Abstract
Many flaviviruses are significant human pathogens. The plus-strand RNA genome of a flavivirus contains a 5′ terminal cap 1 structure (m7GpppAmG). The flavivirus encodes one methyltransferase (MTase), located at the N-terminal portion of the NS5 RNA-dependent RNA polymerase (RdRp). Here we review recent advances in our understanding of flaviviral capping machinery and the implications for drug development. The NS5 MTase catalyzes both guanine N7 and ribose 2′-OH methylations during viral cap formation. Representative flavivirus MTases, from dengue, yellow fever, and West Nile virus (WNV), sequentially generate GpppA → m7GpppA → m7GpppAm. Despite the existence of two distinct methylation activities, the crystal structures of flavivirus MTases showed a single binding site for S-adenosyl-L-methionine (SAM), the methyl donor. This finding indicates that the substrate GpppA-RNA must be repositioned to accept the N7 and 2′-O methyl groups from SAM during the sequential reactions. Further studies demonstrated that distinct RNA elements are required for the methylations of guanine N7 on the cap and of ribose 2′-OH on the first transcribed nucleotide. Mutant enzymes with different methylation defects can trans complement one another in vitro, demonstrating that separate molecules of the enzyme can independently catalyze the two cap methylations in vitro. In the context of the infectious virus, defects in both methylations, or a defect in the N7 methylation alone, are lethal to WNV. However, viruses defective solely in 2′-O methylation are attenuated and can protect mice from later wild-type WNV challenge. The results demonstrate that the N7 methylation activity is essential for the WNV life cycle and, thus, methyltransferase represents a novel and promising target for flavivirus therapy.
Keywords: Flavivirus NS5, RNA cap methylation, methyltransferase, structure and function, inhibitor
1 Introduction
The viral family Flaviviridae includes three genera of enveloped viruses: Flavivirus, Pestivirus, and Hepacivirus (Brinton, 1981; Westaway et al., 1985; Brinton, 2002). About 70 viruses have thus far been found to contain a flavivirus-specific antigen and are classified as belonging to the genus Flavivirus (Westaway et al., 1985). Many flaviviruses are significant human pathogens, including dengue virus serotypes (DENV) 1–4, yellow fever virus (YFV), tick-borne encephalitis complex virus (TBEV), Japanese encephalitis virus (JEV), Murray Valley encephalitis virus (MVEV), St. Louis encephalitis virus (SLEV), and West Nile virus (WNV). The spectrum of diseases caused by flaviviruses ranges from a mild febrile illness to hepatitis, hemorrhagic syndromes, and encephalitis and can be fatal (Asnis et al., 2000; Asnis et al., 2001; Kramer and Bernard, 2001; Shi et al., 2001; Shi et al., 2002a, 2002b).
Among flaviviruses, DENV has spread throughout the tropical and subtropical regions, mainly Southeast and South Asia, Central and South America, and the Caribbean (WHO, 2009a). The four serotypes of DENV represent a major, rapidly growing public health problem, with an estimated 2.5 billion people at risk of the life-threatening diseases: severe dengue fever and dengue hemorrhagic fever (DHF)/dengue shock syndrome (DSS). There are about 50 million to 100 million cases of dengue infection each year, with an estimated 500 000 cases of life-threatening disease in the form of severe dengue, including DHF and DSS (WHO, 2009a).
As is true for DENV, WNV occurs over a broad geographical range and in a wide range of vertebrate host and vector species, and has recently emerged as a significant human pathogen by spreading to the Western hemisphere. WNV was originally isolated in 1937 from the blood of a febrile patient from the West Nile Province of Uganda, and was subsequently found to be widely distributed throughout Africa, the Middle East, parts of Europe, Russia, India, and Indonesia. (Smithburn et al., 1940; Brinton, 2002). Transmission of WNV predominantly involves mosquitoes of the genus Culex as vectors, and wild birds as the reservoir host. WNV has been reported to cause significant equine, avian, and in particular, human disease (Bernard et al., 2001; Bernard and Kramer, 2001; Petersen and Roehrig, 2001). In the human cases, infections by WNVas a result of a mosquito-bite can cause severe central nervous system disease, including encephalitis and meningoencephalitis, potentially fatal infections of the brain. Since its first appearance in the northeastern United States in 1999, WNV has quickly spread along the Eastern Seaboard and to the West Coast of the US. WNVas an emerging pathogen will likely be an increasingly severe threat to much of the U.S. population. WNV has been classified as a potential bioterrorism pathogen by the National Institutes of Health (NIH) (2000).
The World Health Organization has estimated annual human cases of more than 200 000, for YFV (WHO, 2009c), and more than 50 000 for JEV (WHO, 2009b), respectively. However, vaccines for humans are currently available only for YFV, JEV, and TBEV (Burke and Monath, 2001). No clinically approved antiviral therapy is available for treatment of flavivirus infections. Therefore, the development of vaccines and antiviral agents for prevention and treatment of flavivirus infections is a clear public health priority (Kramer et al., 2007).
This article will review recent advances in our understanding of flavivirus RNA cap methylation, and the potential exploitation of flavivirus methyltransferase (MTase) as an antiviral target.
2 Characteristics of the flaviviral genome and replication
Flavivirus virions are spherical in shape with a diameter of 50 to 60 nm (Li et al., 2008; Perera and Kuhn, 2008; Yu et al., 2008). The icosahedral nucleocapsid, about 30 nm in diameter, consists of capsid protein and genomic RNA, and is surrounded by a lipid bilayer in which the viral envelope and membrane proteins are embedded.
The flavivirus genome RNA is single-stranded and of positive (i.e., mRNA-sense) polarity. A cap is present at the 5′ end, followed by the conserved dinucleotide sequence 5′-AG-3′ (Cleaves and Dubin, 1979). The 3′ end of the genome terminates with 5′-CUOH-3′ (Wengler and Wengler, 1981) rather than with a poly(A) tract. The viral genome is approximately 11 kb in length, consisting of a 5′ untranslated region (UTR), a single long open reading frame (ORF), and a 3′ UTR (Fig. 1) (Rice et al., 1985; Shi et al., 2001). The single ORF of WNV, for instance, encodes a polyprotein of about 3433 amino acids, and has a gene order of 5′-C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-3′ (Fig. 1A). The polyprotein is co-and post-translationally processed by viral and cellular proteases into three structural proteins (capsid [C], premembrane [prM] or membrane [M], and envelope [E]) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Chambers et al., 1990). The NS proteins are assumed to be involved primarily in the replication of viral RNA as part of a replicase complex. The majority of the WNV NS proteins are multifunctional. NS1, NS3 and NS5 are large and highly conserved, while NS2A, NS2B, NS4A and NS4B are relatively small and hydrophobic. Glycoprotein NS1 and its interaction with NS4A are required for RNA replication (Lindenbach and Rice, 1997; Muylaert et al., 1997; Lindenbach and Rice, 1999). It was recently reported that NS2A functions during assembly and/or release of infectious flavivirus particles (Kummerer and Rice, 2002). NS2B forms a complex with NS3 and is a required cofactor for the protease activity of NS3 (Chambers et al., 1991; Arias et al., 1993; Chambers et al., 1993; Falgout et al., 1993). NS3 is a multi-functional protein with activities of serine protease (with NS2B as a cofactor), 5′-RNA triphosphatase (RTPase), nucleoside triphosphatase (NTPase), and helicase (Wengler and Wengler, 1991; Warrener et al., 1993; Li et al., 1999). The functions of the membrane-associated NS4A and NS4B are not known. NS5 has RNA-dependent RNA polymerase (RdRp) activity (Tan et al., 1996; Ackermann and Padmanabhan, 2001; Guyatt et al., 2001) and has homology with MTases, which are involved in methylation of the 5′ RNA cap structure (Koonin, 1993; Egloff et al., 2002; Ray et al., 2006). Recently, NS5 was also found to carry RNA guanylyltransferase (GTase) activity (Issur et al., 2009). Upon flavivirus infection, the plus-strand genomic RNA is transcribed into a complementary minus-strand RNA, and that, in turn, serves as the template for the synthesis of more plus-strand genomic RNA (Chambers et al., 1990). The synthesis of flavivirus plus-sense and minus-sense RNAs is asymmetric; plus-sense RNA is produced in 10- to 100-fold excess over minus-sense RNA (Muylaert et al., 1996; Diamond et al., 2000).
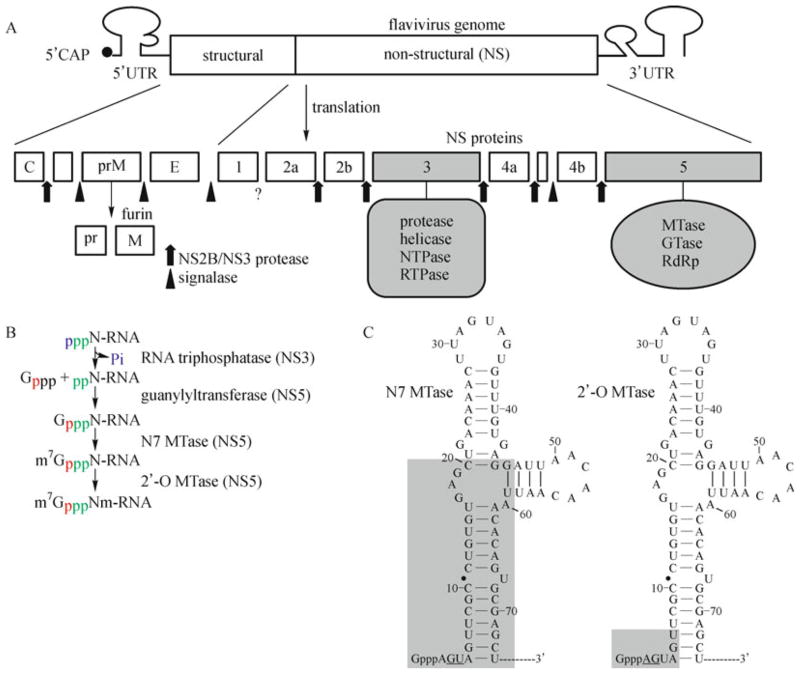
Fig. 1. Schematic representations of flavivirus genome structure (A), RNA cap formation (B), and substrate specificities of the WNV MTase (C).
A: Flavivirus genome structure. Flavivirus genomic RNA consists of a 5′ untranslated region (UTR), a single open reading frame (ORF), and a 3′-UTR. The single ORF encodes three structural and seven nonstructural (NS) proteins. Sites of polyprotein cleavage mediated by the viral NS2B-NS3 and by host signalase and furin are shown, and the enzymatic activities of NS3 and NS5 are also indicated. B: Cap formation of flavivirus RNA. Four enzymatic modifications are required for flavivirus RNA cap formation. Phosphates from different molecules are colored individually to indicate their sources. The putative steps presented here are modified from cellular mRNA cap formation. C: Summary of RNA substrate requirements for the N7 and 2′-O activities of the WNV MTase (Dong et al., 2007). The stem-loop structure formed by the 5′ terminal 74 nt of the WNV genome was predicted by the Mfold program (Zuker, 2003). The shaded regions are important for N7 (left panel) and 2′-O (right panel) methylations. Essential wild-type nucleotides are underlined.
3 Current knowledge of the flaviviral RNA capping enzymes
Eukaryotic mRNAs possess a blocked 5′ end known as the cap structure. Capping occurs on nascent RNA transcripts and is catalyzed by nuclear enzymes. Most animal viruses that replicate in the cytoplasm of infected cells also possess capping activities and produce capped mRNAs. This modification is involved in pre-mRNA processing and transport from the nucleus, in recruitment of mature mRNAs to the ribosomes, and in protection of mRNAs from degradation (Bisaillon and Lemay, 1997; Hodel et al., 1999; Furuichi and Shatkin, 2000). In the cap structure, a guanosine residue blocks the 5′-terminal base of the mRNA or viral RNA via a unique 5′-5′ triphosphate linkage GpppN, where the penultimate “N” nucleoside is derived from the RNA transcript. The blocking guanosine, for almost all cellular and viral mRNAs, contains a methyl group at the N7 position (m7GpppN), generally referred to as cap0 (Furuichi and Shatkin, 2000; Shuman, 2001; Fabrega et al., 2004; Gu and Lima, 2005; De la Pena et al., 2007). In small nuclear RNAs, a m2,7GpppN or m2,2,7GpppN cap structure is often found instead of m7GpppN. The mRNA caps for animal cells and their viruses are generally further modified by methylation at the ribose moiety (2′-OH) of the first, or the first two, transcribed nucleotide(s), respectively, leading to the formation of so-called “cap1” (m7GpppNm) or “cap2” (m7GpppNmNm) structures. When adenosine is found at the penultimate position, the N6 position of adenosine can also be methylated.
The cap structure is generally synthesized by several enzymatic reactions. A common mechanism, entailing either three or four steps, has been proposed for the cap formation (Fig. 1B). In step1, the RNA triphosphatase removes the 5′ γ-phosphate of a primary transcript, and in step 2 RNA GTase then donates a GMP moiety, derived from GTP, to form a 5′-5′ triphosphate linkage, typical for a cap structure. In step 3, the cap is methylated at position 7 of the terminal guanosine by guanine-N7 RNA MTase, using S-adenosyl-L-methionine (SAM or AdoMet) as the methyl donor, and yielding a cap0 and a by-product S-adenosyl-L-homocysteine (SAH or AdoHcy). Quite often, specific RNA (nucleoside-2′-O)-MTase then acts on the riboses of the first nucleotides, respectively giving rise to cap1 in step 4 (Barbosa and Moss, 1978; Furuichi and Shatkin, 2000). Some organisms may also contain cytoplasmic enzymes that can further methylate the RNA molecule to generate the cap2 structure (Furuichi and Shatkin, 2000).
Alternative pathways to generate the cap structure have also been found. For example, the minus-strand RNA vesicular stomatitis virus (VSV) transfers the 5-monophosphate of the RNA to GDP (Abraham et al., 1975; Ogino and Banerjee, 2007); the plus-strand RNA alpha viruses methylate GTP prior to the transfer of m7GMP to the 5-diphosphate of the RNA (Ahola and Kaariainen, 1995). The order of capping and methylation is variable among cellular and viral RNAs (Furuichi and Shatkin, 2000). In the majority of cases, the N7 and 2′-O MTase reactions are catalyzed by two distinct enzymes or two separate domains of one large protein (Cong and Shuman, 1992; Furuichi and Shatkin, 2000; Shuman, 2001; Moure et al., 2006). Flavivirus NS5 protein (Ray et al., 2006) and the L protein of VSV (Li et al., 2006) are the only two known examples of capping MTases that exhibit both N7 and 2′-O-MTase activities but have a single SAM-binding site; that configuration indicates that one active site or one protein domain exists for two enzymatic activities (see below).
In flaviviruses, the NS3 carries RNA triphosphatase activity (Wengler and Wengler, 1991; Warrener et al., 1993; Li et al., 1999); all of the remaining enzymatic activities (RNA GTase, N7 MTase, and 2′-O MTase) have been recently mapped to the N-terminal domain of NS5 protein (Fig. 1B) (Koonin, 1993; Egloff et al., 2002; Ray et al., 2006; Issur et al., 2009).
4 Flaviviral MTase
4.1 Identification of flavivirus MTase
NS5 of flavivirus is located at the C-terminal portion of the viral polyprotein. It is the largest and the most highly conserved of the flaviviral proteins (Chambers et al., 1990; Brinton, 2002). The NS5 C-terminal region has long been known to contain motifs characteristic of RdRps (Kamer and Argos, 1984; Koonin, 1991). The existence of flavivirus NS5 MTase was predicted in 1993, through the identification of a SAM-binding motif that is characteristic of the SAM-dependent MTase, within the N-terminal domain of NS5 (Koonin, 1993).
The MTase activity of an NS5 N-terminal fragment (amino acids (AA) 1–296) of DENV-2 was first biochemically confirmed as an RNA nucleoside 2′-O MTase (Egloff et al., 2002). Egloff et al. reported that the flavivirus NS5 MTase can methylate the nucleoside-2′-O ribose of short artificial capped RNA substrates (GpppAC5 and m7GpppAC5) (Egloff et al., 2002). No N7 MTase activity was observed under their experimental conditions. Recently, both N7 and 2′-O MTase activities were discovered, for both full-length NS5 and an NS5 MTase domain (AA 1–300) of WNV, when an authentic WNV RNA substrate representing the first 190 nucleotides (nt) of the WNV genome was used (Ray et al., 2006; Dong et al., 2007). The dual N7 and 2′-O MTase activities were later confirmed for MTases from other representative flaviviruses such as DENV-1, DENV-2, YFV, and Powassan virus (PWV) (Dong et al., 2007; Zhou et al., 2007; Kroschewski et al., 2008; Milani et al., 2009; Chung et al., 2010). Therefore, it is highly likely that flavivirus encodes the NS5 MTase as a general mechanism for dual methylations of the viral RNA cap.
4.2 Flaviviral MTase substrate specificity
It has been noted that the assay conditions were different between the earlier DENV-2 MTase and the later WNV studies. Zhou et al. sought to optimize conditions for the individual N7 and 2′-O assays, and found that the optimal assay conditions for N7 and 2′-O MTase activities differed (Zhou et al., 2007). The most significant difference is that the optimal pH for the N7 reaction is neutral, pH 7, whereas the optimal pH for 2′-O MTase reaction is pH 10. In contrast to the latter finding, the earlier DENV-2 study has been performed at neutral pH (Egloff et al., 2002). However, this single difference does not suffice to explain the absence of N7 activity in the earlier DENV-2 study (Egloff et al., 2002).
Across various experimental systems that have been used to assay flaviviral MTase activities, the N7 MTase activity was only observed when a long authentic viral RNA substrate was used (Ray et al., 2006; Dong et al., 2007; Zhou et al., 2007; Kroschewski et al., 2008; Milani et al., 2009; Chung et al., 2010). When nonviral short GpppAC5 or m7GpppAC5 substrate or nonviral-specific RNA substrates were used, N7 MTase activity could not be detected (Egloff et al., 2002; Dong et al., 2007; Mastrangelo et al., 2007; Peyrane et al., 2007; Kroschewski et al., 2008; Lim et al., 2008). Indeed, when an authentic RNA substrate representing the first 70 to 211 nt of the DENV-2 genome or the authentic WNV substrate (190-nt) was used, the N7 MTase activity of the DENV-2 or DENV-1 MTase was detected (Dong et al., 2007; Zhou et al., 2007; Kroschewski et al., 2008; Milani et al., 2009; Chung et al., 2010). The observed cross-genus activities of cap methylation between DENV MTases and WNV RNA substrate could result from the conserved 5′ terminal nucleotides and the RNA structure present in the two flaviviruses (Brinton and Dispoto, 1988; Dong et al., 2007). Therefore, the N7 MTase activity appears to be strictly dependent on a virus-specific RNA substrate.
Dong et al. further investigated the details of substrate requirements for both N7 and 2′-O MTase activities of the WNV MTase, by constructing an extensive series of mutations of the 5′-stem loop structure of the 190-nt WNV substrate (Fig. 1C) (Dong et al., 2007). They found that distinct RNA elements are required for the methylations at guanine N7 on the cap and at ribose 2′-OH on the first transcribed nucleotide. In a WNV model, N7 cap methylation requires specific nucleotides at the second (G) and third (U) positions, and a 5′stem-loop structure (although not specificity of the sequence within the stem loop structure); in contrast, 2′-OH ribose methylation requires specific nucleotides at the first (A) and second (G) positions, with a minimum 5′viral RNA length of 20 nt. In addition, Egloff et al. found that the binding of small RNAs to the DENV-2 MTase was higher when the RNA substrates contained the first two authentic viral nucleotides than when non-authentic sequence was used, confirming the important role of the 5′-terminal nucleotides for both N7 and 2′-O-MTase activities (Egloff et al., 2007). Further investigations showed that cap analogues, GpppA and m7GpppA, are not active substrates for the WNV MTase (Dong et al., 2007). For the flavivirus 2′-O MTase activity, many studies have shown that non-viral short RNA can serve as a 2′-O MTase substrate (Egloff et al., 2002; Mastrangelo et al., 2007; Peyrane et al., 2007; Kroschewski et al., 2008; Lim et al., 2008; Selisko et al., 2010). Moreover, Selisko et al. investigated the length requirement for the 2′-O MTase substrate (Selisko et al., 2010). They found that the DENV-2 MTase can perform 2′-O methylation of substrate m7GpppACn or GpppACn when n≥2, and that the 2′-O MTase activity increased with RNA substrate length, reaching a plateau for RNAs containing 6 or 7 nt.
4.3 Flaviviral MTase processivity
Using authentic long WNV GpppA-RNA substrate, Ray et al. performed kinetic analysis of the methylation of substrate GpppA-RNA by the WNV MTase (Ray et al., 2006). The results showed that guanine N7 methylation was first detected and ribose 2′-O methylation only became evident after accumulation of the N7 methylated cap. These observations suggest a sequential methylation of RNA cap in the order of GpppA → m7GpppA → m7GpppAm, indicating that guanine N7 methylation precedes ribose 2′-O methylation. Zhou et al. confirmed the sequential nature of the flaviviral N7 and 2′-O MTase reactions, using MTases from other representative flaviviruses such as DENV-1 and YFV (Zhou et al., 2007).
Recently, Dong et al., using mutants of the WNV MTase defective in either N7 or 2′-O activity, further demonstrated that the two N7 and 2′-O methylation events are independent in vitro; the independence was evidenced by efficient N7 methylation of GpppA-RNA → m7GpppA-RNA and GpppAm-RNA → m7GpppAm-RNA, and by the 2′-O methylation of GpppA-RNA → GpppAm-RNA and m7GpppA-RNA → m7GpppAm-RNA (Dong et al., 2008a). The N7-independent 2′-O methylation was also observed for other flavivirus MTases, when short nonviral RNA substrates were used (Egloff et al., 2002; Mas-trangelo et al., 2007; Selisko et al., 2010). These results suggested two alternative methylation pathways in vitro: GpppA-RNA → m7GpppA-RNA → m7GpppAm-RNA, and GpppA-RNA → GpppAm-RNA → m7GpppAm-RNA.
To investigate the processivity of flavivirus MTase, Dong et al. performed detailed kinetic analysis of the N7 and 2′-O MTase reactions (Dong et al., 2008a). They found that the 2′-O methylation activity prefers substrate m7GpppA-RNA over GpppA-RNA. In contrast, the presence of a 2′-O methyl group in the cap structure was not seen to affect the N7 methylation efficiency. Since the N7 methylation has no preference between substrates GpppA-RNA and GpppAm-RNA, the substrate preference of 2′-O methylation for m7GpppA-RNA could thus be the factor that determines the dominant pathway of WNV cap methylation as GpppA-RNA → m7GpppA-RNA → m7GpppAm-RNA (Dong et al., 2008a).
Recently, Chung et al. reported steady-state kinetic studies of the N7 and 2′-O methylations by the DENV-2 MTase (Chung et al., 2010). They found that N7 methylation precedes 2′-O methylation; the former reaction turns over RNA faster (kcat), resulting in 2.4-fold higher catalytic efficiency. Michaelis constants for SAM in both reactions were about 10-fold lower than the constants for the RNA substrates, suggesting that the rate-limiting steps in the two methylase reactions were associated with RNA templates.
5 Crystal structures of flavivirus MTases
The first crystal structure of flavivirus MTase was determined for the DENV-2 MTase by Egloff et al. in 2002 (Egloff et al., 2002). Since 2006, the recognition of the dual methylation activities of flaviviral MTase makes the enzyme an attractive model in the study of RNA methylation mechanisms. The crystal structures of MTases have since been determined for seven other flaviviruses, namely WNV (Zhou et al., 2007), Meaban virus (MEAV) (Mastrangelo et al., 2007), MVEV (Assenberg et al., 2007), Wesselsbron virus (WESSV) (Bollati et al., 2009c), YFV (Geiss et al., 2009), Yokose virus (YOKV) (Bollati et al., 2009b), and Modoc virus (MODV) (Jansson et al., 2009). In addition, crystal structures of several flaviviral MTases in complex with SAM (co-factor), SAH (byproduct), and/or cap analogs or inhibitors have been determined (Egloff et al., 2002; Benarroch et al., 2004; Assenberg et al., 2007; Egloff et al., 2007; Mastrangelo et al., 2007; Zhou et al., 2007; Bollati et al., 2009b, 2009c; Geiss et al., 2009; Jansson et al., 2009). Although their sequences differ considerably, all of these MTases have similar three-dimensional structures, with overall root-mean-squared deviations within 1.0Å when the structures are superimposed.
The crystal structures of flavivirus MTase domains reveal that these MTases belong to the so-called SAM-dependent MTase superfamily. Members of this class of enzymes utilize the methyl donor SAM to methylate substrates, and the enzymatic reaction generates SAH as a by-product (Fig. 2A). Roughly around 120 types of SAM-dependent MTases have been classified based on their substrate specificity (small molecule, lipid, protein, nucleic acid, etc.) and on the target atom (nitrogen, oxygen, carbon, sulfur, etc.) (Fauman et al., 1999).
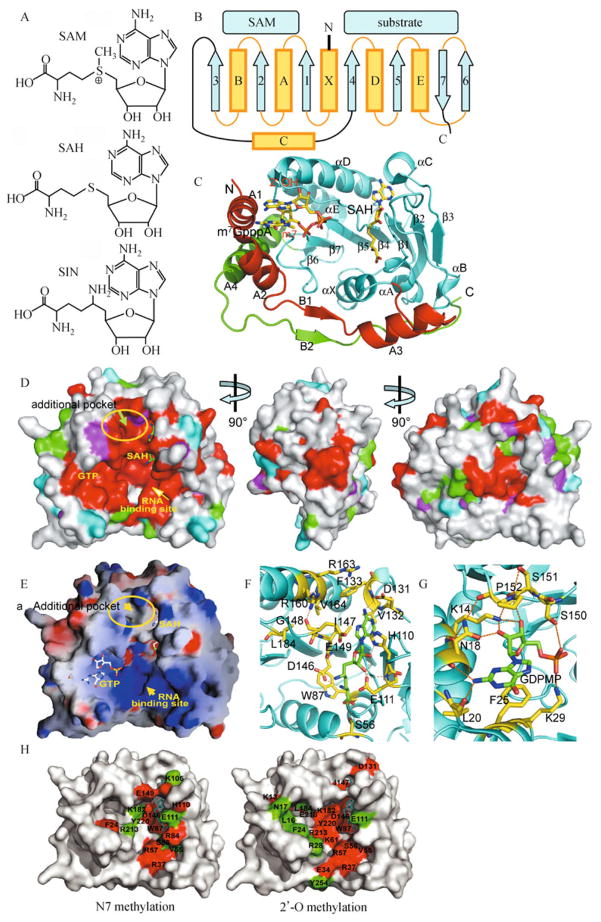
Fig. 2. SAM, SAM analogs, SAM-dependent MTase fold, and critical features of flavivirus MTases.
A: Chemical structure of the methyl donor SAM, the by-product SAH, and the SAM-analog inhibitor sinefungin (SIN); B: Schematic showing the topology of the core fold of the SAM-dependent MTases. The SAM-binding and substrate-binding regions are indicated. Helices are shown as yellow cylinders, and grey arrows denote β-strands. C: Crystal structure of the DENV-2 MTase in complex with m7GpppA and SAH (PDB: 2P3O) (Egloff et al., 2007). The by-product SAH and cap analog m7GpppA are in ball-and-stick presentation. The so-called three domains are colored red, cyan, and green. D: Mapping of sequence conservation to surface of the WNV MTase; the structure is viewed from three angles, differing by two successive rotations of approximately 90° each about the vertical axis. Left-hand image: the face containing the shallow groove where the co-factor, represented in green sticks, and the substrate bind. The side and the back of the molecule are respectively shown in the middle and right-hand images. The surface is colored according to conserved residues, resulting from the multiple alignment of 61 sequences of flaviviral MTases: >85% conserved (red); 70%–85% conservation (green); 60%–70% conservation (magenta); 50%–60% conservation (cyan), and not conserved ( <50%) (white). The bound SAH is depicted in a stick representation in the left-hand image. Three important binding sites and the additional pocket are labeled. E: GRASP (Nicholls et al., 1991) surface representation of the electrostatic potential of the WNV MTase, showing the three flavivirus-conserved binding sites for SAH/SAM, GTP, and RNA. The surface is colored blue for positive (15 kT), red for negative (−15 kT) and white for neutral, where k is the Boltzmann constant and T is the temperature (Nicholls et al., 1991). GTP was modeled according to the DENV-2 MTase-GTP complex (PDB: 2P1D) (Egloff et al., 2002). The bound SAH and GTP molecules are represented in white rods. The putative RNA-binding site is indicated by the area of strongly positively charged residues. F: Close-up view of amino acids involved in the formation of the SAM-binding pocket and the newly identified additional pocket of the WNV MTase. Residues are labeled and shown in a stick representation, with atom colors as follows: carbon, yellow; oxygen, red; and nitrogen, blue. Hydrogen bonds are shown as orange dashed lines. SAH is shown in a green stick representation. G: Close-up view of the GTP-binding pocket of the DENV-2 MTase (PDB: 2P1D) (Egloff et al., 2002). GTP is depicted in a green stick representation. MTase residues are labeled and shown in a yellow stick representation. Atom colors are as in panel (F). H: Residues important for N7 methylation (left) and 2′-O methylation (right) are summarized on the surface of the WNV MTase. Residues whose mutations led to severe reductions in activity to <20% of the WT activity are in red; residues whose mutation led to moderate activity reductions to 20% to 60% of the WT activity are in green. SAM is shown in a cyan stick representation.
To date, crystal or nuclear magnetic resonance (NMR) structures of over one hundred SAM-dependent MTases have been reported (Fauman et al., 1999; Martin and McMillan, 2002). Most of these structures show a common core structure referred to as a “SAM-dependent MTase fold” (Fig. 1B) (Fauman et al., 1999; Martin and McMillan, 2002). The SAM-dependent MTase fold is an open α/β/α sandwich structure. The central part of the core structure is a seven stranded β-sheet. The order of the strands is generally 3214576 (Fig. 1B), with strand 7 antiparallel to all other parallel strands. Helices X, A, and B are located on one side of the β-sheet, and helices C, D, and E on the other. This basic core structure is conserved in all SAM-dependent MTases, despite very low or even completely lack of sequence identity. Modifications usually occur outside the core structure. Some of the proteins have additional domains that play roles in substrate recognition or in separate reactions (Fauman et al., 1999; Martin and McMillan, 2002). Nearly all SAM-dependent MTases display a central cleft between β-strands 1 and 4. This cleft has been described as the active site, where SAM and the substrate bind, and where the methyl transfer occurs (Fig. 2B,C) (Hodel et al., 1996; Fauman et al., 1999; Hodel et al., 1999; Martin and McMillan, 2002). The binding sites for the methyl acceptors vary considerably, because the substrates themselves vary significantly in size and physical characteristics (e.g., protein, DNA, RNA, lipid, small molecule, etc.).
All flaviviral MTase structures are composed of three subdomains. The core subdomain, consisting of a seven-stranded β-sheet surrounded by four α-helices, shows a typical SAM-dependent MTase fold (Fig. 2C, cyan). In addition to the core structure, there are an N-terminal subdomain (Fig. 2C, red) and a C-terminal subdomain (Fig. 2C, green).
Analysis of these structures, coupled with structure-based mutagenesis studies, has led to the definition of three functionally critical regions in the MTase structure: (1) the SAM-binding site; (2) a GTP-binding site for cap-binding; and (3) a highly conserved and positively charged surface groove as a putative RNA-binding site (see below) (Fig. 2C–E) (Egloff et al., 2002; Zhou et al., 2007).
6 Structural and sequential conservations of flavivirus MTases
The flavivirus MTase was first identified through sequence analysis as a sequence motif that was conserved across the SAM-dependent MTases (Koonin, 1993). Further bioin-formatic analysis and structure-based alignment indicated the presence of eight of the nine conserved MTase motifs involved in SAM-binding and catalysis (Malone et al., 1995; Egloff et al., 2002). Bollati et al. analyzed 34 sequences of flavivirus MTases and found three conserved patches on the MTase surface (Egloff et al., 2009b). We extended that analysis by considering all 61 currently available sequences of flavivirus MTases (Supplemental Fig. 1). Among the 265 residues (according to the WNV sequence), for the 61 flavivirus MTases, we found that 25 residues are invariant, 21 have one variation, five have two variations, and four have three variations (Supplemental Fig. 1). When we mapped the sequence-conserved residues on the MTase structures, we found them to form a large continuous surface (with most residues having >85% sequence identity (Fig. 2D, left-hand image; colored red)) covering the SAM-binding site, GTP-binding site, and the putative RNA-binding site (Fig. 2D). In addition, two smaller conserved patches are found on the MTase surface; relative to the surface shown in Fig. 2D (left-hand image), one is to the right side of the larger conserved face (Fig. 2D, middle image) and one is on the opposite (back) face (Fig. 2D, right-hand image). The side patch is composed of residues W121, N122, G263, T264, and R265 (numbering and residue identity are according to the WNV sequence). Although the functional significance of this small conserved patch is unknown, we can speculate that it is important in mediating the intramolecular interactions with the RdRp domain of the flavivirus NS5 protein, giving that the G263T264R265 are the C-termini of the NS5 MTase domain. The small conserved surface on the “back” surface is formed by residues W64, R68, V207, R208, P210, S212, N214, L240, and R243. The functional significance of this surface is also unknown.
7 Structural and functional studies of flaviviral MTases
Since the discovery of flavivirus MTase and the determinations of MTase structures, extensive mutagenesis has been performed on residues considered as potentially essential for the MTase functions (Egloff et al., 2002; Ray et al., 2006; Zhou et al., 2007; Dong et al., 2008a, 2008b; Kroschewski et al., 2008; Geiss et al., 2009).
7.1 The SAM-binding site
Flavivirus MTase, like other SAM-dependent MTases, utilizes SAM as a methyl donor. The majority of crystal structures of flavivirus MTases reveal the presence of a SAM or SAH (the by-product of the methyl transfer reaction) molecule in the SAM-binding site, although neither SAM nor SAH was added during the crystallization, in some of these experiments (Fig. 2F) (Egloff et al., 2002; Zhou et al., 2007; Bollati et al., 2009c; Geiss et al., 2009). Co-crystallization of flavivirus MTases with SAM, SAH, or SAM analogs further confirmed that the cofactor SAM, the by-product SAH, and SAM analogs bind identically in the SAM-binding pocket of the MTase (Assenberg et al., 2007; Mastrangelo et al., 2007; Bollati et al., 2009b, 2009c; Jansson et al., 2009).
Crystal contact analysis of the WNV MTase structure indicated that as many as 20 residues are within 4Å distance to contact the bound SAH molecule (Fig. 2F). These residues are S56, G58, G81, C82, G83, G85, G86, W87, T104, K105, G106, H110, E111, V130, D131, V132, F133, D146, I147, and E149. With the exception of three residues, K105 (30% conserved), V130 (40%), and F133 (61%), all of the residues are well conserved (including eight invariant ones) among flavivirus MTases (Supplemental Fig. 1). Therefore, the SAM/SAH-binding pocket is highly conserved (Fig. 2D).
We recently determined the crystal structure of the WNV MTase in complex with an MTase inhibitor, sinefungin (SIN), which is a SAM analog, at 2.0Å resolution (Liu and Li, unpublished results). Upon examination of the crystal structures of the WNV MTase-SIN and MTase-SAH complexes (Zhou et al., 2007), we observed an additional pocket that is an extension of the cavity holding the adenine base of SAM or one of the SAM analogs (SAH or SIN) (Fig. 2D). The extended pocket is nearly square-shaped with approximate dimensions of 8 Å × 9 Å, and ~ 5 Å deep. Further analysis indicated that a pocket similar to the WNV SAM-pocket extension exists in all known structures of flavivirus MTases (data not shown) (Egloff et al., 2002; Assenberg et al., 2007; Mastrangelo et al., 2007; Zhou et al., 2007; Bollati et al., 2009b, 2009c; Geiss et al., 2009; Jansson et al., 2009). Interestingly, however, upon examination of known structures of SAM-utilizing proteins (over 100 structures available), we found that the extended pocket is specific to the flaviviral MTases, as it has not been seen in structures of other SAM-utilizing proteins (data not shown). These results indicate that the additional pocket is not only a general but also a characteristic feature of flavivirus MTase structures.
The nature of the extended pocket is mainly hydrophobic, and is formed by residues F133, I147, G148, E149, R160, R163, V164, and L184 of the WNV MTase (Fig. 2F). Sequence alignment of flavivirus MTases indicated that residues at positions 133, 147, 148, 149, 160, 163, and 164, and 184 of the WNV MTase are conserved among flaviviruses (Supplemental Fig. 1) (Mastrangelo et al., 2007; Zhou et al., 2007). Among these residues, G148 and E149 are completely conserved. Except for F133 (61% conservation), all of the residues within this pocket are over 90% conserved among 61 flavivirus MTases (Supplemental Fig. 1). In addition, structure-based sequence alignment indicated that the sequence conservation of these residues only occurs among flavivirus MTases, but not among other SAM-dependent MTases, nor, on a broader scale, among other SAM-utilizing proteins (data not shown, and Egloff et al., 2002). Moreover, Dong et al. recently found that mutagenesis of R160 to alanine greatly reduces the N7 MTase activity (Dong and Li, unpublished data). Therefore, the flavivirus-conserved additional pocket is a unique and possibly important feature of flavivirus MTases.
A number of residues (S56, W87, T104, H110, E111, D131, V132, F133, D146, I147, and E149), all of which except F133, are over 90% conservative among flavivirus MTases, making side-chain contacts with the bound SAH/SIN molecule. Although slight differences were observed for binding of the SAM/SAH/SIN molecule to different flavivirus MTases (Davidson, 2009), the bound co-factor/by-product/inhibitor molecule is stabilized by a number of hydrogen bonds, some of which are contributed by side-chain atoms of MTase residues S56 (Oγ), H110 (Nδ1), E111 (Oε2), D131 (Oδ2), and D146 (Oδ1) (Fig. 2F) (Dong et al., 2008b).
Extensive mutagenesis has been performed on the SAM-contacting residues of the WNV and DENV-2 MTases (Ray et al., 2006; Zhou et al., 2007; Dong et al., 2008b; Kroschewski et al., 2008). Those studies showed that mutations of residues predicted to be involved in SAM-binding (such as S56, G81, G83, G85, W87, K105, H110, E111, D131, I147, D146, and E149) reduced, and in some cases abolished, either the N7 or the 2′-O methylation, or both. Collectively, these data confirmed that only one SAM-binding site is required for the methylations of the two N7 and 2′-O positions of the viral RNA cap.
7.2 The GTP-binding site
A novel GTP-binding site was discovered when crystals of the DENV-2 MTase were soaked with a GTP analog GDPMP. The new GTP-binding site was proposed to interact with the cap structure so as to properly position the 2′-OH of the first transcribed adenosine during 2′-O methylation (Egloff et al., 2002). Since that initial recognition, crystal structures of the DENV-2, MVEV, and WESSV MTases in complex with either the GTP analog (ribavirin triphosphate) or cap analogs (m7GTP, GpppA, m7GpppA, GpppG, m7GpppG, and m7GpppGm) have been determined (Benarroch et al., 2004; Assenberg et al., 2007; Egloff et al., 2007; Bollati et al., 2009c; Geiss et al., 2009). Although various conformations have been observed for the second nucleotide (A or G(m)) of the co-crystallized cap analog dinucleotides (such as GpppA, m7GpppA, GpppG, m7GpppG, and m7GpppGm) in the crystal structures of flavivirus MTase-dinucleotide complexes, the guanosine moiety of the cap dinucleotide analogs is always bound in the GTP-binding pocket in identical fashion (Egloff et al., 2002; Benarroch et al., 2004; Assenberg et al., 2007; Egloff et al., 2007; Bollati et al., 2009c; Geiss et al., 2009). Overall, these structures clearly defined the molecular interactions between the MTase and the GTP cap (Fig. 2G).
In contrast to structures of the cellular GTP cap-binding proteins, in which the GTP cap is usually sandwiched between two aromatic residues, the flavivirus MTase binds GTP with only one aromatic residue F25 (equivalent to F24 in WNV) base stacking with the guanine ring (Egloff et al., 2002). Limited polar interactions were also observed between the guanine base and flavivirus MTase. The main-chain hydroxyl oxygens of N18 and Leu20 form hydrogen bonds with the guanine N2 atom. The limited interaction between the guanine base and MTase implies that flavivirus MTase can bind both methylated and unmethylated GTPs (Egloff et al., 2002). This conclusion was confirmed by determinations of the crystal structures of flaviviral MTases in complex with unmethylated GTP analogs (Egloff et al., 2002; Benarroch et al., 2004; Assenberg et al., 2007; Egloff et al., 2007; Bollati et al., 2009c). In addition, Dong et al. demonstrated that the WNV MTase can perform its 2′-O MTase function with either a methylated or an unmethylated RNA substrate (Dong et al., 2008a). However, the 2′-O methylation activity prefers substrate m7GpppA-RNA over GpppA-RNA (Dong et al., 2008a). The structural basis for this selection is unknown, since no specific contact was observed between the N7 methyl group of the guanine cap and the flavivirus MTase, in the structures of the MTase-cap analog complexes. In addition to the MTase-guanine cap interactions, several MTase residues form side-chain hydrogen bonds with the bound GTP analog (Fig. 2G). A side chain oxygen of N18 forms a hydrogen bond with the 2′-OH of the ribose, the side chain of K14 interacts with both 2′-OH and 3′-OH groups through two hydrogen bonds, and the α-phosphate of GTP forms hydrogen bonds with the side chains of K29 and S150 (Fig. 2G). In addition, hydrogen bonds were observed between the YFV MTase residue R213 and the GTP γ-phosphate oxygen, and between the MTase S215 and the GTP α-phosphate oxygen (Geiss et al., 2009).
Sequence alignment shows that the GTP-binding site is conserved among flavivirus MTases, although albeit to a lesser degree than is the case for the SAM-binding site (Fig. 2D and Supplemental Fig. 1). Among the MTase residues contributing specific contacts with the bound GTP or analogs, R213 and S215 are invariant, and K14, N18, F25, K29, and S150 are 89%–95% conserved; however, P152 is less than 40% conserved among flavivirus MTases (Supplemental Fig. 1). Mutagenesis of MTase residues involved in the GTP-binding pocket has been performed for the DENV-2 and WNV MTases (Egloff et al., 2002; Dong et al., 2008b; Geiss et al., 2009). Mutation of F25 to alanine was found to abolish or greatly reduce the GTP binding to the DENV-2 MTase (Egloff et al., 2002; Geiss et al., 2009). Functional studies showed that an F24A mutation of the WNV MTase significantly affected both N7 and 2′-O MTase activities (Dong et al., 2008b). Although point mutations of other GTP-contacting residues, such as N18A and K29A, greatly decreased the GTP-binding ability in the DENV-2 MTase (Egloff et al., 2002; Geiss et al., 2009), mutations of equivalent residues in the WNV MTase did not affect the N7 MTase activity of the enzyme. However, these mutations did reduce the 2′-O activity of the WNV MTase to various degrees (Dong et al., 2008b). Overall, mutations of various GTP-contacting residues (other than F25) did not have any significant effects on the N7 activity, but they all caused various reductions in 2′-O activity (Dong et al., 2008b). Furthermore, Dong et al. showed that GTP and cap analog GpppA specifically inhibited 2′-O methylation in a dose-dependent manner, whereas N7 methylation activity was not affected. Therefore, it has been proposed that the GTP-binding pocket functions only during the 2′-O methylation (Dong et al., 2008b).
7.3 The RNA-binding site
Crystal structures of several flaviviral MTases in complex with cap analog dinucleotides have been determined (Assenberg et al., 2007; Egloff et al., 2007; Bollati et al., 2009c; Geiss et al., 2009). However, these structures provide little information regarding the RNA binding, since the second nucleotide following the GTP cap is distant from the methyl donor SAM, resulting in a conformation for the 2′-OH not suitable for MTase catalysis. Therefore, the conformations may not be physiological ones.
No crystal structure is available for an authentic RNA-MTase complex. The N7 RNA-MTase complex would be particularly difficult to crystallize, since it entails a long authentic RNA substrate with secondary structural elements. However, comparison of the existing structures of flaviviral MTases indicated that a common feature of the flavivirus MTases is a highly positively charged surface adjacent to the SAM- and GTP- binding sites; such strong positively charged surface is not seen in other regions (Fig. 2E) (Egloff et al., 2002; Zhou et al., 2007). The positively charged region is likely the site for binding of the capped RNA substrates. Superposition of the structures of the VP39 2′-O and Encephalitozoon cuniculi (Ecm1) N7 MTases in complex with their RNA substrates (Hodel et al., 1998; Fabrega et al., 2004) onto the WNV MTase structure indicated that the RNA substrates lie along the positively charged region (Dong et al., 2008b).
Sequence alignment indicated that the positively charged putative RNA-binding site is conserved among flavivirus MTases (Fig. 2D and Supplemental Fig. 1). Dong et al. performed extensive mutagenesis of residues within this putative RNA-binding site (Dong et al., 2008b). Together with results of earlier mutagenesis studies (Ray et al., 2006; Zhou et al., 2007), the results showed that two distinct sets of amino acids in the RNA-binding site are required for the N7 and 2′-O methylations. The effects of individual mutations on the two methylation events could be categorized into four groups (Fig. 2H): (1) Residues (R37, R57, W87, and D146) were critical for both methylation activities. Point mutations of these residues reduced both methylation activities to <20% of that of the WT enzyme. (2) Residues (E149 and R84) were selectively important for the N7 methylation activity. Point mutations of these amino acids dramatically suppressed the N7 methylation, but had minor or no effect on the 2′-O methylation activity. (3) Amino acids (L16, E34, K61, K182, L184, E218, and Y254) were selectively important for the 2′-O methylation activity. Point mutations of these residues substantially suppressed the 2′-O methylation, but not the N7 methylation. (4) Residues were not important for either methylation activity.
Overall, the mutagenesis results demonstrated that distinct sets of amino acids are required for the N7 and 2′-O methylations. On the MTase surface, residues critical for the 2′-O methylation are more spread out than those important for the N7 methylation; also, more residues are critically involved in the 2′-O methylation than are involved in the N7 methylation. These results suggest that the RNA substrate binds in different positions on the enzyme surface during the two methylation reactions. In addition, further experiment indicated that point mutation within the RNA-binding site is not sufficient to decrease RNA-MTase complex formation, likely because the complexes are formed through multiple contact sites (Dong et al., 2008b).
8 Mechanism of flavivirus MTase activities
Mechanistic studies on model chemical reactions involving SAM indicated that nucleophilic attack occurs exclusively at the methyl group of SAM; that finding led to the conclusion that methyl transfer from SAM is a classic SN2 reaction requiring a linear arrangement of the nucleophile, methyl carbon, and thioester leaving group in the transition state (Fauman et al., 1999). It was proposed that O-, N-, and S-methylations occur via a straightforward attack of oxygen, nitrogen, or sulfur lone electron pairs on the SAM methyl group (Fauman et al., 1999).
Vaccinia virus protein VP39 is a representative, and the most extensively studied, RNA 2′-O MTase. The crystal structure of the VP39-AdoHcy-m7GpppGA5 complex revealed the geometry of the catalytic center in the context of bound substrates (Hodel et al., 1998). It was proposed that the VP39 MTase utilizes a classic SN2 reaction mechanism for the methylation, possibly involving deprotonation of the target 2′-hydroxyl, so as to generate an attacking nucleophile for the methyl carbon center, followed by a direct SN2 in-line displacement (Hodel et al., 1999).
From sequence and structure comparisons, a putative catalytic tetrad K-D-K-E that is conserved among various 2′-O MTases was proposed to catalyze an SN2-reaction-mediated 2′-O methyl transfer (Schnierle et al., 1994; Hodel et al., 1998; Egloff et al., 2002; Hager et al., 2002). The tetrad is common to several families of site-specific MTases that modify 2′-hydroxyl groups of riboses in mRNA, rRNA, and tRNA. In the DENV-2 MTase, the tetrad is K61-D146-K181-E217 in (Egloff et al., 2002), and in WNV it is K61-D146-K182-E218 (Ray et al., 2006; Zhou et al., 2007). Structural alignment of the flavivirus MTase with the VP39 tertiary complex (MTase, a short RNA with cap, and SAH) (Hodel et al., 1998) showed that the K-D-K-E tetrad is nearly superimposable between the two enzymes. In addition, the side chain of K181(DENV)/K182(WNV) is within hydrogen-bonding distance of the target ribose 2′-hydroxyl of the modeled RNA substrate. K181/K182 of the flavivirus MTase was proposed to be the catalytic center serving as a general base (Egloff et al., 2002; Zhou et al., 2007). Acid/base catalysis has been suggested in the mechanisms of other MTases, for instance, the plant 4-OMTase ChOMTand IOMT (Zubieta et al., 2001) and the N-MTase PRMT3 (Zhang et al., 2000), SET7/9-AdoMet (Kwon et al., 2003), PvuII DNA cytosine N4 MTase (Gong et al., 1997), histamine HNMT (Horton et al., 2001), and gunaidinoacetate MTase (Komoto et al., 2002).
The KDKE model requires that K181/K182 be in the deprotonated form. Interestingly, several studies indicated that the optimal pH for the WNV and DENV-2 2′-O MTase reactions is pH 10 (Lim et al., 2008; Zhou et al., 2007). Since the pKa of Lys is at pH 10, a high pH would facilitate the deprotonation of K182, in turn favoring the deprotonation of the 2′-OH, leading to efficient formation of the SN2-like transition state, so as to accomplish the methyl transfer (Hodel et al., 1998; Hager et al., 2002). However, results of several more recent studies indicated that the 2′-O MTase activity could frequently be detected at neutral pH, when short artificial capped RNA substrates were used (Mastrangelo et al., 2007; Peyrane et al., 2007; Kroschewski et al., 2008; Lim et al., 2008; Selisko et al., 2010). Deprotonation of K182 at neutral pH suggests that the pKa of this amino group is lower than 10. A similar decrease in pKa has previously been suggested for RrmJ, a heat shock-induced RNA 2′-O MTase (Hager et al., 2002). The pKa of a lysine sidechain has been found to be decreased in aldolases, in which the lysine was sited in a hydrophobic microenvironment (Barbas et al., 1997). An alternative mechanism for the perturbation of the pKa of an amine is that chemical tuning of a reactive lysine for Schiff base formation at neutral pH can be accomplished, at least in part, by positioning in proximity to a neighboring protonated lysine residue that electrostatically perturbs the pKa of the amine nucleophile (Frey et al., 1971; Kokesh and Westheimer, 1971; Highbarger et al., 1996). Therefore, the pKa of the catalytic lysine in flavivirus MTase may be decreased as a result of the proximity of the amine to the positive charges from the SAM sulfur atom and the amine group of the other lysine in the catalytic triad.
Biochemical mutagenesis of the WNV MTase showed that 2′-O methylation has a strict requirement of charge and side-chain length for the conserved KDKE motif (Ray et al., 2006; Zhou et al., 2007). All of the KDKE mutations abolished 2′-O methylation, confirming these residues’ essential role in flavivirus 2′-O MTase activity. In contrast, N7 methylation requires only the D146 within the tetrad; all of the other residues facilitate, but are not essential for N7 methylations. These results suggested that the N7 active site is different from the 2′-O site on the MTase (Ray et al., 2006; Zhou et al., 2007).
In contrast to the SN2-reaction mediated mechanism for 2′-O methylation, an in-line mechanism was suggested for N7 methylation by the determination of the structure of Ecm1 N7 MTase (Fabrega et al., 2004): no direct contact is observed between residues of the enzyme and either the attacking nucleophile N7 atom of guanine, the methyl carbon of SAM, or the leaving group sulfur of SAH. Instead, the catalysis is achieved through the close proximity and proper geometry of the two substrates (Fabrega et al., 2004). Superposition of the structures of the WNV and DENV-2 MTases onto that of the Ecm1 N7 MTase-substrate complex (Fabrega et al., 2004) did not suggest any steric hindrance between the guanosine cap analogue and the WNV and DENV-2 MTases (data not shown), providing structural feasibility for a mechanism analog to that for the Ecm1 N7 MTase reaction (Dong et al., 2008b). However, an understanding of the mechanism of flavivirus N7 methylation requires structure determination for the flavivirus MTase in complex with an authentic or N7-reactive RNA substrate.
9 Mechanism of flavivirus processive RNA cap methylation
Biochemical results showed that flavivirus MTase catalyzes cap methylation in a processive manner, with the order of GpppA → m7GpppA → m7GpppAm. However, the crystal structures of flavivirus MTases (Egloff et al., 2002; Assenberg et al., 2007; Mastrangelo et al., 2007; Zhou et al., 2007; Bollati et al., 2009b, 2009c; Geiss et al., 2009; Jansson et al., 2009) have all shown a single SAH-binding site. Alignment of the flaviviral MTase structures with that of the Ecm1 N7 MTase-substrate complex (Fabrega et al., 2004) or with that of the VP39 2′-O MTase-substrate complex (Shiryaev et al., 2006) did not suggest any steric hindrance between the RNA substrate and flavivirus MTase (Mastrangelo et al., 2007; Dong et al., 2008b), providing structural plausibility for the flavivirus MTase to catalyze both methylation reactions. These data, together with the results of mutagenesis and footprinting (Dong et al., 2007; Dong et al., 2008a, 2008b), led us to propose a molecular “repositioning” model for flavivirus RNA cap methylation (Zhou et al., 2007; Dong et al., 2008a, 2008b) (Fig. 3). Under this hypothesis, viral RNA is positioned in distinct ways on the MTase surface during the two methylation reactions. The guanine N7 is first positioned in proximity to the methyl group of SAM, to generate m7GpppA-RNA. Once the guanine N7 has been methylated, the 5′ terminus of m7GpppA-RNA is repositioned. The m7Gppp moiety moves into a GTP-binding pocket, as previously described for the DENV-2 MTase (Egloff et al., 2002). The binding of the m7Gppp moiety in the GTP pocket precisely registers the ribose 2′-OH of the first transcribed adenosine with SAM to generate m7GpppAm-RNA.
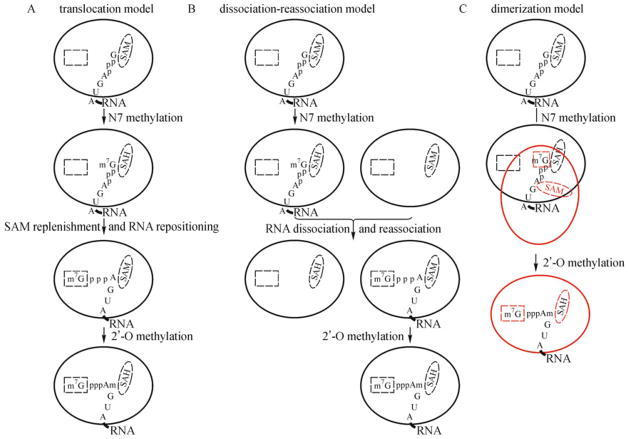
Fig. 3. Models for the sequential N7 and 2′-O methylations of the flavivirus RNA cap.
The SAM-binding and GTP-binding sites are depicted by an oval and rectangle, respectively. In model C, the second MTase molecule and its associated GTP-binding site and SAM are colored in red. See text for details of each of the three models presented.
Our model is consistent with mutagenesis data indicating that distinct sets of amino acids are required for the N7 and 2′-O methylations (Dong et al., 2008b) (Fig. 2H), although currently it is not known how the RNA substrate is re-positioned on the enzyme during the sequential N7 and 2′-OH methylation reactions; three non-mutually exclusive scenarios can be envisaged (Fig. 3). In the first scenario (Fig. 3A), the substrate translocates on the same enzyme molecule, after N7 methylation, to complete the 2′-O methylation (a translocation model) (Zhou et al., 2007). Under this mechanism, the substrate will first bind in the N7 configuration to complete the N7 reaction, and will then translocate, on the same enzyme molecule, to complete the 2′-O methylation. Since SAM, the methyl donor, will be converted to SAH during the N7 MTase reaction, a fresh SAM molecule is required to displace the by-product SAH for the following 2′-O methylation reaction. As an alternative mechanism, in scenario 2 (Fig. 3B), the substrate dissociates from one enzyme molecule after the first N7 methylation, and re-associates with a second enzyme molecule for the second 2′-O methylation (a dissociation and re-association model) (Dong et al., 2008b). This scenario is reminiscent of the RNA capping for the reovirus λ2 (Reinisch et al., 2000) and bluetongue virus (Sutton et al., 2007): the N7 and 2′-O methylations are sequentially executed by two separate MTase domains. In the third scenario (Fig. 3C), the GpppA-RNA substrate binds in one configuration, on one MTase, to catalyze the N7 methylation, and a second MTase dimerizes the MTase-RNA complex to catalyze the 2′-O methylation (a dimerization model).
Recently, using the WNV MTase mutants with defects in either 2′-O or N7 activity, Dong et al. demonstrated that mutant enzymes with different methylation defects can trans complement one another in vitro (Dong et al., 2008a). In addition, sequential treatment of GpppA-RNA with distinct MTase mutants generates fully methylated m7GpppAm-RNA, demonstrating that separate molecules of the enzyme can independently catalyze the two cap methylations in vitro (Dong et al., 2008a). Further studies indicated that N7 methylation of GpppA-RNA → m7GpppA-RNA and of GpppAm-RNA → m7GpppAm-RNA occurred with equal efficiency. However, while the WNV MTase can perform 2′-O methylation of both GpppA-RNA → GpppAm-RNA and m7GpppA-RNA → m7GpppAm-RNA, the 2′-O methylation activity exhibits a preference for substrate m7GpppA-RNA over GpppA-RNA (Dong et al., 2008a). Recently, Chung et al. performed kinetic studies to elucidate the mechanisms of the processive N7 and 2′-O methylations of flavivirus MTase (Chung et al., 2010). Chung et al. demonstrated that sequential N7 to 2′-O methylations occurred via a random bi bi and processive mechanism that does not involve enzyme-RNA dissociation. Analyses of steady state kinetic parameters showed that N7 precedes 2′-O methylation, because it turns over RNA more rapidly. Michaelis constants for SAM in both reactions were about 10-fold lower than those for the respective RNA substrates, suggesting that the rate-limiting steps in methylase reactions are associated with RNA templates. In addition, Assenberg et al. observed a crystallographic dimer of the WVEV MTase in the structure of a flaviviral MTase complexed with GpppA and SAM (Assenberg et al., 2007). Although the GpppA dimer observed in the co-crystal structure is not in a catalytically relevant binding state, the authors proposed that the MTase dimer represents the structure of the methylation complex, supporting a dimerization model (Assenberg et al., 2007).
In addition, Egloff et al. hypothesized that the first step in flavivirus capping entails the binding of a GTP to the GTP pocket of NS5 MTase, followed by binding of an as-yet unidentified GTase, so as to transfer the GTP to nascent diphosphate RNA (Egloff et al., 2007). Then, the capped RNA is repositioned to allow methylation at the guanine-N7 position. Once N7 is methylated, the cofactor SAM is reloaded, and the capped RNA is repositioned such that the methylated cap is in the GTP binding pocket, this registering the 2′-OH of the adenosine ribose for 2′-O methylation. As support for this hypothesis, Issur et al. recently found that the flavivirus NS5 MTase domain also carries RNA GTase activity (Issur et al., 2009).
10 Flaviviral MTase functions
The biological functions of flavivirus MTase have been evaluated through the use of flavivirus replicon and full-length infectious clones of DENV-2 and WNV (Ray et al., 2006; Zhou et al., 2007; Dong et al., 2008b; Kroschewski et al., 2008). Ray et al. first used a luciferase-reporting WNV replicon to demonstrate that (1) the GpppA cap of the WNV replicon is critical for viral translation, and (2) N7 methylation, but not 2′-O methylation, of the cap increases viral translation efficiency, compared to unmethylated cap RNA (Ray et al., 2006).
Further characterization of flaviviral MTase in the context of the viral lifecycle was performed in infectious clones of WNV and DENV-2 (Zhou et al., 2007; Dong et al., 2008b; Kroschewski et al., 2008). A clear correlation was found between the competency of N7 MTase activity and viral replication; also, virus viability required a threshold level of N7 MTase activity. In the context of the full-length virus, a defect in N7 methylation alone or defects in both N7 and 2′-O methylations are lethal to WNV; however, viruses defective solely in 2′-O methylation are only attenuated, and can protect mice from later challenge with the wild-type WNV (Zhou et al., 2007). Similar findings were obtained when the DENV-2 infectious clone was used (Kroschewski et al., 2008). The results demonstrate that the N7, but not 2′-O, methylation activity is essential for the WNV lifecycle (Zhou et al., 2007; Dong et al., 2008b; Kroschewski et al., 2008).
11 Flavivirus MTase inhibitors
Many flaviviruses cause significant human disease. However, currently no clinically approved antiviral therapy is available for treatment of flavivirus-associated diseases. Given that flavivirus MTase has been shown to be essential for the replication of WNV (Dong et al., 2007; Zhou et al., 2007), KUNV (Khromykh et al., 1998), YFV (Bhattacharya et al., 2008), and DENV (Kroschewski et al., 2008), the viral MTase represents a novel and attractive target for flavivirus therapy. In addition, because the N7 MTase activity, but not 2′-O activity, is essential for the virus lifecycle, emphasis should be placed on compounds that can achieve inhibition of N7 methylation activity, when MTase inhibitors are being developed.
Various inhibitors have been found through the use of a variety of techniques including cell-based assay and virtual screening (Benarroch et al., 2004; Luzhkov et al., 2007; Dong et al., 2008b, 2008c; Lim et al., 2008; Bollati et al., 2009a; Milani et al., 2009; Puig-Basagoiti et al., 2009; Sampath and Padmanabhan, 2009; Podvinec et al., 2010; Selisko et al., 2010). Ribavirin 5-triphosphate, a GTP analog, was found to inhibit 2′-O methylation of the DENV-2 MTase by competing with GTP-binding (Benarroch et al., 2004). However, it is not known whether ribavirin inhibits the N7 MTase activity of flavivirus MTase. It has been shown that ribavirin suppresses viral growth through other mechanisms as well (Dong et al., 2008c; Bollati et al., 2009a). When virtual screening was applied to compound libraries, several inhibitors were found to inhibit the 2′-O MTase activity with IC50 (50% inhibition concentration) values in the micromolar range (Luzhkov et al., 2007; Milani et al., 2009; Podvinec et al., 2010). Among these compounds, however, only one compound was test for inhibition of the N7 MTase activity; an IC50 of 127 μmol/L was found for it (Milani et al., 2009). None of the other compounds were tested for their N7 inhibition activity. Using a cell-based assay, Puig-Basagoiti et al. identified a compound that inhibited the WNV N7 MTase activity with an IC50 of 54 μmol/L (Puig-Basagoiti et al., 2009). They also found that the compound inhibited the growth of various flaviviruses with an EC50 (50% effective concentration) in the low micromolar range (Puig-Basagoiti et al., 2009).
In addition, we and others have found that SIN, a SAM analog, is a potent flavivirus MTase inhibitor (Dong et al., 2008b; Lim et al., 2008; Selisko et al., 2010). The crystal structures of the MTases from WESSV (Bollati et al., 2009c) and WNV (Liu and Li, unpublished results) in complex with SIN indicates that SIN binds in the SAM-binding pocket in a manner identical to that of SAM. Dong et al. found that SIN inhibited both the N7 and 2′-O MTase activities with IC50 values of approximately 14 μmol/L (Dong et al., 2008b). SIN was also found to inhibit the 2′-O MTase activity of DENV-2 with an IC50 of less than 0.7 μmol/L (Lim et al., 2008; Selisko et al., 2010). Recently, we found that SIN inhibited the N7 MTase activity of DENV-2 with an IC50 of 0.7 μmol/L (Dong, Liu, Shi and Li, unpublished results). Viral titer reduction assays showed that SIN can suppress WNV replication, with an EC50 of approximately 27 μmol/L, and a therapeutic index of 167 for SIN inhibition against WNV in cell culture (Dong et al., 2008b). These results suggest that SIN analogs have the potential to be developed for flavivirus therapy. The availability of the crystal structure of the NS5 MTase-SIN complexes may promote designing such inhibitor, as well as evaluating the lead compounds identified from the drug screen.
12 Concluding remarks
During the last several years, our understanding of flavivirus capping machinery has grown rapidly, although the answers to many important questions remain elusive. The biochemical characterization of flavivirus NS5 MTase has had a major impact on our understanding of flavivirus capping. Now we know that NS5 MTase plays a major role in RNA cap formation. The MTase domain of NS5 possesses both N7 and 2′-O MTase activities. It can sequentially catalyze the methylations of guanine N7 and ribose 2′-OH to generate N7-methylated and dual (N7 and 2′-O)-methylated RNA molecules. Crystal structures of flaviviral MTases reveal that the MTase uses a single SAM-binding site for both reactions, indicating that the substrate must be repositioned between the dual methylation events. However, the details of the repositioning are currently unknown. Although various models have been proposed for the mechanism of sequential dual methylations, it is currently not known whether one molecule or two of the MTase are involved in cap methylations. It is also unclear whether the substrate dissociates from the MTase upon N7 methylation. In addition, whether the MTase dimerizes during the dual methylations has not been established. Single-molecule fluorescence resonance energy transfer (smFRET) studies may be required to clarify the dual methylation mechanism.
In contrast to the majority of capping enzymes, which lack substrate specificity, flavivirus MTase is now known to require specific viral sequences and structures for the N7 MTase activity and to a lesser extent for the 2′-O MTase activity. Crystal structures of MTases from several flaviviruses and the complexes of these MTases with cap analogs or cap dinucleotide analogs have been determined. These structures have significantly advanced our understanding of the flavivirus MTase, providing a firm basis for functional analysis. However, the cap analogs in the crystals are not in biologically functional configurations. Structures of the MTase in complex with N7 and 2′-O substrates in biologically meaningful configurations will be required to elucidate the structural basis of substrate specificity of the flavivirus MTases.
Functional studies indicated that the N7, but not 2′-O, MTase activity is essential for WNV replication. The N7, but not 2′-O, methylation of the viral RNA cap was also found to increase viral translation efficiency. Mutations with defects in the N7 MTase activity are lethal to WNV. However, thus far the essential role of the N7 MTase has only been evaluated in WNV (Zhou et al., 2007; Dong et al., 2008b), and only limited mutagenesis was performed in a DENV-2 infectious clone (Kroschewski et al., 2008). More experimentation needs to be done to extend the conclusion from WNV to other flaviviruses. In addition, little is known about the role of 2′-O methylation in the viral lifecycle. Viruses defective solely in 2′-O methylation are attenuated and can protect mice from later challenge with wild-type WNV (Zhou et al., 2007). It was found that host innate immune response to WNV infection is dramatically delayed, relative to the response to infection by other RNA viruses (Fredericksen and Gale, 2006). Recent studies have also shown that nucleoside modifications, including 2′-O methylations, could allow the virus to evade RNA-triggered innate immune response (Hornung et al., 2006). Whether 2′-O MTase activity is involved in perturbing the host immune response requires further investigation.
The essential role of flavivirus N7 MTase makes the enzyme an attractive anti-viral drug target. Various promising inhibitors have been found. Among these inhibitors, SIN, a SAM analog, has been extensively investigated. However, since SAM is also a methyl donor for host RNA and protein methylations, its analogs would nonspecifically suppress host MTases, resulting in toxicity. Therefore, identification of features unique to the flaviviral MTase is critical for the design of specific inhibitors of the viral MTase. Indeed, through analysis of crystal structures of the MTase in complex with SAM/SAH/SIN, we found a pocket extension from the SAM-binding pocket (Fig. 2D) (Liu and Li, unpublished results). The pocket is structurally and sequentially conserved among flaviviral MTases, but not among the broader grouping of host SAM-utilizing enzymes. Preliminary studies indicated that the pocket is functionally important for both MTase functions and viral replication (Dong, Liu, Shi and Li, unpublished results). Therefore, SIN analogs bearing modifications that allow the analogs to interact with the additional pocket may display flaviviral specificity, making them potential specific inhibitors of the flaviviral MTase.
Supplementary Material
Acknowledgments
This research was partially supported by grants from the National Institute of Health (NIH) (No. AI07079201A1) to H.L. We thank A. Verschoor for a critical reading of the manuscript.
References
- Abraham G, Rhodes DP, Banerjee AK. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975;5(1):51–58. doi: 10.1016/0092-8674(75)90091-4. [DOI] [PubMed] [Google Scholar]
- Ackermann M, Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J Biol Chem. 2001;276(43):39926–39937. doi: 10.1074/jbc.M104248200. [DOI] [PubMed] [Google Scholar]
- Ahola T, Kääriäinen L. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc Natl Acad Sci USA. 1995;92(2):507–511. doi: 10.1073/pnas.92.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CF, Preugschat F, Strauss JH. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193(2):888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- Asnis DS, Conetta R, Teixeira AA, Waldman G, Sampson BA. The West Nile Virus outbreak of 1999 in New York: the Flushing Hospital experience. Clin Infect Dis. 2000;30(3):413–418. doi: 10.1086/313737. [DOI] [PubMed] [Google Scholar]
- Asnis DS, Conetta R, Waldman G, Teixeira AA. The West Nile virus encephalitis outbreak in the United States (1999–2000): from Flushing, New York, to beyond its borders. Ann N Y Acad Sci. 2001;951:161–171. doi: 10.1111/j.1749-6632.2001.tb02694.x. [DOI] [PubMed] [Google Scholar]
- Assenberg R, Ren J, Verma A, Walter TS, Alderton D, Hurrelbrink RJ, Fuller SD, Bressanelli S, Owens RJ, Stuart DI, Grimes JM. Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J Gen Virol. 2007;88(Pt 8):2228–2236. doi: 10.1099/vir.0.82757-0. [DOI] [PubMed] [Google Scholar]
- Barbas CF, 3rd, Heine A, Zhong G, Hoffmann T, Gramatikova S, Björnestedt R, List B, Anderson J, Stura EA, Wilson IA, Lerner RA. Immune versus natural selection: antibody aldolases with enzymic rates but broader scope. Science. 1997;278(5346):2085–2092. doi: 10.1126/science.278.5346.2085. [DOI] [PubMed] [Google Scholar]
- Barbosa E, Moss B. mRNA(nucleoside-2′-)-methyltransferase from vaccinia virus. Characteristics and substrate specificity. J Biol Chem. 1978;253(21):7698–7702. [PubMed] [Google Scholar]
- Benarroch D, Egloff MP, Mulard L, Guerreiro C, Romette JL, Canard B. A structural basis for the inhibition of the NS5 dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphate. J Biol Chem. 2004;279(34):35638–35643. doi: 10.1074/jbc.M400460200. [DOI] [PubMed] [Google Scholar]
- Bernard KA, Kramer LD. West Nile virus activity in the United States, 2001. Viral Immunol. 2001;14(4):319–338. doi: 10.1089/08828240152716574. [DOI] [PubMed] [Google Scholar]
- Bernard KA, Maffei JG, Jones SA, Kauffman EB, Ebel G, Dupuis AP, 2nd, Ngo KA, Nicholas DC, Young DM, Shi PY, Kulasekera VL, Eidson M, White DJ, Stone WB, Kramer LD and the NY State West Nile Virus Surveillance Team. West Nile virus infection in birds and mosquitoes, New York State, 2000. Emerg Infect Dis. 2001;7(4):679–685. doi: 10.3201/eid0704.010415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Hoover S, Falk SP, Weisblum B, Vestling M, Striker R. Phosphorylation of yellow fever virus NS5 alters methyl-transferase activity. Virology. 2008;380(2):276–284. doi: 10.1016/j.virol.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaillon M, Lemay G. Viral and cellular enzymes involved in synthesis of mRNA cap structure. Virology. 1997;236(1):1–7. doi: 10.1006/viro.1997.8698. [DOI] [PubMed] [Google Scholar]
- Bollati M, Alvarez K, Assenberg R, Baronti C, Canard B, Cook S, Coutard B, Decroly E, de Lamballerie X, Gould EA, Grard G, Grimes JM, Hilgenfeld R, Jansson AM, Malet H, Mancini EJ, Mastrangelo E, Mattevi A, Milani M, Moureau G, Neyts J, Owens RJ, Ren J, Selisko B, Speroni S, Steuber H, Stuart DI, Unge T, Bolognesi M. Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antiviral Res. 2009a Nov 27; doi: 10.1016/j.antiviral.2009.11.009. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati M, Milani M, Mastrangelo E, de Lamballerie X, Canard B, Bolognesi M. Crystal structure of a methyltransferase from a no-known-vector Flavivirus. Biochem Biophys Res Commun. 2009b;382 (1):200–204. doi: 10.1016/j.bbrc.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Bollati M, Milani M, Mastrangelo E, Ricagno S, Tedeschi G, Nonnis S, Decroly E, Selisko B, de Lamballerie X, Coutard B, Canard B, Bolognesi M. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: implications for RNA-capping mechanisms in Flavivirus. J Mol Biol. 2009c;385(1):140–152. doi: 10.1016/j.jmb.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Brinton MA. Isolation of a replication-efficient mutant of West Nile virus from a persistently infected genetically resistant mouse cell culture. J Virol. 1981;39(2):413–421. doi: 10.1128/jvi.39.2.413-421.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton MA. The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu Rev Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology. 1988;162(2):290–299. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- Burke DS, Monath TP. Flaviviruses. Philadelphia, PA: Lippincott William & Wilkins; 2001. [Google Scholar]
- Centers for Disease Control and Prevention (CDC) . Guidelines for surveillance, prevention, and control of West Nile virus infection—United States. MMWR Morb Mortal Wkly Rep. 2000;49(2):25–28. [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Grakoui A, Rice CM. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67(11):6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KY, Dong H, Chao AT, Shi PY, Lescar J, Lim SP. Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2′-O methyltransferase activity in dengue virus. Virology. 2010;402(1):52–60. doi: 10.1016/j.virol.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Cleaves GR, Dubin DT. Methylation status of intracellular dengue type 2 40 S RNA. Virology. 1979;96(1):159–165. doi: 10.1016/0042-6822(79)90181-8. [DOI] [PubMed] [Google Scholar]
- Cong P, Shuman S. Methyltransferase and subunit association domains of vaccinia virus mRNA capping enzyme. J Biol Chem. 1992;267 (23):16424–16429. [PubMed] [Google Scholar]
- Davidson AD. Chapter 2. New insights into flavivirus nonstructural protein 5. Adv Virus Res. 2009;74:41–101. doi: 10.1016/S0065-3527(09)74002-3. [DOI] [PubMed] [Google Scholar]
- De la Peña M, Kyrieleis OJ, Cusack S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 2007;26(23):4913–4925. doi: 10.1038/sj.emboj.7601912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Edgil D, Roberts TG, Lu B, Harris E. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J Virol. 2000;74(17):7814–7823. doi: 10.1128/jvi.74.17.7814-7823.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Ray D, Ren S, Zhang B, Puig-Basagoiti F, Takagi Y, Ho CK, Li H, Shi PY. Distinct RNA elements confer specificity to flavivirus RNA cap methylation events. J Virol. 2007;81(9):4412–4421. doi: 10.1128/JVI.02455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Ren S, Li H, Shi PY. Separate molecules of West Nile virus methyltransferase can independently catalyze the N7 and 2′-O methylations of viral RNA cap. Virology. 2008a;377(1):1–6. doi: 10.1016/j.virol.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Ren S, Zhang B, Zhou Y, Puig-Basagoiti F, Li H, Shi PY. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol. 2008b;82(9):4295–4307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhang B, Shi PY. Flavivirus methyltransferase: a novel antiviral target. Antiviral Res. 2008c;80(1):1–10. doi: 10.1016/j.antiviral.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 2002;21(11):2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol. 2007;372(3):723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell. 2004;13(1):77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- Falgout B, Miller RH, Lai CJ. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J Virol. 1993;67(4):2034–2042. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauman EB, Blumenthal RM, Cheng XD. Structure and evolution of AdoMet-dependent methyltransferases. World Scientific Publishing Co; Singapore: 1999. [Google Scholar]
- Fredericksen BL, Gale M., Jr West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80(6):2913–2923. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey PA, Kokesh FC, Westheimer FH. A reporter group at the active site of acetoacetate decarboxylase. I. Ionization constant of the nitrophenol. J Am Chem Soc. 1971;93(26):7266–7269. doi: 10.1021/ja00755a024. [DOI] [PubMed] [Google Scholar]
- Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss BJ, Thompson AA, Andrews AJ, Sons RL, Gari HH, Keenan SM, Peersen OB. Analysis of flavivirus NS5 methyltransferase cap binding. J Mol Biol. 2009;385(5):1643–1654. doi: 10.1016/j.jmb.2008.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W, O’Gara M, Blumenthal RM, Cheng X. Structure of pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25(14):2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Lima CD. Processing the message: structural insights into capping and decapping mRNA. Curr Opin Struct Biol. 2005;15(1):99–106. doi: 10.1016/j.sbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Guyatt KJ, Westaway EG, Khromykh AA. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J Virol Methods. 2001;92 (1):37–44. doi: 10.1016/s0166-0934(00)00270-6. [DOI] [PubMed] [Google Scholar]
- Hager J, Staker BL, Bugl H, Jakob U. Active site in RrmJ, a heat shock-induced methyltransferase. J Biol Chem. 2002;277(44):41978–41986. doi: 10.1074/jbc.M205423200. [DOI] [PubMed] [Google Scholar]
- Highbarger LA, Gerlt JA, Kenyon GL. Mechanism of the reaction catalyzed by acetoacetate decarboxylase. Importance of lysine 116 in determining the pKa of active-site lysine 115. Biochemistry. 1996;35(1):41–46. doi: 10.1021/bi9518306. [DOI] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Shi X, Quiocho FA. The 1.85 A structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell. 1996;85(2):247–256. doi: 10.1016/s0092-8674(00)81101-0. [DOI] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol Cell. 1998;1(3):443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- Hodel AE, Quiocho FA, Gershon PD. VP39–an mRNA cap-specific 2′-o-methyltransferase. In: Cheng XD, Blementhal RM, editors. S-Adenosylmethionine-dependent methyltransferase: structures and functions. 1999. pp. 255–282. [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Horton JR, Sawada K, Nishibori M, Zhang X, Cheng X. Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons. Structure. 2001;9(9):837–849. doi: 10.1016/s0969-2126(01)00643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issur M, Geiss BJ, Bougie I, Picard-Jean F, Despins S, Mayette J, Hobdey SE, Bisaillon M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA. 2009;15(12):2340–2350. doi: 10.1261/rna.1609709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson AM, Jakobsson E, Johansson P, Lantez V, Coutard B, de Lamballerie X, Unge T, Jones TA. Structure of the methyltransferase domain from the Modoc virus, a flavivirus with no known vector. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 8):796–803. doi: 10.1107/S0907444909017260. [DOI] [PubMed] [Google Scholar]
- Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khromykh AA, Kenney MT, Westaway EG. Trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol. 1998;72(9):7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokesh FC, Westheimer FH. A reporter group at the active site of acetoacetate decarboxylase. II. Ionization constant of the amino group. J Am Chem Soc. 1971;93(26):7270–7274. doi: 10.1021/ja00755a025. [DOI] [PubMed] [Google Scholar]
- Komoto J, Huang Y, Takata Y, Yamada T, Konishi K, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Crystal structure of guanidinoacetate methyltransferase from rat liver: a model structure of protein arginine methyltransferase. J Mol Biol. 2002;320(2):223–235. doi: 10.1016/S0022-2836(02)00448-5. [DOI] [PubMed] [Google Scholar]
- Koonin EV. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72(Pt 9):2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- Koonin EV. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993;74(Pt 4):733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- Kramer LD, Bernard KA. West Nile virus infection in birds and mammals. Ann N Y Acad Sci. 2001;951:84–93. doi: 10.1111/j.1749-6632.2001.tb02687.x. [DOI] [PubMed] [Google Scholar]
- Kramer LD, Li J, Shi PY. West Nile virus. Lancet Neurol. 2007;6(2):171–181. doi: 10.1016/S1474-4422(07)70030-3. [DOI] [PubMed] [Google Scholar]
- Kroschewski H, Lim SP, Butcher RE, Yap TL, Lescar J, Wright PJ, Vasudevan SG, Davidson AD. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J Biol Chem. 2008;283(28):19410–19421. doi: 10.1074/jbc.M800613200. [DOI] [PubMed] [Google Scholar]
- Kümmerer BM, Rice CM. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol. 2002;76(10):4773–4784. doi: 10.1128/JVI.76.10.4773-4784.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee J, Cho Y. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 2003;22(2):292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Clum S, You S, Ebner KE, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73(4):3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang JT, Whelan SP. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci USA. 2006;103(22):8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319(5871):1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- Lim SP, Wen D, Yap TL, Yan CK, Lescar J, Vasudevan SG. A scintillation proximity assay for dengue virus NS5 2′-O-methyltransferase-kinetic and inhibition analyses. Antiviral Res. 2008;80 (3):360–369. doi: 10.1016/j.antiviral.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71 (12):9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73(6):4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzhkov VB, Selisko B, Nordqvist A, Peyrane F, Decroly E, Alvarez K, Karlen A, Canard B, Qvist J. Virtual screening and bioassay study of novel inhibitors for dengue virus mRNA cap (nucleoside-2′O)-methyltransferase. Bioorg Med Chem. 2007;15(24):7795–7802. doi: 10.1016/j.bmc.2007.08.049. [DOI] [PubMed] [Google Scholar]
- Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253(4):618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12(6):783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Mastrangelo E, Bollati M, Milani M, Selisko B, Peyrane F, Canard B, Grard G, de Lamballerie X, Bolognesi M. Structural bases for substrate recognition and activity in Meaban virus nucleoside-2′-O-methyltransferase. Protein Sci. 2007;16(6):1133–1145. doi: 10.1110/ps.072758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani M, Mastrangelo E, Bollati M, Selisko B, Decroly E, Bouvet M, Canard B, Bolognesi M. Flaviviral methyltransferase/RNA interaction: structural basis for enzyme inhibition. Antiviral Res. 2009;83 (1):28–34. doi: 10.1016/j.antiviral.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moure CM, Bowman BR, Gershon PD, Quiocho FA. Crystal structures of the vaccinia virus polyadenylate polymerase hetero-dimer: insights into ATP selectivity and processivity. Mol Cell. 2006;22(3):339–349. doi: 10.1016/j.molcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Muylaert IR, Chambers TJ, Galler R, Rice CM. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology. 1996;222(1):159–168. doi: 10.1006/viro.1996.0406. [DOI] [PubMed] [Google Scholar]
- Muylaert IR, Galler R, Rice CM. Genetic analysis of the yellow fever virus NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol. 1997;71(1):291–298. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell. 2007;25(1):85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Perera R, Kuhn RJ. Structural proteomics of dengue virus. Curr Opin Microbiol. 2008;11(4):369–377. doi: 10.1016/j.mib.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen LR, Roehrig JT. West Nile virus: a reemerging global pathogen. Emerg Infect Dis. 2001;7(4):611–614. doi: 10.3201/eid0704.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrane F, Selisko B, Decroly E, Vasseur JJ, Benarroch D, Canard B, Alvarez K. High-yield production of short GpppA- and 7MeGpppA-capped RNAs and HPLC-monitoring of methyltransfer reactions at the guanine-N7 and adenosine-2′O positions. Nucleic Acids Res. 2007;35(4):e26. doi: 10.1093/nar/gkl1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podvinec M, Lim SP, Schmidt T, Scarsi M, Wen D, Sonntag LS, Sanschagrin P, Shenkin PS, Schwede T. Novel inhibitors of dengue virus methyltransferase: discovery by in vitro-driven virtual screening on a desktop computer grid. J Med Chem. 2010;53(4):1483–1495. doi: 10.1021/jm900776m. [DOI] [PubMed] [Google Scholar]
- Puig-Basagoiti F, Qing M, Dong H, Zhang B, Zou G, Yuan Z, Shi PY. Identification and characterization of inhibitors of West Nile virus. Antiviral Res. 2009;83(1):71–79. doi: 10.1016/j.antiviral.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006;80(17):8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 A resolution. Nature. 2000;404(6781):960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Sampath A, Padmanabhan R. Molecular targets for flavivirus drug discovery. Antiviral Res. 2009;81(1):6–15. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnierle BS, Gershon PD, Moss B. Mutational analysis of a multifunctional protein, with mRNA 5′ cap-specific (nucleoside-2′-O-)-methyltransferase and 3′-adenylyltransferase stimulatory activities, encoded by vaccinia virus. J Biol Chem. 1994;269(32):20700–20706. [PubMed] [Google Scholar]
- Selisko B, Peyrane FF, Canard B, Alvarez K, Decroly E. Biochemical characterization of the (nucleoside-2′O)-methyltransferase activity of dengue virus protein NS5 using purifled capped RNA oligonucleotides (7Me)GpppAC(n) and GpppAC(n) J Gen Virol. 2010;91(Pt 1):112–121. doi: 10.1099/vir.0.015511-0. [DOI] [PubMed] [Google Scholar]
- Shi PY, Kauffman EB, Ren P, Felton A, Tai JH, Dupuis AP, 2nd, Jones SA, Ngo KA, Nicholas DC, Maffei J, Ebel GD, Bernard KA, Kramer LD. High-throughput detection of West Nile virus RNA. J Clin Microbiol. 2001;39(4):1264–1271. doi: 10.1128/JCM.39.4.1264-1271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi PY, Tilgner M, Lo MK. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology. 2002a;296(2):219–233. doi: 10.1006/viro.2002.1453. [DOI] [PubMed] [Google Scholar]
- Shi PY, Tilgner M, Lo MK, Kent KA, Bernard KA. Infectious cDNA clone of the epidemic west nile virus from New York City. J Virol. 2002b;76(12):5847–5856. doi: 10.1128/JVI.76.12.5847-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev SA, Ratnikov BI, Chekanov AV, Sikora S, Rozanov DV, Godzik A, Wang J, Smith JW, Huang Z, Lindberg I, Samuel MA, Diamond MS, Strongin AY. Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem J. 2006;393(Pt 2):503–511. doi: 10.1042/BJ20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- Smithburn BC, Hughes TP, Burke AW, Paul JH. A neurotropic virus isolated from the blood of a native Uganda. Am J Trop Med Hyg. 1940;20:471–492. [Google Scholar]
- Sutton G, Grimes JM, Stuart DI, Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nat Struct Mol Biol. 2007;14(5):449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- Tan BH, Fu J, Sugrue RJ, Yap EH, Chan YC, Tan YH. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216(2):317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- Warrener P, Tamura JK, Collett MS. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67(2):989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G, Wengler G. Terminal sequences of the genome and replicative-from RNA of the flavivirus West Nile virus: absence of poly(A) and possible role in RNA replication. Virology. 1981;113(2):544–555. doi: 10.1016/0042-6822(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Wengler G, Wengler G. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology. 1991;184(2):707–715. doi: 10.1016/0042-6822(91)90440-m. [DOI] [PubMed] [Google Scholar]
- Westaway EG, Brinton MA, Gaidamovich SY, Horzinek MC, Igarashi A, Kaariainen L, Lvov DK, Porterfield JS, Russell PK, Trent DW. Flaviviridae Intervirol. 1985;24:183–192. doi: 10.1159/000149642. [DOI] [PubMed] [Google Scholar]
- WHO. Dengue factsheet. 2009a http://www.who.int/mediacentre/factsheets/fs117/en/
- WHO. Immunization, vaccines and biologicals: Japanese encephalitis. 2009b http://www.who.int/nuvi/je/en/
- WHO. Yellow fever factsheet. 2009c http://www.who.int/mediacentre/factsheets/fs100/en/
- Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 2000;19(14):3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81(8):3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, He XZ, Dixon RA, Noel JP. Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat Struct Biol. 2001;8(3):271–279. doi: 10.1038/85029. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.