Abstract
A small number of major regulatory (selector) genes have been identified in animals that control the development of particular organs or complex structures. In Drosophila, the vestigial gene is required for wing formation and is able to induce wing-like outgrowths on other structures. However, the molecular function of the nuclear Vestigial protein, which bears no informative similarities to other proteins, was unknown. Here, we show that Vestigial requires the function of the Scalloped protein, a member of the TEA family of transcriptional regulators, to directly activate the expression of genes involved in wing morphogenesis. Genetic and molecular analyses reveal that Vestigial regulates wing identity by forming a complex with the Scalloped protein that binds sequence specifically to essential sites in wing-specific enhancers. These enhancers also require the direct inputs of signaling pathways, and the response of an enhancer can be switched to another pathway through changes in signal-transducer binding sites. Combinatorial regulation by selector proteins and signal transducers is likely to be a general feature of the tissue-specific control of gene expression during organogenesis.
Keywords: Organogenesis, signal transduction, selector gene, cis-regulatory elements, wing development
Induction of gene expression by signaling molecules is a central mechanism controlling the patterning of developmental fields. Several short- and long-range signaling proteins have been identified and it has become increasingly clear that their signaling pathways are required for the development of most body structures. However, we know little about the molecular mechanisms that govern the competence of a target tissue to respond to a signal in a particular way. This raises the question of the identity and function of regulatory molecules that may work in conjunction with common signaling pathways to activate tissue-specific patterns of gene expression.
The development of the Drosophila wing is organized by two orthogonal systems of short- and long-range signaling proteins that regulate growth and gene expression (Brook et al. 1996; Neumann and Cohen 1997; Serrano and O’Farrell 1997). The Decapentaplegic (Dpp) and Wingless (Wg) long-range signaling proteins are produced by cells along the anteroposterior and dorsoventral compartment boundaries, and induce the expression of several target genes in the developing wing disc. Dpp induces the spalt (sal), spalt related (salr), and optomotor blind (omb) genes in a nested pattern centered on the Dpp stripe (de Celis et al. 1996a; Grimm and Pflugfelder 1996; Lecuit et al. 1996; Nellen et al. 1996; Sturtevant et al. 1997). The vestigial (vg) gene is even more broadly induced by Dpp and its expression pattern fills the entire wing field (Williams et al. 1991, 1993; Kim et al. 1996). Activation of vg also requires input from the dorsoventral organizing signals Wg and Serrate (Ser). Similarly, the Serum Response Factor (SRF or blistered) gene is regulated by signals from both axes and is expressed in the subregions of the developing wing that become the intervein tissue (Montagne et al. 1996). Other genes have expression patterns that are determined by the signals that organize the dorsoventral axis, for example cut is expressed in a stripe along the dorsoventral compartment boundary and regulated by Notch signaling (de Celis et al. 1996b; Neumann and Cohen 1996; Micchelli et al. 1997).
Recent studies have shown that the expression patterns of these target genes in the wing field are controlled by discrete cis-regulatory elements that respond to particular signaling proteins. Activation of vg is regulated by two separate enhancers that are induced by different signaling pathways. The boundary enhancer is activated along the dorsoventral boundary through direct input of the Notch (N) pathway (Williams et al. 1994; Kim et al. 1996), whereas the quadrant enhancer is activated in the remainder of the growing wing pouch by Dpp and Wg signaling (Kim et al. 1996, 1997; Zecca et al. 1996; Neumann and Cohen 1997). Similarly, the SRF gene has two enhancers, one expressed in the intervein C region along the anteroposterior compartment boundary that is regulated by Hh, and one expressed in adjacent intervein regions (U. Nussbaumer, J. Montagne, J. Groppe, and M. Affolter, in prep.). Finally, enhancers were identified that drive sal and cut expression in the wing pouch (Jack et al. 1991; Kuhnlein et al. 1997). Importantly, all six enhancers are expressed exclusively in the wing (and haltere) pouch, and are not active in leg or eye-antennal discs, even though the inducing signals are active in other tissues.
How are the cut, sal, SRF, and vg enhancers induced specifically in the developing wing pouch but not in other tissues in which these signaling pathways are also active? Apparently, other factors must exist that promote or restrict the responses of cells to signaling inputs. One candidate for such a role is the Vg protein because it is able to reprogram some cells in leg and other imaginal discs to adopt a wing fate (Kim et al. 1996). Vg is required in all wing cells and loss of Vg function eliminates wing formation (Williams et al. 1991, 1993). Vg is expressed in the developing wing discs, but not in leg or eye antennal imaginal discs. Thus, Vg is a selector protein for wing identity and development. However, the molecular function of the Vg protein is not known, although its nuclear localization (Williams et al. 1991) suggests that it could be involved in regulating gene expression.
Here we show that Vg induces wing development and wing-specific gene expression by cooperating with the Scalloped (Sd) protein, a member of the TEA family of DNA-binding proteins (for review, see Jacquemin and Davidson 1997) that is also expressed in the developing wing pouch (Williams et al. 1991, 1993; Campbell et al. 1992). We found that Sd function is required in parallel to Vg to induce wing development. Moreover, Sd and Vg act synergistically to induce several wing-specific enhancers in tissue culture cells. A functional analysis of the SRF intervein C enhancer revealed that this element contains several binding sites for Sd that are essential for enhancer activity in tissue culture and in vivo. Our results, together with the finding that Vg and Sd interact physically (Simmonds et al. 1998), support a model in which Vg and Sd form a complex that directly regulates gene expression and limits the induction of their target enhancers to the developing wing. Thus, it appears that the combinatorial control of cis-regulatory elements by signal transducers and selector proteins is an important mechanism to evoke tissue-specific responses to common signals.
Results
Vg cell autonomously induces wing-specific gene expression
Because ectopic expression of Vg in many imaginal discs induces the outgrowth of wing tissue (Kim et al. 1996), we examined whether the expression of various wing patterning genes was induced in these ectopic growths. Vg is expressed in the entire developing wing pouch (Fig. 1A) whereas Sal and SRF have specific expression patterns within this domain (Fig. 1B,C) but are not expressed in wild-type leg discs (Fig. 1D). Targeted expression of Vg with the Gal4–UAS system induced ectopic expression of Sal and SRF in developing leg imaginal discs (Fig. 1E,F) (Weatherbee et al. 1998). Similarly, the nubbin (nub) gene, which is also expressed and required during wing development (Ng et al. 1995), was ectopically induced in leg discs by Vg expression (Klein and Martinez Arias 1998; data not shown). In each case, only a subset of the cells expressing Vg activated the target gene, which suggests that additional factors control the expression pattern of each gene.
Figure 1.
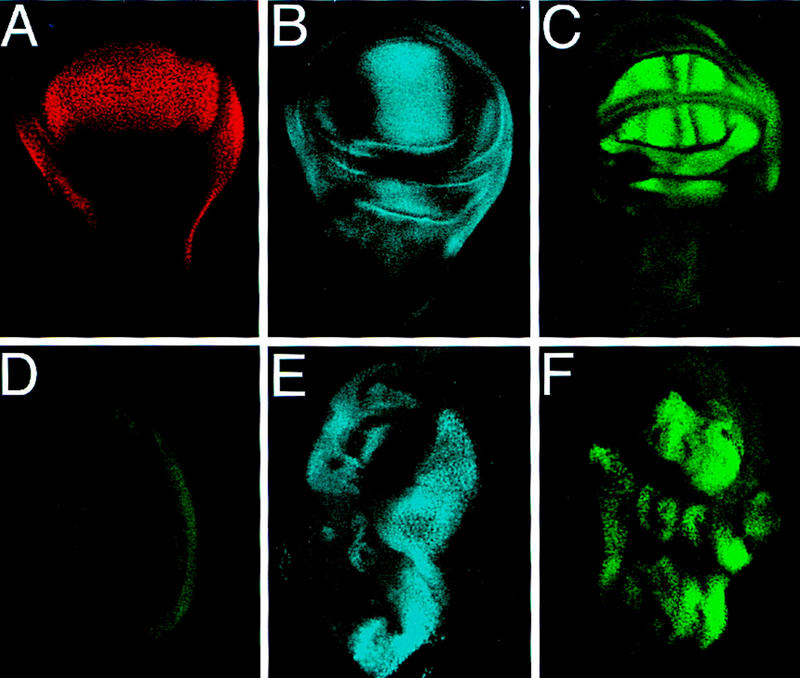
Targeted expression of Vg ectopically induces wing patterning genes. (A–C), Endogenous expression patterns of Vg, Sal, and SRF in third instar wing discs. (A) Vg expression marks the entire developing wing field. (B) Sal is expressed in a band straddling the A/P boundary within the developing wing pouch. (C) SRF is expressed in future intervein tissue whose proximal borders are defined by Vg expression. Both Sal and SRF are also expressed in regions surrounding the wing pouch. (D) Sal (blue) and SRF (green) are not expressed in wild-type leg discs. (E) Sal and (F) SRF, are ectopically induced in leg discs in response to targeted expression of Vg by a Distal-less (Dll)–Gal4 driver. Lower levels of these genes are induced in the legs by use of the dpp–Gal4 driver (not shown). All discs are anterior to the left, in wing discs ventral is at the top, in leg discs dorsal is at the top.
In a first step toward elucidating the molecular mechanism by which Vg regulates gene expression, we studied the response of wing-specific enhancers to ectopic Vg expression. We focused our attention on the boundary and quadrant enhancers of the vg gene (Fig. 2A,D) and the enhancer from the SRF gene that drives expression specifically in the intervein region between veins three and four (intervein C, Fig. 2G) (U. Nussbaumer, J. Montagne, J. Groppe, and M. Affolter, in prep.). All three enhancers were induced by ectopic Vg expression in leg and other imaginal discs. Importantly, ectopic expression of Vg in clones of cells induced the enhancers only within the clones (Fig. 2B,C,E,F,H,I). However, gene expression was not induced in all cells within clones nor in all clones. In addition, each individual enhancer was expressed in different regions of these discs that appear to correlate with the spatial distribution of the different signaling inputs known to be required for activation of these enhancers.
Figure 2.
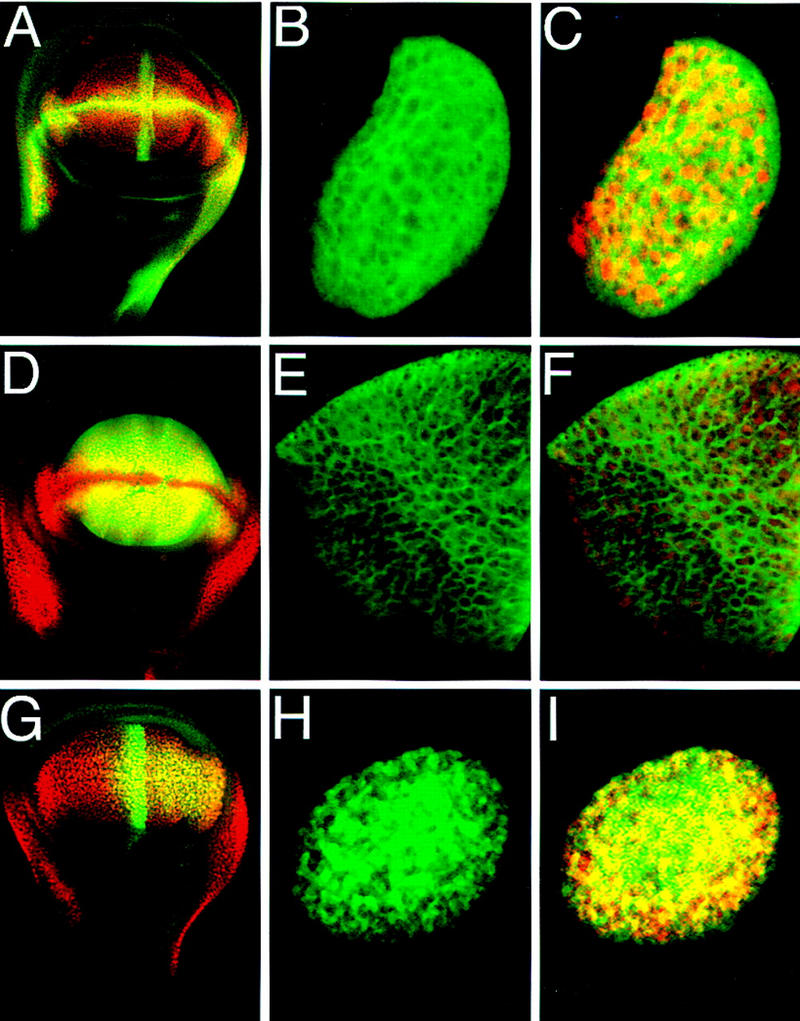
Cell autonomous activation of wing-specific enhancers by Vg. (A) Wing disc double stained for β-gal (green) driven by the vg boundary enhancer and for Vg protein (red). (B) Close-up of a clone of cells in an eye disc that ectopically expressed Vg (red in C) and autonomously induced expression of the vg boundary enhancer reporter construct (green). (D) Wing disc double stained for β-gal (green) driven by the vg quadrant enhancer and Vg protein (red). (E,F) A clone of cells in a leg imaginal disc in which β-gal expression driven by the quadrant enhancer (E, green) is induced by ectopic Vg expression (red in F). (G) Wing disc double stained for β-gal expression (green) driven by the SRF intervein C element and Vg protein (red). (H,I) Clone of cells in the eye in which the intervein C element is activated (H, green) by ectopic Vg expression (red in I).
Sd is required in parallel for Vg function
In the notum primordia of the wing disc, the vg enhancers, as well as the sal, SRF, and nub genes were not induced by ectopic Vg even though the known required extracellular signals were present (Fig. 3A–C; data not shown). Target gene activation could depend then on the function of another gene(s). One candidate for such a factor is the product of the sd gene, which is expressed in a pattern similar to Vg in the wing disc (Fig. 3D) (Campbell et al. 1992) and is required for wing formation (Campbell et al. 1991; Williams et al. 1993) and the proper expression of Vg (Williams et al. 1993) and other genes (Morcillo et al. 1996). In other discs, such as the leg and eye discs, sd is endogenously expressed and is upregulated wherever ectopic Vg was able to induce wing-specific gene expression and trigger wing development (data not shown). We note, however, that a sd enhancer trap line and the SRF-intervein C enhancer transgene were also ectopically induced by Vg in the presumptive notum, although at levels lower than those observed in the wing pouch (Fig. 3E,F). This is consistent with the inability of Vg to trigger wing development and induce other wing patterning genes in the developing notum. On the basis of the correlation between sd expression and activation of Vg target genes, we investigated whether Sd function was required in parallel to Vg to exert its wing inducing activity.
Figure 3.
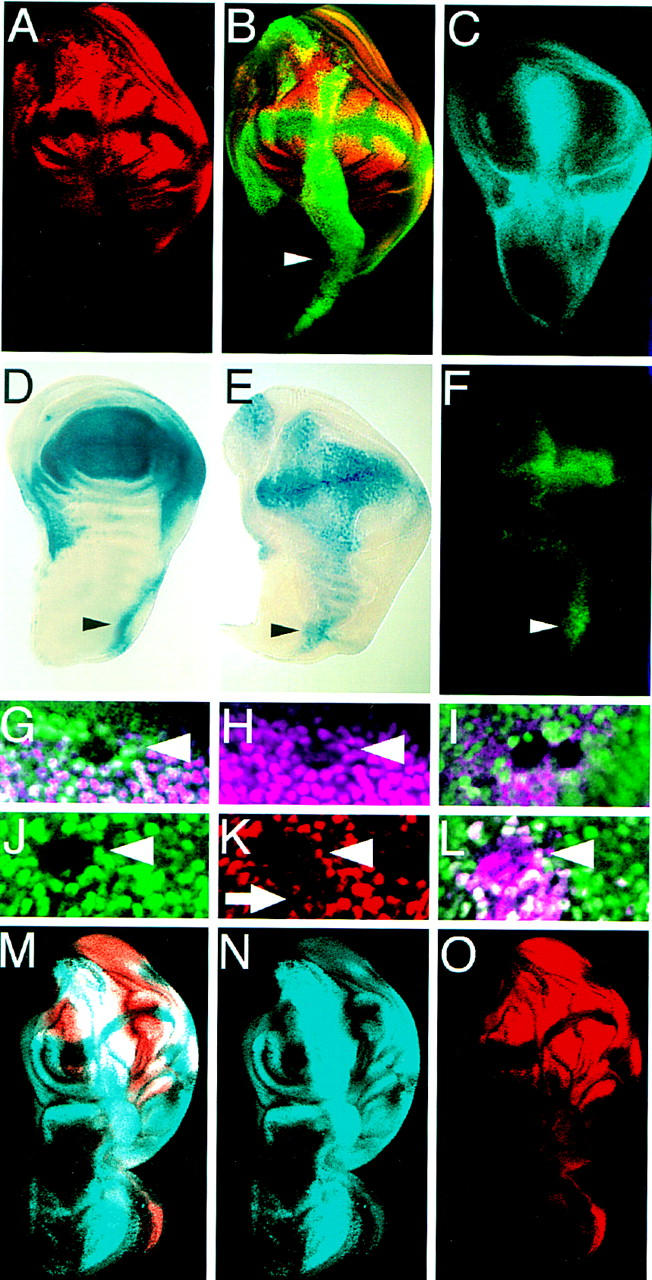
The activation of downstream genes, and autoregulation of vg requires Sd function in parallel to Vg. (A–C) Wing discs in which Vg was expressed along the A/P boundary driven by dpp–Gal4. (A) SRF expression. (B) same disc as in A stained for both SRF (red) and Vg (green). Note that SRF expression does not extend into the notum portion (arrowhead). (C) Sal expression also does not extend into the notum. (D) sd expression pattern in a wing disc revealed by X-gal staining of larvae carrying a lacZ insertion into the sd gene. (E) Ectopic Vg expression driven by dpp–GAL4 induces sd expression at low levels in the notum. (F) Induction of the SRF intervein C element by ectopic Vg expression is highest where endogenous sd expression is highest in the notum (arrowheads in D–F). (G,H) In sd47m mutant clones in the wing disc, Vg expression (purple) is lost in the clone. Mutant clones are illustrated by loss of Myc staining (green). (I) sd47m mutant clones in a wing disc ectopically expressing Vg along the A/P boundary. The clones are situated along the A/P boundary and are marked by loss of Myc staining (green). (J–L) Expression of a lacZ reporter gene driven by the vg quadrant enhancer (purple) is lost in sd47m mutant clones, even in the presence of ectopic Vg. Homozygous sd47m mutant clones in a leg disc in which Vg is ectopically expressed. (J) The mutant clones are illustrated by the lack of Myc expression (green). SRF expression (red) is lost within the clones (K) even though Vg (purple) is still expressed (L). Just outside of the clone, ectopic Vg expression down-regulates SRF (arrow, this down-regulation is also apparent in A). (M–O) Wing discs that ectopically expressed Vg as in A–C with Sd ubiquitously expressed by a heat-inducible transgene. (M) Doble staining for Sal (blue) and SRF (red) (N,O) expression patterns. Coexpression of Vg with Sd induces gene expression in the notum portion of the disc.
To determine if Sd is required in vivo for Vg to induce the expression of wing patterning genes, we first examined expression of target genes in clones that were homozygous for a strong mutation in the sd gene. Expression of the SRF and vg genes was lost in these clones (Fig. 3G,H). The loss of SRF gene expression could have been due directly to the loss of Sd or indirectly to Sd effects on vg expression. To distinguish between these possibilities, we generated sd mutant clones in imaginal discs in which Vg was ectopically expressed via the Gal4–UAS system, thus bypassing the requirement of Sd for vg expression. In these sd mutant clones, SRF (Fig. 3J–L) and vg quadrant enhancer expression (Fig. 3I) was not induced, revealing that Sd function is required for Vg to regulate gene expression.
To distinguish whether Sd was required downstream of or in parallel to Vg, we provided Sd activity in conjunction with Vg in ectopic expression experiments. High levels of ectopic Sd expression produced by the Gal4–UAS system with a dpp–GAL4 driver had a dominant-negative effect, suppressing the ectopic wing phenotypes caused by Vg expression, and repressing endogenous Sal expression in the wing disc (not shown). It appears then that the concentration of Sd is critical for gene expression, a parameter that is also critical for target gene activation in tissue culture by Sd (see below) and the human Sd homolog transcriptional enhancer factor-1 (TEF-1) (Xiao et al. 1991; Hwang et al. 1993). To better control the level of Sd expression, we used a heat-inducible transgene. Coexpression of Sd (by heat shock) and Vg (with a dpp–Gal4 driver) resulted in ectopic outgrowths in the notum and induction of Sal expression along the entire A/P boundary and SRF expression within the ectopic outgrowth (Fig. 3M–O). These phenotypes were never observed by expressing Vg or Sd alone. We conclude that Sd and Vg are required in parallel to induce wing development.
Vg and Sd act synergistically to activate target genes
These genetic experiments, and the discovery that the Vg and Sd proteins interact physically (Simmonds et al. 1998), led us to investigate whether the activation of target genes might occur through a direct interaction between Vg and Sd and cis-regulatory sequences of target genes. We found that the three wing-specific enhancers from the SRF and vg genes were activated synergistically when Sd and Vg were coexpressed in Drosophila S2 cells (Fig. 4). Although each individual protein had some effect on reporter gene expression, this was always significantly less than that observed in the presence of both Vg and Sd (Fig. 4). Titration of the amounts of transfected Vg and Sd plasmids with all enhancers showed that the relative concentration of the two factors was critical and, at any given Vg concentration, high levels of Sd reduced activation (Fig. 4B).
Figure 4.
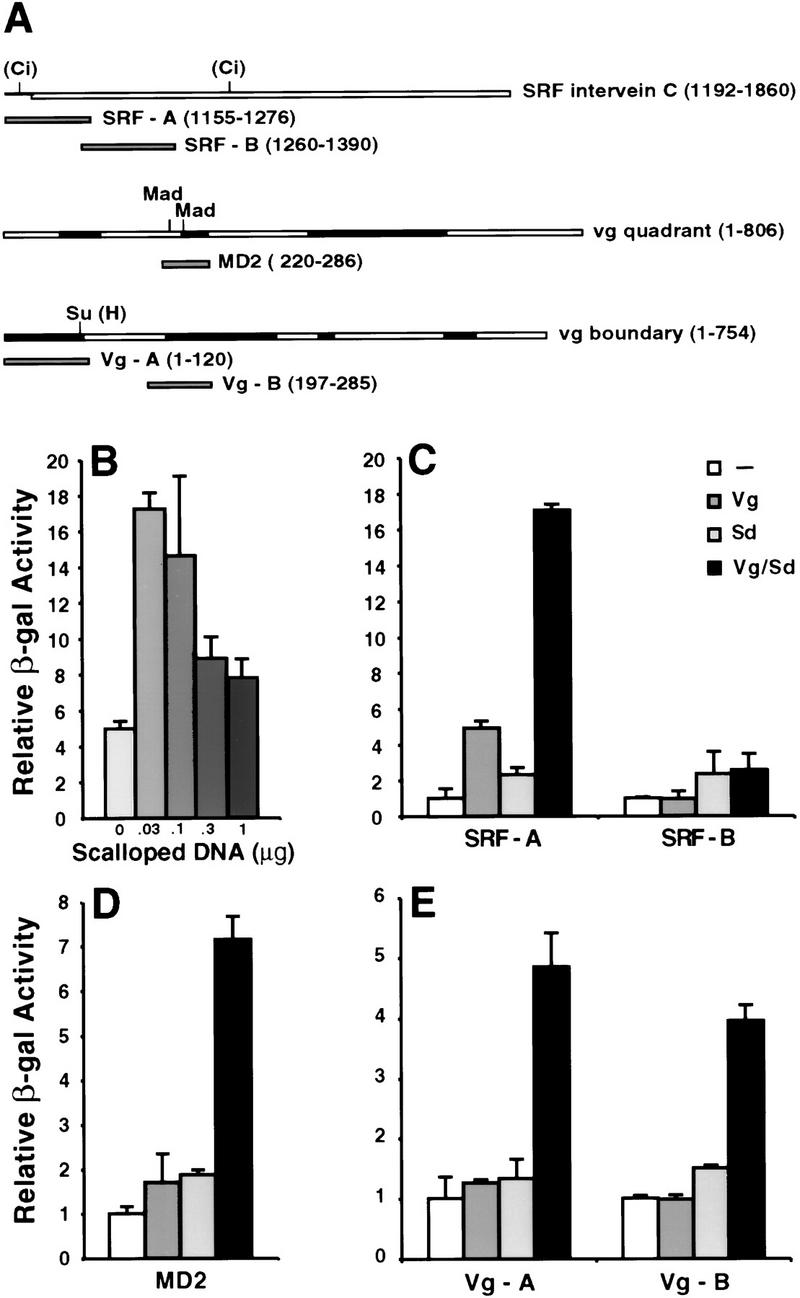
Synergistic activation of cis-regulatory elements of wing-patterning genes by cotransfection of vg and sd. (A) Schematic of Vg-responsive enhancers and fragments analyzed in tissue culture and in vivo. The binding sites for transcriptional regulators of signaling pathways are indicated. Evolutionarily conserved regions of the vg enhancers are shaded. (B) The induction of the SRF-A element by Vg and Sd. Synergistic activation diminishes with increasing Sd concentration. (C) The SRF-A element is activated 17-fold by cotransfection of optimal amounts of Vg and Sd expression vectors, whereas either Vg or Sd alone induce five- and twofold, respectively. The SRF-B element is not activated significantly. (D) MD2, and (E) Vg-A and Vg-B are also activated synergistically by cotransfection of Sd and Vg. Error bars represent 1 s.d. of relative β-gal activity.
To define the sequences of the enhancers that respond to Vg/Sd, we analyzed the activation of smaller fragments from the 704-bp SRF intervein C enhancer, the 806-bp vg quadrant enhancer, and the 754-bp vg boundary enhancer in tissue culture (Fig. 4A). We found that a 125-bp fragment (SRF-A) derived from the 5′ end of the SRF enhancer was activated, whereas an adjacent 131-bp fragment (SRF-B) was not activated (Fig. 4C). A 65-bp fragment from the vg quadrant enhancer (MD2) has been identified that, when multimerized, produces an expression pattern very similar to the full-length enhancer in wing discs (A. Hudson and S. Carroll, unpubl.). When assayed in tissue culture, MD2 was activated by Vg and Sd (Fig. 4D). Within the vg boundary enhancer, a 120-bp fragment sufficient to drive reporter gene expression in the wing pouch (vg-A) as well as a nonoverlapping 90-bp fragment (vg-B) were also activated synergistically by cotransfection of Vg and Sd (Fig. 4E).
Sd affects Vg localization to nuclei
One possible reason for the importance of the concentration of Sd on Vg function concerns the localization of the Vg protein. We observed that in S2 cells transfected with the vg expression plasmid alone, the Vg protein appears to be localized to both the cytoplasm and the nucleus (Fig. 5A–C). In contrast, in cells cotransfected with the Vg and Sd expression plasmids, Vg is clearly localized to nuclei (Fig. 5D–F). Furthermore, we note that Vg localization is more diffuse in sd mutant clones than in sd+ cells; this is also true of ectopic Vg localization in regions of imaginal discs that lack endogenous Sd expression (not shown). Furthermore, deletion of the Sd interaction domain of Vg results in cytoplasmic accumulation of Vg in vivo (Simmonds et al. 1998). Thus, Sd may facilitate the transport or retention of Vg protein in the nucleus and, coupled with the concentration-dependent, synergistic effects of Vg and Sd on target gene expression, these results suggest that the proteins form a complex in vivo.
Figure 5.
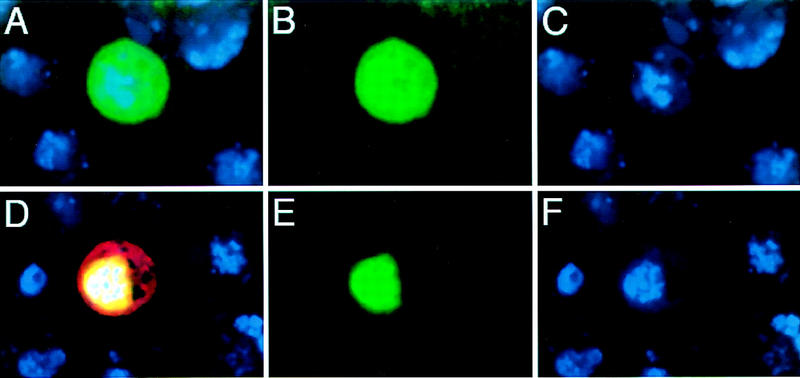
Sd affects the nuclear localization of Vg. Drosophila S2 cells transfected with SRF-A lacZ reporter gene (β-gal in red) and Vg (green) (A–C) or the reporter, Vg, and Sd (D–F). Cotransfection of Sd localizes Vg to the nucleus (E), and induces expression of the SRF-A lacZ reporter gene (D). All cells are counterstained with Topro to reveal DNA in nuclei (blue).
Sd binds specifically to essential sites for target gene activation
Because Sd is a member of the TEA family of transcriptional regulators that contain the highly conserved TEA DNA-binding domain (Jacquemin and Davidson 1997; Davidson et al. 1988), we investigated whether Sd could bind to the regulatory elements of the SRF and vg genes in vitro. We used purified TEA domains in gel mobility shift experiments, as the TEA domain alone is able to bind to DNA cooperatively and with high affinity (Hwang et al. 1993; Jacquemin et al. 1996). The Sd, as well as the TEA domain of its human homolog TEF-1, bound to the SRF-A, but not to the SRF-B fragment with high affinity, comparable to that of both proteins binding to the Sph and GT-IIC TEF-1 binding sites identified in the SV40 enhancer (Fig. 6B,C; data not shown) (Davidson et al. 1988; data not shown). Sd and TEF-1 TEA domains also bound specifically to the vg quadrant MD2 element but with a lower affinity than to SRF-A (data not shown). Neither protein recognized a mutated GT-IIC site shown previously not to be bound by TEF-1 (Davidson et al. 1988). The Sd and TEF-1 TEA domains recognized all sequences tested in a similar manner, indicating that the two TEA domains have virtually identical DNA-binding specificities (Fig. 6C). This is consistent with the finding that TEF-1 can rescue sd mutant flies (Deshpande et al. 1997). The pattern of mobility shifts of the Sd TEA domain bound to the SRF-A fragment closely resembles the pattern of complexes obtained by shifting the Sph fragment (Fig. 6C). The Sph fragment contains two tandem TEF-1 binding sites that are bound cooperatively by the TEF-1 TEA domain (Davidson et al. 1988). Inspection of the SRF-A sequence revealed two tandem sets of potential binding sites that resemble those in the Sph enhancer (A1 and A2; Fig. 6A,B).
Figure 6.
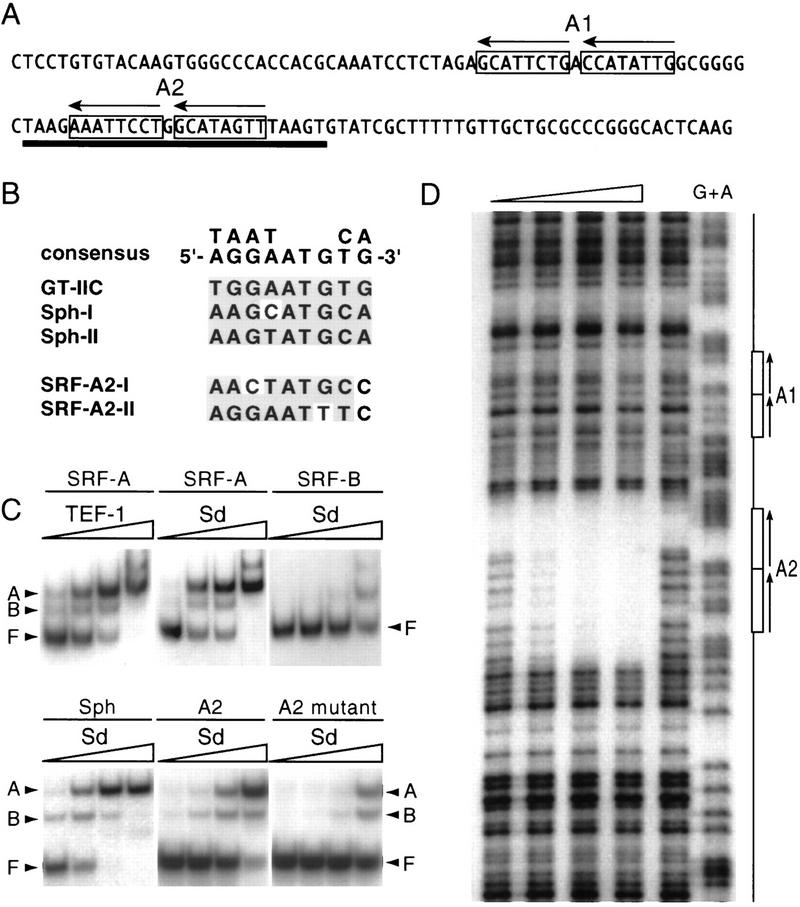
The Scalloped protein binds sequence specifically to the SRF-A element. (A) Sequence of the SRF-A element. Two sets of putative Sd tandem binding sites are boxed (A1, A2) and the orientation of the binding sites indicated by arrows. The extent of the region protected from DNase I digestion by the Sd–TEA domain is underlined. (B) Alignment of the Sd binding sites in the SRF enhancer with known TEF-1 binding sites. Shaded bases fit the TEF-1 consensus. (C) Gel mobility-shift experiments with the SRF-A and SRF-B fragments, the Sph fragment from the SV40 enhancer, and the wild-type and mutated SRF-A2 fragment by use of increasing amounts of purified 6-histidine-tagged Sd or TEF-1 TEA domains. Concentrations are 0.3, 1, 3, and 10 ng/25 μl, respectively. (F) Free probe; (B) complex with one TEA domain bound; (A) complex with two proteins bound. The Sd and TEF-1 TEA domains bind with high affinity to the SRF-A fragment but not to SRF-B. Both proteins bind with similar affinity to the Sph fragment (only the Sd shift is shown). Two molecules of Sd TEA domain bind to the SRF-A2 element but with much lower affinity (comparable to nonspecific binding) when the A2 sites are mutated. (D) DNase I protection assay on the SRF enhancer with increasing amounts of purified Sd TEA domain (1.7, 5, 15, 45 ng from left to right). The lane directly to the left of the G+A sequence ladder contained no Sd TEA protein. The A1 and A2 regions are shown schematically at right. Only the A2 region is protected by Sd protein.
To identify the specific sequences to which Sd bound, we performed DNase I footprinting with the Sd protein on the SRF-A and SRF-B regions (Fig. 6D). The Sd TEA domain protected one of the two tandem sets (A2), whereas neither the other set (A1) nor any other regions were protected, even at the highest protein concentration tested (Fig. 6D). As expected, SRF-B showed no footprint (not shown). To test whether the A2 binding sites were required for activation of SRF-A in tissue culture, we mutated these sites to sequences that were no longer bound specifically by Sd in gel mobility shift assays (Fig. 6C). When transfected into S2 cells, the activation of the mutant reporter plasmid by Vg and Sd was severely reduced (Fig. 7A). Similarly in Drosophila, although the SRF-A element was sufficient to drive expression specifically in the wing pouch (Fig. 7B), the mutated SRF-A lacking Sd binding sites was inactivated (Fig. 7C). Therefore, the sequence-specific binding of Sd to the SRF cis-regulatory element is required for enhancer activation.
Figure 7.
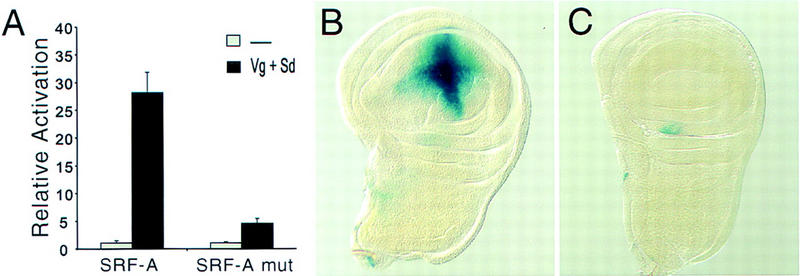
Sd binding sites are required for target gene expression. (A) Activation of the SRF-A reporter by Vg and Sd was drastically reduced by mutating the footprinted A2 sequence to sequences no longer bound by Sd. Error bars represent 1 s.d. of relative β-gal activation. (B) The SRF-A fragment is sufficient to drive β-gal expression specifically in the developing wing pouch. (C) The activity of the SRF-A fragment is abolished when the Sd binding sites are mutated.
Combinatorial control of wing-specific gene expression patterns by Vg/Sd and signaling inputs
Our results demonstrate that the activation of several genes in the wing field requires Vg/Sd function. It is also known that for each of the cis-regulatory elements analyzed here, direct input(s) of particular signaling pathways are also required. Specifically, the activation of the SRF intervein C element requires both Vg/Sd and Hh signaling (U. Nussbaumer, J. Montagne, J. Groppe, and M. Affolter, in prep.), the activation of the vg boundary enhancer requires Vg/Sd and N signaling (Kim et al. 1996), and the activation of the vg quadrant enhancer requires Vg/Sd and Dpp signaling (Kim et al. 1996). Because these regulatory elements are not expressed in all tissues in which the signals are active, nor in all wing cells in which Vg/Sd are active, we deduce that neither the input of various signals nor of Vg/Sd alone are sufficient for gene activation in vivo. Rather, our results suggest that the various wing-specific cis-regulatory elements require a combination of direct inputs, comprising the Vg/Sd selector function, which restricts expression to the wing field, and at least one signal transducer that mediates signaling inputs and hence, the pattern of gene expression within the wing field. One prediction of this model is that gene expression patterns within the wing field may be changed by altering the signal-transducer binding sites within a cis-response element. To test this, we changed the Suppressor of Hairless [Su(H)] binding site that mediates the N input in the vg boundary enhancer (Kim et al. 1996) to sites for the Cubitus interruptus (Ci) protein that transduces Hh signaling. This switches the pattern of gene expression from a N-induced dorsoventral stripe to a Hh-induced anteroposterior stripe while retaining the restriction of gene activation to the wing disc (Fig. 8A).
Figure 8.
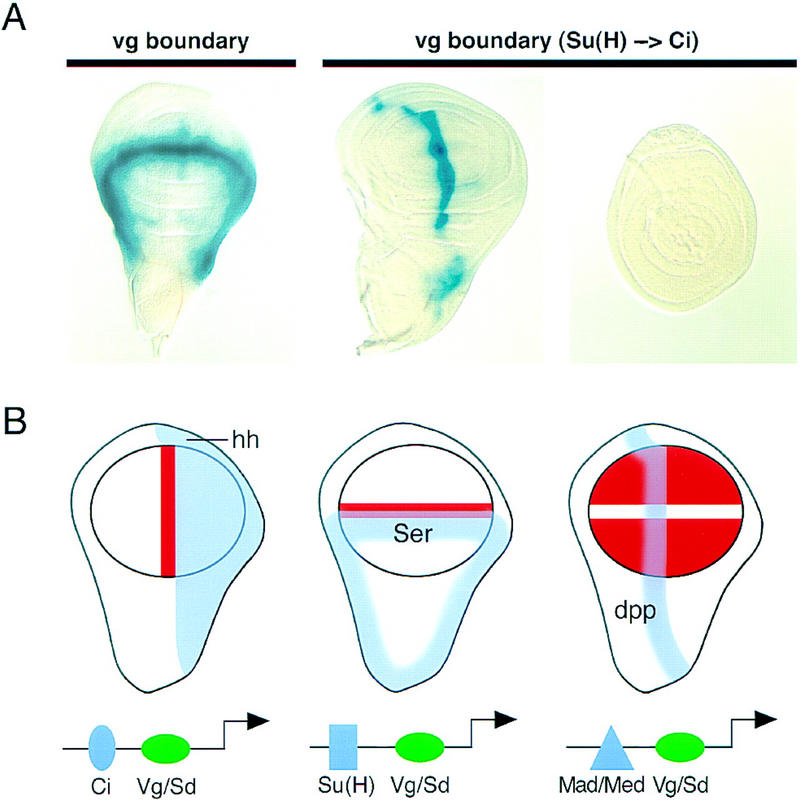
Vg and Sd control the wing-specific responses of target genes to signaling proteins. (A) Switching the specificity of a cis-response element. The vg boundary enhancer is activated through the N pathway by the Su(H) protein along the D/V boundary of the wing disc (left; Kim et al. 1996). Replacing the Su(H) site with two binding sites for the Ci protein changes the response of the enhancer from the N to the Hh pathway and the lacZ reporter gene is now expressed along the anterior side of the A/P boundary in the wing (middle), but not in the leg (right). (B) The selector and signal model for combinatorial control of gene expression in the wing field. Schematics of wing imaginal discs showing the expression patterns of the Dpp, Ser, and Hh signaling proteins (blue). Target gene responses (orange) are restricted to the circular wing field (outlined in black) by the Vg/Sd selector function, which acts together with the DNA-binding transducers of respective signaling pathways on cis-regulatory elements.
Discussion
Vg interacts with Sd to regulate wing-specific gene expression
Our genetic and biochemical results suggest that Vg regulates wing identity by acting together with the Sd protein to control wing-specific patterns of gene expression. The requirement of Sd function for Vg activity and nuclear localization in vivo, the synergy of Vg and Sd in cotransfection experiments, the direct binding of Sd to cis-regulatory sequences required for target gene activation, and the demonstration of a physical interaction between Vg and Sd (Simmonds et al. 1998) all support a model whereby the two proteins form a transcriptional activator in which Sd is the sequence-specific DNA binding component.
The vertebrate Sd homologs, the TEF proteins (for review, see Jacquemin and Davidson 1997) also require specific coactivators for their function (Xiao et al. 1991; Ishiji et al. 1992; Hwang et al. 1993), and one of them was identified as the bHLH protein Max (Gupta et al. 1997). Interestingly, these coactivators have a tissue specific distribution, whereas the TEFs are widely expressed. Similarily, in Drosophila, Sd is expressed and required in several imaginal discs, whereas Vg functions specifically in the developing wing disc. Thus, it is the spatial restriction of Vg expression that determines the boundaries of the wing field.
Tissue-specific responses to signaling pathways: the role of selector genes
Our results demonstrate that the role of the Vg/Sd selector function is to directly regulate wing-specific cis-regulatory elements that also require particular signaling inputs. The patterns of gene expression induced in the wing disc are limited to cells in which both the selector genes and specific signaling pathways are active. The response of the SRF-A, vg boundary, and vg quadrant enhancers to Hh, N, and Dpp signaling are limited to the wing pouch by Vg/Sd and occur in different patterns because of their direct regulation by the Ci, Su(H), and Mad proteins, respectively (Fig. 8B). Furthermore, the finding that the changing of the Su(H) binding site into a Ci binding site in the vg boundary enhancer switches the pattern from a wing-specific dorsoventral N-regulated stripe to a wing-specific anteroposterior Hh-regulated stripe suggests that spatial expression patterns are determined by the sites for individual DNA-binding signal transducers.
One corollary of this model is that for any given signaling protein, different selector proteins may be involved in directing tissue-specific responses in different organs and tissues. It has recently been shown that tissue-specific enhancers in the embryo that are regulated by Dpp also require the action of the Labial/Extradenticle (Grieder et al. 1997) or Tinman (Xu et al. 1998) selector proteins to limit expression to the endoderm or mesoderm, respectively. We suggest that, in general, combinatorial control by selector proteins and common signal transducers at a cis-regulatory level is required for the tissue- and organ-specific responses of target genes to widely deployed signaling systems.
Materials and methods
Ectopic expression of Vg and Sd
Ectopic expression of Vg was driven by the Gal4–UAS expression system (Brand and Perrimon 1993). dpp(4A.3)–Gal4 (Morimura et al. 1996) and Dll–Gal4 (Calleja et al. 1996) drivers were used to activate UAS–Vg (Kim et al. 1996) in a specific spatiotemporal pattern. dpp–Gal4 expresses along the A/P compartment boundary of leg and wing imaginal discs. Dll–Gal4 is expressed in the distal portions of leg and the wing pouch imaginal tissues. Clones of ectopic Vg were generated by mating hs–FLPase; UAS–Vg flies with flies carrying an actin promoter–FRT–CD2–FRT–Gal4 transgene (Pignoni and Zipursky 1997). Heat shock at 34°C for 30 min at 40–60 hr after egg laying induced expression of FLPase catalyzing the site specific recombination between the FRT sites, resulting in Gal4 expression in clones. Larvae were aged 2 days after heat shock and dissected at the late third instar stage. The hs–sd transgene was induced by four heat shocks at 37°C for 45 min every 4 hr. Heat shocks were started at 70–80 hr after egg laying and crawling third instar larvae were dissected for disc strainings.
Sd mutant clones
Mitotic recombination to generate sd mutant clones was induced by γ-rays in progeny from a cross between sd47m/FM7 flies and flies carrying a heat-inducible 18M myc (Bailey and Posakony 1995). Larvae were aged at 25°C an additional 48 hr after irradiation, heat-shocked for 2 hr at 36.5°C to induce Myc expression, and then allowed to recover for 45–120 min at 25°C prior to dissection.
Immunohistochemistry
Antibody and X-gal stainings of imaginal discs were done as described (Halder et al. 1998). Immunohistochemistry of transfected Drosophila S2 cells involved spotting cells onto a slide, fixing in 4% paraformaldehyde followed by a −20°C methanol wash, blocking with 3% BSA, incubating with mouse anti-β-gal (Promega, 1:100) and rabbit anti-Vg (1:100) for 1 hr and then with fluorescent secondary antibodies (Jackson Laboratories) for 1 hr. Topro counterstaining was performed in the final washes. Antibodies to Sal were provided by Reinhard Schuh (Max-Planck Institute für Biophysikalische Chemie, Goettingen, Germany) and to SRF by Markus Affolter (Biocenter, Basel, Switzerland).
Transient transfection of Drosophila S2 cells
vg and sd cDNAs were cloned into the pPacPL expression vector (Koelle and Hogness 1992). Drosophila S2 cells were maintained in M3 medium with 12.5% heat-inactivated fetal bovine serum (FBS) and 1% Pen-Strep (PS) at an average density of 6 × 106 cells/ml with the medium replaced every 2 days. For transfection, cells were seeded at 2 × 106 the night before, washed, and resuspended in M3 without FBS and PS at 3.75 × 106/ml. A mixture of lipofectin, DNA, and M3 was incubated 30 min at room temperature before adding to S2 cells in M3. Following incubation for 6 hr at 25°C, M3 with FBS and PS was added to the cells, which were then incubated for an additional 36 hr at 25°C. lacZ reporter gene expression was assayed with chlorophenol red-β-d-galactopyranoside as a substrate as described (Rouet et al. 1992) except that cell lysis was achieved by use of a hypotonic buffer (20 mm Tris, 10mm NaCl, 20% glycerol, 0.2 mm EDTA, 0.1% NP-40) in place of quick freeze–thaws. This colorometic assay was quantified at 595 nm by a standard 96-well plate reader. Relative activation values are normalized to β-Gal activity of the reporter gene alone in S2 cells. Reported activation of MD2, Vg-A, and Vg-B was with 0.1 μg of Vg and 0.1 μg of Sd; SRF-A, SRF-A mutant, and SRF-B was with 0.1 μg of Vg and 0.03 μg of Sd expression plasmid. All transfections were performed in triplicate in at least three independent experiments. Transfection efficiency ranged from 5% to 15% in independent experiments as assayed by antibody staining for Vg expression. The efficiency was not significantly different between reporter genes or in the presence/absence of Sd and was consistent among triplicate samples. Representative data from a single experiment is shown.
Reporter constructs
Reporter constructs were created by cloning subfragments of the vg quadrant enhancer (Kim et al. 1996; MD2), the vg boundary enhancer (Vg-A and Vg-B), and the SRF Intervein C enhancer (SRF-A and SRF-B) (U. Nussbaumer, J. Montagne, J. Groppe, and M. Affolter, in prep.) into the hsp–lacZ–CaSpeR plasmid (Nelson and Laughon 1993). In the mutant version of the SRF-A reporter construct, the sequence of the A2 binding site was changed to 5′-AAAAATTAGTGGAATAAGT-3′ (mutated bases underlined; the same mutations abolish binding of TEF-1 to high affinity sites in the GT-IIC and Sph enhancers (Davidson et al. 1988)). To replace the Su(H) site in the vg boundary enhancer, the sequence ctagTGGGTGGTCgatccaaagTGGGTGGTCgtaccg, which contains two tandem Ci-binding sites (shown in uppercase), was inserted into the Su(H) site (Kim et al. 1996).
Purification of His-tagged TEA domains
Construction of the TEF-1 TEA domain expression plasmid was described previously (Jacquemin et al. 1996). The Sd TEA domain expression plasmid was constructed by mutagenizing the codon that encodes the one amino acid that differs between the Sd and TEF-1 TEA domains with PCR. Both plasmids express TEA domains tagged with six histidines at their carboxyl terminus. Expression was induced for 2 hr in BL21 Lys S cells and was purified on nickel chelate columns (Novagen). Proteins were dialyzed against buffer containing 100 mm KCl, 10 mm Tris (pH 7.5), 250 μm EDTA, 1 mm DTT, frozen, and stored at −80°C in binding buffer (12% glycerol, 15 mm HEPES at pH 7.9, 50 mm KCl, 4 mmMgCl2, 100 μg/ml BSA, 1 mm DTT). The TEA domains were nearly homogeneous as judged by SDS-PAGE.
Electrophoretic mobility-shift assays
Electrophoretic mobility shift assays were performed by incubating 1–3 fmoles of labeled template with 0.1–10 ng TEA domain and 5 ng of poly [d(AT)] in 25 μl of binding buffer. After 30 min at room temperature, complexes were separated on 6% polyacrylamide gels and standard 0.5× TBE buffer. Templates were annealed oligonucleotides that were radioactively labeled by fill in with Klenow on EcoRI overhangs on either side. Sequences of the upper strand oligonucleotides were Sph (5′-AATTCGCAGAAGTATGCAAAGCATGCATCTC-3′), GT-IIC (5′-AATTCAGCTGTGGAATGTGTGTC-3′), GT-IIC mutant (5′-AATTCACCTTTCTAATTTGTGTC-3′). SRF-A and SRF-B templates: The corresponding regions were PCR amplified with primers that contained EcoRI sites and subcloned into pCRII (Invitrogen). Fragments were then excised with EcoRI, gel purified, and radioactively labeled by fill-in reaction.
DNase I footprinting
A fragment spanning the SRF-A and SRF-B regions was PCR amplified and subcloned into pCRII (Invitrogen). The insert was then cut on one side with XhoI, radioactively labeled by fill-in reaction, and gel purified after complete removal of the insert with HindIII. A total of 30,000 cpm of this fragment was incubated for 30 min together with the Sd TEA domain at room temperature in incubation buffer containing 100 mm KCl, 15 mm HEPES (pH 7.9), 12% glycerol, 4 mm MgCl2, 1 mm CaCl2, 1 mm DTT and 100 μg/ml BSA. For digestion, 5 μl of DNase solution (1:10 dilution of Promega’s RQ1 DNase I in binding buffer without BSA) was added and the reaction stopped after 1 min by adding 140 μl of stop solution (final concentrations: 0.5% SD, 15 mm EDTA, 100 mm NaCl, 25 μg/ml tRNA). After phenol/chloroform extraction and ethanol precipitation, the samples were separated on a 6% polyacrylamide sequencing gel. G+A sequencing reactions were done according to Maxam and Gilbert (1980).
Acknowledgments
We thank John Bell, Ken Irvine, and Alfonso Martinez-Arias for communication of results prior to publication; Ute Nussbaumer, Jacques Montagne, and Markus Affolter for detailed information and valuable materials concerning the SRF enhancer; Ken Irvine for the UAS–sd stock; Irwin Davidson for provision of the TEF-1 expression construct; Scott Weatherbee for the image in Figure 1C; Mike Hoffmann for Drosophila S2 cells; Rosa Barrio, Reinhard Schuh, Markus Affolter, and William Chia for antibodies; Jim Williams for the hs–sd construct; Kathy Vaccaro for technical assistance and Jamie Wilson for help with manuscript preparation. G.H. holds a long-term fellowship from the European Molecular Biology Organization, P.P. is funded by a WARF fellowship, and MEK is a National Institutes of Health postdoctoral fellow. This work was supported by a grant from the National Science Foundation (A.S.L.) and by the Howard Hughes Medical Institute (S.B.C.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sbcarrol@facstaff.wisc.edu; FAX (608) 262-9343.
References
- Bailey A, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brook W, Diaz-Benjumea F, Cohen S. Organizing spatial pattern in limb development. Annu Rev Cell Dev Biol. 1996;12:161–180. doi: 10.1146/annurev.cellbio.12.1.161. [DOI] [PubMed] [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–254. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Campbell S, Duttaroy A, Katzen A, Chovnick A. Cloning and characterization of the scalloped region of Drosophila melanogaster. Genetics. 1991;127:367–380. doi: 10.1093/genetics/127.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes & Dev. 1992;6:367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- Davidson I, Xiao J, Rosales R, Staub A, Chambon P. The HeLa cell protein TEF-1 binds specifically and cooperatively to two SV40 enhancer motifs of unrelated sequence. Cell. 1988;54:931–942. doi: 10.1016/0092-8674(88)90108-0. [DOI] [PubMed] [Google Scholar]
- de Celis J, Barrio R, Kafatos F. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature. 1996a;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- de Celis J, Garcia-Bellido A, Bray S. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 1996b;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- Deshpande N, Chopra A, Rangarajan A, Shashidhara L, Rodrigues V, Krishna S. The human transcription enhancer factor-1, TEF-1, can substitute for Drosophila scalloped during wingblade development. J Biol Chem. 1997;272:10664–10668. doi: 10.1074/jbc.272.16.10664. [DOI] [PubMed] [Google Scholar]
- Grieder N, Marty T, Ryco H, Mann R, Affolter M. Synergistic activation of a Drosophila enhancer by HOM/EXD and DPP signaling. EMBO J. 1997;16:7402–7410. doi: 10.1093/emboj/16.24.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S, Pflugfelder G. Control of the gene optomotor-blind in Drosophila wing development by decapentaplegic and wingless. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- Gupta M, Amin C, Gupta M, Hay N, Zak R. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of cardiac α-Myosin heavy-chain gene expression. Mol Cel Biol. 1997;17:3924–3936. doi: 10.1128/mcb.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring W. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Hwang J-J, Chambon P, Davidson I. Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J. 1993;12:2337–2348. doi: 10.1002/j.1460-2075.1993.tb05888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiji T, Lace M, Parkkinen S, Anderson R, Haugen T, Cripe T, Xiao J-H, Davidson I, Chambon P, Turek L. Transcriptional enhancer factor (TEF-1) and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. EMBO J. 1992;11:2271–2281. doi: 10.1002/j.1460-2075.1992.tb05286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack J, Dorsett D, Delotto Y, Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 1991;113:735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Davidson I. The role of the TEF transcription factors in cardiogenesis and other developmental processes. Trends Cardiovasc Med. 1997;7:192–197. doi: 10.1016/S1050-1738(97)00052-2. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Hwang J-J, Martials J, Dollé P, Davidson I. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J Biol Chem. 1996;271:21775–21785. doi: 10.1074/jbc.271.36.21775. [DOI] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch J, Kraus M, Vorwerk K, Magee J, Carroll S. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll SB, Laughon A. MAD binds to DNA and directly mediates activation of vestigial by DPP. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Klein T, Martinez Arias A. Different spatial and temporal interactions between Notch, wingless, and vestigial specify proximal and distal pattern elements of the wing in Drosophila. Dev Biol. 1998;194:196–212. doi: 10.1006/dbio.1997.8829. [DOI] [PubMed] [Google Scholar]
- Koelle, M. and D. Hogness. 1992. Technical notes: Plasmid vectors for expressing proteins in tissue culture cells or studying enhancer function. Dros. Inf. Newsl. 8.
- Kuhnlein R, Bronner G, Taubert H, Schuh R. Regulation of Drosophila spalt gene expression. Mech Dev. 1997;66:107–118. doi: 10.1016/s0925-4773(97)00103-2. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Brook W, Ng M, Calleja M, Sun H, Cohen S. Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing. Nature. 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- Maxam A, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Meth Enzymol. 1980;65:499–525. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Micchelli C, Rulifson E, Blair S. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Winglesss and a dominant nagative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- Montagne J, Groppe J, Guillemin K, Krasnow M, Gehring W, Affolter M. The Drosophila serum response factor gene is required for the formation of intervein tissue of the wing and is allelic to blistered. Development. 1996;122:2589–2597. doi: 10.1242/dev.122.9.2589. [DOI] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Dorsett D. Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics. 1996;144:1153–1154. doi: 10.1093/genetics/144.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura S, Maves L, Chen Y, Hoffmann F. decapentaplegic overexpression affects Drosophila wing and leg imaginal disc development and wingless expression. Dev Biol. 1996;10:136–151. doi: 10.1006/dbio.1996.0151. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range actions of a Dpp morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Nelson H, Laughon A. The DNA binding specificity of the Drosophila fushi tarazu protein: A possible role for DNA bending in homeodomain recognition. Wilhelm Roux’s Arch Dev Biol. 1993;202:341–354. [PubMed] [Google Scholar]
- Neumann C, Cohen S. A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development. 1996;122:3477–3485. doi: 10.1242/dev.122.11.3477. [DOI] [PubMed] [Google Scholar]
- ————— Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- Ng M, Diaz-Benjumea F, Cohen S. nubbin encodes a POU-domain protein required for proximal-distal patterning in the Drosophila wing. Development. 1995;121:589–599. doi: 10.1242/dev.121.2.589. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky S. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Rouet P, Raguenez G, Salier J. Optimized assays for quantifying transient expressions of co-transfected beta-galactasidase and CAT reporter genes. BioTechniques. 1992;13:700–701. [PubMed] [Google Scholar]
- Serrano N, O’Farrell P. Limb morphogenesis: Connections between patterning and growth. Curr Biol. 1997;7:R186–R195. doi: 10.1016/s0960-9822(97)70085-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds, A., X. Liu, K. Soanes, H. Krause, K. Irvine, and J. Bell. 1998. Molecular interactions between Vestigial and Scalloped to promote wing formation in Drosophila. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Sturtevant M, Biehs B, Marin E, Bier E. The spalt gene links the A/P compartment boundary to a linear adult structure in the Drosophila wing. Development. 1997;124:21–32. doi: 10.1242/dev.124.1.21. [DOI] [PubMed] [Google Scholar]
- Weatherbee S, Halder G, Hudson A, Kim J, Carroll S. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes & Dev. 1998;10:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Bell J, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes & Dev. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Carroll SB. Pattern formation in a secondary field: A hierarchy of regulatory genes subdivides the developing Drosophila wing disc into discrete sub-regions. Development. 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Vorwerk K, Carroll SB. Organization of wing formation and induction of a wing-patterning gene at a compartment boundary. Nature. 1994;368:299–305. doi: 10.1038/368299a0. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Davidson H, Matthes JM, Garn Cloning, expression and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- Xu X, Yin Z, Hudson J, Ferguson E, Frasch M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes & Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long-range action of a wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]


