Abstract
Scalloped (Sd) and Vestigial (Vg) are each needed for Drosophila wing development. We show that Sd is required for Vg function and that altering their relative cellular levels inhibits wing formation. In vitro, Vg binds directly to both Sd and its human homolog, Transcription Enhancer Factor-1. The interaction domains map to a small region of Vg that is essential for Vg-mediated gene activation and to the carboxy-terminal half of Sd. Our observations indicate that Vg and Sd function coordinately to control the expression of genes required for wing development, which implies that Vg is a tissue-specific transcriptional intermediary factor of Sd.
Keywords: Vestigial, Scalloped, Wing, Drosphila, transcription, development
The Drosophila vestigial (vg) and scalloped (sd) genes are expressed in similar patterns during wing development, and mutations in either gene lead to loss of wing tissue (Campbell et al. 1991, 1992; Williams et al. 1991, 1993). Vg is a developmentally regulated nuclear protein of previously unknown function and is required principally for the development of the wing and haltere (Williams et al. 1991). Sd is part of a highly conserved family of transcription factors, the TEA/ATTS domain proteins and is an essential protein with a wider developmental role (Campbell et al. 1991, 1992).
Expression of vg in cells of the developing wing primordia is established by a number of conserved signaling pathways and is required for subsequent cell proliferation and patterning. Expression of wingless (wg), as well as interactions between dorsal and ventral cells that activate the Notch receptor, initially directs limited vg expression along the dorsal–ventral (D/V) wing boundary (Williams et al. 1994; Kim et al. 1995, 1996; for review, see Irvine and Vogt 1997). Subsequent vg expression in the wing primordia occurs in response to both the D/V Wg signal and Decapentaplegic (Dpp), a member of the Transforming Growth Factor-β (TGF-β) protein family, secreted by cells along the anterior–posterior (A/P) border. (Blair 1994; Kim et al. 1996, 1997; Zecca et al. 1996; Neumann and Cohen 1997). By the late third larval instar, maximal amounts of Vg are seen in cells at the D/V wing disc boundary, whereas cells located farther from this border produce progressively less Vg (Williams et al. 1991). vg is also required to maintain sd expression in the wing progenitor cells, and sd is similarly required for the maintenance of elevated vg expression (Williams et al. 1993).
A cellular role for Sd can be inferred from studies of its human homolog Transcription Enhancer Factor-1 (TEF-1). TEF-1 binds to SV40 enhancer sequences via a TEA/ATTS class DNA-binding domain, and has been shown to require transcriptional intermediary factors (TIFs) for proper function (Xiao et al. 1991; Ishiji et al. 1992; Hwang et al. 1993; Gupta et al. 1997). Interestingly, it has been reported that the Sd TEA/ATTS domain does not bind the same enhancer DNA sequences in vitro as TEF-1, although TEF-1 can substitute for Sd during Drosphila wing development (Hwang et al. 1993; Deshpande et al. 1997). This suggests that other factors within Drosphila wing cells interact with and modify the specificity of both Sd and TEF-1 in a similar fashion.
Results and Discussion
Previous analysis has shown that ectopic expression of Vg, under control of a dpp enhancer, can induce transformation of some cells in the eye, antenna, leg, and genital imaginal discs into wing-specific fates, as well as causing tissue overgrowth (Fig. 1b; Kim et al. 1996). Importantly, whereas Vg expression is normally restricted to the wing and haltere imaginal discs, a subset of cells within almost all imaginal discs normally express sd (Campbell et al. 1992). Thus, when Vg is ectopically produced a supply of Sd is already present in those tissues. As Sd is required for formation of the normal wing, we tested whether there is a similar requirement for Sd in the formation of Vg-induced ectopic wings. The induction of wing tissue overgrowths by ectopic Vg was partially suppressed in animals heterozygous for a strong viable allele of sd (Fig. 1d), sd58, and was completely suppressed in sd58 hemizygotes (Fig. 1e). These observations demonstrate that Vg requires Sd to transform cells to wing fates. This requirement does not appear to reflect a role for Sd as a downstream effector of Vg function, as expression of Sd alone, whether under the control of dpp or other promoters, does not induce the formation of ectopic wing tissue (Fig. 1c; data not shown). Instead, these observations suggest that Sd and Vg could act in parallel to induce wing cell fates.
Figure 1.
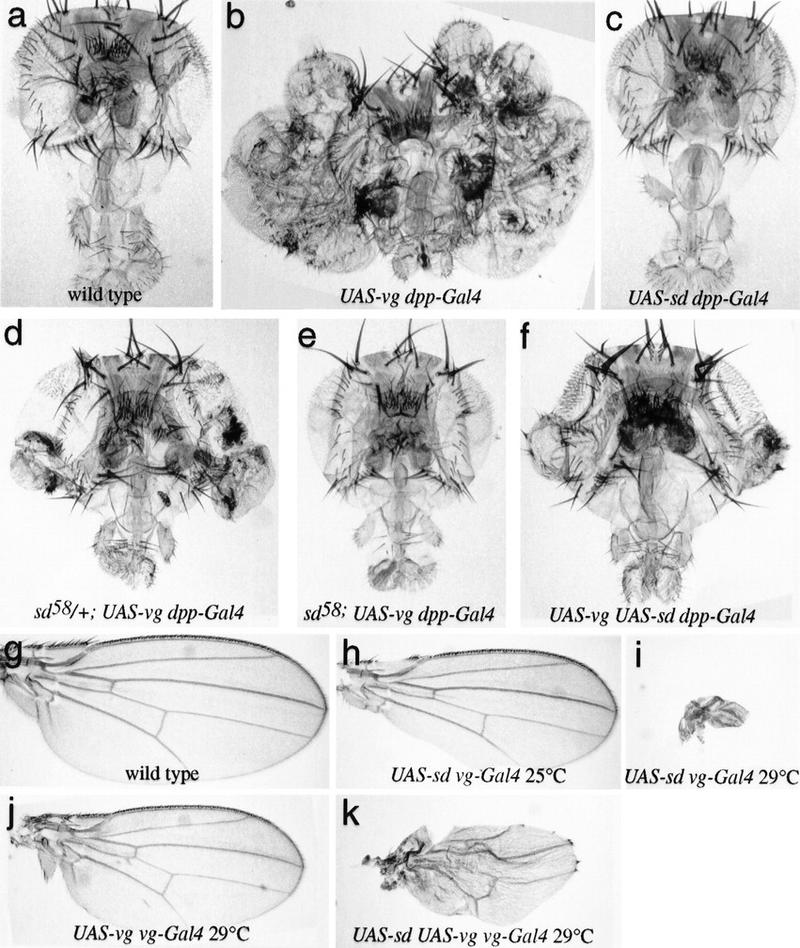
Vg requires Sd and is sensitive to Sd levels in vivo. (a–f) Drosphila heads; (g–k) Drosphila wings, in which the Vg and/or Sd proteins have been ectopically expressed using the UAS–Gal4 system (Brand and Perrimon 1993). (a) Wild type. (b) UAS–vg dpp–GAL4. A massive outgrowth of wing tissue from the eye occurs. (c) UAS–sd dpp–GAL4. No wing tissue is induced, and higher levels of expression (e.g., using ptc–GAL4) result in loss of head tissue. (d) sd58/+; UAS–vg dpp–GAL4. The sd58 mutation reduces the amount of Sd produced, and formation of ectopic wing tissue is partially suppressed. (e) sd58; UAS–vg dpp–GAL4. The formation of wing tissue is completely suppressed when no wild-type copy of sd is present. (f) UAS–vg UAS–sd dpp–GAL4. The formation of wing tissue is also partially suppressed when levels of Sd are increased. Raising or lowering the levels of Sd relative to that of Vg within the developing wing disc produces corresponding phenotypes. (g) Wild type. (h) UAS–sd vg–GAL4 at 25°C. Elevating Sd levels in wing progenitor cells leads to incomplete formation of the wing margin. (i) UAS–sd vg–GAL4 at 29°C. This effect becomes more severe in flies raised at 29°C, where almost no wing tissue is formed. (j) UAS–vg vg–GAL4 at 29°C. The elevated Vg levels in the wings of these animals leads to a slight reduction in wing size. (k) UAS–sd UAS–vg vg–GAL4 at 29°C. The loss of wing tissue induced by excess Sd (i) is partially suppressed by the simultaneous increase in Vg expression.
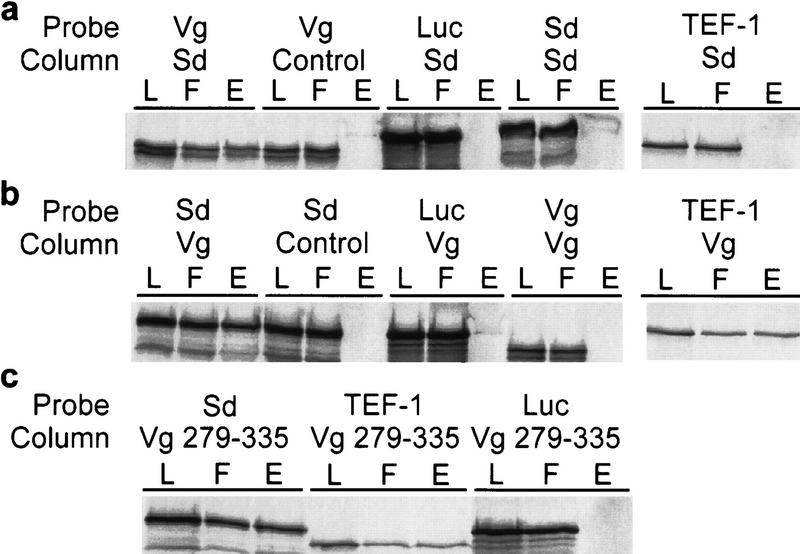
The possibility that coordinate action of Sd and Vg is effected via a direct protein–protein interaction was examined by in vitro-binding experiments. Microaffinity columns bound with bacterially expressed Sd protein selectively retain in vitro-translated Vg (Fig. 2a). Similarly, Vg columns show specific retention of Sd (Fig. 2b). TEF-1, the vertebrate homolog of Sd, was retained on Vg columns with similar affinity (Fig. 2b). Control columns containing the homeodomain proteins Engrailed (En) and Fushi tarazu (Ftz; not shown) did not retain Vg or Sd. Additionally, Luciferase (Luc) did not bind to Vg or Sd columns, confirming that the Sd–Vg interaction is specific (Fig. 2a–c). Notably, neither Vg nor Sd bound to themselves either, suggesting that they do not form homomultimers (Fig. 2a,b).
Figure 2.
Vg and Sd exhibit a strong protein–protein interaction in vitro. (a) Affinity chromatography was performed using bound bacterially expressed Sd protein [(L) column load; (F) column flowthrough; (E) column eluate]. Sd columns selectively retain Vg and not Sd or TEF-1. As a control for nonspecific binding, Luc was expressed in the same in vitro system and shown to not bind to the Sd column. Similarly, the column matrix alone does not retain significant amounts of labeled Vg. (b) Vg columns specifically bind Sd protein but not Vg or Luc. The human TEF-1 protein, a homolog of Drosphila Sd, also shows a similar specific affinity for Vg. (c) Affinity columns bound with a protein consisting of Vg amino acids 279–335 (see Fig. 3) can selectively retain Sd or TEF-1 but not Luc.
To map Vg–Sd and Vg–TEF-1 interaction domains, a Far Western blotting assay was used to screen 15 deleted proteins that remove terminal or internal regions of Vg (Fig. 3a–c). Only Vg proteins that contain amino acids 279–335 have any significant affinity for Sd (Fig. 3b,c). The Vg–Sd interaction appears to be limited to this 56-amino-acid domain, as Sd does not bind to a deleted Vg protein missing only these amino acids, and a construct encoding only this portion of the protein will still bind to Sd. Significantly, a duplicate panel of Vg deletion proteins probed with TEF-1 (Fig. 3b,c) shows that TEF-1 interacts with Vg via the same protein domain. Affinity columns containing this protein fragment of Vg bind Sd and TEF-1 protein as well as full-length Vg does (Fig. 2c). This Sd/TEF-1-binding domain of Vg is serine rich and includes putative phosphorylation sites (Williams et al. 1991). Phosphorylation of Vg at these sites may potentially modify the Vg–Sd interaction. This region is highly conserved in Vg proteins from Drosphila virilis (Fig. 3d) and Aedes aegypti (S. Carroll, pers. comm.). Similar sequences also occur in mammalian genomic and expressed sequence tag databases (Fig. 3d). The amino- and carboxy-terminal portions of Sd were also tested to map which region of Sd interacts with Vg. Previous studies with TEF-1 demonstrated that regions mediating interaction with cell-specific TIFs were separable from the DNA-binding TEA/ATTS domain (Xiao et al. 1991). The Vg-binding region of Sd maps to the carboxy-terminal half of the protein, separable from the TEA/ATTS domain in the amino-terminal half (Fig. 3e). The carboxy-terminal portion of Sd is also highly similar to TEF-1 (Campbell et al. 1992; Hwang et al 1993; Despande et al. 1997), which is consistent with the observation (Fig. 3c) that TEF-1 binds to Vg with the same affinity as Sd does. To confirm the direct protein–protein interaction between Vg and Sd in a cellular environment, a yeast two-hybrid assay was used. In yeast, Vg and Sd proteins show a specific and reciprocal interaction when fused to either Gal4-binding domain (pGBDU) or Gal4 activation domain (pACT) fusion constructs (not shown; James et al. 1996). Each construct alone or paired with nonspecific (pSNF1BD) bait sequences does not activate a lacZ reporter.
Figure 3.
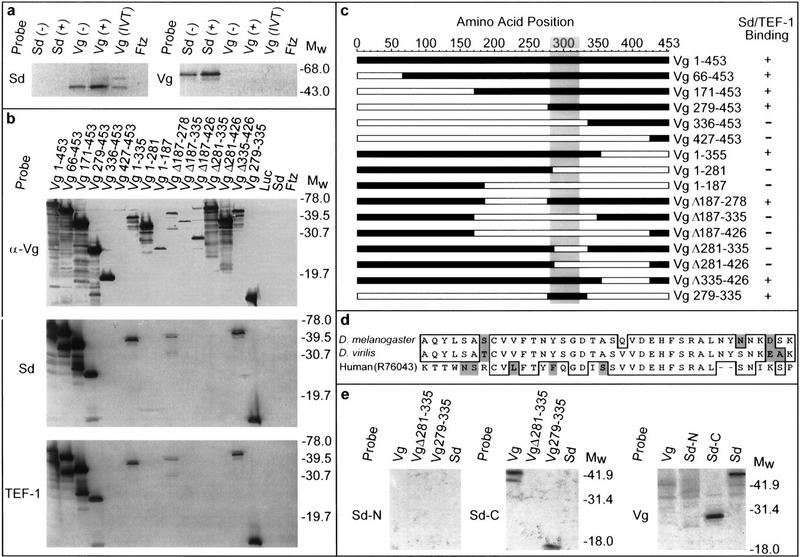
Mapping the Vg–Sd interaction domain. (a) Far Western blots of bacterial or in vitro-translated (IVT) Vg or Sd were probed with labeled Vg or Sd probes. Ftz protein expressed under similar conditions was also included on each blot to detect nonspecific probe binding. Sd selectively binds bacterially produced and IVT Vg, whereas a corresponding blot probed with Vg shows binding only to Sd. As the detected band is more intense in cell extracts after induction (+) of the expression plasmid than before (−), this confirms that each is plasmid and not endogenously encoded. (b) All but one of the specific deletions of Vg (c) were recognized by anti-Vg antibody (α-Vg), confirming that Vg is present in each lane. Expression of the Vg 427–453 protein was verified by Coomassie staining. When these deleted proteins were probed with Sd or TEF-1 under conditions similar to those above, only proteins retaining amino acids 279–335 of Vg were detected. Luc, Sd, or Ftz control lanes did not show any signals. (c) A map summarizing the position and relative size of the deletions (open bars) within Vg assayed for binding (+ or −) to Sd or TEF-1. The shaded area denotes a region that is highly similar to Vg from D. virilis and A. aegypti (S. Carroll, pers. comm.). (d) This region is also highly similar to sequences identified in mammalian genomic and expressed sequence tag databases (GenBank/EST accession nos. Z798880, Z97632, AA474871, AA571483, W81241, R65857, R73306, R76043). In addition to the 38 amino acids indicated, the smallest tested portion of Vg that has Sd-binding ability includes an amino-terminal Q and amino acids (ESSSPMSSRNFPPSFWN) carboxy-terminal to this homologous region. (e) Probes corresponding to the amino-terminal half of Sd (amino acids 1–242, Sd-N) show no interaction with Vg. The carboxy-terminal half of Sd (amino acids 240–440, Sd-C) binds to Vg protein containing only amino acids 279–335 (Sd-binding domain). A reciprocal blot using full-length Vg (Vg) as a probe shows interaction only with the carboxy-terminal half of Sd (Sd-C).
We employed the UAS–Gal4 system (Brand and Perrimon 1993) to further examine the in vivo significance of the Sd–Vg interaction. As vg is normally required for sd expression in wing imaginal discs (Williams et al. 1993), we examined the influence of ectopic Vg expression on sd. Normally, sd expression appears confined to the wing pouch region of the wing imaginal disc, although very low levels can sometimes be detected elsewhere. Strong sd expression is induced outside of the wing pouch in wing disc cells where vg is expressed ectopically under the control of the patched (ptc) promoter (UAS–vg ptc–GAL4) (Fig. 4 a–d). Activation of a downstream target gene by ectopic expression of Vg and Sd was observed using expression of the cut (ct) gene as an assay. ct is likely a direct Vg–Sd target in Drosphila, as sd, vg, and ct interact genetically, and the Sd TEA/ATTS domain has been shown to bind to ct wing margin enhancer DNA sequences (Morcillo et al. 1996). Consistent with our observation of ectopic sd expression and the hypothesis that Sd and Vg function coordinately to regulate gene expression, ectopic expression of Vg in the wing under ptc–GAL4 control also produces activation of ct (Fig. 4e). However, when a UAS–vg construct that is missing the 56-amino-acid Sd binding domain is expressed, sd and ct are not induced (Fig. 4g,h). Thus, activation of both target genes, sd and ct, by Vg requires the Sd–Vg interaction domain identified in vitro, implying that this is Sd dependent. Moreover, the UAS–vg construct with the Sd-binding domain deleted is also unable to induce the formation of ectopic wing tissue (results not shown), consistent with the observation that this induction is sd dependent (Fig. 1e). Interestingly, Vg protein deleted for the Sd-binding domain is found in the cytoplasm rather than in the nucleus (Fig. 4f),which implies that association with Sd is required for nuclear localization of Vg.
Figure 4.
Influence of ectopic Vg on gene expression. (a) Third instar larval wing discs of genotype sd–lacZ; ptc–GAL4 showing a normal pattern of high levels of sd–lacZ expression in all cells of the presumptive wing pouch (Campbell et al. 1992). Using in situ hybridization to sd mRNA, low levels of sd expression are also detected outside of the wing pouch (not shown). A localized depression of sd activity is usually seen where the D/V intersects the A/P boundary. (b) ptc–GAL4 activates a UAS–lacZ reporter along the A/P axis of the wing disc. Note that the highest levels of activation occur in cells immediately adjacent to the border (arrow); lower levels of lacZ expression are seen in more anterior cells. (c) A ptc–GAL4; UAS–vg wing imaginal disc shows a similar graded pattern of ectopic vg expression along the A/P margin (arrow). (d) Third instar wing discs of genotype sd–lacZ; ptc–GAL4; UAS–vg. The ectopic vg expression activates sd along the A/P margin, causing a nongraded level of activation of the sd–lacZ reporter gene in vg-expressing cells (arrow). (e) A wing imaginal disc of the same genotype as c stained with anti-ct antibody. Ectopic activation of ct occurs in some cells that express ectopic Vg and Sd along the A/P margin. (f) VgΔ281–335 protein (driven by ptc–GAL4) is found primarily in the cytoplasm [arrow and inset (high magnification)] compared to the nuclear localization of endogenous wild-type Vg (arrowhead). (g) In discs containing sd–lacZ, ptc–GAL4 mediated expression of a UAS–vgΔ281–335 construct (missing the Sd-binding domain) does not induce sd expression along the A/P margin (arrow). (h) Where high levels of ectopic VgΔ281–335 and Sd overlap, ct expression remains in the wild-type pattern along the D/V border.
Further support of a direct Sd–Vg interaction in vivo comes from the observation that elevated amounts of Sd protein can also result in programmed cell death and wing tissue loss (Fig. 1g–i; data not shown). These phenotypes are very similar to those of sd or vg loss-of-function mutants (Williams et al. 1991, 1993; Campbell et al. 1992). The apparent dominant-negative effect of Sd overexpression suggests that a Vg–Sd heterodimer is functional in vivo and that Sd alone can compete with the functional heterodimer for binding to DNA or other essential cofactors. This suggestion is consistent with studies of the transcriptional activity of TEF-1 and Sd in cultured cell lines, in which dominant-negative effects (squelching) appear to occur when they are overexpressed (Xiao et al. 1991; Ishiji et al. 1992; Hwang et al. 1993; Halder et al. 1998). Direct in vivo support for this hypothesis comes from the observation that overexpression of Sd is able to suppress the consequences of ectopic Vg expression almost as efficiently as loss of sd function (Fig. 1f). Moreover, overexpression of Vg in the wing is able to partially suppress the tissue loss otherwise associated with Sd overexpression (Fig. 1, cf. k and i). Together, these observations argue that balanced levels of Sd and Vg are essential for normal wing development and further support the conclusion that the specific interaction between Vg and Sd identified in vitro is essential for function in vivo. The exquisite sensitivity of wing tissue growth to the Sd/Vg ratio also raises the possibility that variation of this balance could be a mechanism of growth control during normal development.
A wide variety of studies have suggested that TEA/ATTS domain proteins require tissue-specific TIFs, although relatively little progress has been made toward identifying and characterizing these TIFs (Xiao et al. 1987, 1991; Ishiji et al. 1992; Hwang et al. 1993; Chen et al. 1994; Farrance and Ordahl 1996; Gavrias et al. 1996; Jacquemin et al. 1996; Stewart et al. 1996; Gupta et al. 1997). According to the definitions established by the analysis of TEF-1 (Xiao et al. 1987, 1991; Ishiji et al. 1992), a TIF for Sd would be expected to bind directly to Sd, to show a restricted pattern of expression, and to be required for Sd function in vivo. Previous studies have shown that sd and vg have similar mutant phenotypes and patterns of expression in the developing wing (Williams et al. 1991; Kim et al. 1996, 1997). The results presented here demonstrate that Sd and Vg proteins bind to each other and that the cooperative action of Sd and Vg is required in vivo for Drosphila wing development. These observations, together with the analysis of the coordinate regulation of downstream target genes by Sd and Vg (Halder et al. 1998), argue that Vg functions as a tissue-specific TIF for Sd. Although it is possible that Vg interacts with proteins other than Sd, genetic studies argue against this, because all vg mutant phenotypes are shared by sd. In contrast, sd is required for the development of other tissues in which vg is not required (Campbell et al. 1991; Williams et al. 1991). Thus, it is likely that there are other trans-acting factors in Drosphila that interact with Sd.
Although the DNA target sequence of Sd is as yet uncharacterized, one target of a yeast TEA/ATTS domain protein (TEC-1) is an element in the TEC-1 promoter (Madhani and Fink 1997). Likewise, in flies, one target of Sd–Vg is likely to be the sd promoter itself, as activation of sd during early development is dependent on Vg (Williams et al. 1993), and ectopic Vg induces elevated expression of sd (Fig. 4). This suggests a model whereby low levels of Sd expression within wing imaginal discs are elevated in the presence of Vg by positive autoregulation. The dependence of elevated levels of Vg in the wing disc on sd suggests that vg is also a target of positive autoregulation, and direct evidence for this now been obtained (Halder et al. 1998).
The TEA/ATTS domain protein family is involved in developmental processes as diverse as mammalian neuronal and cardiac muscle development (Chen et al. 1994; Jacquemin et al. 1996; Yockey et al. 1996) to conidial formation in Aspergillus (Gavrias et al. 1996) and pseudohyphal growth of Saccharomyces cerevisiae (Madhani and Fink 1997). Although Vg homologs have not yet been identified in these organisms, we have found that genes containing sequences related to the Sd/TEF-1 interaction domain of Vg are conserved in mammals; these genes are thus candidate TIFs for mammalian TEF-1-related proteins (Fig. 3). One of these candidate TIFs is expressed in fetal heart tissue, which is intriguing given that gene-targeted mutations in TEF-1 result in cardiac defects (Chen et al. 1994). Future challenges will be to determine whether genes encoding these putative Sd-interacting domains actually function as TIFs for TEF-1 or related proteins, and whether distinct regulatory TIFs have evolved that adapt the transcriptional activities of conserved Sd/TEF-1 homologs to specific functions in different tissues in their respective organisms.
Materials and methods
For the in vitro protein interaction assays, PCR fragments containing either full-length sd or vg coding region were cloned into pET16b (Novagen) and expressed in Escherichia coli (BL21–DE3 pLysE) to produce 6× His-tagged Vg or Sd protein. Inverse PCR, using primers that amplified specific portions of the vg or sd coding region shown in Figure 3, was used to create deleted Vg and Sd expression constructs. Recombinant proteins containing the amino-terminal histidine tag were purified on Ni2+ resin following the manufacturer’s directions (Invitrogen). Affinity chromatography, Western, and Far Western blotting were performed as described previously (Guichet et al. 1997), with the following modifications. For microcolumn affinity chromatography, 500 μl of a 200 μg/ml solution of each protein to be tested was coupled to 100 μl of Ni2+ resin in binding buffer (20 mm NaH2PO4 at pH 7.8, 0.5 m NaCl) and rewashed according to the manufacturer’s directions (Invitrogen). Columns with Ni2+ resin alone served as controls for nonspecific binding of the labeled probe. Each column was then equilibrated in 100 mm NaCl, 0.1 m Tris (pH 7.6), and 10% glycerol. Columns were subsequently blocked using a solution of 0.5% skim milk powder in equilibrium solution. 35S-Labeled probes were made by cloning the relevant coding region of each protein to be tested into pBluescript SK(−) and then expressed using the TnT in vitro-coupled rabbit reticulocyte system (Promega). The resulting probes were purified using Sepharose G25 spin columns, and a 2-μl aliquot of each was then quantified using a PhosphorImager. Approximately 40–45 μl of each labeling reaction was combined with 80 μl of blocking solution and passed over the respective affinity column. Any bound protein was eluted using 100 μl of the equilibration solution plus 2% SDS. For Far Western blotting, ∼10 μl of total lysate from bacteria expressing Vg, Sd, or control protein per lane was separated on a 13% SDS–polyacrylamide gel and blotted to nitrocellulose. Proteins on the resulting blot were serially renatured in a series of 6, 3, 1, and 0.1 m guanidine–HCl solutions containing 20 mm Tris (pH 7.6), 100 mm NaCl, 0.1% Tween 20, 2% skim milk powder, 10% glycerol, and 1 mm EDTA. Probes prepared as described were incubated with each blot for 2 hr at 4°C in blocking solution plus 1 mm DTT. Antibody staining of imaginal discs was performed as described previously (Simmonds et al. 1995). Rabbit anti-Vg was used at a dilution of 1/50 on imaginal discs and at 1/200 for Western blotting (Williams et al. 1991). Mouse anti-β-galactosidase (Promega) and mouse anti-Ct 2B10 (Blochlinger et al. 1990) were used at 1/500. sd–lacZ refers to the sdETX4 line described by Campbell et al. (1992). UAS–sd and UAS–vgΔ281–335 transgenes were constructed by cloning a 2.05-kb XmnI–ClaI fragment of a sd cDNA (Campbell et al. 1992) or a PCR fragment consisting of the coding region of the VgΔ281–335 expression construct into pUAST (Brand and Perrimon 1993).
Acknowledgments
We thank Sean Carroll for communication of results prior to publication. A.J.S. was supported by the Alberta Heritage Foundation for Medical Research (AHMFR) and a Charles H. Best postdoctoral fellowship. Research in the laboratory of J.B.B. is supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada. Research in K.D.I.’s laboratory was supported by a grant from the New Jersey Commission on Cancer Research. The anti-Ct 2B10 antibody was obtained from the Developmental Studies Hybridoma Bank (University of Iowa). We acknowledge Garry Ritzel for the yeast two-hybrid assays, Shelagh Campbell and Sarah Hughes for suggestions and critical reading of this manuscript, and Shelagh Campbell, Arthur Chovnick, Sean Carroll, I. Davidson, and P. Chambon for reagents.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jbell@gpu.srv.ualberta.ca; FAX (403) 492-9234.
References
- Blair SS. A role for the segment polarity gene shaggy-zeste white 3 in the specification of regional identity in the developing wing of Drosphila. Dev Biol. 1994;162:229–244. doi: 10.1006/dbio.1994.1081. [DOI] [PubMed] [Google Scholar]
- Blochlinger KR, Bodmer R, Jan LY, Jan YN. Patterns of expression of cut, a protein required for external sensory organ development in wild type and mutant Drosphila embryos. Genes & Dev. 1990;4:1322–1331. doi: 10.1101/gad.4.8.1322. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Campbell SD, Duttaroy A, Katzen AL, Chovnick A. Cloning and characterization of the scalloped region of Drosphila melanogaster. Genetics. 1991;127:367–380. doi: 10.1093/genetics/127.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SD, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosphila. Genes & Dev. 1992;6:367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- Chen Z, Friedrich GA, Soriano P. Transcriptional enhancer factor 1 disruption by a retroviral gene trap leads to heart defects and embryonic lethality in mice. Genes & Dev. 1994;8:2293–2301. doi: 10.1101/gad.8.19.2293. [DOI] [PubMed] [Google Scholar]
- Deshpande N, Chopra A, Rangarajan A, Shashidhara LS, Rodrigues V, Krishna S. The human transcription enhancer factor-1, TEF-1, can substitute for Drosphila scalloped during wingblade development. J Biol Chem. 1997;272:10664–10668. doi: 10.1074/jbc.272.16.10664. [DOI] [PubMed] [Google Scholar]
- Farrance IK, Ordahl CP. The role of transcription enhancer factor-1 (TEF-1) related proteins in the formation of M-CAT binding complexes in muscle and non-muscle tissues. J Biol Chem. 1996;271:8266–8274. doi: 10.1074/jbc.271.14.8266. [DOI] [PubMed] [Google Scholar]
- Gavrias V, Andrianopoulos A, Gimeno CJ, Timberlake WE. Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- Guichet A, Copeland JW, Erdelyi M, Hlousek D, Zavorszky P, Ho J, Brown S, Percival-Smith A, Krause HM, Ephrussi A. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- Gupta MP, Amin CS, Gupta M, Hay N, Zak R. Transcription enhancer factor 1 interacts with a basic helix-loop-helix zipper protein, Max, for positive regulation of Cardiac a-myosin heavy-chain gene expression. Mol Cell Biol. 1997;17:3924–3936. doi: 10.1128/mcb.17.7.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder, G., P. Polaczyk, M.E. Kraus, A. Hudson, J. Kim, A. Laughon, and S. Carroll. 1998. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in response to signaling proteins. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Hwang JJ, Chambon P, Davidson I. Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J. 1993;12:2337–2348. doi: 10.1002/j.1460-2075.1993.tb05888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine KD, Vogt TF. Dorsal-ventral signaling in limb development. Curr Opin Cell Biol. 1997;9:867–876. doi: 10.1016/s0955-0674(97)80090-7. [DOI] [PubMed] [Google Scholar]
- Ishiji T, Lace MJ, Parkkinen S, Anderson RD, Haugen TH, Cripe TP, Xiao JH, Davidson I, Chambon P, Turek LP. Transcriptional enhancer factor (TEF)-1 and its cell-specific co-activator activate human papillomavirus-16 E6 and E7 oncogene transcription in keratinocytes and cervical carcinoma cells. EMBO J. 1992;11:2271–2281. doi: 10.1002/j.1460-2075.1992.tb05286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemin P, Hwang JJ, Martial JA, Dolle P, Davidson I. A novel family of developmentally regulated mammalian transcription factors containing the TEA/ATTS DNA binding domain. J Biol Chem. 1996;271:21775–21785. doi: 10.1074/jbc.271.36.21775. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Irvine KD, Carroll SB. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosphila wing. Cell. 1995;82:795–802. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosphila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosphila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Dorsett D. Genes regulating the remote wing margin enhancer in the Drosphila cut locus. Genetics. 1996;144:1143–1154. doi: 10.1093/genetics/144.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CJ, Cohen SM. Long-range action of Wingless organizes the dorsal-ventral axis of the Drosphila wing. Development. 1997;124:871–880. doi: 10.1242/dev.124.4.871. [DOI] [PubMed] [Google Scholar]
- Simmonds AJ, Brook WJ, Cohen SM, Bell JB. Distinguishable functions for engrailed and invected in anterior-posterior patterning in the Drosphila wing. Nature. 1995;376:424–427. doi: 10.1038/376424a0. [DOI] [PubMed] [Google Scholar]
- Stewart AF, Richard CW, III, Suzow J, Stephan D, Weremowicz S, Morton CC, Adra CN. Cloning of human RTEF-1, a transcriptional enhancer factor-1-related gene preferentially expressed in skeletal muscle: Evidence for an ancient multigene family. Genomics. 1996;36:68–76. doi: 10.1006/geno.1996.0522. [DOI] [PubMed] [Google Scholar]
- Williams JA, Bell JB, Carroll SB. Control of Drosphila wing and haltere development by the nuclear vestigial gene product. Genes & Dev. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Carroll SB. Pattern formation in a secondary field: A hierarchy of regulatory genes subdivides the developing Drosphila wing disc into discrete subregions. Development. 1993;117:571–584. doi: 10.1242/dev.117.2.571. [DOI] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Vorwerk K, Carroll SB. Organization of wing formation and induction of a wing-patterning gene at the dorsal/ventral compartment boundary. Nature. 1994;368:299–305. doi: 10.1038/368299a0. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Davidson I, Ferrandon D, Rosales R, Vigneron M, Macchi M, Ruffenach F, Chambon P. One cell-specific and three ubiquitous nuclear proteins bind in vitro to overlapping motifs in the domain B1 of the SV40 enhancer. EMBO J. 1987;6:3005–3013. doi: 10.1002/j.1460-2075.1987.tb02606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- Yockey CE, Smith G, Izumo S, Shimizu N. cDNA cloning and characterization of murine transcriptional enhancer factor-1-related protein 1, a transcription factor that binds to the M-CAT motif. J Biol Chem. 1996;271:3727–3736. doi: 10.1074/jbc.271.7.3727. [DOI] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long range action of a wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]