Abstract
Gentle touch sensation in Caenorhabditis elegans is mediated by the MEC-4/MEC-10 channel complex, which is expressed exclusively in six touch receptor neurons (TRNs). The complex contains two pore-forming subunits, MEC-4 and MEC-10, as well as the accessory subunits MEC-2, MEC-6, and UNC-24. MEC-4 is essential for channel function, but beyond its role as a pore-forming subunit, the functional contribution of MEC-10 to the channel complex and to touch sensation is unclear. We addressed this question using behavioral assays, in vivo electrophysiological recordings from TRNs, and heterologous expression of mutant MEC-10 isoforms. Animals with a deletion in mec-10 showed only a partial loss of touch sensitivity and a modest decrease in the size of the mechanoreceptor current (MRC). In contrast, five previously identified mec-10 alleles acted as recessive gain-of-function alleles that resulted in complete touch insensitivity. Each of these alleles produced a substantial decrease in MRC size and a shift in the reversal potential in vivo. The latter finding indicates that these mec-10 mutations alter the ionic selectivity of the transduction channel in vivo. All mec-10 mutant animals had properly localized channel complexes, indicating that the loss of MRCs was not attributable to a dramatic mislocalization of transduction channels. Finally, electrophysiological examination of heterologously expressed complexes suggests that mutant MEC-10 proteins may affect channel current via MEC-2.
Introduction
The degenerin/epithelial Na+ channel (DEG/ENaC) proteins comprise a diverse family of ion channel proteins found in invertebrates and vertebrates (for review, see Kellenberger and Schild, 2002). DEG/ENaC proteins are involved in diverse physiological processes, such as sodium transport, mechanosensation, and salt and water taste, but share a common structure with two transmembrane domains, intracellular termini, and a large extracellular loop. Both homomeric and heteromeric channels can be formed.
Mutations in the genes for two DEG/ENaC proteins, MEC-4 and MEC-10, disrupt gentle touch sensation in the nematode Caenorhabditis elegans (Chalfie and Sulston, 1981; Driscoll and Chalfie, 1991; Huang and Chalfie, 1994). These two proteins and at least three accessory subunits (the prohibitin-domain proteins MEC-2 and UNC-24 and the paraoxonase-like protein MEC-6) form a channel complex that transduces touch (Huang et al., 1995; Chelur et al., 2002; Zhang et al., 2004; O'Hagan et al., 2005).
Regions of MEC-4 and MEC-10 both contribute to the pore of the channel complex, as evidenced by the ability of mutations in either protein to affect the ion selectivity of the mechanoreceptor current (MRC) (O'Hagan et al., 2005). The two proteins share extensive sequence similarity, yet the two genes encoding them show dramatically different genetics. This difference may reflect the different roles that the two proteins play in the mechanosensory channel complex. Saturation mutagenesis screens for touch-insensitive animals identified 59 mec-4 mutant alleles compared with only six mec-10 mutant alleles (Chalfie and Sulston, 1981; Chalfie and Au, 1989). Furthermore, although the different mec-4 alleles have mutations scattered throughout the gene, all but one of five mec-10 mutations are clustered within a 25 nt stretch (the defect in the sixth allele has not been identified; Huang and Chalfie, 1994). These observations suggest that MEC-10 may play a minor role in touch sensation. This idea is supported by the finding that no MRC is generated in mec-4 null animals with a wild-type mec-10 allele (O'Hagan et al., 2005), suggesting that MEC-10 is not sufficient for generation of MRCs in the absence of MEC-4. Furthermore, a gain-of-function mutation in mec-10 that causes the degeneration of the touch receptor neurons (TRNs) requires wild-type mec-4, but similar mutations in mec-4 do not require wild-type mec-10 (Chalfie and Wolinsky, 1990; Huang and Chalfie, 1994). However, all six of the previously characterized mec-10 mutations result in complete touch insensitivity. If these mutations cause the loss of MEC-10 activity, this finding implies that MEC-10 plays a critical role in mechanosensation.
In this paper, we resolve the apparent paradox of MEC-10 function by showing that the protein is not required for a behavioral or electrophysiological response to touch. Instead, MEC-10 plays a regulatory role in the channel complex and is essential for full sensitivity to gentle touch. In addition, we show that five previously identified mec-10 mutant alleles are not loss-of-function mutations but recessive gain-of-function alleles. Finally, by recording currents from heterologously expressed channel complexes, we show that the gain-of-function effect of the mec-10 mutations may be mediated via the MEC-2 accessory subunit.
Materials and Methods
Worm strains.
Wild type (N2) and strains with the mutations mec-3(e1338), mec-4(u253), mec-10(e1515), mec-10(u20), mec-10(u390), mec-10(u332), mec-10(e1715), lin-35(n745), and mec-4(u339) have been described previously (Brenner, 1974; Chalfie and Au, 1989; Huang and Chalfie, 1994; Lu and Horvitz, 1998) or constructed genetically (Brenner, 1974). In addition, we used TU2769, a strain with an integrated array containing mec-17::gfp and lin-15(+) in a lin-15(n765ts) background (O'Hagan et al., 2005), to generate strains with labeled TRNs. mec-10(ok1104) was obtained from the C. elegans Gene Knockout Consortium at Oklahoma Medical Research Foundation (Oklahoma City, OK). mec-10(tm1552) was obtained from the National Bioresource Project for the Nematode at Tokyo Women's Medical University (Tokyo, Japan). For visualization of MEC-4::YFP, an extrachromosomal array of Pmec-4 mec-4::yfpPmec-4cfp (Chelur et al., 2002) was integrated using gamma irradiation (Mello and Fire, 1995) and outcrossed seven times to wild type before crossing into the mec-10 backgrounds.
mec-10(ok1104) sequencing. The following primer pairs were used for PCR amplification of overlapping 1–2 kb regions of the mec-10 gene for sequencing (Genewiz): 5′-GAAGGAATTTTTTGGGATGGGG and 5′-ACGGGTTCAAATTGCAAAGA; 5′-CACGGATATACAATTGAAGTTTGAC and 5′-CACTATCGCCAAAGTATTCCC; 5′-GATCGGAACTCAAGAAGGAG and 5′-CGACACTTGAATGATCCGTG; and 5′-GTTAGGAACATTTGATACGGTTTC and 5′-CAAAAAAAAAATGCAAAAGTGTGTACCC. cDNA was sequenced from products amplified from mRNA isolated from ok1104 animals by RT-PCR using the following primer pairs: 5′-CGTAGTCGCAGTCGATTTCA and 5′-CGACACTTGAATGATCCGTG; and 5′-CGTAGTCGCAGTCGATTTCA and 5′-CACTATCGCCAAAGTATTCCC.
Touch assays.
Animals were assayed for response to gentle touch as described by Chalfie and Sulston (1981) and scored as described by Hobert et al. (1999). Each animal was usually touched 10 times, alternating between touch to the anterior and the posterior part of the animal. In experiments to test whether mec-10 mutations resulted in differential responses along the TRN processes, we touched individual animals at specific locations only once. All touch assays were performed as blind tests.
To determine the touch sensitivity of animals that had a mec-10 mutant allele and the mec-10(ok1104) deletion allele, we mated mec-10(ok1104) hermaphrodites with males hemizygous for the mec-10 mutations and homozygous for the mec-17::gfp transgene from TU2769. Animals with this transgene express GFP in the TRNs without any significant effect on the touch sensitivity of the animal (O'Hagan et al., 2005). GFP-positive F1 progeny were picked from the crosses and tested for response to touch as described above.
Harsh touch was tested by prodding 20 wild-type, mec-4(u253), mec-3(e1338), and mec-10(tm1552) mec-4(u253) animals near the vulva with a platinum wire.
RNA interference.
RNA interference (RNAi) was performed by feeding, essentially as described by Kamath and Ahringer (2003), using three different RNAi sensitizing backgrounds: lin-35(n745) (Lehner et al., 2006), Punc-119sid-1Punc-119gfpPmec-6mec-6; lin-15b(n744), and Punc-119 sid-1Punc-119gfpPmec-6mec-6 (Calixto et al., 2010). The latter was used for knockdown of mec-10 in ok1104 animals, because mec-10 and lin-15b are located in close proximity to each other on chromosome X. Worms were fed Escherichia coli expressing double-stranded RNA against either mec-10 or mec-4 [Geneservice RNAi library, ID number X-4G16 (mec-10) or X-7D15 (mec-4) (Fraser et al., 2000); primer pairs for the mec-10 clone: forward, ATCGGAAAACCAACACTTGC and reverse, CGTAGTCGCAGTCGATTTCA; and for the mec-4 clone: forward, TACCTGCAACGGAAAGATCC and reverse, ATACAACGGAAAGACGCCAC]. To control for nonspecific effects of RNAi, we compared the touch response of test animals with that of animals fed bacteria expressing double-stranded RNA against GFP [pPD128.110 from the Fire Lab Vector kit, Addgene plasmid 1649 (Addgene); sequence available at http://www.addgene.org]. The bacteria was grown in liquid at 37°C for ∼12 h before seeding onto nematode growth medium–isopropyl-thio-β-d-galactoside (NGM–IPTG) plates and then grown at room temperature for 24 h before use. Eggs were harvested by bleaching gravid adult animals in 0.1 m KOH, 10% bleach solution for 5 min. The eggs were washed three times in M9 buffer and then placed on seeded NGM–IPTG plates and grown at 20°C (lin-35 background) or 15°C [Punc-119sid-1 Punc-119gfpPmec-6mec-6; lin-15b(n744) and Punc-119sid-1Punc-119gfpPmec-6 mec-6 backgrounds]. Touch assays were as described above and were performed blind, with respect to both the strain and the bacteria on which it was raised.
Whole-mount immunochemistry.
Animals were fixed with 1% formaldehyde for 30 min and permeabilized with β-mercaptoethanol, DTT, and H2O2 for antibody staining as described (Miller and Shakes, 1995). Fixed animals were stained with a rabbit polyclonal antibody against the N terminus of MEC-2 (1:200) (Zhang et al., 2004) overnight at 4°C and Alexa Fluor 488-conjugated goat anti-rabbit antibody (1:2000) (Invitrogen) for 2 h at room temperature. Fluorescent micrographs were collected at 100× and used to compute inter-punctum intervals as follows: line segments tracing PLM neurons were straightened using the “straighten selection” function in Fiji (http://pacific.mpi-cbg.de/wiki/index.php?title=Fiji&oldid=4701), and straightened segments were used to compute intensity line scans from which inter-punctum intervals were measured from the distance between intensity peaks.
In vivo electrophysiology.
Electrical responses to a mechanical stimulus were recorded at a holding potential of −74 mV from the cell body of a PLM touch receptor neuron that was exposed in the tail of an immobilized worm. Whole-cell recordings and data analysis were performed as described by O'Hagan et al. (2005), except data were collected using a HEKA EPC-10 amplifier. Recordings with a holding current of less than −15 pA and a series resistance of <100 MΩ were used for analysis (average series resistance was 64 MΩ). External saline contained the following (in mm): 145 NaCl, 5 KCl, 5 MgCl2, 1 CaCl2, 10 KHEPES, pH 7.2 (adjusted to ∼320 mOsm with d-glucose). Internal saline contained the following (in mm): 125 Kgluconate, 22 NaCl, 1 MgCl2, 0.6 CaCl2, 10 NaHEPES, and 10 K2EGTA, pH 7.2. Sulforhodamine 101 (20 μm) was added to the internal saline to visualize the touch receptor neurite and confirm connection to the cell after recording. Holding potential was corrected for liquid junction potentials. For current–voltage relations, the membrane potential at the stimulus site was adjusted for attenuation because of the distance between the point of stimulus and the recording electrode (Vm), using the following equation (O'Hagan et al., 2005): Vm = Vh cosh(ln/λn − ds/λn)/cosh(ln/λn), where Vh is the holding potential, ln is the length of the neurite, λn is the length constant, and ds is the distance between stimulus and electrode. The length of the neurite was estimated from the length of the body of the worm [measured from a video recording using NIH ImageJ (http://rsbweb.nih.gov/ij/)] using the following relationship (O'Hagan et al., 2005): ln = 123 + 0.34L, where L is the body length (in micrometers). ds was measured from a video recording, and λn was estimated as described by O'Hagan et al. (2005).
Pressure dependence of MRCs was calculated as described by O'Hagan et al. (2005). The average P½ of wild-type MRCs we found in these experiments was similar but not identical to the previously published value. This difference is likely attributable to the errors associated with estimating the spring constants of the reference probe and the stimulus probe, as well as the area of stimulus probe that contacted the cuticle of the animal. However, this systematic difference had no effect on the interpretation of results, as differences in P½ values between wild type and mutants were highly consistent.
Single-channel conductance of the mechanoreceptor channel in wild-type and mec-10 deletion animals was determined using nonstationary noise analysis (Heinemann and Conti, 1992) as described by O'Hagan et al. (2005).
Heterologous expression, electrophysiology, coimmunoprecipitation, and detection of surface-expressed channels.
Expression constructs for MEC-4d (A713T), MEC-2, MEC-6, MEC-10, myc::MEC-4d (A713T), and MEC-10::GFP have all been described previously (Chelur et al., 2002; Goodman et al., 2002). Point mutations corresponding to mec-10 mutations were generated using the QuikChange site-directed mutagenesis kit (Stratagene).
cRNA was synthesized using the T7 mMESSAGE mMACHINE kit (Applied Biosystems/Ambion). Ten nanograms of MEC-4d, MEC-2, MEC-10 and 1 ng of MEC-6 cRNA were injected into Xenopus laevis oocytes. Oocytes were cultured in ND-96 with penicillin–streptomycin and 300 μm amiloride at 17°C and recorded from or used for biochemical experiments, 5 d after injection. Two-electrode voltage-clamp recordings, coimmunoprecipitation, and surface expression experiments were performed as described previously (Chelur et al., 2002; Goodman et al., 2002).
For two-electrode voltage-clamp recordings, normal bath solution contained the following (in mm): 100 Na-gluconate, 2 KCl, 1 CaCl2, 2 MgCl2, and 10 NaHEPES, pH 7.2. Pipettes were filled with 3 m KCl. Amiloride was added from a stock solution (0.1 m in DMSO) to a final concentration of 300 μm to the bath solution to record amiloride-insensitive currents.
For coimmunoprecipitation experiments, oocytes were injected with cRNAs producing myc::MEC-4d, MEC-2, and wild-type or mutant forms of MEC-10::GFP. An agarose-conjugated polyclonal antibody against c-myc (A-14; Santa Cruz Biotechnology) was used to precipitate myc::MEC-4d from the lysate, followed by staining of Western blots with an HRP-conjugated monoclonal antibody against GFP (B-2; Santa Cruz Biotechnology) to detect GFP-tagged wild-type or mutant forms of MEC-10.
Surface expression was tested essentially as described (Goodman et al., 2002). In brief, oocytes were injected with cRNAs for myc:: MEC-4d, MEC-2, and wild-type or mutant forms of MEC-10. Five days after injection, healthy and intact cells were selected and incubated in EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific Pierce Protein Research Products). Unbound biotin was quenched with free glycine, and the oocytes were washed several times before lysis. Lysates were incubated with agarose-conjugated avidin beads to precipitate biotinylated surface proteins. Samples were then run on SDS-PAGE, and Western blots were stained with HRP-conjugated c-myc antibody (9E10; Santa Cruz Biotechnology) to detect surface-expressed myc::MEC-4d.
Results
Decreasing or eliminating mec-10 results in a partial defect in touch sensation
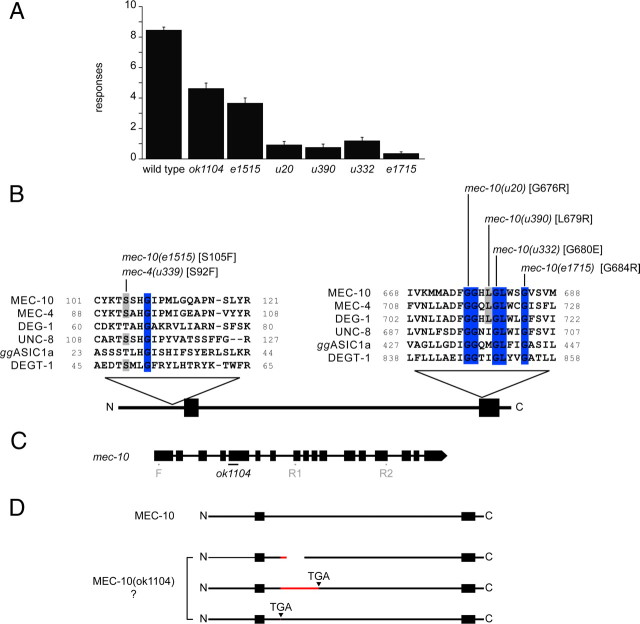
Saturation mutagenesis screens for touch-insensitive animals identified five mec-10 alleles (all recessive) with confirmed mutations: e1515, u20, u390, u332, and e1715 (Chalfie and Sulston, 1981; Chalfie and Au, 1989). On average, mec-10 mutant animals respond only to 1–4 touches of 10, whereas wild-type animals respond to 9 touches of 10 (Fig. 1A). Each of the mec-10 alleles has a single missense mutation (Huang and Chalfie, 1994) that affects a protein domain that is conserved within the DEG/ENaC protein family (Fig. 1B). Therefore, these mutations may affect critical functions not only of MEC-10 but also of DEG/ENaC channel proteins more generally.
Figure 1.
mec-10 mutant alleles. A, Touch sensitivity of mec-10 animals. Each animal was touched 10 times, as described in Materials and Methods (mean ± SEM, n = 30 for each). B, Location of missense mutations in mec-10 mutant alleles within conserved regions of DEG/ENaC proteins. Protein sequences were aligned using COBALT (Papadopoulos and Agarwala, 2007). Except ASIC1a (Gallus gallus), all sequences are from C. elegans. Gray shading indicates conserved residues at sites mutated in mec-10. Residues conserved among all the proteins are highlighted in blue. Transmembrane domains are represented by black boxes. C, The ok1104 allele carries a 143 bp deletion (black bar) at the junction of the fourth intron and the fifth exon in the mec-10 gene. Location of primer pairs used for RT-PCR amplification is indicated in gray (F, R1, and R2). D, Putative protein products in ok1104 animals as derived from conceptual translation of three different RT-PCR products (see Results). Transmembrane domains are represented by black boxes, foreign amino acid residues by red bars, and a deletion by a white bar.
To determine whether loss of mec-10 also results in a touch-insensitive phenotype, we characterized a candidate null allele, mec-10(ok1104). The ok1104 allele was identified by PCR-based screening of a deletion library, rather than a phenotype-based screen. ok1104 animals had a partial defect in response to gentle touch, a phenotype that was weaker than any of the five previously identified mec-10 alleles (Fig. 1A).
ok1104 animals have a 143 bp deletion in the mec-10 gene starting at the junction between the fourth intron and the fifth exon. To learn how this deletion affects MEC-10 expression, we used two primer pairs for RT-PCR to compare mec-10 mRNA in wild-type and ok1104 animals (Fig. 1C). One primer pair (F-R1) resulted in multiple products from ok1104 animals and a single band from wild-type animals (data not shown). A single RT-PCR product was detected in ok1104 and wild-type animals with a second primer pair (F-R2) that spans a region including that covered by the first primer pair. We sequenced the RT-PCR product from ok1104 animals from the F-R2 primer pair and two of the products from the F-R1 primer. Conceptual translation of these three gene products revealed the following (Fig. 1D): a transcript encoding an aberrant MEC-10 product with a deletion of 47 aa (L190 to G236) and two transcripts encoding premature stop codons. This analysis indicates that the ok1104 deletion allele disrupts transcription of the mec-10 gene but leaves open the possibility that ok1104 animals might express a mutant MEC-10 protein.
We further tested the effects of reduced MEC-10 activity on touch sensitivity in vivo using RNAi. Initially, we used strains carrying lin-35(n745), which increases the efficiency of RNAi in the TRNs (Lehner et al., 2006). RNAi against mec-10 in these animals had no effect on touch sensitivity (Fig. 2A), whereas RNAi against mec-4 reduced touch sensitivity as found previously (Lehner et al., 2006).
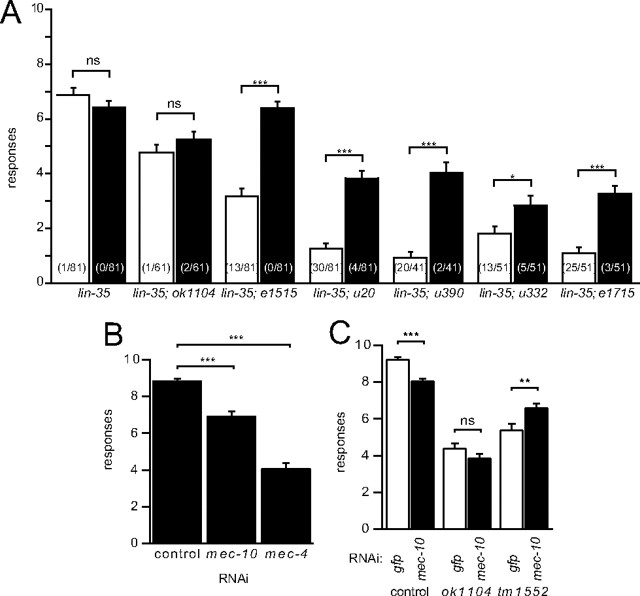
Figure 2.
Effects of mec-10 RNAi. A, RNAi against mec-10 in lin-35(n745) has no effect on animals with wild-type mec-10 or animals with the ok1104 deletion allele and enhances the touch sensitivity of animals with missense mutations in mec-10. Animals were fed bacteria expressing dsRNA against either GFP (white bars) or mec-10 (black bars) and assayed for touch response as described in Materials and Methods. Bars are mean ± SEM for n = 41–81 animals. Significance in all panels from Student's t test: ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.0001. Numbers in parentheses represent the number of animals that were completely unresponsive. B, RNAi against mec-10 results in a partial loss of touch sensitivity in a strain that is hypersensitive to RNAi [Punc-119sid-1 Punc-119gfp Pmec-6mec-6; lin-15b(n744)], whereas RNAi against mec-4 produces a more dramatic loss of touch sensitivity. Animals were fed bacteria expressing dsRNA against either GFP (control), mec-10, or mec-4 as indicated and assayed for touch response as described in Materials and Methods (mean ± SEM, n = 60 for each). C, RNAi against mec-10 in a hypersensitive RNAi background (Punc-119sid-1 Punc-119gfp Pmec-6mec-6) reduces the touch sensitivity of animals with wild-type mec-10 (control) and does not affect the touch response of animals with a deletion in mec-10 (ok1104 or tm1552). Animals were fed bacteria expressing dsRNA against either GFP (white bars) or mec-10 (black bars) and assayed for touch response as described in Materials and Methods (mean ± SEM, n = 90–100 for each).
If MEC-10 were required for touch sensitivity, we would expect animals treated with mec-10 RNAi to have touch sensitivity similar to that of animals carrying the ok1104 deletion allele (Fig. 1A). RNAi against mec-10 in lin-35(n745) animals, however, had no detectable effect on the touch response. Because this discrepancy could be attributable to incomplete knockdown of mec-10, we retested mec-10 RNAi in animals whose neurons were made very sensitive to RNAi by neuronal expression of the gene sid-1 (Calixto et al., 2010). RNAi against mec-10 in these animals produced partially touch-insensitive animals (Fig. 2B). However, RNAi against mec-4 produced an even more dramatic reduction in touch sensitivity in sid-1(+) transgenic animals. These results mirror the more severe touch-insensitive phenotype of mec-4 null mutants compared with mec-10 deletion animals. The phenotype of ok1104 animals and the effects of knockdown of mec-10 suggest that MEC-10 is not essential for touch sensation. This result could explain why no null alleles of mec-10 were identified in previous screens for touch-insensitive animals.
These RNAi results suggest that the ok1104 deletion causes a loss of functional MEC-10 protein. If, however, ok1104 resulted in a polypeptide that interfered with proper function of the channel complex, then knockdown of mec-10 expression by RNAi in ok1104 animals should rescue their touch response (the ok1104 allele has >90% of the region targeted by mec-10 RNAi). In contrast, we found that RNAi against mec-10 had no effect on the touch response in ok1104 mutants with either the lin-35 mutation (Fig. 2A) or the sid-1(+) transgene (Fig. 2C). These results suggest that either a truncated protein is not made in ok1104 animals, or if it is made, it does not interfere with the function of the touch receptor complex.
We also tested the behavioral response of trans-heterozygotes with one copy of e1515, u20, u390, u332, or e1715 and one copy of ok1104. The touch responses of these animals were indistinguishable from homozygous mec-10 mutant animals (supplemental Fig. 1A,B, available at www.jneurosci.org as supplemental material). This result also suggests that ok1104 is a loss-of-function allele.
Chatzigeorgiou et al. (2010a,b) reported that mec-10(tm1552), an allele with a deletion in the region of the gene that encodes the extracellular domain [as with mec-10(ok1104)], also produced partial touch insensitivity. Therefore, both ok1104 and tm1552 are probably similar loss-of-function alleles. Moreover, a similarly deleted region in mec-4(u253) results in a loss-of-function allele (Hong et al., 2000). We used ok1104 in our experiments because RNAi against mec-10 did not alter touch sensitivity. In contrast, RNAi for mec-10 increased the touch sensitivity of tm1552 animals (Fig. 2C).
Chatzigeorgiou et al. (2010a) also reported that the deletion in mec-10(tm1552) results in differential touch insensitivity along the length of the TRN processes. In contrast, we found that wild-type, mec-10(ok1104), and mec-10(tm1552) animals all showed a reduced response to touch near the ALM cell body compared with touch near the second pharyngeal bulb (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). No difference in response was detected from the two ends of the PLM neurons.
Chatzigeorgiou et al. (2010b) have proposed that MEC-10 is essential for harsh touch sensitivity. Neither animals homozygous for missense mutations in mec-10 (Huang and Chalfie, 1994) nor animals with both a mec-4 null mutation (u253) and a mec-10 deletion (tm1552) were insensitive to harsh touch (this study). All wild-type animals responded to prodding by a platinum wire, whereas only 15 ± 8% (mean ± SEM, n = 20 for all) of mec-3(e1338) animals, which are defective in sensing harsh touch (Way and Chalfie, 1989), responded. However, 85 ± 8% of mec-4(u253) and 80 ± 9% of mec-10(ok1104)mec-4(u253) double-mutant animals responded to harsh touch stimuli.
mec-10 alleles that result in touch insensitivity contain gain-of-function mutations
The results presented thus far suggest that loss of mec-10 causes only mild touch insensitivity (Figs. 1A, 2B), whereas several mutant mec-10 alleles result in greater loss of touch sensitivity (Fig. 1A). To test whether this more severe defect is caused by gain-of-function mutations in the mec-10 alleles, we used RNAi to knock down expression of mutant mec-10. In contrast to the effects of RNAi against mec-10 in animals carrying either wild-type mec-10 or the ok1104 allele, RNAi against mec-10 reduced the penetrance and expressivity of touch insensitivity of mec-10 mutant animals (Fig. 2A). Thus, decreasing expression of mutant MEC-10 proteins resulted in animals that responded more often to repeated stimuli and fewer animals completely failed to respond to touch (Fig. 2A). These results support the conclusion that the mec-10 mutations are gain-of-function mutations.
Transduction channel subunits are positioned normally in mec-10 mutant animals
The MEC-4/MEC-10 channel complex is localized in puncta along the process of the TRNs (Chelur et al., 2002; Zhang et al., 2004). The partial or complete loss of touch sensitivity in mec-10 animals could be a result of inefficient or improper localization of transduction channel subunits in the TRNs. To test for defects in the localization of the receptor complex in mec-10 deletion and mutant animals, we assayed the localization of two components of the channel complex, MEC-4 and MEC-2. Using MEC-4::YFP and an anti-MEC-2 antibody, we found that the transduction channel in mec-10 mutant animals was localized in a punctate pattern similar to that seen in wild-type animals (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). This result indicates that the touch insensitivity seen in mec-10 animals is likely a result of interference with the function and not a gross mislocalization of the transduction channel.
Loss of MEC-10 decreases MRC amplitude but not single-channel conductance or pressure sensitivity
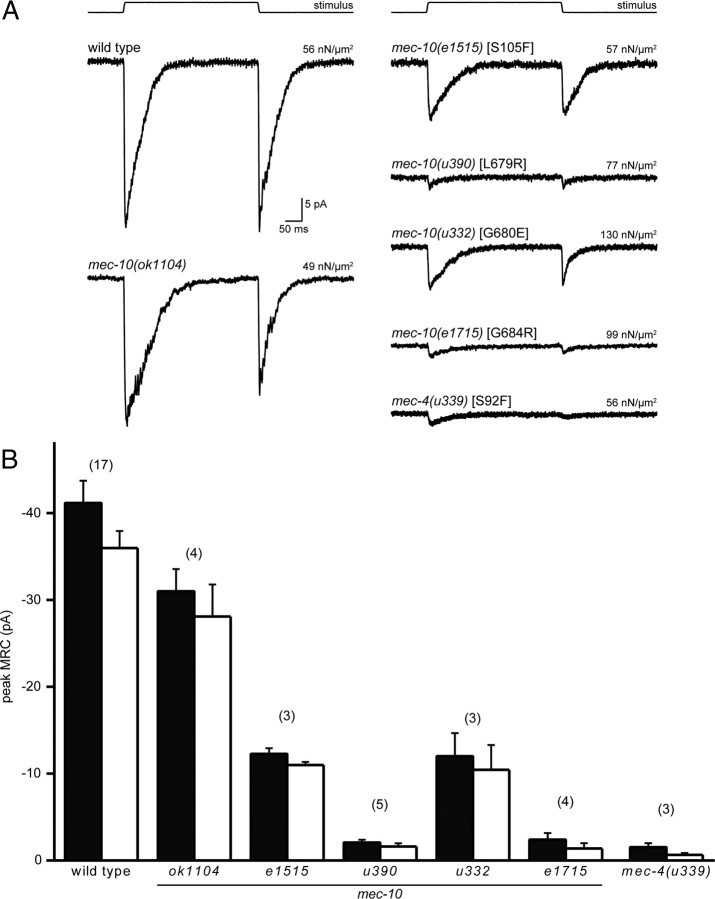
We recorded MRCs from PLM touch receptor neurons in live animals to determine how loss of MEC-10 affects channel function. Application and removal of a mechanical stimulus elicits large inward MRCs in both wild-type and mec-10(ok1104) deletion mutants (Fig. 3A). Loss of mec-10 did not detectably affect the latency of MRC activation at either the onset or offset of a touch but slowed the rate of MRC activation and adaptation of the onset response (Table 1). Specifically, rates for activation (τ1) and adaptation (τ2) of “on” MRCs were τ1 = 2.6 ± 0.2 ms and τ2 = 51 ± 3 ms for wild-type animals but were τ1 = 4.2 ± 0.3 ms and τ2 = 67 ± 3 ms in mec-10(ok1104) animals (p < 0.05 by one-way ANOVA). Thus, MEC-10 is required for normal MRC kinetics in vivo. TRNs from animals with the ok1104 mutation also had smaller peak MRC amplitudes, ∼75% that of wild-type animals.
Figure 3.
mec-10 and mec-4 mutants have MRCs with decreased amplitude. A, Examples of in vivo recordings of MRCs from PLM touch receptor neurons in wild-type, mec-10, and mec-4 animals (at −74 mV) in response to a saturating mechanical stimulus (top). Each trace is an average of 20 sweeps. Stimulus pressure is indicated above each trace. B, Maximum peak amplitude of MRC recorded from PLM touch receptor neurons (at −74 mV), at the onset (black bars) and offset (white bars) of a response to a saturating mechanical stimulus (mean ± SEM, numbers in parentheses indicate number of cells tested).
Table 1.
Kinetics of “on” and “off” MRCs
| Genotype | On |
Off |
||||
|---|---|---|---|---|---|---|
| Latency (ms) | t1 (ms) | τ2 (ms) | Latency (ms) | τ1 (ms) | τ2 (ms) | |
| Wild type | 0.7 ± 0.1 | 2.6 ± 0.2 | 51 ± 3 | 1.9 ± 0.1 | 2.0 ± 0.2 | 49 ± 6 |
| (n = 23) | (n = 12) | (n = 12) | (n = 23) | (n = 12) | (n = 12) | |
| ok1104 | 0.7 ± 0.1 | 4.2 ± 0.3* | 67 ± 3* | 1.8 ± 0.1 | 1.6 ± 0.1 | 41 ± 3 |
| (n = 10) | (n = 10) | (n = 10) | (n = 10) | (n = 10) | (n = 10) | |
| e1515 | 1.0 ± 0.2 | 2.7 ± 0.5 | 48 ± 6 | 3.0 ± 0.3* | 2.3 ± 0.2 | 47 ± 3 |
| (n = 5) | (n = 5) | (n = 5) | (n = 5) | (n = 5) | (n = 5) | |
| u332 | 0.8 ± 0.1 | 2.9 ± 0.2 | 49 ± 4 | 2.7 ± 0.2* | 1.5 ± 0.1 | 20 ± 2* |
| (n = 8) | (n = 7) | (n = 7) | (n = 8) | (n = 7) | (n = 7) | |
Average latency, activation (τ1), and adaptation (τ2) time constants were calculated from MRC responses to stimuli of at least 40 nN/μm2 [mean ± SEM, n is number of recordings;
*p < 0.05, one-way ANOVA and subsequent pairwise comparisons (Tukey's test)].
We tested whether the change in MRC size reflected a decrease in single-channel conductance. Using noise analysis to estimate the apparent single-channel conductance, we found that it was similar in wild-type and ok1104 mutants: 15.6 ± 0.8 pS for wild type (n = 25) and 15.0 ± 1.8 pS for ok1104 (n = 9). Thus, the effect of the ok1104 deletion on MRC amplitude is not attributable to a decrease in single-channel conductance. This in vivo observation is consistent with previous work showing that coexpressing MEC-10 with MEC-4 in Xenopus oocytes has no effect on the single-channel conductance measured directly in outside-out membrane patches (Brown et al., 2008).
Next, we investigated whether the decrease in MRC size in mec-10(ok1104) mutants was attributable to a loss of pressure sensitivity and found that normalized pressure–response curves were similar in wild-type and mec-10(ok1104) mutants (supplemental Fig. 3A,B, available at www.jneurosci.org as supplemental material). Collectively, these results indicate that MEC-10 is not essential for the formation of pressure-sensitive ion channels in the PLM neurons but fine tunes the time course of activation and adaptation. Additionally, the results suggest that the reduction in MRC size reflects a decrease in either the number of channels available to be opened by external force in vivo, a decrease in the peak open probability, or a combination of both factors.
Missense mutations in mec-10 and mec-4 decrease MRC amplitude
In contrast to TRNs in mec-10(ok1104) mutants, which have nearly wild-type MRCs, TRNs from animals carrying mec-10 missense alleles exhibited a dramatic reduction in peak MRC size. For instance, mec-10(e1515) animals had MRCs with a maximum amplitude that was ∼30% of that in wild-type animals. Because the e1515 mutation replaces a conserved serine in the N terminus of MEC-10 with a phenylalanine (S105F), this result suggests that the N terminus is essential for normal DEG/ENaC function in vivo. To learn whether this effect was specific to MEC-10 or a more general feature of DEG/ENaC channels, we recorded from mec-4(u339) animals in which the homologous residue in MEC-4 is identically changed (S92F) (Fig. 1B). We found that MRCs in mec-4(u339) animals were reduced to 4% of their wild-type amplitude. MRC amplitude was also dramatically decreased by the u332, u390, and e1715 alleles, which affect conserved residues in the second transmembrane domain (TM2) of MEC-10 (Fig. 3). The effect of mec-10 mutations on MRC amplitude mirrored the severity of the behavioral defects such that mutations that give rise to mild defects in behavior such as e1515 also retain larger-amplitude MRCs.
As in wild-type animals, MRC amplitude increased with applied pressure in e1515 and u332 mutants. Thus, mutant channel complexes retain their ability to detect external force. Consistent with this idea, neither e1515 nor u332 showed an increase in the stimulus pressure required to fully saturate MRC amplitude (supplemental Fig. 3C,D, available at www.jneurosci.org as supplemental material). (The MRCs in u390 and e1715 animals were too small to analyze.) The pressure–response curve for e1515 was slightly shifted to the right (supplemental Fig. 3C, available at www.jneurosci.org as supplemental material): P½ values were 15 ± 4 nN/μm2 (n = 3) and 8 ± 1 nN/μm2 (n = 17) in e1515 and wild-type animals, respectively. Thus, larger pressures were required for half-maximal amplitude MRC activation in e1515 animals, suggesting that the mutation in e1515 in the N terminus of the protein influences channel gating. Pressure–response curves in u332 animals, in contrast, were indistinguishable from wild type (supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
We also examined the latencies for MRC activation at the onset and the offset of the mechanical stimulus in e1515 and u332 mutant animals (Table 1; MRCs in u390 and e1715 were too small to analyze). The measured responses latencies are <1 ms and represent an upper bound on the time elapsed between the delivery (or removal) of the mechanical stimulus and activation of channels in vivo. Latencies for channel activation in response to stimulus onset were similar to wild type in both e1515 and u332 mutants. In contrast, latencies for the “off” response were increased. As in wild-type animals, MRCs in mec-10 missense mutants activated rapidly and adapted during continuous stimulation (Fig. 3A). The time course of MRCs in e1515 mutants was similar to that recorded in wild-type PLM neurons (Table 1). MRCs in u332, however, showed a 2.5-fold increase in the adaptation rate (τ2) of the off response. Previous work also showed an increased rate of adaptation and response latency in MRCs recorded from mec-10(u20) animals (O'Hagan et al., 2005).
To determine whether mutations in mec-10 and mec-4 specifically affect MRCs or they affect the general excitability of TRNs, we recorded the response of voltage-gated currents to a series of voltage steps. These currents are generated in response to voltage pulses and are independent of the mechanosensitive response. Peak and steady-state voltage-gated currents recorded from all mutant animals were qualitatively similar to those recorded from wild-type animals and from previous studies (O'Hagan et al., 2005) (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). Hence, these mutations appear to affect mechanotransduction specifically.
Missense mutations in mec-10, but not loss of MEC-10, alter MRC ion selectivity
Recordings of MRCs in wild-type animals demonstrate that the MEC-4/MEC-10 transduction channel is Na+ selective (O'Hagan et al., 2005). Such selectivity is preserved when MEC-4/MEC-10 channels are expressed in Xenopus oocytes and is insensitive to the presence or absence of MEC-10 (Goodman et al., 2002; Brown et al., 2007). Consistent with the properties of channels expressed in oocytes, MRCs reversed polarity near +20 mV in both wild-type and ok1104 animals (Fig. 4A). Thus, loss of MEC-10 does not appear to affect ion selectivity in vivo. In contrast, mutations in MEC-10 shifted the reversal potential for MRCs by −40 mV or more (Fig. 4B,C). The negative reversal potential of the MRC in mec-10 mutant animals suggests that mutant channels have increased permeability to K+ ions. A similar effect was reported previously for MRCs recorded from mec-10(u20) and mec-4(u2) animals (O'Hagan et al., 2005). This result suggests that the shift in ion selectivity is caused by gain-of-function mutations in mec-10 rather than a nonfunctional MEC-10.
Figure 4.
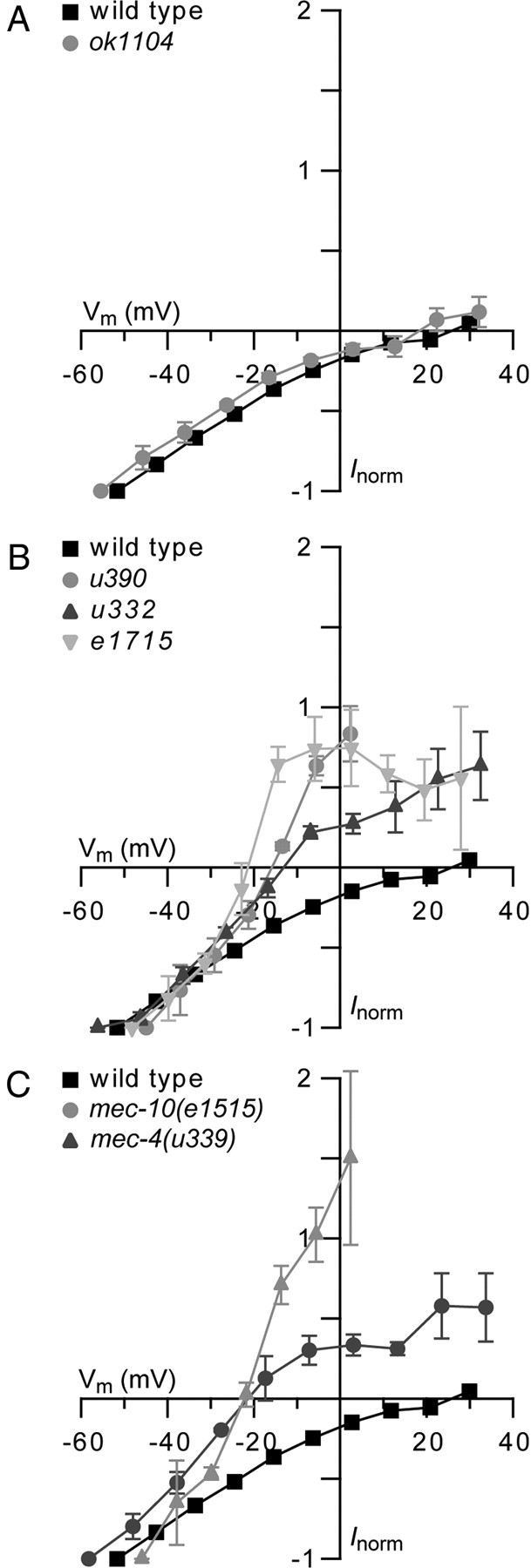
Point mutations in mec-10 and mec-4 alter the reversal potential of the MRC, whereas a deletion in mec-10 does not affect the reversal potential. Current–voltage relations for MRCs recorded from PLM cells at the onset of a saturating mechanical stimulus in mutant (gray) and wild-type (black) animals (mean ± SEM). MRCs were recorded at −114 to +66 mV in 20 mV increments. Current was normalized (Inorm) to the current recorded at a holding potential of −114 mV, and membrane potential (Vm) was adjusted for voltage attenuation as described in Materials and Methods. A, mec-10(ok1104) (n = 4) compared with wild-type (n = 7). B, mec-10(u390) (n = 3), mec-10(u332) (n = 4), and mec-10(e1715) (n = 4) compared with wild-type (n = 7). C, mec-10(e1515) (n = 3) and mec-4(u339) (n = 3) compared with wild type (n = 7).
To test this interpretation, we measured the MRC–voltage relationship in mec-4(u339) animals. If the gain-of-function mutation in e1515 is responsible for the shift in ion selectivity, a mutation in the conserved S92 residue in mec-4(u339) should cause a similar shift in the reversal potential of the MRCs. We found that this was the case: MRCs recorded in both mec-10(e1515) and mec-4(u339) animals reversed polarity near −20 mV (Fig. 4C).
Mutant MEC-10 requires MEC-2
In addition to MEC-4 and MEC-10, several non-pore-forming accessory subunits form part of the mechanosensory channel complex. These accessory subunits include MEC-2, a PHB-domain protein, and MEC-6, a paraoxonase-like protein. These proteins colocalize with the channel along the process of the TRN and interact with MEC-4 and MEC-10 (Chelur et al., 2002; Goodman et al., 2002; Zhang et al., 2004). Expression of MEC-2 and MEC-6 in Xenopus oocytes affects both the size and the properties of the MEC-4 current. To investigate how mutant MEC-10 proteins interact with other subunits of the channel complex, we expressed wild-type and mutant forms of MEC-10 along with MEC-4, MEC-2, and MEC-6 in Xenopus oocytes. As in previous studies, we used a constitutively active MEC-4 isoform [MEC-4d also known as MEC-4(A713T)] (Goodman et al., 2002).
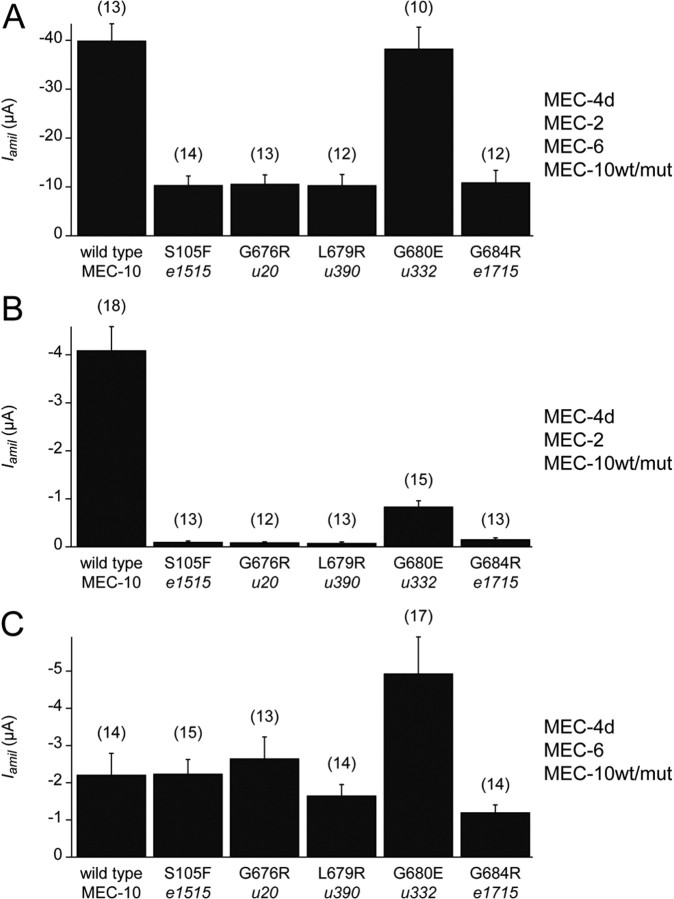
Coexpression of MEC-10 with MEC-4d, MEC-2, and MEC-6 reduced the amiloride-sensitive current recorded from Xenopus oocytes compared with current recorded from oocytes expressing MEC-4d, MEC-2, and MEC-6 (our unpublished observation). We found that mutant MEC-10 proteins further reduced the current. Current recorded from oocytes expressing MEC-4d, MEC-2, MEC-6 and mutant MEC-10 was as low as 25% of the current recorded from oocytes expressing MEC-4d, MEC-2, MEC-6 and wild-type MEC-10 (Fig. 5A). Therefore, oocytes expressing MEC-4d/MEC-10/MEC-2/MEC-6 channel complexes recapitulate the gain-of-function effects of the mec-10 mutations in vivo. The one exception was MEC-10(G680E), corresponding to the u332 allele, which resulted in an amiloride-sensitive current with an amplitude comparable with that recorded from oocytes expressing channel complexes with wild-type MEC-10. The mutant phenotype produced by the u332 allele may result from interactions with other proteins of the channel complex in vivo.
Figure 5.
MEC-10 mutant proteins inhibit the amiloride-sensitive current in Xenopus oocytes in a MEC-2-dependent manner. Amiloride-sensitive current (Iamil) recorded at −85 mV from the MEC-4d channel complex expressed with wild-type and mutant forms of MEC-10 as indicated beneath each bar (mean ± SEM, numbers in parentheses represent number of cells tested). A, Oocytes expressing MEC-4d, MEC-2, MEC-6, and wild-type and mutant forms of MEC-10 as noted. Except for the G680E variant, currents recorded from channels with MEC-10 variants were significantly different from currents from channels with wild-type MEC-10 (p < 0.00001, Student's t test). B, Oocytes expressing MEC-4d, MEC-2, and wild-type and mutant forms of MEC-10 as noted. Currents recorded from channels with MEC-10 variant proteins were significantly different from those from channels with wild-type MEC-10 (p < 0.00001, Student's t test). C, Oocytes expressing MEC-4d, MEC-6, and wild-type and mutant forms of MEC-10 as noted. Currents recorded from channels with G680E were significantly larger than currents from channels with wild-type MEC-10 (p < 0.05, Student's t test). No significant difference in current between channels with any of the other MEC-10 variants and those with wild-type MEC-10 (p = 0.12–0.97, Student's t test).
The reduction of the amiloride-sensitive current by mutant versions of MEC-10 did not require MEC-6. The current was decreased by >95% relative to channel complexes with wild-type MEC-10 for all the MEC-10 mutant proteins coexpressed with MEC-4d and MEC-2 in the absence of MEC-6 except MEC-10(G680E), which decreased the current by 80% (Fig. 5B).
In contrast, the reduction of the amiloride-sensitive current by mutant MEC-10 proteins required MEC-2. Coexpression of MEC-10 mutant proteins with MEC-4d and MEC-6 produced currents that were similar to channel complexes with wild-type MEC-10 (Fig. 5C). The exception was MEC-10(G680E), which produced more than twofold larger current than wild-type MEC-10 when coexpressed with MEC-4d/MEC-6.
The effect of the mutation in the u332 allele [MEC-10(G680E)] was anomalous to the other mec-10 alleles in heterologously expressed channels. MEC-10(G680E) decreased the current recorded from the MEC-4d/MEC-2/MEC-10 channel complexes less, relative to wild-type MEC-10, than any of the other four MEC-10 mutant proteins. MEC-10(G680E) also increased the current from the MEC-4d/MEC-6/MEC-10 complex relative to wild type, whereas the other mutant proteins functioned similarly to wild-type MEC-10. The net effect of the MEC-10(G680E) mutation, however, was similar to the effect of wild-type MEC-10 in MEC-4d/MEC-2/MEC-6 channel complexes. However, both behavioral responses of u332 animals and in vivo recordings from these animals were similar to the other mec-10 mutant animals. Differences in heterologously expressed channels could be attributable to an interaction between the residue mutated in u332 with the activating mutation in MEC-4d, which is in close proximity to the site of the mec-10 mutations. Conversely, these differences could be caused by additional proteins that interact with the MEC-4/MEC-10 complex in vivo but are not present in the heterologous system.
Next we tested whether the mutations in the mec-10 alleles decrease the association between mutant MEC-10 proteins and MEC-4d in Xenopus oocytes. We found that all mutant forms of MEC-10 coimmunoprecipitated with MEC-4d when coexpressed with MEC-2 (supplemental Fig. 5A, available at www.jneurosci.org as supplemental material). We also tested whether coexpression of mutated MEC-10 proteins resulted in fewer MEC-4d channel complexes expressed at the surface of the cell. We found that MEC-4d is expressed at the surface of the oocyte when coexpressed with either wild-type or mutant forms of MEC-10 and MEC-2 (supplemental Fig. 5B, available at www.jneurosci.org as supplemental material). Together, these results indicate that the effects of the MEC-10 mutations on the current recorded from heterologously expressed MEC-4d channels are likely attributable to an effect on the function of the channel complex and not interactions between the channel subunits or trafficking to the surface of the oocyte. These findings are in agreement with in vivo recordings of MRCs.
Discussion
Functional diversity of DEG/ENaC channel subunits
Although the amino acid sequences of MEC-4 and MEC-10 are 54% identical, these pore-forming channel subunits are not interchangeable. mec-4 null animals, which retain the wild-type mec-10 gene, lack MRCs (O'Hagan et al., 2005), and overexpression of one gene cannot rescue the touch insensitivity caused by mutation of the other gene (Huang and Chalfie, 1994). Because mec-10(ok1104) animals have substantial MRCs, MEC-10 is not essential to form channels in the PLM neurons. MEC-10 is needed, however, for optimal ion channel activity in vivo. MRCs generated in response to saturating mechanical stimuli in mec-10 deletion mutants are smaller than in wild-type PLM neurons, and these animals show reduced touch sensitivity in behavioral assays. Previous observations had also suggested a minor role for MEC-10 based on the differential sensitivity of the TRNs to activating d mutations in mec-4 and mec-10 (Huang and Chalfie, 1994). Thus, as with β or γENaC (Canessa et al., 1993, 1994; Lingueglia et al., 1993) and ASIC2b (Lingueglia et al., 1997; Hesselager et al., 2004), MEC-10 cannot form functional channels on its own and plays a regulatory role in MEC-4 mechanotransduction complexes.
The mec-10 gene is expressed in the gentle touch-sensitive TRNs and the harsh touch-sensitive PVD and FLP neurons (Huang and Chalfie, 1994). However, only the TRNs that sense gentle touch coexpress mec-4. In PLM, loss of MEC-10 decreases, but does not eliminate, the maximal MRC evoked by low-intensity or “gentle” stimuli. Recently, Chatzigeorgiou et al. (2010b) reported that the response to harsh touch in the PVD neurons and in the TRNs absolutely requires MEC-10 and another DEG/ENaC protein, DEGT-1. [Harsh touch sensitivity in FLP requires MEC-10 but not DEGT-1 (Chatzigeorgiou and Schafer, 2011).] Our experiments lead us to question the role of MEC-10 in the PVD neurons. First, using the same deletion (tm1552) used by Chatzigeorgiou et al. (2010b), we could not detect a significant loss of harsh touch sensitivity in animals lacking mec-4. Second, if mec-10 were essential for the harsh touch channel of the PVD neurons and acted similarly to its role in the MEC-4 channel complex, one might expect that animals with the recessive gain-of-function mutations in mec-10 would also be harsh touch insensitive, but they are not (Huang and Chalfie, 1994). We propose that mec-10 is not likely to be essential in either the TRNs or the PVD neurons.
Interactions between subunits in the mechanoreceptor channel complex
MEC-2 and MEC-6 synergistically increase the current from the MEC-4d/MEC-10 channel complex in vitro by increasing the number of channels in an active state (Chelur et al., 2002; Brown et al., 2008). We show that heterologously expressed mutant MEC-10 proteins decrease currents from the MEC-4d/MEC-10 channel, analogous to the in vivo effects of mutations in mec-10. This effect is dependent on the presence of MEC-2 but not MEC-6. Previous work showed that MEC-2 decreases the amiloride sensitivity of the channel and increases single-channel conductance, suggesting that MEC-4 and MEC-10 together form an amiloride-binding site that can be modified by MEC-2 (Chelur et al., 2002; Brown et al., 2008). The MEC-2 dependence of the gain-of-function effects of the MEC-10 missense mutations may indicate that the interaction between MEC-2 and MEC-4 causes a conformational change. This conformational change could render the channel complex susceptible to interference by the MEC-10 mutations. Alternatively, or additionally, MEC-2 could directly affect MEC-10 conformation.
Structural basis of ion selectivity in DEG/ENaC channels
Five mec-10 alleles (u20, u390, u332, e1715, and e1515) and two mec-4 alleles (u2, u339) encode point mutations that alter the reversal potential of MRCs in vivo (this study and O'Hagan et al., 2005). Whereas wild-type MRCs reverse polarity near the equilibrium potential for Na+ ions, mutant MRCs reverse polarity 40 mV negative to this value, on average (Fig. 4) (O'Hagan et al., 2005, their Fig. 6C). The simplest explanation for this change in reversal potential is that point mutations encoded by mec-10 and mec-4 missense alleles increase K+ permeability. Such an increase in permeability could arise from an increase in the diameter of the mutant pore, because the selectivity sequence of wild-type DEG/ENaC channels mirrors ionic size (Kellenberger and Schild, 2002).
Five of the seven missense mec-10 and mec-4 alleles analyzed in vivo result in amino acid substitutions of conserved residues in TM2 of MEC-10 and MEC-4 (Fig. 1B). A high-resolution crystal structure of a related DEG/ENaC channel, chicken ASIC1a, reveals that TM2 forms a long α helix, tilted at 50° with respect to the plane of the membrane (Gonzales et al., 2009). All of the affected TM2 residues are predicted to lie on the same side of the TM2 helix and localize to the cytoplasmic side of the putative gate. Four of five mutations affect conserved glycines (the fifth is a lysine residue that is conserved in many but not all family members). Our experimental data support the idea that main-chain carbonyl oxygen atoms at such glycine residues coordinate permeant ions in the open-channel conformation (Gonzales et al., 2009). Indeed, two of the glycine residues proposed to coordinate ions in the open-channel state by Gonzales et al. (2009) correspond to the residues affected in u332 and e1715. Mutations at the position equivalent to the site mutated in e1715 affect ion selectivity in vertebrate ENaC channels (for review, see Kellenberger and Schild, 2002). A third glycine, which is mutated in both mec-10(u20) and mec-4(u2) and conserved across phyla, was proposed to form a desensitization gate in cASIC1a (Gonzales et al., 2009). Our current and previous results (O'Hagan et al., 2005) demonstrate that this glycine has an additional role in regulating ion selectivity and may also coordinate permeant ions in the open channel.
Loss of MEC-10 has little or no effect on MRC reversal potential (Fig. 4A), implying that homomeric MEC-4 channels (formed in mec-10 deletion mutants) and heteromeric MEC-4/MEC-10 channels have identical ion selectivity profiles. Similar results were seen when these channel proteins were expressed in Xenopus oocytes (Goodman et al., 2002; Brown et al., 2007) and underscores the importance of the glycine surface in TM2, which is identical in MEC-4 and MEC-10, for regulating pore size and coordinating permeant ions.
The mec-10 gain-of-function alleles affect more than a common surface on TM2; the e1515 and u339 mutations affect a conserved serine residue in the N terminus of MEC-10 and MEC-4, respectively (Fig. 1B). These mutations reduce the peak amplitude of the MRCs and change the ion selectivity of the transduction channel. Thus, the N terminus of MEC-4 and MEC-10 is involved in ion selectivity in vivo. The contribution of the N terminus to selectivity is shared by vertebrate DEG/ENaC proteins because mutating the homologous position in ASIC2 and ASIC3 also increases K+ permeability (Coscoy et al., 1999).
In principle, such a selectivity change could offset the effect of degeneration-inducing mutations in MEC-4 by limiting the depolarization induced by constitutive channel activation in the mutants. Consistent with this idea, transgenic expression of a u339 [MEC-4(S92F)] mutant isoform suppresses degeneration induced by the toxic MEC-4(A713V) protein in vivo (Hong et al., 2000). The structural basis for the effect of the N terminus on selectivity is unknown but could involve direct interactions with the pore-lining TM2 in the open-channel state.
In this study and that of O'Hagan et al. (2005), we identified five residues (in TM2 and the cytoplasmic N terminus) that can mutate to alter selectivity. These data imply that the structural basis of ion selectivity in DEG/ENaC is distinct from the well-characterized selectivity filter of K+-selective ion channels (Armstrong, 2003). In particular, K+-selective channels rely on a highly conserved 3 aa sequence oriented such that backbone carbonyls form closely spaced binding sites for K+ ions in the pore. The Na+ selectivity of DEG/ENaC channels, in contrast, appears to rely on residues distributed across at least three helical turns in TM2 and extending into the N terminus. Thus, the body plan of the selectivity filter of DEG/ENaC channels is distinct from that of ion channels that contain a pore-loop motif similar to that found in bacterial and eukaryotic K+ and Na+ channels. Because DEG/ENaC channels are conserved in animals and absent from both plant and bacterial genomes (Goodman and Schwarz, 2003; Hunter et al., 2009), this type of ion selectivity filter may represent an animal-specific motif in ion channels.
Footnotes
This work was supported by National Institutes of Health Grants GM30997 (M.C.) and NS047715 (M.B.G.) and a Howard Hughes Medical Institute Predoctoral Fellowship (R.O.). We thank C. A. Yao for helpful comments on this manuscript, I. Topalidou for help with the touch sensitivity studies, and A. L. Eastwood for generating and providing the mec-4 mec-10 double mutant.
References
- Armstrong CM. Voltage-gated K channels. Sci STKE. 2003;2003:re10. doi: 10.1126/stke.2003.188.re10. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Fernandez-Illescas SM, Liao Z, Goodman MB. Gain-of-function mutations in the MEC-4 DEG/ENaC sensory mechanotransduction channel alter gating and drug blockade. J Gen Physiol. 2007;129:161–173. doi: 10.1085/jgp.200609672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AL, Liao Z, Goodman MB. MEC-2 and MEC-6 in the Caenorhabditis elegans sensory mechanotransduction complex: auxiliary subunits that enable channel activity. J Gen Physiol. 2008;131:605–616. doi: 10.1085/jgp.200709910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canessa CM, Horisberger JD, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature. 1993;361:467–470. doi: 10.1038/361467a0. [DOI] [PubMed] [Google Scholar]
- Canessa CM, Schild L, Buell G, Thorens B, Gautschi I, Horisberger JD, Rossier BC. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989;243:1027–1033. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Wolinsky E. The identification and suppression of inherited neurodegeneration in Caenorhabditis elegans. Nature. 1990;345:410–416. doi: 10.1038/345410a0. [DOI] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Schafer WR. Lateral facilitation between primary mechanosensory neurons controls nose touch perception in C. elegans. Neuron. 2011;70:299–309. doi: 10.1016/j.neuron.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Grundy L, Kindt KS, Lee WH, Driscoll M, Schafer WR. Spatial asymmetry in the mechanosensory phenotypes of the C. elegans DEG/ENaC gene mec-10. J Neurophysiol. 2010a;104:3334–3344. doi: 10.1152/jn.00330.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzigeorgiou M, Yoo S, Watson JD, Lee WH, Spencer WC, Kindt KS, Hwang SW, Miller DM, 3rd, Treinin M, Driscoll M, Schafer WR. Specific roles for DEG/ENaC and TRP channels in touch and thermosensation in C. elegans nociceptors. Nat Neurosci. 2010b;13:861–868. doi: 10.1038/nn.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelur DS, Ernstrom GG, Goodman MB, Yao CA, Chen L, O'Hagan R, Chalfie M. The mechanosensory protein MEC-6 is a subunit of the C. elegans touch-cell degenerin channel. Nature. 2002;420:669–673. doi: 10.1038/nature01205. [DOI] [PubMed] [Google Scholar]
- Coscoy S, de Weille JR, Lingueglia E, Lazdunski M. The pre-transmembrane 1 domain of acid-sensing ion channels participates in the ion pore. J Biol Chem. 1999;274:10129–10132. doi: 10.1074/jbc.274.15.10129. [DOI] [PubMed] [Google Scholar]
- Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MB, Schwarz EM. Transducing touch in Caenorhabditis elegans. Annu Rev Physiol. 2003;65:429–452. doi: 10.1146/annurev.physiol.65.092101.142659. [DOI] [PubMed] [Google Scholar]
- Goodman MB, Ernstrom GG, Chelur DS, O'Hagan R, Yao CA, Chalfie M. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- Heinemann SH, Conti F. Nonstationary noise analysis and application to patch clamp recordings. Methods Enzymol. 1992;207:131–148. doi: 10.1016/0076-6879(92)07009-d. [DOI] [PubMed] [Google Scholar]
- Hesselager M, Timmermann DB, Ahring PK. pH dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J Biol Chem. 2004;279:11006–11015. doi: 10.1074/jbc.M313507200. [DOI] [PubMed] [Google Scholar]
- Hobert O, Moerman DG, Clark KA, Beckerle MC, Ruvkun G. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Mano I, Driscoll M. In vivo structure–function analyses of Caenorhabditis elegans MEC-4, a candidate mechanosensory ion channel subunit. J Neurosci. 2000;20:2575–2588. doi: 10.1523/JNEUROSCI.20-07-02575.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Chalfie M. Gene interactions affecting mechanosensory transduction in Caenorhabditis elegans. Nature. 1994;367:467–470. doi: 10.1038/367467a0. [DOI] [PubMed] [Google Scholar]
- Huang M, Gu G, Ferguson EL, Chalfie M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature. 1995;378:292–295. doi: 10.1038/378292a0. [DOI] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 2002;82:735–767. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- Lehner B, Calixto A, Crombie C, Tischler J, Fortunato A, Chalfie M, Fraser AG. Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol. 2006;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingueglia E, Voilley N, Waldmann R, Lazdunski M, Barbry P. Expression cloning of an epithelial amiloride-sensitive Na+ channel. A new channel type with homologies to Caenorhabditis elegans degenerins. FEBS Lett. 1993;318:95–99. doi: 10.1016/0014-5793(93)81336-x. [DOI] [PubMed] [Google Scholar]
- Lingueglia E, de Weille JR, Bassilana F, Heurteaux C, Sakai H, Waldmann R, Lazdunski M. A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J Biol Chem. 1997;272:29778–29783. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- O'Hagan R, Chalfie M, Goodman MB. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat Neurosci. 2005;8:43–50. doi: 10.1038/nn1362. [DOI] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Way JC, Chalfie M. The mec-3 gene of Caenorhabditis elegans requires its own product for maintained expression and is expressed in three neuronal cell types. Genes Dev. 1989;3:1823–1833. doi: 10.1101/gad.3.12a.1823. [DOI] [PubMed] [Google Scholar]
- Zhang S, Arnadottir J, Keller C, Caldwell GA, Yao CA, Chalfie M. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr Biol. 2004;14:1888–1896. doi: 10.1016/j.cub.2004.10.030. [DOI] [PubMed] [Google Scholar]