Abstract
The Elongin BC complex was identified initially as a positive regulator of RNA polymerase II (Pol II) elongation factor Elongin A and subsequently as a component of the multiprotein von Hippel-Lindau (VHL) tumor suppressor complex, in which it participates in both tumor suppression and negative regulation of hypoxia-inducible genes. Elongin B is a ubiquitin-like protein, and Elongin C is a Skp1-like protein that binds to a BC-box motif that is present in both Elongin A and VHL and is distinct from the conserved F-box motif recognized by Skp1. In this report, we demonstrate that the Elongin BC complex also binds to a functional BC box present in the SOCS box, a sequence motif identified recently in the suppressor of cytokine signaling-1 (SOCS-1) protein, as well as in a collection of additional proteins belonging to the SOCS, ras, WD-40 repeat, SPRY domain, and ankyrin repeat families. In addition, we present evidence (1) that the Elongin BC complex is a component of a multiprotein SOCS-1 complex that attenuates Jak/STAT signaling by binding to Jak2 and inhibiting Jak2 kinase, and (2) that by interacting with the SOCS box, the Elongin BC complex can increase expression of the SOCS-1 protein by inhibiting its degradation. These results suggest that Elongin BC is a multifunctional regulatory complex capable of controlling multiple pathways in the cell through interaction with a short degenerate sequence motif found in many different proteins.
Keywords: Elongin BC complex, SOCS box, SOCS-1, Jak/STAT signaling, protein degradation
The cytokine-inducible suppressors of cytokine signaling (SOCS) proteins (also referred to as JABs, SSIs, or CISs) are negative regulators of cytokine-induced Jak/STAT signaling and inhibit phosphorylation of STATs by binding to and inhibiting Jak kinases (Endo et al. 1997; Ki et al. 1997; Naka et al. 1997; Starr et al. 1997) or through interactions with receptor tyrosine kinases (Yoshimura et al. 1995; Matsumoto et al. 1997). Several members of the cytokine superfamily, including interleukin-6, leukemia-inhibitory factor, erythropoietin, growth hormone, and leptin, induce transcriptional activation of one or more of the SOCS genes (Yoshimura et al. 1995; Endo et al. 1997; Masuhara et al. 1997; Naka et al. 1997; Starr et al. 1997; Adams et al. 1998; Auerenhammer et al. 1998; Bjorbaek et al. 1998), suggesting that the SOCS proteins function as part of a negative feedback loop that dampens or attenuates cytokine signal transduction.
The SOCS family presently includes eight proteins that all share an amino-terminal SH2 domain and a novel ∼50 amino acid carboxy-terminal domain referred to as the SOCS box (Endo et al. 1997; Minamoto et al. 1997; Starr et al. 1997; Hilton et al. 1998). In addition, Hilton et al. (1998) recently obtained evidence for an expanded role for SOCS-box proteins in cellular regulation by searching DNA databases and identifying a large number of additional SOCS-box proteins, including previously uncharacterized members of the ras, WD-40 repeat, ankyrin repeat, and SPRY domain families. Although the function(s) of the SOCS box has not yet been elucidated, it has been proposed that it might function directly in inhibition of Jak or receptor tyrosine kinases, in light of evidence indicating that the SH2 domain of the SOCS-1 protein is necessary but not sufficient for preventing Jak2 kinase-dependent activation of STATs (Endo et al. 1997). Data addressing the role of the SOCS box in inhibition of Jak or receptor tyrosine kinases have not yet been reported, however, and SOCS-box interacting proteins have not been identified.
The Elongin BC complex was initially identified as a potent activator of RNA polymerase II (Pol II) elongation factor Elongin A (Bradsher et al. 1993; Aso et al. 1995), which is one of several transcription factors that stimulate the rate of elongation by RNA Pol II by suppressing transient pausing by polymerase at many sites along the DNA. Elongin B is a 118-amino-acid ubiquitin-like protein composed of an 84-amino-acid amino-terminal ubiquitin–homology (UbH) domain fused to a 34-amino-acid carboxy-terminal tail (Garrett et al. 1995). Elongin C is a 112-amino-acid protein (Garrett et al. 1994) similar in sequence to the multifunctional SKP1 protein (Zhang et al. 1995; Bai et al. 1996; Connelly and Hieter 1996; Feldman et al. 1997; Skowyra et al. 1997; Patton et al. 1998b), which has been shown to bind to the conserved F-box motif present in a large number of proteins and to participate in regulation of diverse cellular processes including the cell cycle, transcription, and development.
The Elongin BC complex is also a component of the multiprotein von Hippel–Lindau (VHL) tumor suppressor complex (Duan et al. 1995b; Kibel et al. 1995). The VHL gene on chromosome 3p25.5 is mutated in the majority of sporadic clear cell renal carcinomas and in VHL disease, an autosomal dominant familial cancer syndrome that predisposes affected individuals to a variety of tumors including clear cell renal carcinomas, cerebellar hemangioblastomas and hemangiomas, retinal angiomata, and pheochromocytomas (Latif et al. 1993; Gnarra et al. 1996a).
Further investigation of the function of the Elongin BC complex revealed that VHL is a potent inhibitor of Elongin BC-dependent activation of Elongin A in vitro. Interaction of Elongin BC with Elongin A and VHL is mutually exclusive and depends on binding of Elongin C to a short ∼12-amino-acid BC-box motif that is evolutionarily conserved in Elongin A and VHL proteins and that is a site of frequent mutations in VHL disease (Duan et al. 1995b; Kibel et al. 1995; Aso et al. 1996; Gnarra et al. 1996a). On the basis of these findings, it was initially proposed that VHL may function in cells as a negative regulator of Elongin A transcriptional activity by competing with Elongin A for binding to the Elongin BC complex. One prediction of this simple competition model is that the Elongin BC complex is limiting in cells.
In the process of investigating this model, we fractionated liver extracts and quantitated the relative amounts of Elongin A, VHL, and Elongin BC. Unexpectedly, we found (1) that Elongin BC is much more abundant in cells than either Elongin A or VHL, and (2) that Elongin BC is present in multiple chromatographically distinct complexes. Taken together with recent evidence indicating that Elongin BC functions in VHL-dependent negative regulation of expression of hypoxia-inducible genes like VEGF and GLUT1 by controlling the stability of their mRNAs (Gnarra et al. 1996b; Iliopoulos et al. 1996; Lonergan et al. 1998), these findings suggest that like SKP1 (Bai et al. 1996; Feldman et al. 1997; Skowyra et al. 1997; Patton et al. 1998b), the Elongin BC complex could interact with and regulate multiple proteins that play roles in distinct cellular processes.
In this report, we present evidence consistent with this hypothesis. We identify the SOCS-box proteins as a new class of Elongin BC binding proteins. We show that Elongin BC interacts with a conserved BC box located in the amino terminus of the SOCS-box motif found in members of the SOCS, ras, WD-40 repeat, ankyrin repeat, and SPRY domain families. In addition, we show that (1) the Elongin BC complex is a component of a multiprotein SOCS-1 complex that binds to and inhibits Jak kinase and activation of STATs, (2) that the SOCS box is not responsible for SOCS-1 inhibition of Jak kinase, but (3) that by interacting with the SOCS box, Elongin BC can regulate SOCS-1 expression by inhibiting its degradation. These results suggest that Elongin BC participates in regulation of multiple cellular pathways through interaction with a short degenerate sequence motif found in a diverse collection of proteins.
Results
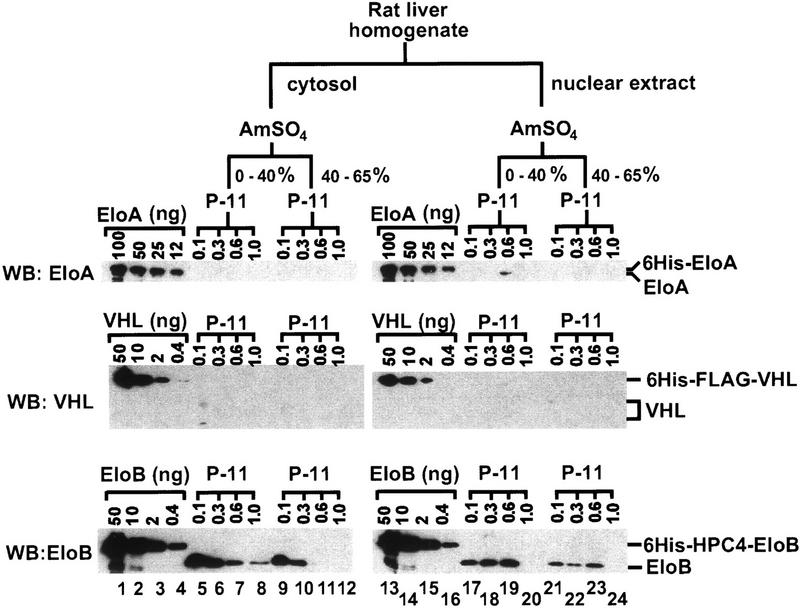
To determine the relative cellular concentrations and distributions of the Elongin A, VHL, and Elongin BC proteins, we systematically fractionated ∼150 grams of rat liver and measured the levels of Elongin A, VHL, and Elongin B by quantitative Western blotting. As shown in Figure 1, nearly all of the detectable VHL (∼1 μg) was found in the 0.1 m KCl phosphocellulose flowthrough fraction derived from a 0%–40% ammonium sulfate precipitation of cytosol, and nearly all of the detectable Elongin A (∼5 μg) was found in the 0.6 m KCl phosphocellulose fraction derived from a 0%–40% ammonium sulfate precipitation of nuclear extract. In contrast, a substantially greater quantity of Elongin B (∼150 μg) was found distributed in multiple phosphocellulose fractions derived from both cytosol and nuclear extract. Because our anti-Elongin C antibodies were not of sufficient quality, we were unable to obtain reliable estimates of the levels of Elongin C protein in the relatively crude phosphocellulose fractions. Further purification of Elongin B-containing species in each of the phosphocellulose fractions examined, however, has revealed the presence of equimolar amounts of Elongins B and C in more highly purified fractions, suggesting that the majority of Elongin B is associated with Elongin C (data not shown). Taken together, these results raised the possibility that Elongin BC may be present in cells in association with additional proteins besides Elongin A and VHL.
Figure 1.
Partial purification of Elongin BC-containing complexes from rat liver. Homogenate was prepared from the livers of 10 male Sprague–Dawley rats (200–250 grams) and subjected to subcellular fractionation, ammonium sulfate precipitation, and phosphocellulose chromatography as described (Conaway et al. 1996). Ten microliters of each fraction was subjected to SDS-PAGE along with the indicated amounts of recombinant histidine-tagged Elongin A (6His–EloA), histidine- and FLAG-tagged VHL (6His–FLAG–VHL), or histidine- and HPC4-tagged Elongin B (6His–HPC4–EloB) and then immunoblotted with anti-Elongin A, anti-VHL, and anti-Elongin B antibodies. The 0.1, 0.3, 0.6, and 1 m KCl eluates from phosphocellulose are indicated as 0.1, 0.3, 0.6, and 1.0, respectively. The volumes of the fractions shown in lanes 5–8 were 50, 25, 25, and 20 ml; fractions in lanes 9–12 were 200, 150, 120, and 120 ml; fractions in lanes 17–20 were 15, 10, 10, and 10 ml; and fractions in lanes 21–24 were 40, 20, 15, and 15 ml. (AmSO4) Ammonium sulfate; (EloA) Elongin A; (EloB) Elongin B; (P-11) phosphocellulose.
Sequence alignments of Elongin A and VHL proteins from mammalian cells, Caenorhabditis elegans, and Saccharomyces cerevisiae reveal a single region that is conserved between Elongin A and VHL and that has the consensus sequence T/SL/MxxxC/SxxxV/L/I (Fig. 2A). Several lines of evidence argue that this conserved region defines a consensus Elongin BC binding site or BC box. First, Elongin A and VHL mutants lacking this region are unable to bind to the Elongin BC complex, and mutation of conserved residues within this region interferes with Elongin transcriptional activity and with binding of Elongin BC to the VHL protein (Aso et al. 1995, 1996; Duan et al. 1995a,b; Kibel et al. 1995; Kishida et al. 1995). Second, a GST–VHL (157–172) fusion protein, which includes the conserved region, is sufficient to bind Elongin BC (Kibel et al. 1995). Finally, peptides corresponding to VHL (157–172) inhibit the binding of Elongin BC to VHL and to Elongin A, whereas point-mutant derivatives altered at the conserved residues do not (Kibel et al. 1995; data not shown).
Figure 2.
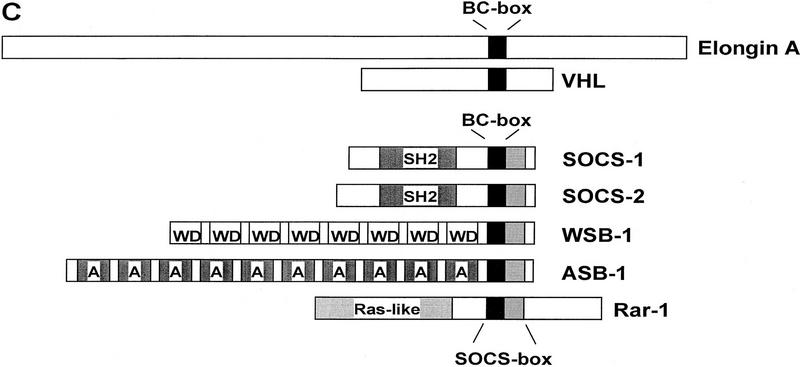
The SOCS box includes a consensus Elongin BC binding site. (A) Sequence alignment of the Elongin BC binding site regions of VHL and Elongin A proteins from mammalian cells, C. elegans, and S. cerevisiae. (B) The SOCS-box regions of several SOCS-box proteins (Hilton et al. 1998) are aligned beneath Elongin BC binding sites from VHL and Elongin A proteins. (C) Diagrammatic representation of Elongin A, VHL, and the SOCS-box proteins used in this study.
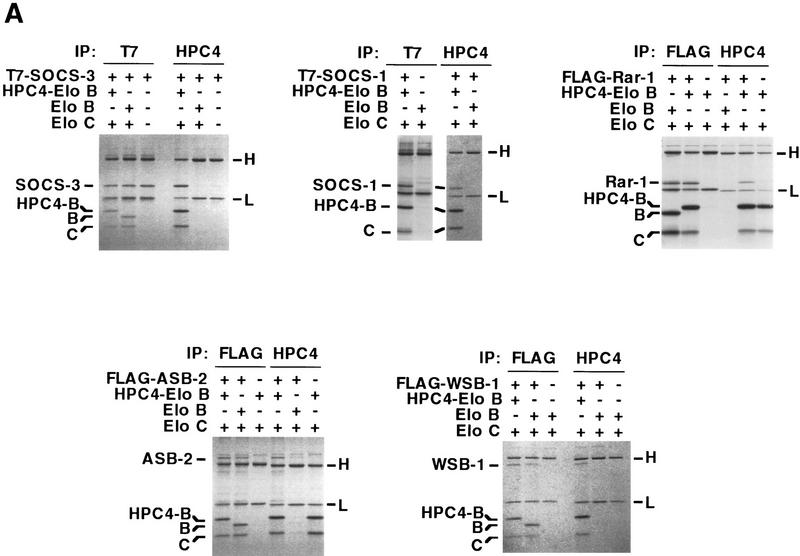
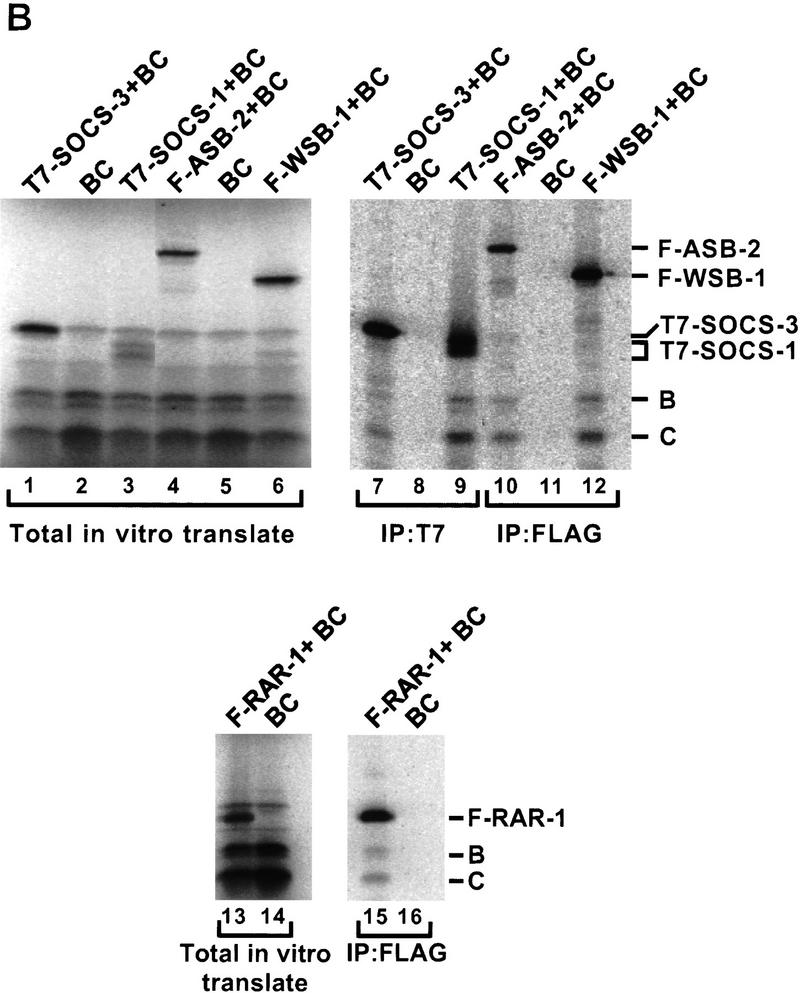
Recently, Hilton et al. (1998) identified a large family of SOCS-box proteins belonging to the SOCS, Ras, WD-40 repeat, ankyrin repeat, and SPRY domain families. As shown in Figure 2B, the amino-terminal half of the SOCS box found in each of these proteins includes a potential Elongin BC binding site. To determine whether SOCS-box proteins bind the Elongin BC complex, we tested the abilities of representative members of four of the five SOCS-box containing protein families to bind Elongin BC in vitro. ORFs encoding full-length SOCS-1, SOCS-3, and the WD-40 repeat-containing protein WSB-1, all but the first 9 and final 34 amino acids of the ras-related protein Rar-1, and the 545 carboxy-terminal amino acids of the ankyrin repeat-containing protein ASB-2 (Fig. 2C) were expressed in Escherichia coli with amino-terminal 6-histidine and epitope tags, purified by Ni2+-agarose affinity chromatography, and tested in coimmunoprecipitation assays for their abilities to bind Elongin BC. As shown in Figure 3A, all five of the recombinant SOCS-box proteins tested could be specifically coimmunoprecipitated with Elongin BC at high ionic strength (0.5 m NaCl) and in the presence of detergent. We note that a super-stoichiometric amount of bacterially expressed Elongin BC was sometimes immunoprecipitated with epitope-tagged SOCS-box proteins (e.g., Flag–Rar-1 and Flag–ASB2), most likely due to the tendency of Elongin BC to form multimers (Takagi et al. 1997). We also assessed the binding of in vitro-translated SOCS-1, SOCS-3, WSB-1, ASB-2, and RAR-1 to in vitro-translated Elongin BC. As shown in Figure 3B, in vitro-translated Elongin BC was specifically coimmunoprecipitated with each of the in vitro-translated SOCS-box containing proteins.
Figure 3.
Binding of the Elongin BC complex to SOCS-box proteins. (A) Recombinant proteins were expressed in E. coli and purified from guanidine-solubilized inclusion bodies by Ni2+–agarose affinity chromatography. Untagged or epitope-tagged SOCS-1, SOCS-3, Rar-1, WSB-1, ASB-1, Elongin B, and Elongin C were mixed together in the combinations indicated, renatured by dilution and dialysis, and immunoprecipitated with the antibodies indicated. Following SDS-PAGE, immunoprecipitated proteins were visualized by Coomassie Blue staining. (B) T7-tagged SOCS-1 and SOCS-3, Flag-tagged WSB-1, ASB-2, and RAR-1 were in vitro transcribed and translated together with HPC-4 tagged Elongin B and HSV-tagged Elongin C by use of the Promega TNT/T7 rabbit reticulocyte lysate kit with [35S]methionine according to the manufacturer’s instructions. In vitro-translated proteins were immunoprecipitated with the antibodies indicated, subjected to SDS-PAGE, and visualized with a PhosphorImager (Molecular Dynamics). (H,L) Immunoglobulin heavy and light chains, respectively. (IP) Immunoprecipitate; (WB) Western blot; (F) Flag epitope; (B) Elongin B; (C) Elongin C.
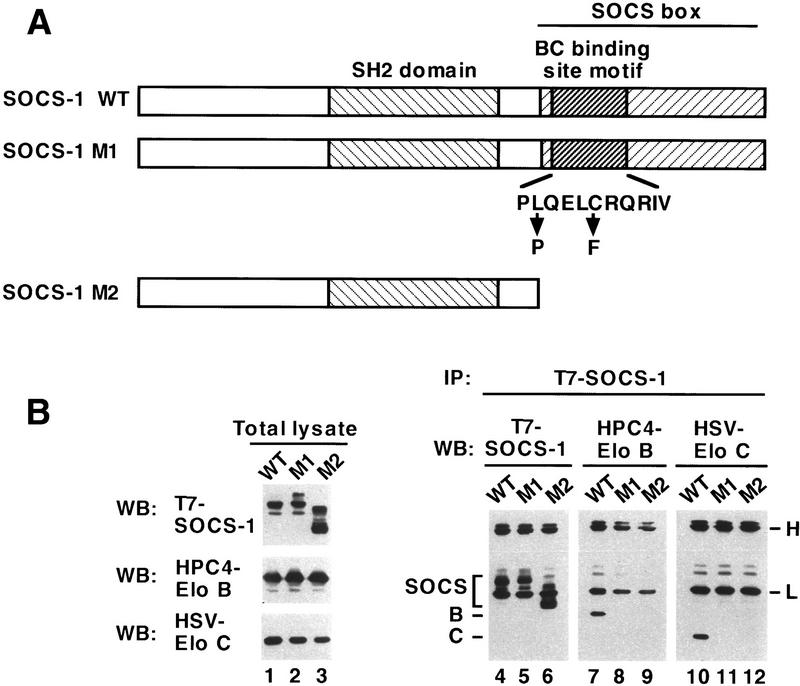
To determine whether the BC box motif in the amino terminus of the SOCS box is necessary for binding of Elongin BC to a SOCS-box protein, we compared the abilities of wild-type SOCS-1 and SOCS-1 mutants to bind Elongin BC. Baculovirus and mammalian expression vectors encoding wild-type epitope-tagged SOCS-1 and two SOCS-1 derivatives containing mutations of the potential Elongin BC binding site were constructed (Fig. 4A). One SOCS-1 mutant (M1) contained point mutations at positions 2 (L → P) and 6 (C → F) of the consensus BC-box motif shown in Figure 2A; point mutations at the corresponding positions of Elongin A and VHL disrupt Elongin BC binding. The other SOCS-1 mutant (M2) was a carboxy-terminal deletion mutant that lacked the entire SOCS-box, including the amino-terminal BC-box motif. T7-tagged SOCS-1 and SOCS-1 mutants M1 and M2 were coexpressed with epitope-tagged Elongins B and C in Sf21 insect cells and in mammalian 293T cells and their abilities to bind Elongin BC were assessed in coimmunoprecipitation experiments. As shown in Figure 4B, coimmunopreciptiation of the Elongin BC complex with SOCS-1 in Sf21 cells was strongly dependent on the presence of an intact BC box. Elongins B and C were both coimmunoprecipitated with wild-type SOCS-1 (lanes 7,10). In contrast, Elongins B and C did not coimmunoprecipitate with either of the SOCS-1 mutants M1 (lanes 8,11) or M2 (lanes 9,12), even though approximately equivalent levels of the wild-type and mutant SOCS-1 proteins were expressed in baculovirus-infected cells (lanes 1–3). Similarly, coimmunoprecipitation of the Elongin BC complex with SOCS-1 in 293T cells was strongly dependent on the presence of an intact BC box (Fig. 5A, top and middle panels).
Figure 4.
Binding of the Elongin BC complex to SOCS-1 is disrupted by mutation of the SOCS-box Elongin BC binding site motif. (Lanes 1–3) Twenty-five micrograms of total cell lysate from Sf21 cells infected with the baculovirus vectors encoding HPC4-tagged Elongin B, HSV-tagged Elongin C, and the indicated wild-type or mutant T7-tagged SOCS-1 proteins was immunoblotted with anti-T7, anti-HPC4, or anti-HSV antibodies. (Lanes 4–12) Three hundred micrograms of total cell lysate protein from baculovirus-infected Sf21 cells was immunopreciptiated with antibodies specific for T7 epitope (IP: T7-SOCS-1), HPC4 epitope (IP: HPC4-EloB), or HSV epitope (IP: HSV-EloC) and then immunoblotted with anti-T7, anti-HPC4, or anti-HSV antibodies. (H,L) Immunoglobulin heavy and light chains, respectively.
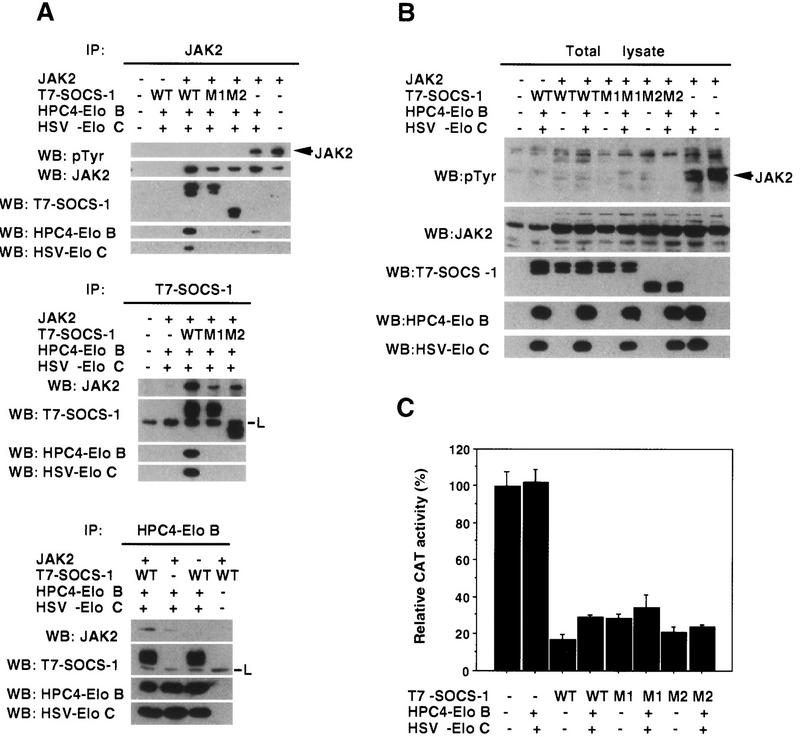
Figure 5.
Elongins B and C are components of the SOCS-1 complex but are not required for inhibition of Jak activity. 293T cells were transiently transfected with the indicated combinations of Jak2, wild-type or mutant T7–SOCS-1, HPC4–Elongin B, and HSV–Elongin C expression vectors. The amount of the T7–SOCS-1 expression vectors used in each transfection was adjusted so that comparable levels of wild-type SOCS-1 and SOCS-1 mutants would be expressed. (A) Three hundred micrograms of total cell lysate protein from transfected cells was immunopreciptiated with antibodies specific for Jak2 (IP: Jak2), T7 epitope (IP: T7-SOCS-1), or HPC4 epitope (IP: HPC4-EloB) and then immunoblotted with anti-pTyr, anti-Jak2, anti-T7, anti-HPC4, or anti-HSV antibodies. (L) Immunoglobulin light chain. (B) Twenty-five micrograms of the indicated total cell lysates was immunoblotted with the antibodies indicated. (C) Hep3B cells were transiently transfected in 35-mm dishes with 1 μg each of the IL6-inducible STAT-dependent CAT reporter construct TIK (gift of W. Liao, M.D. Anderson Cancer Center, Houston, TX) and the β-galactosidase control reporter pCMVβ (Clontech), with or without 1 μg of expression vectors encoding T7-tagged SOCS-1, HPC4-tagged Elongin B, and HSV-tagged Elongin C. After 40 hr, cells were stimulated with 100 ng/ml IL-6 for 3 hr, and cell extracts were assayed for CAT and β-galactosidase activities with the CAT and β-galactosidase assay systems from Promega. CAT activity, normalized to β-galactosidase activity, is expressed as the percent of that observed in lysates from cells not transfected with SOCS-1, Elongin B, and Elongin C expression vectors.
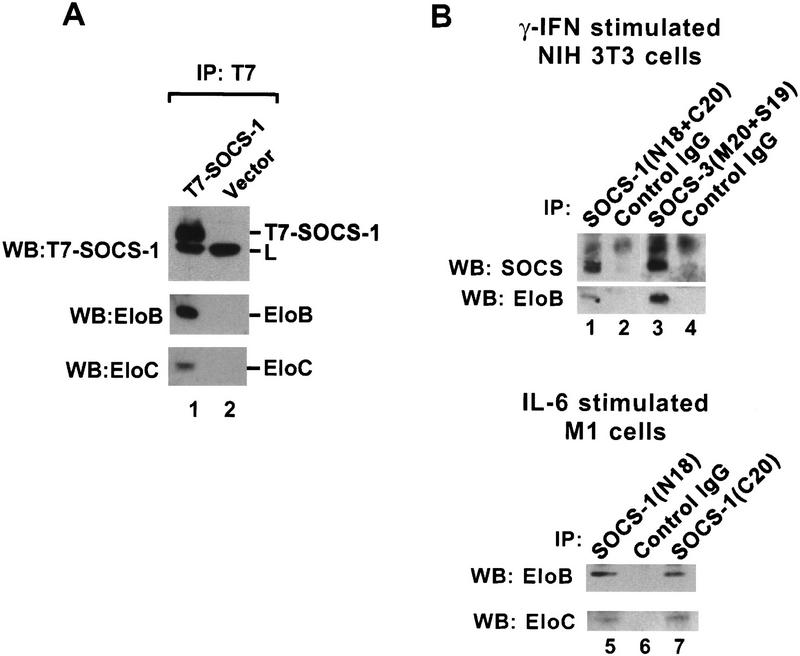
Two lines of evidence indicate that Elongin BC expressed at physiological concentrations is capable of interacting with SOCS-box proteins. First, endogenous Elongins B and C could be immunoprecipitated with anti-T7 epitope antibody from lysates of Hep3B cells stably transfected with a vector encoding T7-tagged SOCS-1, but not from lysates of cells stably transfected with a control vector (Fig. 6A) or with a vector encoding the T7-tagged SOCS-1 M1 double point mutant (Fig. 7B, below; T. Kamura, J.W. Conaway, and R.C. Conaway, unpubl.). Second, endogenous Elongins B and C could be immunoprecipitated from lysates of γ-interferon-stimulated NIH-3T3 cells and IL-6-stimulated M1 cells with goat polyclonal antibodies raised against an amino-terminal peptide of SOCS-1 (N18), a carboxy-terminal peptide of SOCS-1 (C20), and amino- and carboxy-terminal peptides of SOCS-3 (S19 and M20) but not with control goat IgG (Fig. 6B).
Figure 6.
Interaction of SOCS-Box proteins with Elongin BC. (A) Hep3B cells stably transformed with T7-SOCS-1 (lane 1) or empty control vector (lane 2) were incubated for 9 hr with 100 μm ZnCl2 in DMEM, 10% fetal calf serum prior to harvesting and cell lysis. Two-hundred micrograms of total cell lysate protein was immunoprecipitated with anti-T7 antibodies and then immunoblotted with anti-T7, anti-Elongin B, or anti-Elongin C antibodies. (B) NIH-3T3 cells were stimulated with 1000 units of γ-interferon for 5 hr before harvesting and cell lysis (lanes 1–4). M1 cells were stimulated with 50 ng/ml IL-6 for 5 hr before harvesting and cell lysis (lanes 5–7). Approximately 5 mg of total cell lysate protein was immunoprecipitated with the indicated antibodies and then immunoblotted with anti-SOCS-1 (N18) (lanes 1–2), anti-SOCS-3 (S19) (lanes 3–4), and anti-Elongin B and anti-Elongin C antibodies as indicated.
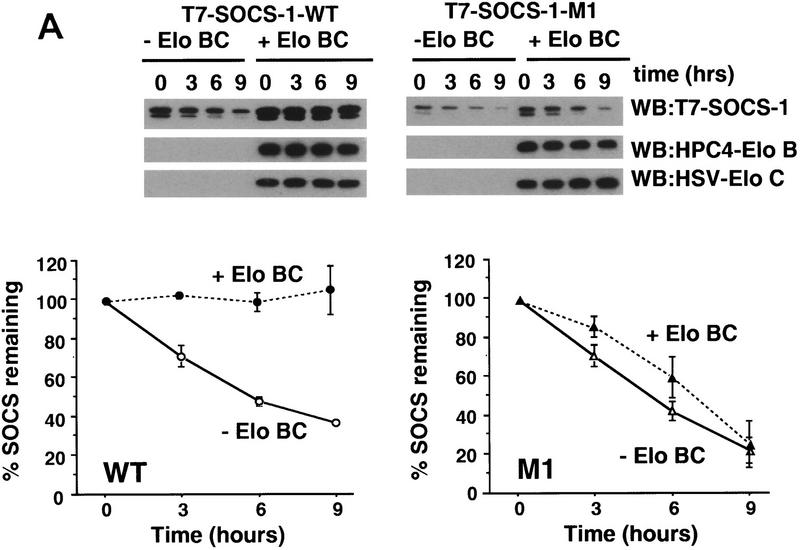
Figure 7.
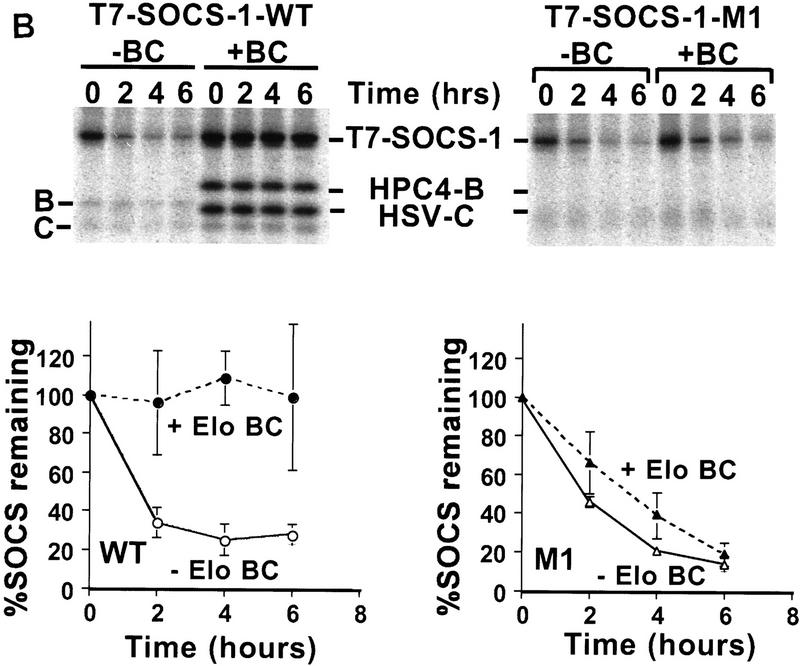
Interaction of Elongin BC with SOCS-1 increases its stability. 293T cells were transiently transfected in 35-mm dishes with 1 μg of the HPC4-Elongin B expression vector, 1 μg of the HSV-Elongin C expression vector, and 1 μg of the wild-type T7-SOCS-1 expression vector or 3 μg of the T7-SOCS-1 M1 expression vector as indicated. (A) Cycloheximide block. Twenty-four hours after transfection, 100 μg/ml cycloheximide was added to culture dishes. Cells were harvested and lysed after an additional incubation for the indicated times. Twenty-five micrograms of total cell lysate protein was immunoblotted with the indicated antibodies. SOCS-1 protein was quantitated by densitometry of Western blots using NIH Image 1.60/ppc. (B) [35S]methionine pulse chase. Cultures were washed twice with PBS and incubated with methionine-free medium containing 10% dialyzed fetal calf serum for 1 hr prior to addition of [35S]methionine (0.5 mCi/ml of medium; EXPRE35S35S protein labeling mix, NEN), followed by an additional 2 hr incubation. The cells were then washed twice with PBS and cultured in DMEM with 10% fetal calf serum. Cells were harvested and lysed after an additional incubation for the indicated times. Approximately 200 μg of total cell lysate protein was immunoprecipitated with T7 antibody and analyzed by SDS-PAGE. Radioactive proteins were detected and quantitated by PhosphorImager analysis. The data plotted represents the mean of three independent experiments. Error bars indicate the standard error of the mean.
Although results of initial studies on SOCS-box proteins provided little insight into the function of the SOCS box (Endo et al. 1997; Naka et al. 1997; Starr et al. 1997), it had been suggested that the SOCS box might play a role in suppressing signal transduction, perhaps by inhibiting protein kinase activities, or, alternatively, that it might regulate aspects of protein metabolism such as protein stability (Hilton et al. 1998). SOCS-1 has been shown to bind to and inhibit tyrosine phosphorylation by multiple kinases including Jak2 (Endo et al. 1997; Naka et al. 1997; Starr et al. 1997). To address the role of the SOCS box and the Elongin BC complex in regulation of Jak2 kinase by SOCS-1, epitope-tagged Elongins B and C, Jak2, and T7-tagged wild-type SOCS-1 or the M1 or M2 mutants were coexpressed in 293T cells. Coimmunoprecipitation experiments revealed that Elongins B and C are components of a multiprotein SOCS-1 complex that binds to Jak2. As shown in Figure 5A, Elongins B and C could be coimmunoprecipitated by anti-Jak2 antibodies in the presence of wild-type SOCS-1 but not the M1 or M2 mutants. Furthermore, Elongins B and C could be coimmunoprecipitated by anti-T7 antibodies in the presence of wild-type SOCS-1 but not the SOCS-1 M1 or M2 mutants, and Jak2 was coimmunoprecipitated with SOCS-1 and Elongins B and C by antibodies against the HPC4 epitope on Elongin B or by anti-T7 antibodies; the small amount of Jak2 coimmunoprecipitated by anti-HPC4 from cells not transfected with SOCS-1 was most likely due to the presence of endogenous SOCS proteins. Consistent with the previous observation that the SOCS-1 SH2 domain is sufficient for binding to Jak2 (Endo et al. 1997), wild-type SOCS-1 and both SOCS-1 mutants could be coimmunoprecipitated with Jak2. In addition, we observed that all three SOCS-1 proteins were capable of inhibiting tyrosine phosphorylation of Jak2 (Fig. 5A, top, and B) and of inhibiting IL-6-induced activation of a STAT-dependent CAT reporter (Fig. 5C), indicating that an intact BC box is not essential for these SOCS-1 functions.
In the course of these experiments, we observed that wild-type SOCS-1 was expressed at significantly higher levels than SOCS-1 mutants M1 or M2 following transfection of 293T cells with equal concentrations of expression vectors (data not shown). To determine whether this increased expression of wild-type SOCS-1 could be attributed at least in part to stabilization of SOCS-1 by Elongin BC, wild-type SOCS-1 or the M1 double point mutant was transiently expressed in 293T cells with or without Elongins B and C. Twenty-four hours after transfection, cycloheximide was added to cultures to block further protein synthesis, and the rate of decay of wild-type and mutant SOCS-1 proteins was measured by Western blotting. As shown in Figure 7A, the level of wild-type SOCS-1 protein remained constant for at least 9 hr after addition of cycloheximide to cells cotransfected with SOCS-1 and Elongins B and C. In the absence of cotransfected Elongins B and C, however, 50% of SOCS-1 protein was lost within ∼5 hr. Thus, wild-type SOCS-1 protein is stabilized by coexpression of Elongins B and C. In contrast to wild-type SOCS-1, the M1 mutant was unstable in both the presence and absence of cotransfected Elongins B and C.
Similar results were obtained when the decay rates of wild-type SOCS-1 and the M1 double point mutant were measured in [35S]methionine pulse-chase experiments. In these experiments, T7-tagged SOCS-1 proteins were transiently expressed in 293T cells with or without Elongins B and C. Twenty-four hours after transfection, cells were pulse-labeled with [35S]methionine and then grown for varying lengths of time in culture medium containing unlabeled methionine. Radioactively labeled SOCS-1 proteins were then immunoprecipitated from cell lysates with anti-T7 epitope antibody, and the amounts of SOCS-1 proteins remaining after the chase were quantitated by analysis with a PhosphorImager. As shown in Figure 7B, wild-type SOCS-1 protein was very stable in cells cotransfected with SOCS-1 and Elongins B and C, but decayed rapidly in the absence of cotransfected Elongins B and C. In addition, the M1 mutant decayed rapidly in both the presence and absence of cotransfected Elongins B and C. Taken together, these results indicate that stabilization of SOCS-1 by the Elongin BC complex depends on an intact Elongin BC binding site and strongly suggest that stabilization of SOCS-1 requires interaction of the Elongin BC complex with the SOCS box.
Discussion
The Elongin BC complex was identified initially as a positive regulator of RNA Pol II elongation factor Elongin A and subsequently as a component of the multiprotein VHL tumor-suppressor complex. Interaction of the Elongin BC complex with Elongin A and VHL depends on binding of Elongin C to an ∼12 amino acid BC-box motif shared by Elongin A and VHL. In the process of investigating the functional relationship between Elongin A, VHL, and the Elongin BC complex, we discovered that Elongin BC is much more abundant in cell extracts than either Elongin A or VHL and, further, that Elongin BC is present in extracts in multiple, chromatographically distinct species. In light of these findings, we considered the possibility that the Elongin BC complex might play multiple roles in cellular regulation by controlling the activities of Elongin A, VHL, and perhaps other proteins in distinct cellular processes. As part of our effort to identify the repertoire of cellular Elongin BC-interacting proteins, we sought to identify additional BC box proteins. In the course of this work, we discovered that a functional BC-box is a component of the SOCS-box motif described recently (Starr et al. 1997; Hilton et al. 1998; Nicholson and Hilton 1998).
The ∼50 amino acid SOCS-box motif is composed of two well-conserved blocks of sequence, which are separated by 2–10 nonconserved residues. The carboxy-terminal-conserved region is an L/P-rich sequence of unknown function, and, as we have shown here, the amino-terminal conserved region is a consensus BC box. Each modular SOCS box protein includes a SOCS-box at its extreme carboxyl terminus and, depending on the protein family, either an SH2, Ras-like, WD40 repeat, ankyrin repeat, or SPRY domain at its amino terminus (Hilton et al. 1998). In this report we have tested representative members of four of the five SOCS-box protein families for their abilities to bind Elongin BC. Our results indicate that the Elongin BC complex interacts not only with Elongin A and VHL, but also with all SOCS-box proteins tested, including members of the SOCS, Ras, WD-40 repeat, and ankyrin repeat families. In addition, we have made findings that shed light on the roles of the SOCS-box and the Elongin BC–SOCS-box interaction in regulating the activity of SOCS-1, the best characterized member of the SOCS-box family.
SOCS-1 was shown previously to attenuate Jak/STAT signaling by binding to and inhibiting Jak kinase-dependent phosphorylation and activation of STATs (Endo et al. 1997; Naka et al. 1997; Starr et al. 1997). These studies demonstrated that the SH2 domain of SOCS-1 is critical for binding to Jak, but is not sufficient for inhibition of Jak kinase, raising the possibility that the role of the SOCS box in SOCS-box proteins might be to control directly the activities of target protein kinases or other regulatory molecules, whereas the amino-terminal SH2, Ras, WD-40 repeat, and ankyrin repeat domains might be responsible for dictating the specificity of interaction of SOCS-box proteins with their targets (Hilton et al. 1998). We observe, however, that SOCS-1 mutants lacking the SOCS-box or containing BC-box mutations that disrupt interaction with Elongin BC still retain their ability to inhibit Jak kinase, arguing that, at least in the case of SOCS-1, neither the SOCS box nor Elongin BC is directly involved in regulating the target protein. On the other hand, we observe that interaction of Elongin BC with the SOCS box markedly increases the stability of the SOCS-1 protein. Consistent with our findings, while this paper was under revision, Narazaki et al. (1998) reported that although the SOCS-1 SOCS box is not essential for inhibition of Jak kinase activities or for suppression of IL-6 signaling in cells, it does serve to protect SOCS-1 from degradation by the proteasome. Although free wild-type SOCS-1 or SOCS-1 lacking a functional Elongin BC binding site may be rapidly degraded simply because it is normally part of a multiprotein complex, our results raise the possibility that the Elongin BC–SOCS-box interaction could function at least in part to fine tune the concentration of SOCS-1 in cells and thus indirectly regulate the activity of the target protein. The potential importance of properly regulating cellular concentrations of SOCS proteins is underscored by recent results suggesting that the leptin resistance and consequent obesity of Ay/a mice may be due in part to inappropriately elevated SOCS-3 levels (Bjorbaek et al. 1998).
The observation that the Elongin BC complex binds to and can regulate the activity or stability of multiple proteins brings to light a striking parallel between Elongin C and and its structural homolog Skp1. Similar to Elongin C, which interacts with the conserved BC-box motif present in Elongin A, VHL, and the SOCS-box proteins, Skp1 interacts with the conserved F-box motif present in multiple families of proteins and thereby participates in regulation of diverse cellular processes (Zhang et al. 1995; Bai et al. 1996; Connelly and Hieter 1996; Feldman et al. 1997; Skowyra et al. 1997; Patton et al. 1998b). Skp1 is a component of the S. cerevisiae CBF3 kinetochore complex, in which it binds to and mediates phosphorylation-dependent activation of the F-box protein Ctf13, perhaps by bridging Ctf13 to the kinase(s) responsible for its phosphorylation (Connelly and Hieter 1996; Kaplan et al. 1997). Mammalian Skp1 has been shown to bind to an F box in cyclin F, although the functional consequences of this interaction are unknown (Bai et al. 1996). Skp1 is also a component of several Skp1–Cdc53–F box (SCF) E3 ubiquitin ligase complexes, in which it functions as an adapter that bridges Cdc53 and F-box proteins like the WD-40 repeat proteins Cdc4p and Met30p and the leucine-rich repeat (LRR) protein Grr1 (Bai et al. 1996; Feldman et al. 1997; Skowyra et al. 1997; Patton et al. 1998a,b). Notably, the Cdc53-like mammalian Cul2 protein is associated with the VHL-Elongin BC complex, and Elongin BC appears to bridge Cul2 to VHL in a manner analogous to the SCF configuration (Pause et al. 1997; Lonergan et al. 1998). There is presently no evidence, however, that the VHL tumor-suppressor complex is an E3 ubiquitin ligase.
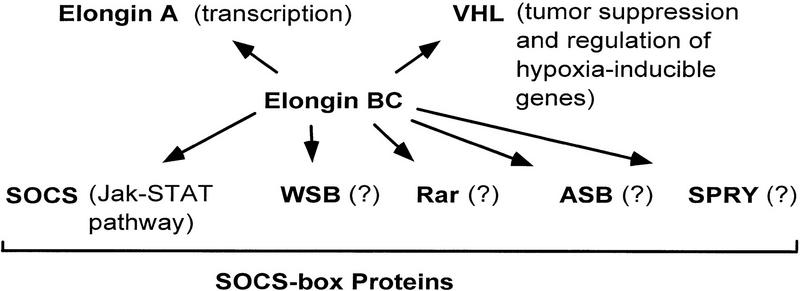
In summary, our findings are consistent with the model that Elongin BC is a multifunctional regulatory complex that is capable of controlling multiple pathways in the cell through its interaction with a short degenerate sequence motif found in many proteins, including Elongin A, VHL, and the SOCS-box family of proteins (Fig. 8). Elongins B and C and many BC-box proteins are highly conserved throughout evolution (Aso et al. 1996; Aso and Conrad 1997; Takagi et al. 1997; Nicholson and Hilton 1998). Thus, it is reasonable to anticipate that Elongin BC regulatory pathways are evolutionarily conserved. Given their high degree of conservation and their involvement in multiple, apparently distinct biochemical pathways, further analysis of the Elongin BC complex and BC-box proteins are likely to reveal new and important modes of regulation.
Figure 8.
Multiple functions for the Elongin BC complex.
Materials and methods
Expression of recombinant proteins in E. coli
Full-length mouse SOCS-1 was expressed in pRSET B (Invitrogen) with amino-terminal 6-histidine, T7, and Xpress tags. Full-length human SOCS-3 was expressed in pRSET B with amino-terminal 6-histidine, T7, and Xpress tags. Mouse Rar-1 (EST accession no. AA241036) containing sequences homologous to residues 9–244 of human RAR-1 was expressed in pRSET B with amino-terminal 6-histidine and Flag tags. Full-length mouse WSB-1 was expressed in pRSET B with amino-terminal 6-histidine and Flag tags. Mouse ASB-2 (EST accession no. AA104067) containing the carboxy-terminal 545 amino acids of the ORF was expressed in pRSET B with amino-terminal 6-histidine and Flag tags. Full-length rat Elongin B was expressed in pET 16b (Novagen) with an amino-terminal 6-histidine tag and in pRSET B with an amino-terminal 6-histidine tag and a carboxy-terminal HPC4 tag (Stearns et al. 1988). Full-length rat Elongin C was expressed in pET 16b with an amino-terminal 6-histidine tag. Proteins were purified from inclusion bodies by Ni2+-agarose affinity chromatography and renatured in various combinations as described (Aso et al. 1995).
Expression of recombinant proteins in insect cells
cDNAs encoding full-length mouse SOCS-1 containing amino-terminal 6-histidine, T7, and Xpress tags, mouse SOCS-1 mutants M1 and M2 containing amino-terminal 6-histidine, T7, and Xpress tags, full-length rat Elongin B containing an amino-terminal 6-histidine tag and a carboxy-terminal HPC4 tag, and full-length rat Elongin C containing amino-terminal 6-histidine and HSV tags were subcloned into pBacPAK 8, and recombinant baculoviruses were generated by the BacPAK baculovirus expression system (Clontech). Sf21 cells were cultured in Sf-900 II SFM with 5% fetal calf serum at 27°C and infected with appropriate recombinant baculoviruses. Sixty hours after infection, cells were collected and lysed by gentle vortexing in ice-cold buffer containing 40 mm HEPES–NaOH (pH 7.9), 150 mm NaCl, 1 mm DTT, 0.5% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin. Lysates were centrifuged at 10,000g for 20 min at 4°C. The supernatants were used for immunoprecipitations.
Expression of recombinant proteins in mammalian cells
cDNAs encoding full-length SOCS-1 and SOCS-1 mutants M1 and M2 containing amino-terminal 6-histidine, T7, and Xpress tags, full-length rat Elongin B containing a carboxy-terminal HPC4 tag, and full-length Elongin C containing an amino-terminal HSV tag were subcloned into the pCI–neo vector (Promega). The Jak2 Prk5 expression construct was a gift from J. Ihle (HHMI and St. Jude Children’s Hospital, Memphis, TN). Twenty-four hours after transfection of 293T cells, cells were collected and lysed in ice-cold buffer containing 40 mm HEPES–NaOH (pH 7.9), 150 mm NaCl, 1 mm DTT, 0.5% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 1 mm Na orthovanadate, 5 μg/ml leupeptin, 5 μg/ml anti-pain, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin. Lysates were centrifuged at 10,000g for 20 min at 4°C. To prepare Hep3B derivatives stably expressing SOCS-1 or the SOCS-1 M1 mutant, SOCS-1 wild type or M1 were subcloned into p1093 (Sompayrac and Danna 1994) under the control of a zinc-inducible metallothionein promoter. Stable transformants were selected in 500 μg/ml G418 in DMEM with 10% fetal calf serum and maintained.
Immunoprecipitations and Western blotting
Anti-T7 and anti-HSV antibodies were from Novagen. Anti-HPC4 monoclonal antibody (Stearns et al. 1988) was provided by C. Esmon (HHMI and Oklahoma Medical Research Foundation); anti-Elongin C monoclonal antibody was from Transduction Laboratories; and anti-VHL monoclonal antibody (Ig32) was from Pharmingen. Elongin A was detected by a monoclonal antibody (ELA-4) that was raised against rat Elongin A and recognizes an epitope between residues 481 and 520 of Elongin A. Anti-Jak2 antibody was from Santa Cruz Biotechnology, Inc. To measure binding of Elongins B and C to SOCS-box proteins from insect cells or E. coli, baculovirus-infected cell lysates or purified, renatured bacterially expressed proteins were incubated with antibody for 1 hr at 4°C and then with protein A/G agarose (Santa Cruz) for 1 hr. Protein A/G agarose was washed three times in buffer containing 40 mm HEPES–NaOH (pH 7.9), 500 mm NaCl, 1 mm DTT, and 0.5% (vol/vol) Triton X-100. To measure protein–protein interactions in mammalian cells, cell lysates were incubated with antibody for 1 hr at 4°C and then with protein A–Sepharose CL-4B for 1 hr or, in experiments with goat polyclonal antibodies, protein A/G agarose. Protein A or protein A/G beads were washed three times in buffer containing 40 mm HEPES–NaOH (pH 7.9), 150 mm NaCl, 1 mm DTT, and 0.5% (vol/vol) Triton X-100. Immunoprecipitated proteins were analyzed by electrophoresis through SDS–polyacrylamide gels. Proteins were visualized by silver or Coomassie Blue staining or transferred to nitrocellulose membranes (Micron Separations, Inc.) and visualized by Western blotting with the chemiluminescence reagent (NEN).
Acknowledgments
We are grateful to Winship Herr for helpful comments on the manuscript. We thank J. Bradsher and K. Smith for sharing results prior to publication, C. Esmon for anti-HPC4 monoclonal antibody, J. Ihle for the mammalian expression vector encoding Jak2, W. Liao for the STAT-dependent CAT reporter, and K. Jackson of the Molecular Biology Resource Center at the Oklahoma Center for Molecular Medicine for oligonucleotide synthesis. This work was supported in part by National Institutes of Health grant GM41628 and funds provided to the Oklahoma Medical Research Foundation by the H.A. and Mary K. Chapman Charitable Trust. J.W.C. and W.G.K. are investigators of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL conawayj@omrf.ouhsc.edu; FAX (405) 271-1580.
References
- Adams TE, Hansen JA, Starr R, Nicola NA, Hilton DJ, Billestrup N. Growth hormone preferentially induces the rapid, transient expression of SOCS-3, a novel inhibitor of cytokine receptor signaling. J Biol Chem. 1998;273:1285–1287. doi: 10.1074/jbc.273.3.1285. [DOI] [PubMed] [Google Scholar]
- Aso T, Conrad MN. Molecular cloning of DNAs encoding the regulatory subunits of elongin from Saccharomyces cerevisiae and Drosophila melanogaster. Biochem Biophys Res Commun. 1997;241:334–340. doi: 10.1006/bbrc.1997.7819. [DOI] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): A multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Aso T, Haque D, Barstead RJ, Conaway RC, Conaway JW. The inducible elongin A elongation activation domain: Structure, function, and interaction with the elongin BC complex. EMBO J. 1996;15:5557–5566. [PMC free article] [PubMed] [Google Scholar]
- Auerenhammer CJ, Chesnokova V, Bousqet C, Melmed S. Pituitary corticotroph SOCS-3: A novel intracellular regulation of leukemia-inhibitory factor-mediated proopiomelanocortin gene expression and adrenocorticotropin secretion. Mol Endocrinol. 1998;12:954–961. doi: 10.1210/mend.12.7.0140. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bjorbaek C, Elmquist JK, Frantz JD, Shoelson SE, Flier JS. Identification of SOCS-3 as a potential mediator of central leptin resistance. Mol Cell. 1998;1:619–625. doi: 10.1016/s1097-2765(00)80062-3. [DOI] [PubMed] [Google Scholar]
- Bradsher JN, Jackson KW, Conaway RC, Conaway JW. RNA polymerase II transcription factor SIII: I. Identification, purification, and properties. J Biol Chem. 1993;268:25587–25593. [PubMed] [Google Scholar]
- Conaway RC, Reines D, Garrett KP, Powell W, Conaway JW. Purification of RNA polymerase II general transcription factors from rat liver. Methods Enzymol. 1996;273:194–207. doi: 10.1016/s0076-6879(96)73020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan DR, Humphrey JS, Chen DYT, Weng Y, Sukegawa J, Lee S, Gnarra JR, Linehan WM, Klausner RD. Characterization of the VHL tumor suppressor gene product: Localization, complex formation, and the effect of natural inactivating mutations. Proc Natl Acad Sci. 1995a;92:6459–6463. doi: 10.1073/pnas.92.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan DR, Pause A, Burgess WH, Aso T, Chen DYT, Garrett KP, Conaway RC, Conaway JW, Linehan WM, Klausner RD. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science. 1995b;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- Endo T, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- Feldman RMR, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/Cullin catalyzes ubiqutination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Garrett KP, Tan S, Bradsher JN, Lane WS, Conaway JW, Conaway RC. Molecular cloning of an essential subunit of RNA polymerase II elongation factor SIII. Proc Natl Acad Sci. 1994;91:5237–5241. doi: 10.1073/pnas.91.12.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett KP, Aso T, Bradsher JN, Foundling SI, Lane WS, Conaway RC, Conaway JW. Positive regulation of general transcription SIII by a tailed ubiquitin homolog. Proc Natl Acad Sci. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra JR, Duan DR, Weng Y, Humphrey JS, Chen DY, Lee S, Pause A, Dudley CF, Latif F, Kuzmin I, Schmidt L, Duh FM, Stackhouse T, Chen F, Kishida T, Wei MH, Lerman MI, Zbar B, Klausner RD, Linehan WM. Molecular cloning of the von Hippel-Lindau tumor suppressor gene and its role in renal carcinoma. Biochim Biophys Acta. 1996a;1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Zhou S, Merrill MJ, Wagner JR, Krumm A, Papavassiliou E, Oldfield EH, Klausner RD, Linehan WM. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci. 1996b;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a carboxy-terminal SOCS box form five structural classes. Proc Natl Acad Sci. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos O, Levy AP, Jiang C, Kaelin WG, Goldberg MA. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Hyman AA, Sorger PK. Regulating the yeast kinetechore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- Ki O, Kajigaya S, Yamashita Y, Miyazoto A, Hatake K, Miura Y, Ikeda U, Shimada K, Ozawa K, Mano H. SOCS-1/JAB/SSI-1 can bind to and suppress Tec protein-tyrosine kinase. J Biol Chem. 1997;272:27178–27182. doi: 10.1074/jbc.272.43.27178. [DOI] [PubMed] [Google Scholar]
- Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- Kishida T, Stackhouse TM, Chen F, Lerman MI, Zbar B. Cellular proteins that bind the von Hippel-Lindau disease gene product: Mapping of binding domains and the effect of missense mutations. Cancer Res. 1995;20:4544–4548. [PubMed] [Google Scholar]
- Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L, Schmidt L, Zhou F, Li H, Wei MH, Chen F, Glenn G, Choyke P, Walther MM, Weng Y, Duan DR, Dean M, Glavac K, Richards FM, Crossey PA, Ferguson-Smith MA, Le Paslier D, Chumakov I, Cohen D, Chinault AC, Maher ER, Linehan WM, Zbar B, Lerman MI. Identification of the von Hippel-Lindau Disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- Lonergan KM, Iliopoulos O, Ohh M, Kamura T, Conaway RC, Conaway JW, Kaelin WG. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing Elongins B/C and Cul2. Mol Cell Biol. 1998;18:732–741. doi: 10.1128/mcb.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuhara M, Sakamoto H, Matsumoto A, Suzuki R, Yasukawa H, Mitsui K, Wakioka T, Tanimura S, Sasaki A, Misawa H, Yokouchi M, Ohtsubo M, Yoshimura A. Cloning and characterization of novel CIS family genes. Biochem Biophys Res Commun. 1997;239:439–446. doi: 10.1006/bbrc.1997.7484. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- Minamoto S, Ikegami K, Ueno K, Narazaki M, Naka T, Yamamoto H, Matsumoto T, Saito H, Hosoe S, Kishimoto T. Cloning and functional analysis of new members of STAT induced STAT inhibitor (SSI) family: SSI-2 and SSI-3. Biochem Biophys Res Commun. 1997;237:79–83. doi: 10.1006/bbrc.1997.7080. [DOI] [PubMed] [Google Scholar]
- Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, Akira S, Kishimoto T. Structure and function of a new STAT-induced inhibitor. Nature. 1997;387:924–928. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- Narazaki M, Fujimoto M, Matsumoto T, Morita Y, Saito H, Kajita T, Yoshizaki K, Naka T, Kishimoto T. Three distinct domains of SSI-1 / SOCS-1 / JAB protein are required for its suppression of interleukin 6 signaling. Proc Natl Acad Sci. 1998;95:13130–13134. doi: 10.1073/pnas.95.22.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson SE, Hilton DJ. The SOCS proteins: A new family of negative regulators of signal transduction. J Leukoc Biol. 1998;63:665–668. doi: 10.1002/jlb.63.6.665. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complex that regulate cell division and methionine biosynthesis in yeast. Genes & Dev. 1998a;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Biochem Sci. 1998b;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Pause A, Lee S, Worrell RA, Chen DYT, Burgess WH, Linehan WM, Klausner RD. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Sompayrac L, Danna KJ. An amino-terminal fragment of SV40 antigen induces cellular DNA synthesis in quiescent rat cells. Virology. 1994;200:849–853. doi: 10.1006/viro.1994.1255. [DOI] [PubMed] [Google Scholar]
- Starr R, Willson TA, Viney EM, Murray LJL, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- Stearns DJ, Kurosawa S, Sims PJ, Esmon NL, Esmon CT. The interaction of a Ca2+-dependent monoclonal antibody with the protein C activation peptide region. Evidence for obligatory Ca2+ binding to both antigen and antibody. J Biol Chem. 1988;263:826–832. [PubMed] [Google Scholar]
- Takagi Y, Pause A, Conaway RC, Conaway JW. Identification of Elongin C sequences required for interaction with the Von-Hippel Lindau tumor suppressor protein. J Biol Chem. 1997;272:27444–27449. doi: 10.1074/jbc.272.43.27444. [DOI] [PubMed] [Google Scholar]
- Yoshimura A, Ohkubo T, Kiguchi T, Jenkins NA, Gilbert DJ, Copeland NG, Hara T, Miyajima A. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK S-phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]