Abstract
Saccharomyces cerevisiae carries ∼150 copies of rDNA in tandem repeats. It was found that the absence of an essential subunit of RNA polymerase I (Pol I) in rpa135 deletion mutants triggers a gradual decrease in rDNA repeat number to about one-half the normal level. Reintroduction of the missing RPA135 gene induced a gradual increase in repeat number back to the normal level. Gene FOB1 was shown to be essential for both the decrease and increase of rDNA repeats. FOB1 was shown previously to be required for replication fork blocking (RFB) activity at RFB site in rDNA and for recombination hot-spot (HOT1) activity. Thus, DNA replication fork blockage appears to stimulate recombination and play an essential role in rDNA expansion/contraction and sequence homogenization, and possibly, in the instability of repeated sequences in general. RNA Pol I, on the other hand, appears to control repeat numbers, perhaps by stabilizing rDNA with the normal repeat numbers as a stable nucleolar structure.
Keywords: Replication fork blocking, RNA polymerase I, gene amplification, recombination hot spot, ribosomal DNA, sequence homogenization
In most eukaryotic organisms, the genes for rRNAs (rDNA) are clustered in long tandem repeats on one or a few chromosomes. The total number of these chromosomal rDNA repeats appears to be maintained at a level appropriate for each organism. However, under certain conditions, alterations of rDNA copy numbers have been observed. Although an increase as extrachromosomal copies was found as a specific developmental event during Xenopus oogenesis (Brown and Dawid 1968; Gall 1968), changes in the number of tandem chromosomal repeats (Ritossa 1968; Russel and Rodland 1986) appear to represent a more general feature of rDNA repeats. For example, variations in rDNA copy number were observed in yeast quite often (see, e.g., Rustchenko et al. 1993), although factors responsible for the variations were not well defined. Other studies focusing on recombination in yeast rDNA were carried out with genetic markers inserted into yeast rDNA repeats and have provided useful information on possible mechanisms involved in sequence homogenization and expansion/contraction of rDNA repeats (Petes 1980; Szostak and Wu 1980; Gangloff et al. 1996). However, factors determining optimal numbers of rDNA repeats as well as those inducing expansion or contraction of rDNA have remained unexplored. In Drosophila, certain bobbed mutations on the X-chromosome are caused by a partial deficiency of rDNA repeats. These mutations have been studied extensively with respect to conditions causing reversion to wild type, which is accompanied by an rDNA copy number increase (called magnification), as well as possible mechanisms involved (Ritossa 1968; Tartof 1974; Hawley and Marcus 1989). However, these studies were done mostly by use of elaborate formal genetics of the bobbed locus, and the system has not been suitable for more direct molecular analyses of the mechanisms involved in rDNA expansion/contraction. In this paper, we describe a yeast system in which expansion and contraction of rDNA repeats can be directly studied, and report some initial results obtained with this system.
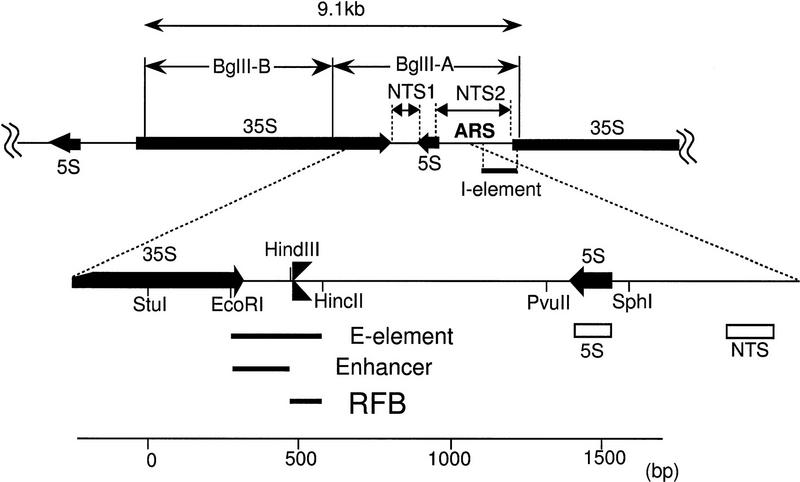
The yeast Saccharomyces cerevisiae carries 100–200 copies of the rDNA unit that are tandemly repeated on chromosome XII (Petes 1979). A single unit consists of two transcribed regions, 35S precursor rRNA and 5S rRNA coding regions, and two nontranscribed regions, NTS1 and NTS2 (Fig. 1). The DNA encoding 35S and 5S rRNA genes is transcribed by RNA polymerases I (Pol I) and III, respectively. The origin of replication (ARS) and the replication fork barrier (RFB) sites are located in NTS2 and NTS1, respectively (Skryabin et al. 1984; Brewer and Fangman 1988; Linskens and Huberman 1988). The RFB site contains a specific nucleotide sequence of ∼100 bp that allows progression of the replication fork in the direction of 35S rRNA transcription, but not in the opposite direction (Brewer et al. 1992; Kobayashi et al. 1992). Two sites analogous to the yeast ARS and the RFB site have been found in the corresponding rDNA regions in several other eukaryotes (Little et al. 1993; Bastia and Mohanty 1996; Gencheva et al. 1996; Gogel et al. 1996; Lopez-Estrano et al. 1998). The RFB site overlaps the E element of HOT1 (Keil and Roeder 1984; Voelkel-Meiman et al. 1987). HOT1 is a DNA element that stimulates mitotic intra- and interchromosomal recombination at nearby regions when inserted at a non-rDNA location (Keil and Roeder 1984). HOT1 consists of two elements: the E element, which overlaps the enhancer for Pol I transcription (Elion and Warner 1984), and the I element, which corresponds to the promoter region for Pol I transcription (Fig. 1). The requirement of these two elements for HOT1 activity suggests that Pol I transcription activity and the stimulation of recombination (HOT1 activity) are causally related, and this suggestion has been supported by the demonstration of a requirement of Pol I for HOT1 activity (Huang and Keil 1995). However, no experimental evidence has been reported to indicate the actual operation of the HOT1 system at the normal rDNA locus.
Figure 1.
Structure of rDNA repeats in S. cerevisiae. Locations of the 35S and 5S rRNA genes (the direction of transcription indicated by arrows), the two nontranscribed spacer regions (NTS1 and NTS2), ARS (replication origin), and the HOT1 I element are shown at top. BglII-A and B DNA fragments are also shown. NTS1 and its surrounding regions are expanded. Three solid bars represent the HOT1 E element, Pol I enhancer, and RFB site (also indicated by  ). Two open rectangles, indicated as 5S and NTS, are DNA regions used for the competitive PCR assay.
). Two open rectangles, indicated as 5S and NTS, are DNA regions used for the competitive PCR assay.
Because of the location of the RFB site near the rDNA transcription termination site and its directionality in function, it has been suggested previously that the RFB site might have evolved to prevent collision between the transcription and replication machineries (Brewer et al. 1992). However, transcription of rDNA itself does not appear to inhibit progression of the DNA fork coming from the opposite direction (Brewer et al. 1992) and the physiological significance of the replication fork block has remained unknown. In Escherichia coli, termination (Ter) sequences similar to RFB sites have been identified. These sequences, together with the Tus protein that binds to the Ter sequences, appear to function to ensure that two replication forks that have initiated bidirectionally from the origin of replication can enter the termination zone of the chromosome but cannot escape it (for review, see Hill 1996). It was shown previously that recombinational hotspots exist near E. coli Ter sites, and that RFB activity at the Ter sites may be causally related to hyper-recombination activity (Horiuchi et al. 1994; Horiuchi and Fujimura 1995). On the basis of the analogy to the E. coli Ter system, we considered the possibility that the RFB function in yeast rDNA might be related to recombinational events in rDNA. To explore this possibility, we previously screened mutants defective in the HOT1 function for simultaneous loss of the RFB function. In this way, we identified the gene FOB1 (fork blocking) whose mutation causes a simultaneous loss of both HOT1 and RFB functions (Kobayashi and Horiuchi 1996). Identification of such a gene strongly supports the suggestion that RFB and HOT1 functions are in fact related. We have now demonstrated that the FOB1 gene is essential for both expansion and contraction of yeast rDNA repeats, and propose a model we call the fork block-dependent recombination model. Additionally, we have found that Pol I plays an important role in determining rDNA repeat number.
Results
Reduction of rDNA copy number in an rpa135 mutant
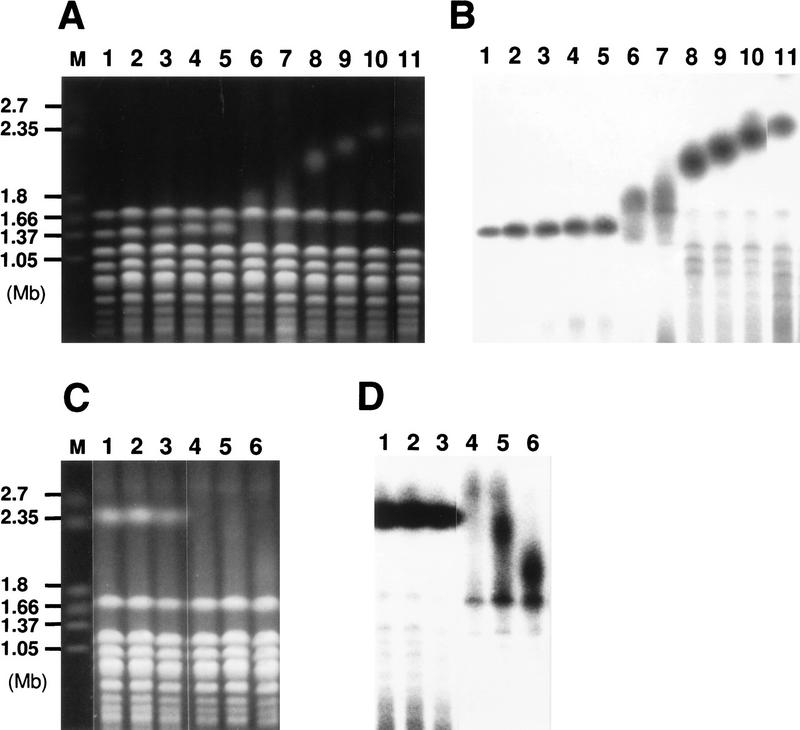
RPA135 is an essential gene encoding the second largest subunit (A135) of yeast Pol I. Strain NOY408-1a has a large deletion in this gene (rpa135Δ::LEU2), but is able to grow on galactose because it carries a multicopy plasmid (pNOY102; helper plasmid) containing the 35S rRNA coding region fused to a strong Pol II promoter, the GAL7 promoter (Nogi et al. 1991). A reduction in rDNA repeats in this strain was first noted by Brewer and coworkers (Brewer et al. 1992). We confirmed this finding. DNA was digested with BglII and subjected to electrophoresis followed by Southern analysis with an rDNA probe specific for chromosomal rDNA repeats (Fig. 1). A single-copy gene, MCM2, was used as an internal control for normalization. As shown in Figure 2, the rDNA copy number in NOY408-1a (lane 1) was significantly reduced relative to that in the parental diploid strain, NOY408, carrying a copy of RPA135 in addition to a copy of rpa135Δ::LEU2 (lane 15) or a sister RPA135 strain, NOY408-1b (lane 16). Quantification showed that copy number was reduced to approximately one-half of that in the control strains (see below).
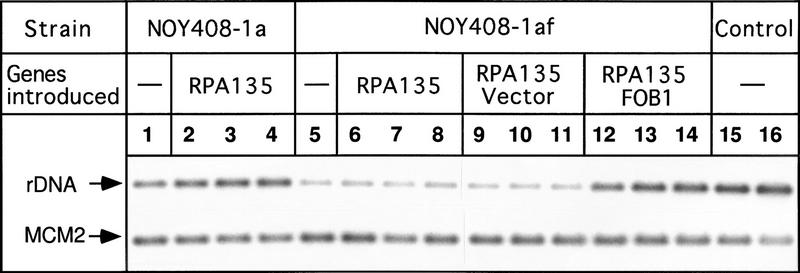
Figure 2.
Southern hybridization analysis of rDNA copy numbers. The RPA135 gene was introduced by transformation into strains NOY408-1a (rpa135), NOY408-1af (rpa135 fob1), and NOY408-1af derivatives carrying FOB1 on a plasmid or control vector plasmid as indicated. DNA samples were prepared at 44 (lanes 2,6,9,12), 80 (lanes 3,7,10,13), and 116 generations (lanes 4,8,11,14) after introduction of RPA135. NOY408-1a, and NOY408-1af, which did not receive RPA135 (lanes 1,5, respectively), were also analyzed together with control strains NOY408 (lane 15) and NOY408-1b (lane 16). In addition to rDNA, a single copy gene, MCM2, was analyzed as a reference.
To determine absolute copy numbers of rDNA in these strains, a competitive PCR assay (Diviacco et al. 1992) was carried out. This assay allows quantification of a target DNA sequence by coamplifying it in the presence of known amounts of a competitor DNA that shares the same primer recognition sites. We selected two rDNA specific sequences, the 5S rRNA gene and a region in NTS2 (open boxes in Fig. 1), and two reference single-copy genes, MCM2 and PPR1. DNA was prepared from NOY408-1a (and NOY408-1af, see below) and the two control strains, NOY408 and NOY408-1b. Known amounts of competitor DNA were added to each of the target DNA samples to be analyzed and PCR reactions were carried out with appropriate primers. Absolute copy numbers of rDNA were calculated by dividing the averages of the values for the two specific rDNA sequences (5S and NTS) by the averages of the values for the two reference single-copy genes (MCM2 and PPR1). Numbers of rDNA copies calculated in this way are shown in Figure 3. The copy number of rDNA in NOY408-1a (rpa135) was ∼80 copies per haploid genome. This value was approximately one-half of those in the control strains NOY408 and NOY408-1b (∼150 copies per haploid genome), confirming the results of Southern blot analysis.
Figure 3.
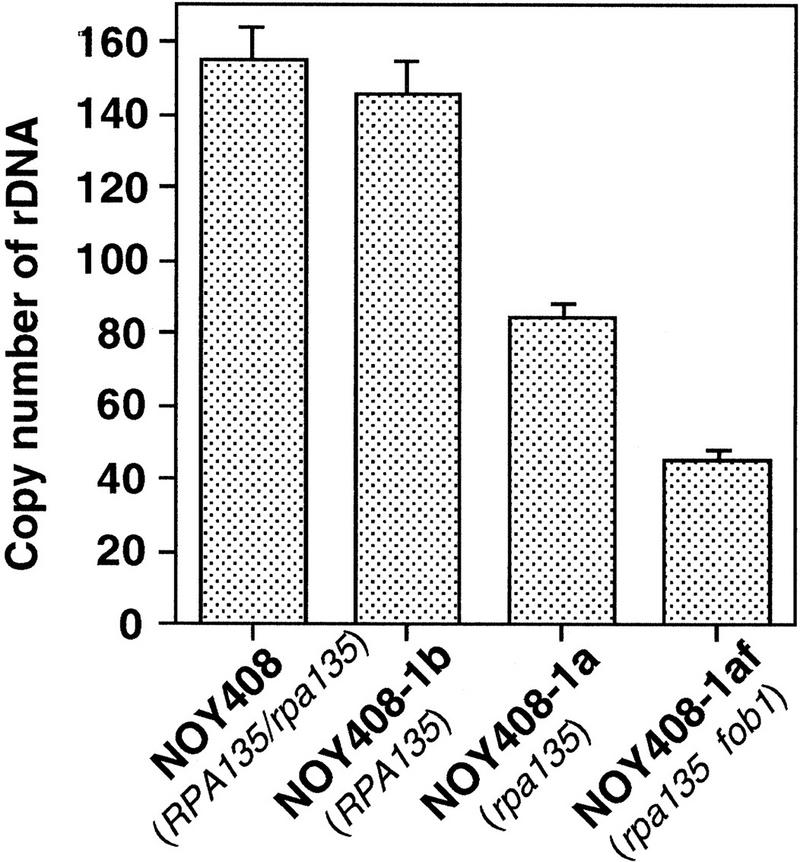
Absolute rDNA copy numbers determined by a competitive PCR assay. Values given are rDNA copy numbers per haploid genome determined as described in Materials and Methods and text.
We repeated tetrad dissection of the parent diploid, NOY408, and followed the kinetics of the decrease in rDNA copy number in freshly germinated rpa135 haploid segregants. We found that the reduction in copy number was slow and required many generations (see below), but the individual rDNA copy numbers of all the rpa135 segregants eventually reached a value similar to that for the original strain, NOY408-1a (data not shown). We conclude that in the absence of Pol I, rDNA copy number decreases to approximately one-half of the normal value in this strain background.
Restoration of rDNA copy number upon introduction of the RPA135 gene into the rpa135 mutant
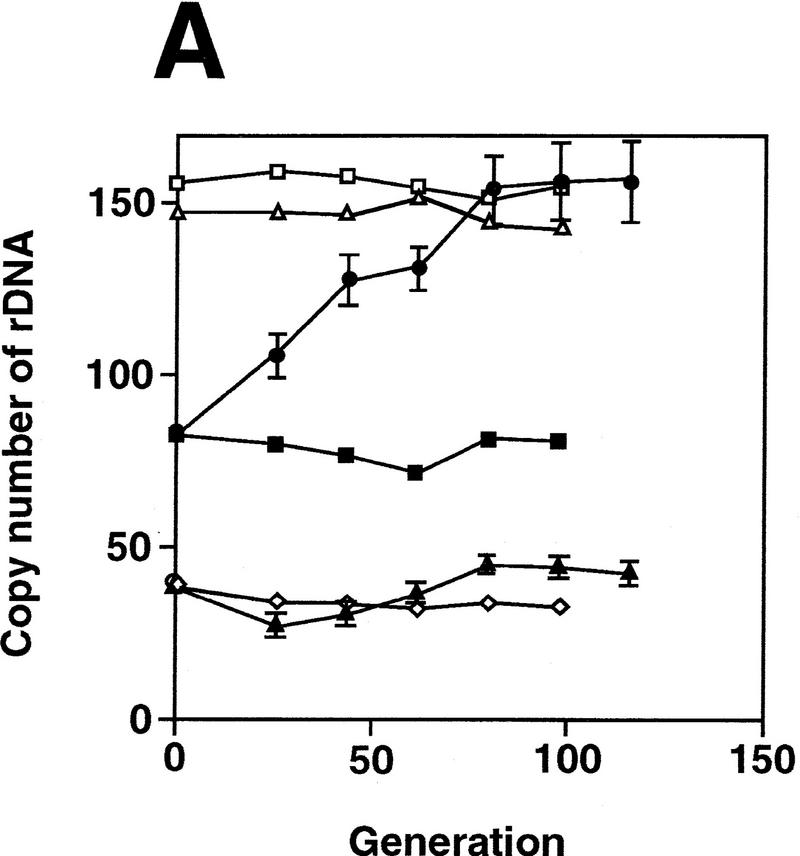
To examine whether the reduction in rDNA copy number in the rpa135 mutant is reversible, we introduced the wild-type RPA135 gene into the rpa135 mutant strain NOY408-1a. The mutant cells were transformed with a CEN plasmid carrying RPA135 (pNOY117). Four independent transformants were isolated and their rDNA copy numbers were determined at times corresponding to 44, 80, and 116 generations in glucose medium after the introduction of RPA135 (see Materials and Methods). Typical Southern hybridization data for one of the transformants are shown in Figure 2 (lanes 2–4 , DNAs obtained from cells at 44, 80, and 116 generations, respectively). Ratios of rDNA to MCM2 were quantified and the absolute rDNA copy numbers were calculated by comparing these ratios (rDNA/MCM2) with the corresponding ratio of a reference strain, NOY408-1b (Fig. 2, lane 16), which has 150 copies of rDNA as described above. The averages of the values obtained for the four independent transformants were calculated and the results are shown in Figure 4A (•). Copy numbers of rDNA in the RPA135 transformants increased gradually during subculture, reaching a value similar to those in control strains after 80 generations, and remained at this value thereafter. In parallel, we subjected the rpa135 mutant strain, NOY408-1a, to the same treatment and measured rDNA copy numbers (Fig. 4A, █). There was no significant change in the copy number over 100 generations in NOY408-1a. (It should be noted that, although rRNA is synthesized from the GAL7–35S rDNA fusion on the helper plasmid by Pol II, 5S rRNA is synthesized from the rDNA repeats on the chromosome by Pol III. Therefore, we thought that the absence of a further decrease in rDNA repeat number in the rpa135 strain might be due to selection against cells with a deficiency in 5S rRNA synthesis. However, replacing the helper plasmid with a similar plasmid carrying an additional 5S rRNA gene and continuing subculture did not lead to any further reduction of rDNA repeats.) These experiments demonstrate that yeast cells have the ability to increase rDNA copy number, and that the absence of intact Pol I in NOY408-1a apparently prevents this increase in rDNA copy number.
Figure 4.
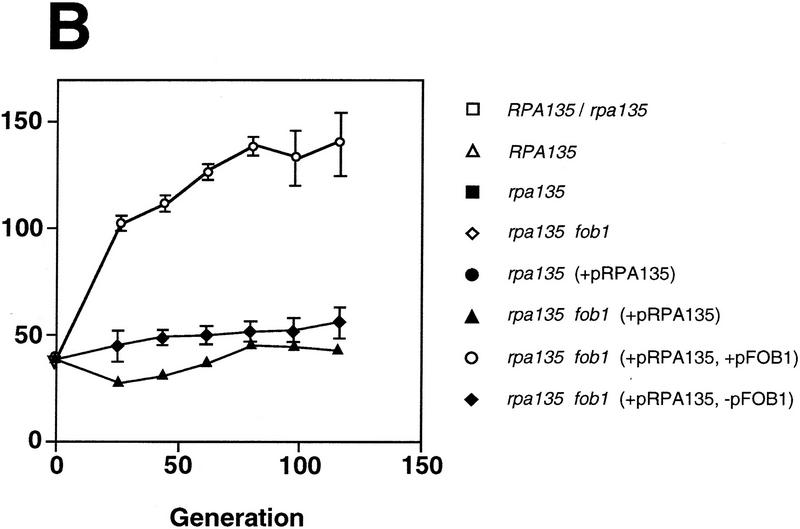
Pol I- and FOB1-dependent increases of rDNA copy numbers. Experiments similar to the ones shown in Fig. 2 were carried out. (A) NOY408-1a (rpa135) and (B) NOY408-1af (rpa135 fob1), as well as their derivatives with (+YEp–FOB1) and without (+YEplac195) the FOB1 gene were analyzed (YEp–FOB1 and YEplac195 are indicated as +pFOB1 and −pFOB1, respectively). Strains that contained RPA135 on plasmid pNOY117 are indicated by the strains’ designation, followed by +pRPA135.NOY408 (RPA135/rpa135) and NOY408-1b (RPA135) were also analyzed. Copy numbers of rDNA relative to the single copy gene MCM2 were analyzed at various generations after introduction of RPA135. Four independent transformants were analyzed. Values shown are the averages obtained for the four cultures (standard deviations are shown). Control cultures, which did not receive RPA135, were also analyzed in parallel (standard deviations are not shown).
Absence of rDNA copy number increase in a fob1 derivative of the rpa135 mutant
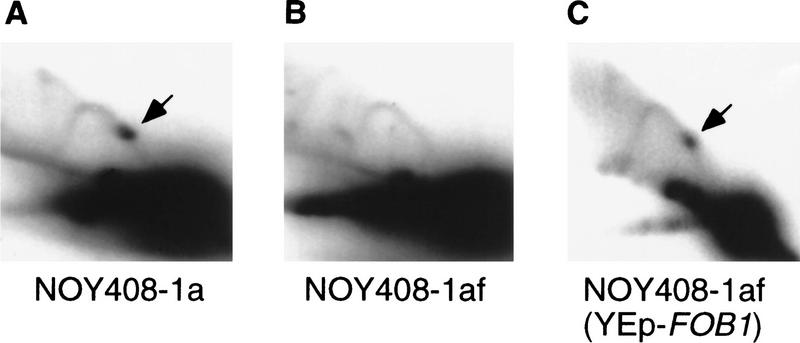
It was shown previously that RFB activity at the RFB site takes place in the absence of rDNA transcription by Pol I (Brewer et al. 1992; Kobayashi et al. 1992). This was confirmed in the experiment shown in Figure 5, which analyzed restriction-enzyme-digested DNA by two-dimensional agarose gel electrophoresis. Accumulation of chromosomal rDNA with replication fork arrest at the RFB site in the rpa135 mutant (NOY408-1a) was indicated by a dense spot (Fig. 5A). We constructed a fob1 derivative of NOY408-1a (NOY408-1af; rpa135Δ::LEU2 fob1Δ::HIS3) by disrupting the FOB1 gene with HIS3. This strain did not show the pertinent spot (Fig. 5B), indicating the absence of the RFB activity. Introduction of the FOB1 gene on a multicopy plasmid (YEp–FOB1) into this fob1 mutant derivative restored the RFB activity (Fig. 5C). These results show that the replication fork block seen under conditions of no rDNA transcription is still FOB1 dependent.
Figure 5.
RFB activity analyzed by two-dimensional gel analysis. DNA was prepared from strains indicated, digested with BglII and SphI and subjected to two-dimensional agarose gel electrophoresis followed by Southern hybridization with a rDNA probe (the HindIII–SphI fragment; see Fig. 1). Spots indicated by arrows show accumulation of Y-form DNA molecules at RFB site.
We determined rDNA copy number in the rpa135 fob1 strain (NOY408-1af) with and without introduction of the RPA135 gene. As shown in Figure 2, the copy number of rDNA in NOY408-1af (lane 5) was even smaller than that in the parental strain NOY408-1a (lane 1; discussed below). This conclusion was also confirmed by the competitive PCR assay described above (Fig. 3). Introduction of the RPA135 gene into NOY408-1af did not lead to any increase in the rDNA copy number during subcultures extending to 116 generations (Fig. 2, lanes 6–8). This result was in clear contrast to the increase seen for the rpa135 FOB1 strain (lanes 2–4). The NOY408-1af strain that contained the missing FOB1 gene on a plasmid regained the ability to increase rDNA copy number on introduction of RPA135 (lanes 12–14). The increase was not observed in a control NOY408-1af strain that contained the plasmid vector without FOB1 (lanes 9–11).
Quantitative data obtained in the above experiments are shown in Figure 4. The copy numbers of rDNA (∼40 copies) in NOY408-1af did not show significant change over 100 generations regardless of whether it carried RPA135 (Fig. 4A,▴) or not (Fig. 4A,⋄). As in the case of NOY408-1a (pNOY117), NOY408-1af (pNOY117, YEp–FOB1) showed a gradual increase in rDNA copy number (Fig. 4B,○), reaching a value similar to the one in control strains after 80 generations. Thus, the rate of increase was approximately one copy per generation in both cases. These results demonstrate that rDNA copy number increase in this system requires not only intact Pol I but also the functional FOB1 gene.
Association of the increased rDNA with chromosome XII
The rDNA repeat is present on chromosome XII. From the sequence information, the size of the sum of non-rDNA regions of this chromosome is ∼1100 kb (Goffeau et al. 1996). According to the present copy number determination (∼150 copies of the 9.1-kb rDNA unit), the total size of the rDNA repeat in the control strains is ∼1400 kb and the size of chromosome XII is ∼2500 kb or 2.5 Mb. Thus, significant changes in the number of repeats should be detectable by analyzing the size of chromosome XII by use of pulse-field gel electrophoresis. Yeast cells used for the Southern hybridization experiments shown in Figure 2 were gently lysed in agarose gel blocks and subjected to pulse-field gel electrophoresis. After the electrophoresis, gels were stained with ethidium bromide (Fig. 6A) followed by hybridization with an rDNA-specific probe and autoradiography (Fig. 6B). It is known that the electrophoretic behavior of chromosome XII is anomalous in such pulse-field gel electrophoresis (Olson 1991). One of the anomalies is that chromosome XII forms a diffuse band presumably due to copy number heterogeneity of the rDNA repeats in the yeast cell population. The broad band of chromosome XII in the control strain NOY408 corresponded to an average size of ∼2.5 Mb (Fig. 6A,B, lane 11) which is consistent with the calculated value for 150 rDNA repeats as mentioned above. Chromosome XII in the strain NOY408-1a showed a heterogeneous size distribution ranging from ∼1.2 to 2.0 Mb (Fig. 6A,B, lanes 6 and 7). This size distribution corresponds, by formal calculation, to chromosome sizes with a very small rDNA copy number to ∼100 rDNA copies (2000 − 1100/9.1 = 99). Calculation of rDNA copy number estimated in this way must undoubtedly involve large errors. Nevertheless, it is clear that the size of rDNA repeats in NOY408-1a is probably very heterogeneous and the value (∼80 copies per genome) obtained by the Southern hybridizaton/competitive PCR assay is an average value for the popultion of NOY408-1a cells. Unexpectedly, strain NOY408-1af, which is a fob1 derivative of NOY408-1a, showed chromosome XII as a clearly defined band (Fig. 6A,B, lanes 1,2), indicating a more homogeneous size distribution in the fob1 cell population (see Discussion).
Figure 6.
Analysis of the size of chromosome XII by pulse-field gel electrophoresis. (A,B). Experiments similar to those in Figs. 2 and 4 were carried out. The RPA135 gene was introduced into NOY408-1a (lanes 8–10) and NOY408-1af (lanes 3–5). Control cultures without RPA135 introduction, NOY408-1a (lanes 6,7) and NOY408-1af (lanes 1,2) were also grown in parallel. Samples were taken at 44 (lanes 1,3,6,8), 80 (lanes 4,9), and 116 generations (lanes 2,5,7,10) after introduction of RPA135. Wild-type control culture, NOY408, was also analyzed (lane 11). (C,D). Haploid strains NOY408-2af (rpa135 fob1) (lanes 1–3) and NOY408-2a (rpa135) (lanes 4–6) were freshly constructed by tetrad dissection of diploid strains NOY408-f and NOY408, respectively. Samples were taken at 44 (lanes 1,4), 80 (lanes 2,5), and 116 generations (lanes 3,6) after germination of spores, and the size of chromosome XII was analyzed by pulse-field gel electrophoresis. (A,C) Chromosome patterns revealed by staining with ethidium bromide. (B,D) Autoradiographs obtained after hybridization with an rDNA probe. Size markers (lane M) are Hansenula wingei chromosomes (Bio-Rad, Hercules, CA). The background cross hybridization of the rDNA probe to the chromosomes without rDNA is not uniform in panels B and D. The reason is not clear, but its pattern was not reproducible. It should also be noted that heterogeneous chromosome XII (in B, lanes 6 and 7, and D lanes 4–6) wih specific hybridization overlapped chromosome IV with nonspecific hybridization, resulting in an artifactual strong band of ∼1.7 Mb in B, lane 7, and D, lanes 4–6.
As mentioned above, NOY408-1a that received RPA135 (as plasmid pNOY117) increased rDNA copy number gradually during subsequent subcultures. The size of chromosome XII increased as shown in Figure 6, A and B, lanes 8–10, which correspond to samples taken at 44, 80, and 116 generations after introduction of RPA135. The size of chromosome XII in the sample at 116 generations (lane 10) was ∼2.4 Mb and was close to that of the control strain NOY408 (lane 11). In contrast to NOY408-1a, introduction of RPA135 into NOY408-1af, which is fob1, did not cause an increase in the size of chromosome XII (Fig. 6A,B, lanes 3–5), as expected from the results of analysis of rDNA copy number by Southern hybridization. Similarly, restoration of the ability to increase rDNA copy number by introducing the FOB1 gene (as YEp–FOB1) into this strain, NOY408-1af (pNOY117), was confirmed by gradual increases in the size of chromosome XII (observed during subcultures of 44, 80, and 116 generations, data not shown). A control strain, which received the vector plasmid without FOB1, did not show such an increase and maintained chromosome XII at the original size of ∼1.5 Mb during subcultures extending to 116 generations (data not shown). These experiments demonstrate that the increase in rDNA copy number observed on introduction of the RPA135 gene into FOB1 strains represents an expansion of tandem rDNA repeats, similar to Drosophila rDNA magnification, rather than amplification as an extrachromosomal plasmid.
Requirement of FOB1 for contraction of rDNA repeats in rpa135 deletion strains
The experiments described above established that FOB1 is required for expansion of rDNA repeats. We then examined the question of whether the FOB1 gene is required for the contraction of rDNA repeats observed in strain NOY408-1a. NOY408-1a was constructed previously by tetrad dissection of the parent diploid strain, NOY408 (RPA135/rpa135, FOB1/FOB1, pNOY102). We first constructed a derivative of NOY408, NOY408-f (RPA135/rpa135, fob1/fob1, pNOY102), by disrupting both copies of FOB1 carried by NOY408. NOY408-f was sporulated, tetrads were dissected, and haploid segregants growing on galactose were analyzed. The size of chromosome XII in haploid segregants carrying rpa135 and fob1 (and pNOY102) was analyzed at 44, 80, and 116 generations after germination of spores. In parallel, NOY408 was sporulated, tetrads were dissected, and a haploid segregant (NOY408-2a) with the same genotype as NOY408-1a (rpa135 FOB1, pNOY102) was similarly analyzed by pulse-field gel electrophoresis. The results are shown in Figure 6, C and D. The size of chromosome XII in NOY 408-2a decreased gradually during subcultures and a heterogeneous size distribution was also evident (lanes 4–6). In contrast, an isogenic fob1 haploid segregant, NOY408-2af, showed no decrease in the size of chromosome XII, and maintained a homogeneous size of ∼2.4 Mb (Fig. 6C,D, lanes 1–3). Thus, we conclude that FOB1 is required not only for expansion, but also for contraction of rDNA repeats.
Discussion
We have shown that the absence of an essential subunit of Pol I (or Pol I transcription of rDNA) triggers a gradual decrease in the number of tandem rDNA repeats and reintroduction of the missing Pol I gene induces a gradual increase of tandem repeats. Using this system, we have demonstrated that the gene FOB1 is required for both the decrease and the increase of rDNA repeats. In contrast, Pol I is required to maintain rDNA repeats at the normal level of ∼150 per cell. Once the RPA135 gene is deleted, rDNA levels decrease but can be restored by introduction of the RPA135 gene (Fig. 7A).
Figure 7.
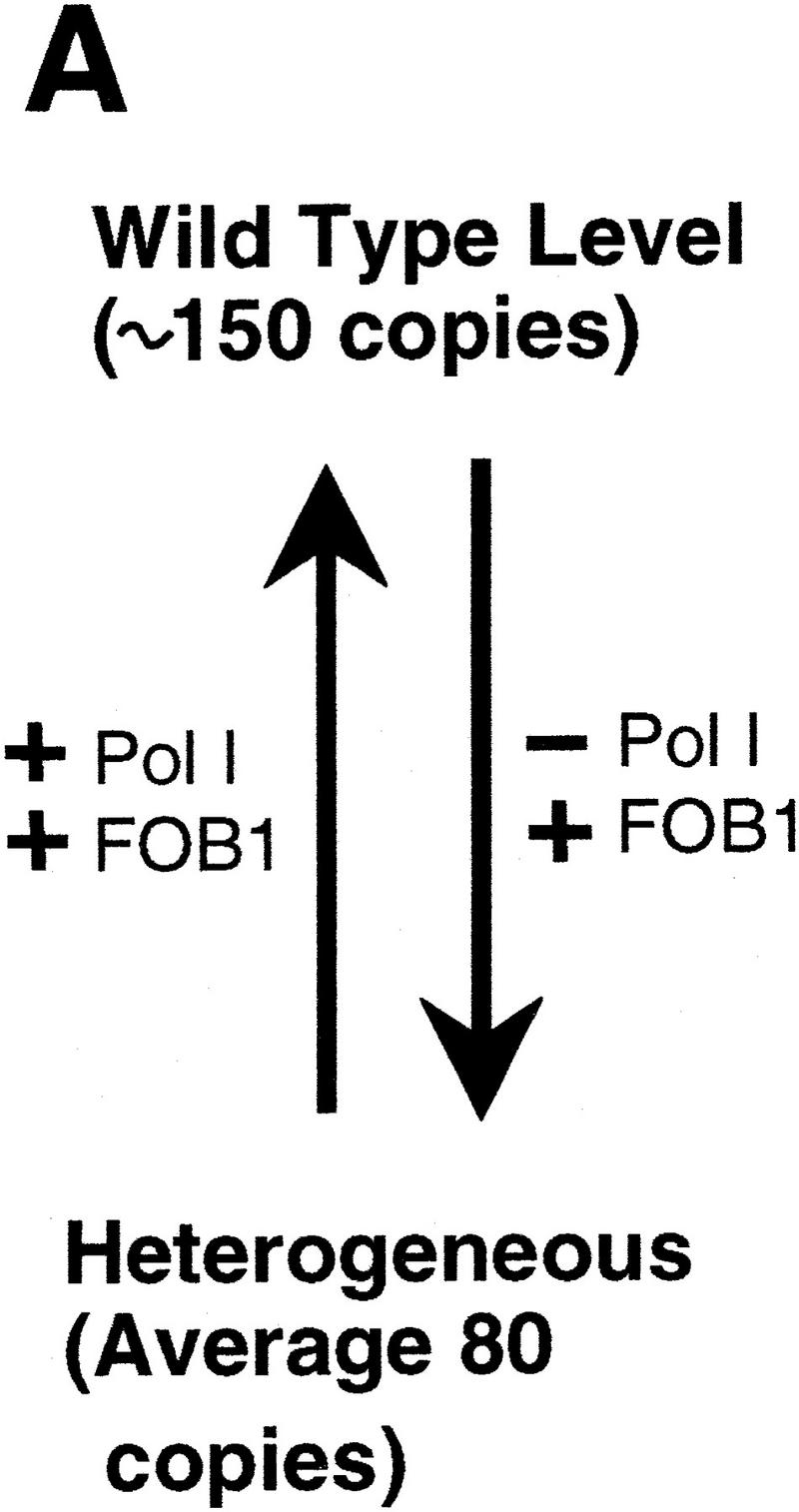
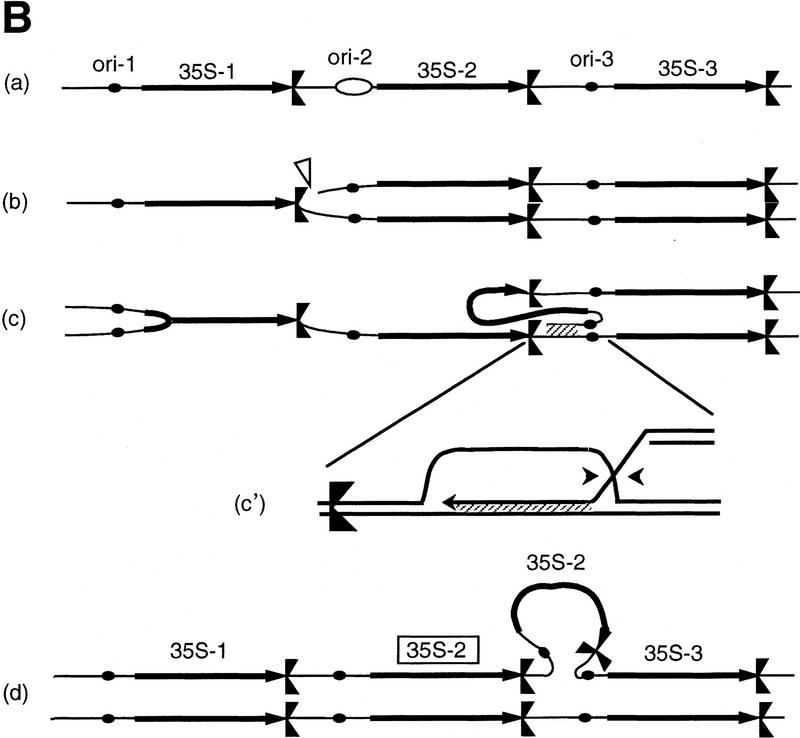
(A) Summary of the rDNA expansion/contraction system studied, and (B) fork block-dependent recombination model for rDNA expansion/contraction. The position of ARS and RFB are shown as filled oval and  , respectively. Individual lines represent chromatids with double-stranded DNA except for c′, where individual single-stranded DNAs are displayed to show detail of the structure formed after strand invasion. In this model, DNA replication starts from one of the ARS (ori-2) bidirectionally (a). In the yeast rDNA repeats, about one in three ARS sites is used as an active origin (Brewer and Fangman 1988; Linskens and Huberman 1988). A leftward replication fork is arrested at the RFB site and an exposed single-stranded region is cleaved by a nuclease (indicated by an open arrowhead in b). A recombination enzyme activates the resulting DSB end, and a strand invasion at a homologous duplex (a downstream sister chromatid near ori-3 in this example) takes place (c and c‘). A new replication fork is formed as a result of resolution of the Holliday junction (arrowheads). The new replication fork meets with the rightward replication fork from upstream, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as boxed 35S-2 (d).
, respectively. Individual lines represent chromatids with double-stranded DNA except for c′, where individual single-stranded DNAs are displayed to show detail of the structure formed after strand invasion. In this model, DNA replication starts from one of the ARS (ori-2) bidirectionally (a). In the yeast rDNA repeats, about one in three ARS sites is used as an active origin (Brewer and Fangman 1988; Linskens and Huberman 1988). A leftward replication fork is arrested at the RFB site and an exposed single-stranded region is cleaved by a nuclease (indicated by an open arrowhead in b). A recombination enzyme activates the resulting DSB end, and a strand invasion at a homologous duplex (a downstream sister chromatid near ori-3 in this example) takes place (c and c‘). A new replication fork is formed as a result of resolution of the Holliday junction (arrowheads). The new replication fork meets with the rightward replication fork from upstream, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as boxed 35S-2 (d).
It was shown previously that blockage of the replication fork at the Ter site in E. coli stimulates homologous recombination at nearby sister chromosomal regions (Horiuchi et al. 1994; Horiuchi and Fujimura 1995). In addition, in the yeast HOT1 recombination system, one of the two DNA elements required for stimulation of recombination, the E element, contains a RFB site and FOB1 was found to be required for the stimulation of recombination (Kobayashi and Horiuchi 1996). Although the HOT1 system is a homologous recombination system artificially constructed outside the rDNA locus, this finding, combined with the results described in this paper, demonstrates that FOB1 is, in fact, important for stimulating recombination within the native rDNA. Thus, in analogy to the E. coli Ter system, we propose that the DNA replication fork block at the RFB site in rDNA repeats stimulates recombination and plays an essential role in rDNA copy number adjustment and sequence homogenization. We specifically suggest that pausing of the DNA replication machinery at the RFB site stimulates double-strand-break (DSB) formation, perhaps in the sister chromatid with newly synthesized-lagging DNA fragments. This DSB then is repaired via gene conversion by a mechanism similar to the DSB repair model for recombination proposed by Szostak et al. (1983). Because of the tandem structure of rDNA repeats, loss or gain as well as no change of rDNA repeats is expected to take place depending on which of the rDNA units is used for gap repair. Figure 7B illustrates how a net gain of one rDNA copy may take place according to the proposed model. Conversely, if the DSB end interacts with an upstream rDNA unit on the sister chromatid (e.g., at the site near ori-1), a net loss of one rDNA copy will be the result of the gap repair. In addition, DSB repair might take place by intrachromatid recombination rather than by the sister-chromatid recombination shown in Figure 7B. Such DSB repair will lead to formation of DNA circles and a net loss of rDNA repeats after segregational loss of the circles, or a net gain of repeats after replication and reinsertion of the circles. We call the model developed here the fork block-dependent recombination model.
It has been argued frequently that unequal crossing over is responsible for maintaining homogeneity of repeat units and adapting the repeat length to the environment in tandemly repeated genes including rDNA repeats (Szostak and Wu 1980; Tartof 1988; Hawley and Marcus 1989). However, by analyzing reporter genes inserted into rDNA repeats of S. cerevisiae, Rothstein and coworkers recently demonstrated that gene conversion, and not unequal sister chromatid exchange, is the predominant mechanism responsible for the expansion/contraction of rDNA repeats (Gangloff et al. 1996). The model proposed by these workers involves DSB and gap repair between nonaligned sister rDNA chromatids. Our model is similar to theirs, but specifically proposes that the DSB is produced at the RFB site of rDNA repeats and the FOB1-encoded protein is required for this initial event. It emphasizes the importance of DNA replication fork arrest and predicts that expansion/contraction of rDNA tandem repeats is coupled with DNA replication. This prediction is consistent with the recent discovery by Zou and Rothstein (1997) that the Holliday junction recombination intermediate, an x-shaped DNA molecule, analyzed by an rDNA probe, is mainly detected at S phase in wild-type cells. They suggested that recombination is stimulated to repair replication-related lesions. In addition, from the rate of loss of a single reporter gene in rDNA repeats, Zou and Rothstein (1997) estimated 0.57 deletion events in rDNA per cell cycle. They also estimated that 3.6 Holliday junctions form in rDNA per cell cycle, and thus suggested that a majority of recombinational repair does not lead to rearrangement of repeat structures. In the present study, the rate of net increase of rDNA repeats observed on introduction of the RPA135 into the rpa135 FOB1 strain was approximately one copy per generation (Fig. 4). A similar rate also was observed for the rate of net decrease of rDNA repeats after inactivation of Pol I (Fig. 6C,D). Althugh the net decrease was analyzed in Pol I disrupted mutants and might not reflect events in normal yeast cells, the net increase was analyzed in yeast cells growing normally by use of Pol I. Thus, the observed rate, one copy increase per generation, may reflect the frequency of unequal sister chromatid gene conversion observed by Zou and Rothstein (1997) using a reporter gene in rDNA. The FOB1-dependent recombinational events responsible for rDNA expansion/contraction are also expected to lead to DSB repair without repeat number change. Therefore, the frequency of recombinational events (3.6 events per cell cycle) estimated by Zou and Rothstein (1997) can be largely accounted for by FOB1-dependent recombinational events as postulated in our model. We suggest that FOB1 plays a major role in normal recombinational events that take place at rDNA repeats in growing yeast cells. Actually, we observed that intra- and inter-sister chromatid recombination was greatly suppressed in normally growing fob1 cells (K. Johzuka and T. Horiuchi, in prep.).
FOB1-dependent replication fork arrest at the RFB site can take place in the absence of Pol I (Brewer et al. 1992; this paper). The FOB1-dependent net decrease of rDNA copy numbers can also take place in the absence of Pol I. Even though the net increase of rDNA repeats apparently required Pol I in our experiments, FOB1 appears to stimulate both the increase and decrease of rDNA repeats in the absence of Pol I. This inference is based on the observed heterogeneity in the size of repeats in NOY408-1a cells. Clones of this strain, which were derived from individual single cells, still showed the same kind of heterogeneity. In contrast, fob1 mutants derived from this strain showed a remarkable homogeneity (Fig. 6) and the size of chromosome XII was different depending on the fob1 mutant isolates from NOY408-1a (T. Kobayashi, unpubl.). It appears that in the absence of Pol I, rDNA repeat numbers continue to fluctuate, but the average copy number is kept at ∼80. Disruption of the FOB1 gene by transformation may prevent the fluctuation by inhibiting FOB1-dependent recombinational events, leading to rDNA copy numbers being fixed at values close to those the individual cells had at the time of gene disruption.
Pol I is required for HOT1 activity (Huang and Keil 1995). It is not clear why stimulation of recombination in the HOT1 assay requires Pol I, whereas recombinational events involved in expansion/contraction of rDNA repeats do not. It should be noted that the HOT1 assay uses artificial recombination systems in non-rDNA chromosomal regions. It is possible that the two cis-acting elements, E and I, together with Pol I (and other protein factors) may form a structure resembling rDNA structures, which might enable FOB1-dependent stimulation of recombination to take place.
What is the role of Pol I (or its absence) in directing rDNA repeat numbers toward a net increase (or a net decrease)? It might be argued that rDNA repeat numbers change as a result of selection of cells with a growth advantage. However, there was no significant change in the growth rate of NOY408-1a in the course of repeat number increase (from ∼80 copies to ∼150 copies; Fig. 4A) after introduction of the RPA135 gene. Similarly, there was no significant change in growth rate of NOY408-2a in the course of repeat number decrease (from ∼150 copies to ∼80 copies; Fig. 6C,D) after disruption of RPA135 gene. Furthermore, there was no significant difference in growth rate between NOY408-1a (rpa135) and NOY408-1af (rpa135 fob1) after introduction of the RPA135 gene and establishment of the final rDNA repeat numbers. The former restored the original repeat number (150 copies) after 80 generations, but the latter maintained its reduced repeat number (40 copies). Therefore, selection of cells with a growth advantage cannot explain expansion/contraction of rDNA repeats, at least in this range. Parenthetically, we note that only a fraction of rRNA genes were found to be transcribed even in actively growing cells in a variety of systems including yeast (Conconi et al. 1989; Dammann et al. 1993). It appears that the total number of rDNA repeats is not directly related to the rate of rRNA synthesis.
One possible explanation for the role of Pol I in controlling rDNA repeat numbers is to hypothesize that nucleolar structures play a role in stabilizing rDNA repeats. Strain NOY408-1a does not have the intact crescent nucleolar structure (Oakes et al. 1993) and Pol I plays a crucial role in the maintenance of nucleolar structure in addition to its functional role in rDNA transcription (Oakes et al. 1993, 1998). Perhaps there are some nucleolar components/substructures that interact with rDNA to form a stable complex. The number of repeats (∼150 copies in our strain) may be determined by the stability of such a structure. In the absence of the intact nucleolar structure, rDNA with the normal repeat length may become unstable and shift to more stable structures consisting of rDNA arrays with smaller repeat numbers and some available components/structures, which might substitute for the proposed intact nucleolar components/substructures. This model can also explain why expansion of the rDNA repeats stops at the original level (∼150) after reintroduction of RPA135 to cells with reduced rDNA repeat numbers.
Structures of rDNA repeats are basically conserved. RFB sites have been identified in rDNA repeats of yeast (Brewer and Fangman 1988; Linskens and Huberman 1988), peas (Hernandez et al. 1988), frogs (Wiesendanger et al. 1994), mouse (Lopez-Estrano et al. 1998) and human cells (Little et al. 1993). All of them are located near the 3′ end of the rRNA gene. This conservation indicates a functional importance of replication fork block. The results presented in this paper suggest that the primary function of replication fork block in yeast is to stimulate recombination to maintain rDNA homogeneity and to allow expansion/contraction of rDNA repeats. We suggest a similar role for RFB sites in other organisms.
Finally, we note that the information obtained in the present work might be relevant to gene amplification in non-rDNA systems, such as the amplification of dihydrofolate reductase gene induced by methotrexate treatment, and that of c-myc during tumorigenesis (Stark and Wahl 1984; Schimke 1988). DSB repair was suggested previously to be involved in the massive expansion of triplet repeats associated with many neurological diseases (e.g., see Gangloff et al. 1996). It remains to be seen whether a mechanism similar to the fork block-dependent expansion of rDNA repeats plays a role in such gene amplification or triplet repeat expansion observed in mammalian cells.
Materials and methods
Strains, plasmids, and media
Yeast strains and plasmids are listed in Table 1. Disruption of FOB1 was carried out in the following way. Plasmid pUC-fob1Δ::HIS3 was constructed from pUC–FOB1 (Kobayashi and Horiuchi 1996) by replacing the NruI–ClaI fragment containing most of the FOB1 coding region with a NruI–ClaI fragment containing HIS3 prepared by PCR. YEp–FOB1 was constructed by inserting the FOB1 gene amplified by PCR (Kobayashi and Horiuchi 1996) into the BamHI site of YEplac195. pNOY117 is a derivative of pRS314 (Sikorski and Hieter 1989) with a 5-kb XhoI–EcoRV fragment carrying RPA135 (Yano and Nomura 1991) inserted between the XhoI and EcoRV sites of the vector. This plasmid was constructed by Hisaho Sonoda (University of California, Irvine). SD is a synthetic glucose medium (Kaiser et al. 1994). SD supplemented with required amino acids and adenine was used to select transformants or strains carrying unstable plasmids. SG and YEP–galactose are the same as SD and YEPD (Yano and Nomura 1991), respectively, except that 2% glucose is substituted by 2% galactose. YEP–galactose was used to grow rpa135 deletion strains carrying helper plasmid pNOY102. The incubation temperature was 30°C.
Table 1.
Yeast strains and plasmids
| Designation
|
Genotypes and comments
|
Source
|
|---|---|---|
| Strain | ||
| NOY408 | MATa/MATα ade2-1/ade2-1 ura3-1/ura3-1 his3-11/his3-11 trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 rpa135Δ∷LEU2/RPA135 pNOY102 | Nogi et al. (1991) |
| NOY408-f | same as NOY408 except for fob1Δ∷HIS3/fob1Δ∷HIS3 | |
| NOY408-1a | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rpa135Δ∷LEU2 pNOY102 | Nogi et al. (1991) |
| NOY408-2a | same as NOY408-1a; freshly constructed from NOY408 | |
| NOY408-1b | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 pNOY102 | Nogi et al. (1991) |
| NOY408-1af | same as NOY408-1a except for fob1Δ∷HIS3 | |
| NOY408-2af | the same genotype as NOY408-1af, but constructed from NOY408-f by tetrad dissection | |
| Plasmid | ||
| pNOY102 | multicopy plasmid carrying GAL7–35S rDNA, 2μ, URA3 | Nogi et al. (1991) |
| pNOY117 | CEN6, ARSH4, TRP1, RPA135 | |
| YEplac195 | multicopy plasmid vector, 2μ, URA3 | Gietz and Sugino (1988) |
| YEp–FOB1 | YEplac195 carrying FOB1 | |
Isolation of DNA and Southern hybridization analysis
The approximate number of generations of yeast cells used for rDNA copy determination was first estimated from the size of colonies formed from single transformants (e.g., rpa135 strains that received gene RPA135 by transformation) or from haploid meiotic segregants. Single colonies with 1-mm diameter contained ∼2 × 105 cells and we assumed that cells in the colonies corresponded to progenies after 18 generations starting from individual ancestor cells. After streaking and taking cells again from colonies with 1-mm diameter (36 generations), subcultures in liquid YEP–galactose medium were initiated and growth was followed by measuring the absorbance at 600 nm. DNA was isolated, digested with BglII, subjected to 1% agarose gel electrophoresis, and analyzed by Southern hybridization using probes for rDNA (SphI–HindIII fragment in Fig. 1) and for MCM2 (1.4-kb fragment prepared by PCR) as described (Sambrook et al. 1989). The amounts of probes hybridized were determined by Fuji Bio-Imaging Analyzer (BAS2000, Fuji film, Japan).
Determination of rDNA copy numbers by a competitive PCR assay
Competitive PCR assays were done according to the method described in Diviacco et al. (1992). Competitor DNA that was added as a reference for target DNA samples was a derivative of pUC18 into which four reference DNA segments were inserted into four separate cloning sites. The reference DNAs were NTS, the 5S gene (Fig. 1, □), MCM2, and PPR1, each carrying an additional 20 nucleotides (derived from phage λ) inserted in the middle. These four reference DNA segments were designed not to have an EcoRI site. Yeast DNA samples were digested with EcoRI and then mixed with known amounts of competitor DNA. PCR reactions were carried out by use of two primers for each segment, yielding PCR products of two different sizes for each segment; competitor (reference) segments gave products with sizes larger than the corresponding target segments by 20 nucleotides. Amplified products were subjected to electrophoresis in 10% polyacrylamide gels, followed by staining with ethidium bromide and quantification by densitometry. The ratio of target to competitor DNA was determined for each segment and absolute numbers of rDNA copies were calculated as explained in the Results section.
Other methods
For pulse-field gel electrophoresis, chromosomal DNA was isolated as described previously (Smith et al. 1988) and subjected to gel electrophoresis in a 0.8% agarose gel, 0.5× TBE buffer, using CHEF–DRII (Bio-Rad, Richmond, CA) with a pulse time of 300–900 sec, 100 CV at 14°C for 68 hr. The gel was then stained with 0.5 mg/ml ethidium bromide for 30 min at room temperature, photographed, and then subjected to Southern hybridization analysis (Sambrook et al. 1989). RFB activity was analyzed by two-dimensional gel electrophoresis as described previously (Brewer and Fangman 1987).
Acknowledgments
We thank Drs. S. Arfin and S. Jinks-Robertson for critical reading of the manuscript. This work was supported in part by grants from the Ministry of Education, Science and Culture, Japan (to T.H.) and a grant from the National Institutes of Health (to M.N.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kishori@nibb.ac.jp; FAX 81-564-55-7690.
References
- Bastia D, Mohanty BK. Mechanisms for completing DNA replication. In: DePamphilis ML, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 177–215. [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- ————— A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Lockshon D, Fangman WL. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- Brown DD, Dawid IB. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science. 1968;160:272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Dammann R, Lucchini R, Koller T, Sogo JM. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviacco S, Norio P, Zentilin L, Menzo S, Clementi M, Biamonti G, Riva S, Falaschi A, Giacca M. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene. 1992;122:313–320. doi: 10.1016/0378-1119(92)90220-j. [DOI] [PubMed] [Google Scholar]
- Elion EA, Warner JR. The major promoter element of rRNA transcription in yeast lies 2kb upstream. Cell. 1984;39:663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Gall JG. Differential synthesis of the genes for ribosomal RNA during amphibian oogenesis. Proc Natl Acad Sci. 1968;60:553–560. doi: 10.1073/pnas.60.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Zou H, Rothstein R. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 1996;15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- Gencheva M, Anachkova B, Russev G. Mapping the sites of initiation of DNA replication in rat and human rRNA genes. J Biol Chem. 1996;271:2608–2614. doi: 10.1074/jbc.271.5.2608. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gogel E, Langst G, Grummt I, Kunkel E, Grummt F. Mapping of replication initiation sites in the mouse ribosomal gene cluster. Chromosoma. 1996;104:511–518. doi: 10.1007/BF00352115. [DOI] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Gailbert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:546–551. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Hawley RS, Marcus CH. Recombinational controls of rDNA redundancy in Drosophila. Annu Rev Genet. 1989;23:87–120. doi: 10.1146/annurev.ge.23.120189.000511. [DOI] [PubMed] [Google Scholar]
- Hernandez P, Lamm SS, Bjerknes CA, Van’t Hof J. Replication termini in the rDNA of synchronized pea root cells (Pisum sativum) EMBO J. 1988;7:303–308. doi: 10.1002/j.1460-2075.1988.tb02813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TM. Features of the chromosomal terminus region. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Vol. 2. Washington, DC.: ASM Press; 1996. pp. 1602–1614. [Google Scholar]
- Horiuchi T, Fujimura Y. Recombinational rescue of the stalled DNA replication fork: A model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol. 1995;177:783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol. 1994;176:4656–4663. doi: 10.1128/jb.176.15.4656-4663.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GS, Keil RL. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics. 1995;141:845–855. doi: 10.1093/genetics/141.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Keil RL, Roeder GS. Cis-acting recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Hidaka M, Nishizawa M, Horiuchi T. Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol & Gen Genet. 1992;233:355–362. doi: 10.1007/BF00265431. [DOI] [PubMed] [Google Scholar]
- Linskens MHK, Huberman JA. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4927–4935. doi: 10.1128/mcb.8.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RD, Platt T H, Schildkraut CL. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Estrano C, Schvartzman JB, Krimer DB, Hernandez P. Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J Mol Biol. 1998;277:249–256. doi: 10.1006/jmbi.1997.1607. [DOI] [PubMed] [Google Scholar]
- Nogi Y, Yano R, Nomura M. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc Natl Acad Sci. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes ML, Nogi Y, Clark MW, Nomura M. Structural alterations of the nucleolus in mutants of Saccharomyces cereviaise defective in RNA polymerase I. Mol Cell Biol. 1993;13:2441–2455. doi: 10.1128/mcb.13.4.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes ML, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MV. Genome structure and organization in Saccharomyces cerevisiae. In: Broach JR, Pringle JR, Jones EW, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 1–39. [Google Scholar]
- Petes TD. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci. 1979;76:410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980;19:765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- Ritossa FM. Unstable redundancy of genes of ribosomal RNA. Proc Natl Acad Sci. 1968;60:509–516. doi: 10.1073/pnas.60.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel PJ, Rodland KD. Magnification of rRNA gene number in a Neurospora crassa strain with a partial deletion of the nucleolus organizer. Chromosoma. 1986;93:337–340. doi: 10.1007/BF00327592. [DOI] [PubMed] [Google Scholar]
- Rustchenko EP, Curran TM, Sherman F. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol. 1993;175:7189–7199. doi: 10.1128/jb.175.22.7189-7199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schimke RT. Gene amplification in cultured cells. J Biol Chem. 1988;263:5989–5992. [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulations of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin KG, Eldarov MA, Larionov VL, Bayev AA, Klootwijk J, de Regt VC, Veldman GM, Planta RJ, Georgiev OI, Hadjiolov AA. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984;12:2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Klco SR, Cantor CR. Pulse-field gel electrophoresis and the technology of large DNA molecules. In: Dacis KE, editor. Genome analysis. Oxford, UK: IRL Press; 1988. pp. 41–112. [Google Scholar]
- Stark GR, Wahl GM. Gene Amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tartof KD. Unequal mitotic sister chromatid exchange as the mechanism of ribosomal RNA gene magnification. Proc Natl Acad Sci. 1974;71:1272–1276. doi: 10.1073/pnas.71.4.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Unequal crossing over then and now. Genetics. 1988;120:1–6. doi: 10.1093/genetics/120.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Keil RL, Roeder GS. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- Wiesendanger B, Lucchini R, Koller T, Sogo JM. Replication fork barriers in the Xenopus rDNA. Nucleic Acids Res. 1994;22:5038–5046. doi: 10.1093/nar/22.23.5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano R, Nomura M. Suppressor analysis of temperature-sensitive mutations of the largest subunit of RNA polymerase I in Saccharomyces cerevisiae: A suppressor gene encodes the second-largest subunit of RNA polymerase I. Mol Cell Biol. 1991;11:754–764. doi: 10.1128/mcb.11.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]