the modification or the charge states of the peptides especially in a complex mixture, which will generate ambiguity in a neutral loss-triggered approach by anticipating the masses for monitoring.
A more detailed analysis on the occurrence of specific signature ions of HexNAc revealed some stable candidates for a product-ion-monitoring model, such as the combination of three ions at m/z 204, m/z 138, and m/z 126, which can be implemented with the HCD-trigger-ETD approach via a decision tree mechanism in the future.
We further demonstrated the capability of the HCD/ETD method in characterizing O-GalNAc and O-Mannose modified peptides as well, suggesting a more general applicability of the future HCD-trigger-ETD approach. Along with the advancement of both hardware and software in mass spectrometry, we anticipate that an HCD-product-ion-triggered-ETD approach on a hybrid linear ion trap/Orbitrap platform will greatly improve the sensitivity of detection and the accuracy in assigning the glycosylation sites on peptides from a complex mixture. Furthermore, this study sheds light on the action of the O-GlcNAc transferase in that it appears to be promiscuous with regards to which Ser/Thr residue is used in clustered regions of hydroxyl-containing amino acids in substrate proteins.
Supplementary Material
Figure S1. Comparison between the extracted ion chromatogram at m/z 204.086 and the total ion chromatogram of the HCD-alone LC-MS/MS experiment on the enriched HEK293T sample.
Figure S2. Comparison between the total ion chromatogram and the respective extracted ion chromatogram at m/z 204.086, m/z 186.076, m/z 168.065, m/z 144.065, m/z 138.054, and m/z 126.054 from the HCD/ETD LC-MS/MS experiment of the enriched HEK293T sample.
Figure S3. Multiple-engine database search strategy for the analysis on the enriched HEK293T sample.
Figure S4. HCD and ETD spectra of O-Mannose and O-GalNAc modified peptides.
Table S1. Fragments of HexNAc and Hexose oxonium ions.
Table S2. List of proteins identified in the enriched HEK293T sample by HCD/ETD approach.
Table S3. List of validated O-GlcNAc modified peptides identified in the enriched HEK293T sample by HCD/ETD approach.
Table S4. List of validated O-GlcNAc modified peptides identified in the enriched HEK293T sample by HCD-alone approach.
Table S5. List of validated O-GlcNAc modified peptides identified in the enriched HEK293T sample by HCD/ETD approach using multiple-engine database searches.
Acknowledgments
This work was supported by a P41 grant from NCRR (P41RR018502, LW, senior investigator) and an R01 grant from NIDDK (R01DK075069, LW). We thank Dr. Gerald W. Hart at Johns Hopkins University School of Medicine for providing the O-GlcNAc modified standard peptides and Dr. David Live at the University of Georgia for the synthesis of O-Mannose and O-GalNAc modified peptides.
Abbreviations
- AGC
automatic gain control
- CID
collision induced dissociation
- DTT
dithiothreitol
- ETD
electron transfer dissociation
- FDR
false discovery rate
- FWHM
full width at half maximum
- GalNAc
N-acetylgalactosamine
- GlcNAc
N-acetylglucosamine
- HCD
high-energy C-trap dissociation
- Hex
hexose
- HexNAc
N-acetylhexosamine
- LC
liquid chromatography
- Man
mannose
- MS
mass spectrometry
- OGA
O-linked β-N-acetylglucosaminidase
- OGT
O-linked β-N-acetylglucosamine transferase
- pepFDR
peptide-level FDR
- TRP domain
tetratricopeptide repeat domain
References
- 1.Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005(312):re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 2.Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673(1–2):13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446(7139):1017–22. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 4.Laczy B, Hill BG, Wang K, Paterson AJ, White CR, Xing D, Chen YF, Darley-Usmar V, Oparil S, Chatham JC. Protein O-GlcNAcylation: a new signaling paradigm for the cardiovascular system. Am J Physiol Heart Circ Physiol. 2009;296(1):H13–28. doi: 10.1152/ajpheart.01056.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copeland RJ, Bullen JW, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: roles in insulin resistance and glucose toxicity. Am J Physiol Endocrinol Metab. 2008;295(1):E17–28. doi: 10.1152/ajpendo.90281.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dias WB, Hart GW. O-GlcNAc modification in diabetes and Alzheimer’s disease. Mol Biosyst. 2007;3(11):766–72. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre T, Guinez C, Dehennaut V, Beseme-Dekeyser O, Morelle W, Michalski JC. Does O-GlcNAc play a role in neurodegenerative diseases? Expert Rev Proteomics. 2005;2(2):265–75. doi: 10.1586/14789450.2.2.265. [DOI] [PubMed] [Google Scholar]
- 8.Hu P, Shimoji S, Hart GW. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 2010;584(12):2526–38. doi: 10.1016/j.febslet.2010.04.044. [DOI] [PubMed] [Google Scholar]
- 9.Haynes PA, Aebersold R. Simultaneous detection and identification of O-GlcNAc-modified glycoproteins using liquid chromatography-tandem mass spectrometry. Anal Chem. 2000;72(21):5402–10. doi: 10.1021/ac000512w. [DOI] [PubMed] [Google Scholar]
- 10.Teo CF, Ingale S, Wolfert MA, Elsayed GA, Not LG, Chatham JC, Wells L, Boons GJ. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol. 2010;6(5):338–43. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal Biochem. 2001;293(2):169–77. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- 12.Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104(5):1143–56. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Identification of protein O-GlcNAcylation sites using electron transfer dissociation mass spectrometry on native peptides. Proc Natl Acad Sci U S A. 2009;106(22):8894–9. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5(5):923–34. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3(104):ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3(6):339–48. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 17.Jebanathirajah J, Steen H, Roepstorff P. Using optimized collision energies and high resolution, high accuracy fragment ion selection to improve glycopeptide detection by precursor ion scanning. J Am Soc Mass Spectrom. 2003;14(7):777–84. doi: 10.1016/S1044-0305(03)00263-0. [DOI] [PubMed] [Google Scholar]
- 18.Chalkley RJ, Burlingame AL. Identification of GlcNAcylation sites of peptides and alpha-crystallin using Q-TOF mass spectrometry. J Am Soc Mass Spectrom. 2001;12(10):1106–13. doi: 10.1016/s1044-0305(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 19.Huddleston MJ, Bean MF, Carr SA. Collisional fragmentation of glycopeptides by electrospray ionization LC/MS and LC/MS/MS: methods for selective detection of glycopeptides in protein digests. Anal Chem. 1993;65(7):877–84. doi: 10.1021/ac00055a009. [DOI] [PubMed] [Google Scholar]
- 20.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4(9):709–12. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 21.Carr SA, Huddleston MJ, Bean MF. Selective identification and differentiation of N- and O-linked oligosaccharides in glycoproteins by liquid chromatography-mass spectrometry. Protein Sci. 1993;2(2):183–96. doi: 10.1002/pro.5560020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim JM, Sherling D, Teo CF, Hausman DB, Lin D, Wells L. Defining the regulated secreted proteome of rodent adipocytes upon the induction of insulin resistance. J Proteome Res. 2008;7(3):1251–63. doi: 10.1021/pr7006945. [DOI] [PubMed] [Google Scholar]
- 23.Scherl A, Shaffer SA, Taylor GK, Hernandez P, Appel RD, Binz PA, Goodlett DR. On the benefits of acquiring peptide fragment ions at high measured mass accuracy. J Am Soc Mass Spectrom. 2008;19(6):891–901. doi: 10.1016/j.jasms.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764(12):1811–22. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JE, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal Chem. 2007;79(2):477–85. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterman SM, Mulholland JJ. A novel approach for identification and characterization of glycoproteins using a hybrid linear ion trap/FT-ICR mass spectrometer. J Am Soc Mass Spectrom. 2006;17(2):168–79. doi: 10.1016/j.jasms.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Zachara NE, Hart GW. O-GlcNAc modification: a nutritional sensor that modulates proteasome function. Trends Cell Biol. 2004;14(5):218–21. doi: 10.1016/j.tcb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Zachara NE, Molina H, Wong KY, Pandey A, Hart GW. The dynamic stress-induced “O-GlcNAc-ome” highlights functions for O-GlcNAc in regulating DNA damage/repair and other cellular pathways. Amino Acids. 2011;40(3):793–808. doi: 10.1007/s00726-010-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeidan Q, Wang Z, De Maio A, Hart GW. O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol Biol Cell. 2010;21(12):1922–36. doi: 10.1091/mbc.E09-11-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zachara NE, Cheung WD, Hart GW. Nucleocytoplasmic glycosylation, O-GlcNAc: identification and site mapping. Methods Mol Biol. 2004;284:175–94. doi: 10.1385/1-59259-816-1:175. [DOI] [PubMed] [Google Scholar]
- 31.Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1(10):791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci U S A. 2008;105(37):13793–8. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Park K, Comer F, Hsieh-Wilson LC, Saudek CD, Hart GW. Site-specific GlcNAcylation of human erythrocyte proteins: potential biomarker(s) for diabetes. Diabetes. 2009;58(2):309–17. doi: 10.2337/db08-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boutilier K, Ross M, Podtelejnikov AV, Orsi C, Taylor R, Taylor P, Figeys D. Comparison of different search engines using validated MS/MS test datasets. Analytica Chimica Acta. 2005;534:10. [Google Scholar]
- 35.Yu W, Taylor JA, Davis MT, Bonilla LE, Lee KA, Auger PL, Farnsworth CC, Welcher AA, Patterson SD. Maximizing the sensitivity and reliability of peptide identification in large-scale proteomic experiments by harnessing multiple search engines. Proteomics. 2010;10(6):1172–89. doi: 10.1002/pmic.200900074. [DOI] [PubMed] [Google Scholar]
- 36.Carrascal M, Gay M, Ovelleiro D, Casas V, Gelpi E, Abian J. Characterization of the human plasma phosphoproteome using linear ion trap mass spectrometry and multiple search engines. J Proteome Res. 2010;9(2):876–84. doi: 10.1021/pr900780s. [DOI] [PubMed] [Google Scholar]
- 37.Searle BC, Turner M, Nesvizhskii AI. Improving sensitivity by probabilistically combining results from multiple MS/MS search methodologies. J Proteome Res. 2008;7(1):245–53. doi: 10.1021/pr070540w. [DOI] [PubMed] [Google Scholar]
- 38.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 39.Nolte D, Muller U. Human O-GlcNAc transferase (OGT): genomic structure analysis of splice variants, fine mapping in Xq13.1. Mamm Genome. 2002;13(1):62–4. doi: 10.1007/s00335-001-2108-9. [DOI] [PubMed] [Google Scholar]
- 40.Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97(11):5735–9. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275(15):10983–8. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- 42.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J Biol Chem. 2008;283(49):33935–41. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer SP, Hart GW. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J Biol Chem. 2003;278(27):24608–16. doi: 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 44.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274(45):32015–22. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 45.Segu ZM, Mechref Y. Characterizing protein glycosylation sites through higher-energy C-trap dissociation. Rapid Commun Mass Spectrom. 2010;24(9):1217–25. doi: 10.1002/rcm.4485. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Ficarro SB, Li S, Marto JA. Optimized Orbitrap HCD for quantitative analysis of phosphopeptides. J Am Soc Mass Spectrom. 2009;20(8):1425–34. doi: 10.1016/j.jasms.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 47.Boja ES, Phillips D, French SA, Harris RA, Balaban RS. Quantitative mitochondrial phosphoproteomics using iTRAQ on an LTQ-Orbitrap with high energy collision dissociation. J Proteome Res. 2009;8(10):4665–75. doi: 10.1021/pr900387b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carr SA, Annan RS, Huddleston MJ. Mapping posttranslational modifications of proteins by MS-based selective detection: application to phosphoproteomics. Methods Enzymol. 2005;405:82–115. doi: 10.1016/S0076-6879(05)05005-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison between the extracted ion chromatogram at m/z 204.086 and the total ion chromatogram of the HCD-alone LC-MS/MS experiment on the enriched HEK293T sample.
Figure S2. Comparison between the total ion chromatogram and the respective extracted ion chromatogram at m/z 204.086, m/z 186.076, m/z 168.065, m/z 144.065, m/z 138.054, and m/z 126.054 from the HCD/ETD LC-MS/MS experiment of the enriched HEK293T sample.
Figure S3. Multiple-engine database search strategy for the analysis on the enriched HEK293T sample.
Figure S4. HCD and ETD spectra of O-Mannose and O-GalNAc modified peptides.
Table S1. Fragments of HexNAc and Hexose oxonium ions.
Table S2. List of proteins identified in the enriched HEK293T sample by HCD/ETD approach.
Table S3. List of validated O-GlcNAc modified peptides identified in the enriched HEK293T sample by HCD/ETD approach.
Table S4. List of validated O-GlcNAc modified peptides identified in the enriched HEK293T sample by HCD-alone approach.
Table S5. List of validated O-GlcNAc modified peptides identified in the enriched HEK293T sample by HCD/ETD approach using multiple-engine database searches.