Abstract
To directly determine whether recombinational repair of double-strand breaks (DSBs) can occur between heterologous chromosomes and lead to chromosomal rearrangements in mammalian cells, we employed an ES cell system to analyze recombination between repeats on heterologous chromosomes. We found that recombination is induced at least 1000-fold following the introduction of a DSB in one repeat. Most (98%) recombinants repaired the DSB by gene conversion in which a small amount of sequence information was transferred from the unbroken chromosome onto the broken chromosome. The remaining recombinants transferred a larger amount of information, but still no chromosomal aberrations were apparent. Thus, mammalian cells are capable of searching genome-wide for sequences that are suitable for DSB repair. The lack of crossover events that would have led to translocations supports a model in which recombination is coupled to replication.
Keywords: DNA double-strand break, recombination, translocation, ES cells
The faithful repair of DNA damage such as chromosomal double-strand breaks (DSBs) is necessary for the maintenance of genomic integrity. Aberrantly repaired DSBs are expected to result in chromosomal rearrangements, including translocations, which promote mutagenesis, transformation, or even cell lethality. Multiple mechanisms have evolved to ensure proper repair of DSBs, details of which are now being elucidated. Historically, the repair of DSBs in mammalian cells was presumed to occur almost exclusively by nonhomologous end joining (Jackson and Jeggo 1995), a process that is integral to V(D)J recombination during lymphocyte development (Taccioli et al. 1993) and possibly isotype switching as well (Casellas et al. 1998). Aberrations of these break repair processes are known to contribute to multiple lymphoid malignancies (see Hiom et al. 1998 and references therein). Direct examination of normal DSB repair in cultured cells, however, has recently contrasted with this view by revealing that homologous recombination is also a major pathway of DSB repair in mammalian cells (Liang et al. 1998). In fact, chromosome breaks are the most potent inducers of recombination known in mammalian cells, increasing homologous recombination three orders of magnitude or more (Rouet et al. 1994; Choulika et al. 1995; Sargent et al. 1997; Liang et al. 1998). The centrality of homologous recombination to genomic integrity is strongly supported by mouse knockouts of Rad51, a homolog to the prokaryotic strand exchange protein RecA, as loss of Rad51 function results in early embryonic lethality (Lim and Hasty 1996; Tsuzuki et al. 1996).
Although homologous recombination is a major DNA repair pathway in mammalian cells, large fractions of mammalian genomes are composed of repetitive elements. Thus, an interesting paradox exists: Mammalian cells appear to be potentially at high risk for genome rearrangements, yet such rearrangements are not generally seen. In humans repetitive elements constitute fully one-third of the genome, with Alu elements, the most abundant class of dispersed repeat sequences, estimated at 106 copies (for discussion, see Schmid 1996 and references therein). Alu elements, each composed of two similar 280-bp monomers, are highly homologous to each other, and 70%–98% homologous to the Alu consensus sequence. It has been suggested that the common 26-bp core sequence is a recombinational hot spot with sequence similarity to the prokaryotic recombination hot spot chi. A few cases of repetitive element-mediated recombination in human cells, and its deleterious consequences, have been reported (Cooper et al. 1998), and some, including complex rearrangements in chronic myelogenous leukemia (CML) patients, may involve Alu–Alu-mediated ectopic recombination (Morris et al. 1996; Jeffs et al. 1998).
Given the quantity of repetitive elements in mammalian genomes, no cell would escape genomic rearrangements if recombination, in particular events involving crossing-over, occurred at ectopic sites with significant frequency. Thus, cells with complex genomes must possess mechanisms that limit recombination, or the outcomes of recombination, between a large fraction of the genome. It has been suggested that sequence divergence between repeats plays a role in this suppression (Elliott et al. 1998; Waldman and Liskay 1988), yet mismatch repair mutants in which recombination between diverged sequences is elevated do not exhibit obvious genome rearrangements. Ectopic position has also been predicted to play a role in suppressing homologous recombination. Yet nonhomologous joining of different chromosomes has been documented in translocations, suggesting that DNA sequences at ectopic sites can interact with each other (e.g., Wang et al. 1997). In addition, at least in some instances in the mouse, ectopic recombination can occur at a detectable frequency (Baker and Read 1992; Murti et al. 1994).
Because DSBs are potent inducers of recombination, we directly determined the potential of homologous DSB repair to lead to chromosomal translocations in mammalian cells. For this, we developed a system to analyze recombination between repeats located on two heterologous chromosomes. Specifically, chromosomes 14 and 17 in mouse embryonic stem (ES) cells were marked with two defective neomycin phosphotransferase (neo) genes, one of which contains the cleavage site for the rare-cutting endonuclease, I-SceI. Recombination between the neo gene sequences can result in a neo+ gene. Although spontaneous recombination was undetectable, a DSB was found to induce interchromosomal recombination >1000-fold. However, when recombinants were examined, no chromosomal aberrations, including translocations, were detected. Almost 98% of recombinants were found to have undergone gene conversions unassociated with crossing-over. In these cases, a small amount of information was transferred at the break site from the unbroken chromosome. In the remaining recombinants, a larger amount of sequence information was transferred. These results clearly indicate that following a DSB, the cell is capable of searching the genome and finding homologous sequences suitable for repair, even when these sequences reside on otherwise nonhomologous chromosomes. However, crossover events that would result in genome rearrangements are suppressed. The lack of crossover events, as well as the recovery of recombinants in which a larger amount of sequence is transferred during recombination, supports a model in which recombination is coupled to replication.
Results
Interchromosomal recombination substrates
To examine homologous recombination between two different chromosomes, gene targeting constructs were designed to introduce either of two defective neo genes to mouse chromosomes 17 and 14 (Fig. 1). The defective neo genes, S2neo and Pneo, differ in the type and position of the mutation. As described previously (Moynahan and Jasin 1997), Pneo contains a PacI recognition site in the 5′ portion of the neo coding region to render the gene nonfunctional. Using a linked hprt gene, Pneo has been targeted to the retinoblastoma (Rb) locus of chromosome 14 in E14TG2a mouse ES cells to create the N cell line (Moynahan and Jasin 1997). The second defective neo gene S2neo is nonfunctional because of the replacement of a NcoI site in the 3′ coding region with the recognition site for the rare-cutting I-SceI endonuclease (Smih et al. 1995). The targeting construct for the S2neo gene contains a linked promoterless hygromycin (hyg) gene flanked by pim-1 sequences from chromosome 17 (te Riele et al. 1990). Because we do not know the relative orientation of the pim-1 and Rb genes on the two chromosomes, we constructed two pim-1 targeting vectors (Fig. 1A). The F (forward) construct has the S2neo gene in the same transcriptional orientation as the pim-1/hyg gene fusion. The R (reverse) construct has the S2neo gene in the reverse orientation. With this design, reciprocal recombination leading to translocation of chromosome arms should be selectable with one of the two orientations, without leading to acentric or dicentric chromosomes (Fig. 1B).
Figure 1.
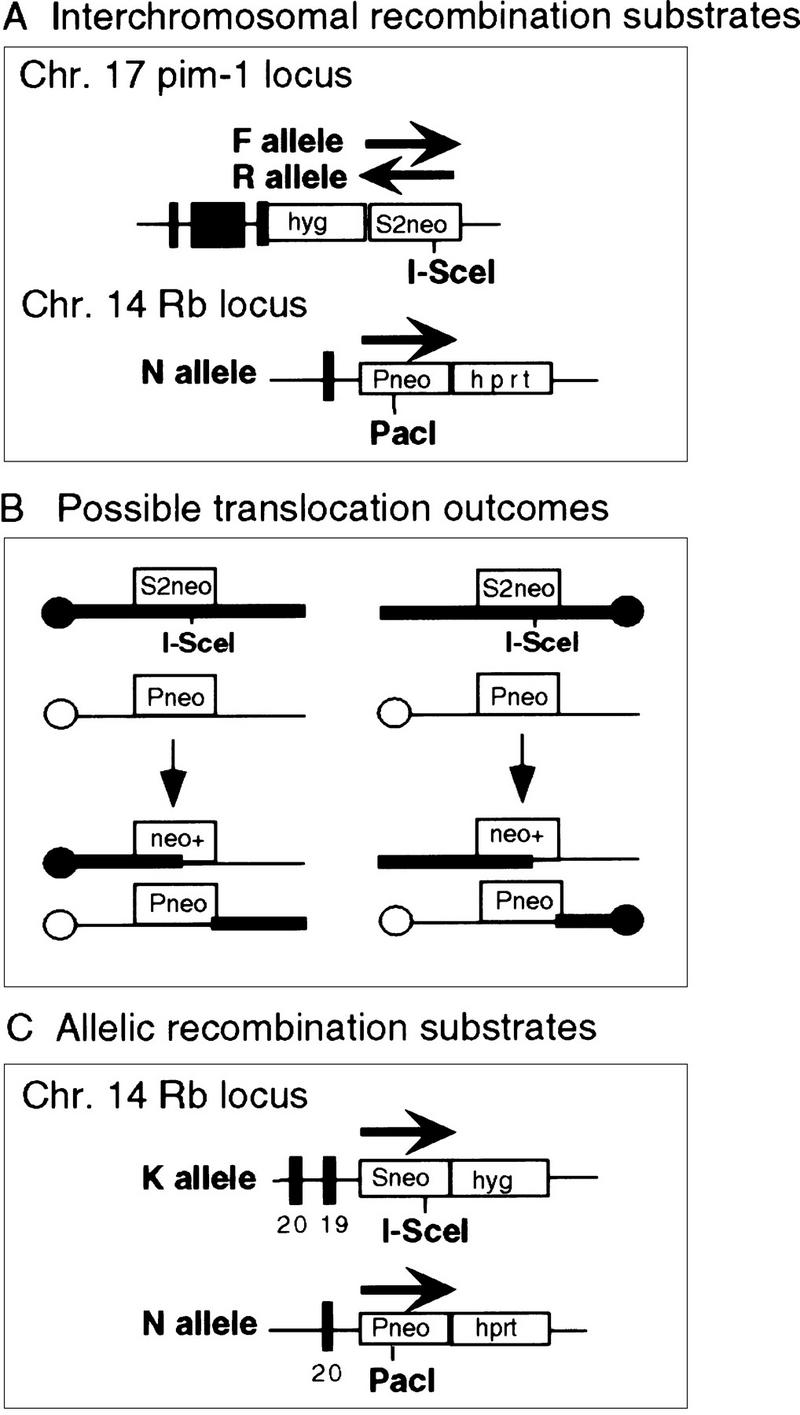
Substrates for DSB-induced interchromosomal and allelic recombination. (A) Two defective neo genes, S2neo, and Pneo, were targeted to mouse chromosomes 17 (F or R allele) and 14 (N allele). The forward orientation (F allele) of the S2neo gene is present in the FN cell lines and the reverse orientation (R allele) is present in the RN cell lines. Conversion with an associated crossover would result in reciprocal translocation products (B). In one orientation of the S2neo gene, monocentric chromosomes would result. In the opposite orientation, an acentric/dicentric pair would result. On chromosome 17, which has a genetic length of ∼60 cM, pim-1 is located ∼16 cM from the centromere (Cuypers et al. 1984). On chromosome 14, which has a genetic length of ∼70 cM, Rb is located ∼41 cM from the centromere (Stone et al.1989). After a reciprocal translocation, the neo genes would be centrally located on both translocation chromosomes. Substrates to examine recombination at allelic positions on homologs were designed similarly to those used to examine recombination between nonhomologous chromosomes (C). In the allelic substrates, Sneo and Pneo were targeted to the two Rb alleles on chromosome 14. The N allele (Pneo) is the same as that used in the interchromosomal recombination experiments (see A). The K allele contains the Sneo gene integrated in the other Rb allele at a similar position, except that the integration did not delete Rb exon 19. Thus, the K allele did not disrupt the Rb gene. Allelic recombination events were scored by conversion of the I-SceI site in the K allele accompanied by loss of heterozygosity at the downstream hyg gene.
The F and R targeting vectors were introduced into the N cell line. Of the 29 hygromycin-resistant colonies obtained with transfection of the F construct, 6 (21%) were correctly targeted to one of the pim-1 alleles (Fig. 2); these cell lines were termed FN. Of the 19 hygromycin-resistant colonies obtained with transfection of the R construct, 11 (58%) were correctly targeted (Fig. 2); these cell lines were termed RN. No additional random integrations were detected in any of the targeted cell lines used in subsequent experiments. As a control, an F cell line was also created by introducing the F targeting construct into E14TG2a cells and screening for targeted integration into the pim-1 locus.
Figure 2.
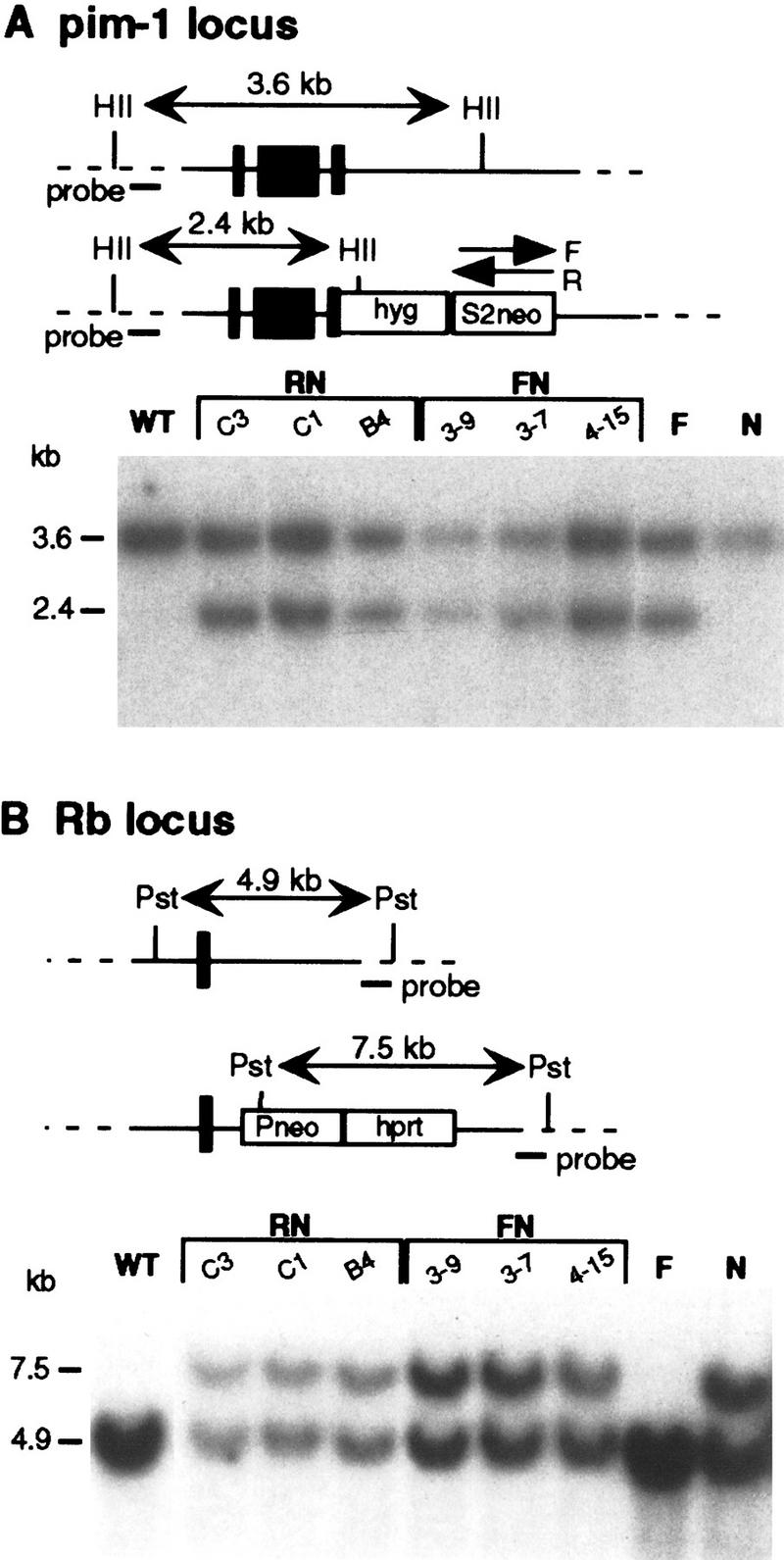
Targeting the recombination reporter genes to chromosomes 17 and 14. (A) The S2neo gene was targeted to the pim-1 locus of chromosome 17 in the N cell line using a coselectable hyg gene (te Riele et al. 1990). A HincII (HII) digest was diagnostic for successful gene targeting. Southern blot analysis of the RN and FN parental cell lines used in the subsequent recombination experiments is shown, as is the analysis of the control F and N cell lines, which are singly targeted. (Black vertical bars) First 3 exons of the pim-1 locus. (B) The presence of the Pneo N allele at the Rb locus on chromosome 14 as described previously (Moynahan and Jasin 1997), was verified using the diagnostic PstI digest. (Black vertical bar) Exon 20 of the Rb locus, with exon 19 of Rb being deleted during the targeting event.
Homologous recombination between the two different mouse chromosomes at the neo loci may restore a functional neo gene. The total length of homology is ∼1.1 kb with the two mutations, the PacI and I-SceI recognition sites, being separated by 526 bp of perfect homology. Downstream of the I-SceI (NcoI) recognition site, there is 297 bp of homology before the termination of either neo gene. A simple gene conversion without crossing-over will maintain the parental configuration of alleles with loss of either the PacI or I-SceI site in one of the alleles (see below). In contrast, gene conversion with crossing-over will result in a chromosomal translocation event (Fig. 1B). If a translocation occurs in G1, reciprocal translocation products will be present in all daughter cells after the next S phase. If a translocation occurs in G2, reciprocal translocation products can segregate together or away from each other at mitosis. We attempted to detect spontaneous recombinants of either type by plating 1 × 109 FN cells in G418. No colonies survived indicating that spontaneous interchromosomal recombination between the two loci is extremely rare, <1 × 10−9.
DSB induction of interchromosomal recombination
Because DSBs are potent inducers of homologous recombination in mammalian cells (e.g., Rouet et al. 1994), DSB-induced recombination between the heterologs was examined even though spontaneous recombination had been undetectable. An I-SceI expression vector, pCBASce, was electroporated into three independently derived FN cell lines and three independently derived RN cell lines. Because the I-SceI endonuclease has an 18-bp recognition site (Colleaux et al. 1988) cleavage of the genome in cells transfected with the I-SceI expression vector is highly restricted to the S2neo gene (Rouet et al. 1994). neo+ colonies were selected in G418. Following induction of a DSB, G418R colonies were readily obtained from all FN and RN cell lines at a similar level (Table 1). The average frequency of recombination was 3.8 × 10−6 in the FN and 3.2 × 10−6 in the RN cell lines (Table 1; data not shown). In contrast, electroporation of the I-SceI expression vector into the singly targeted N and F cell lines did not give rise to any neo+ colonies because of the lack of a homologous donor substrate for recombination.
Table 1.
Induction of interchromosomal recombination by DSBs
| Cell line
|
DNA
|
No. of neo+ colonies
|
Recombination frequency
|
|---|---|---|---|
| N | pCBASce | 0 | <2.6 × 10−7 |
| pneo-WT | 1a | 2.6 × 10−7 | |
| pCBASce+pneo-WT | 0 | <2.6 × 10−7 | |
| F | pCBASce | 0 | <4.7 × 10−7 |
| pneo-WT | 0 | <4.7 × 10−7 | |
| pCBASce+pneo-WT | 2,118 | 1.0 × 10−3 | |
| FN | pCBASce | 129 | 2.0 × 10−6 |
| 3-7 | pneo-WT | 0 | <1.5 × 10−8 |
| pCBASce+pneo-WT | 13,400 | 2.1 × 10−4 | |
| FN | pCBASce | 67 | 3.5 × 10−6 |
| 4-15 | pneo-WT | 0 | <6.7 × 10−8 |
| pCBASce+pneo-WT | 6,932 | 4.1 × 10−4 | |
| FN | pCBASce | 109 | 7.3 × 10−6 |
| 3-9 | pneo-WT | 0 | <6.7 × 10−8 |
| pCBASce+pneo-WT | 4,388 | 2.9 × 10−4 | |
| RN | pCBASce | 101 | 3.4 × 10−6 |
| B4 | pneo-WT | 0 | <3.4 × 10−8 |
| pCBASce+pneo-WT | 14,940 | 3.6 × 10−4 | |
| RN | pCBASce | 82 | 2.6 × 10−6 |
| C1 | pneo-WT | 0 | <3.2 × 10−8 |
| pCBASce+pneo-WT | 6,540 | 2.1 × 10−4 | |
| RN | pCBASce | 40 | 3.7 × 10−6 |
| C3 | pneo-WT | 0 | <9.2 × 10−8 |
| pCBASce+pneo-WT | 2,640 | 2.5 × 10−4 |
Three independently derived FN and RN cell lines were electroporated with the I-SceI endonuclease expression vector pCBASce, a plasmid containing a neo gene fragment pneo-WT, or both plasmids, as indicated. Cell lines containing either the F allele or N allele alone were similarly electroporated as controls. Cells (1 × 108) of the FN and RN cell lines and cells (2 × 107) of the F and N cell lines were electroporated. The recombination frequency was calculated as the number of G418R clones per number of cells surviving electroporation (as counted 20 hr postelectroporation).
This single clone was observed after fixation and therefore could not be confirmed to be a true recombinant.
For comparison, each of the cell lines was also examined for the ability to undergo DSB-promoted gene targeting. We have shown previously that a DSB in a chromosomally integrated S2neo gene stimulates gene targeting 1000-fold, to >10−4 of the transfected population, when the donor substrate is an extrachromosomal plasmid (Elliott et al. 1998). This plasmid, pneo-WT, contains an internal neo gene fragment of 745 bp that can correct the I-SceI recognition site mutation in the chromosome but not the PacI site mutation in Pneo. In the absence of a DSB at the chromosomal locus, gene targeting is observed only rarely with this donor substrate, if at all (Elliott et al. 1998). Similar results were observed here. Electroporation of pneo-WT alone rarely resulted in any G418R colonies; electroporation of both pCBASce and pneo-WT into cell lines containing the S2neo gene (the F, FN, and RN cell lines) gave G418R colonies at a frequency of 1 × 10−3 to 4 × 10−4. This indicates that the lower frequency of DSB-induced heterolog recombination is not caused by chromosomal position of the S2neo gene but, instead, to the type of recombination being promoted. Colonies were not obtained from electroporation of the N cell line with pneo-WT and pCBASce, as a DSB is not induced in the Pneo gene and the homology in pneo-WT cannot correct the PacI site mutation.
These data demonstrate that a single DSB can stimulate recombination between loci on nonhomologous chromosomes at least three orders of magnitude, from <1 × 10−9 to >1 × 10−6. Gene targeting is also stimulated at least three orders of magnitude by a DSB. However, the absolute frequency of recombination with gene targeting is 100-fold higher than recombination between chromosomes.
Short tract gene conversion events
We used diagnostic enzymatic digests and Southern blotting of individual G418R clones to characterize the recombination products. Almost all of the clones had undergone short tract gene conversion (STGC) events using the small region of homology from the N allele to repair the broken F or R allele (Table 2). In fact, this type of event was observed in 97% and 99% of the FN and RN clones, respectively.
Table 2.
Summary of DSB-induced interchromosomal recombination products
|
|
No. of clones
|
|
|---|---|---|
| FN-derived
|
RN-derived
|
|
| STGC | 94 | 86 |
| LTGC | 3 | 1 |
| Reciprocal or nonreciprocal translocation | 0 | 0 |
The major interchromosomal recombination product was a STGC product, observed in 97% of the FN clones and 99% of the RN clones. In the remaining clones, LTGC events were seen in which at least a few kilobases downstream of the break site on chromosome 17 was converted to sequences from chromosome 14. There was no evidence for translocations in any of the clones.
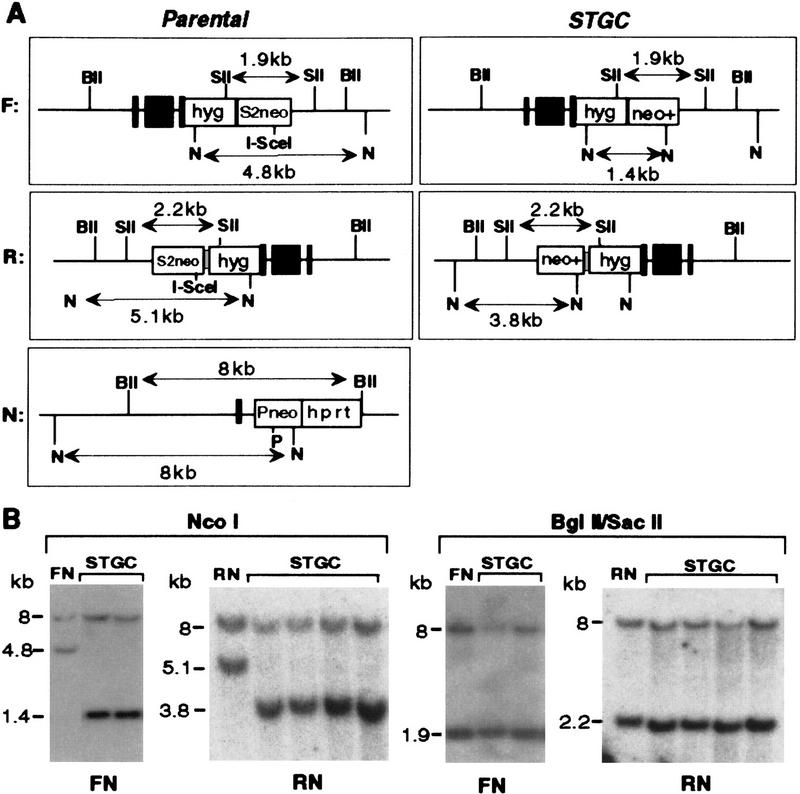
The deduced band fragments and representative Southern blots of this event are shown in Figure 3. For the FN and RN parental cell lines, NcoI-digested genomic DNA gives 4.8- and 5.1-kb bands for the F and R alleles, respectively, when hybridized to a 5′ neo gene probe, and the N allele gives an 8-kb band. During a gene conversion event, the cleaved I-SceI site in the F or R allele is repaired from the wild-type N allele sequence to become an NcoI site. Gene conversions were confirmed by the loss of the parental F or R allele and the appearance of a 1.4-kb band in the FN clones or a 3.8-kb band in the RN clones (Fig. 3). As also expected for a gene conversion event, the N allele on the unbroken chromosome remained unchanged in each of the G418R clones.
Figure 3.
STGC products. The large majority of interchromosomal recombination products were from STGC events. (A) Restriction maps of parental and STGC recombinant alleles. NcoI digestion is diagnostic for homologous repair of the I-SceI-generated DSB. The 4.8- and 5.1-kb NcoI fragments of the parental FN and RN cell lines, respectively, become 1.4 kb in the FN neo+ clones and 3.8 kb in the RN neo+ clones. The 8-kb fragment of the N allele remains unchanged. A BglII–SacII digest is used to characterize the type of recombination event, as the sites flank the neo homology. For the STGC events, the BglII–SacII fragments of the neo+ allele are 1.9 and 2.2 kb, the same size as observed in the parental FN and RN cell lines, respectively. As with the NcoI digest, the 8-kb BglII–SacII band of the N allele remains unchanged. (BII) BglII; (N) NcoI; (P) PacI; and (SII) SacII. (B) Southern blot analysis of representative STGC clones. DNA was probed with a 5′ neo gene fragment.
That loss of the I-SceI site and gain of the NcoI site was the only apparent change within the pim-1 locus of the STGC clones was demonstrated using the BglII and SacII restriction endonucleases whose sites are in chromosomal sequences that flank the neo homology (Fig. 3). For the FN and RN parental lines the S2neo gene of the F and R alleles is contained within 1.9- and 2.2-kb SacII fragments, respectively, and the Pneo gene is within an 8-kb BglII fragment. These fragments remained unchanged in all of the clones designated as arising from STGC events. When these blots were reprobed with an hprt fragment, hybridization was specific to the 8-kb N allele (data not shown), with no indication that hprt sequences had been incorporated into the F or R alleles during DSB repair, as predicted for a STGC.
Long tract gene conversion events
Although STGC was the major observed recombination event, a small portion of clones contained repair bands in Southern blots that were consistent with gene conversion extending beyond the 3′ end of the Pneo gene into a region of heterology between the two chromosomes (Fig. 4). We designated these as long tract gene conversion (LTGC) events. Although conversion can extend any distance 3′ of the I-SceI site, the presence of the 5′ PacI mutation restricted our ability to score LTGC events that extended >526 bp 5′ of the I-SceI site. Three of 97 G418R clones from the FN parental lines and 1 of 87 G418R clones from the RN parental lines had gene structures consistent with this type of event (Table 2).
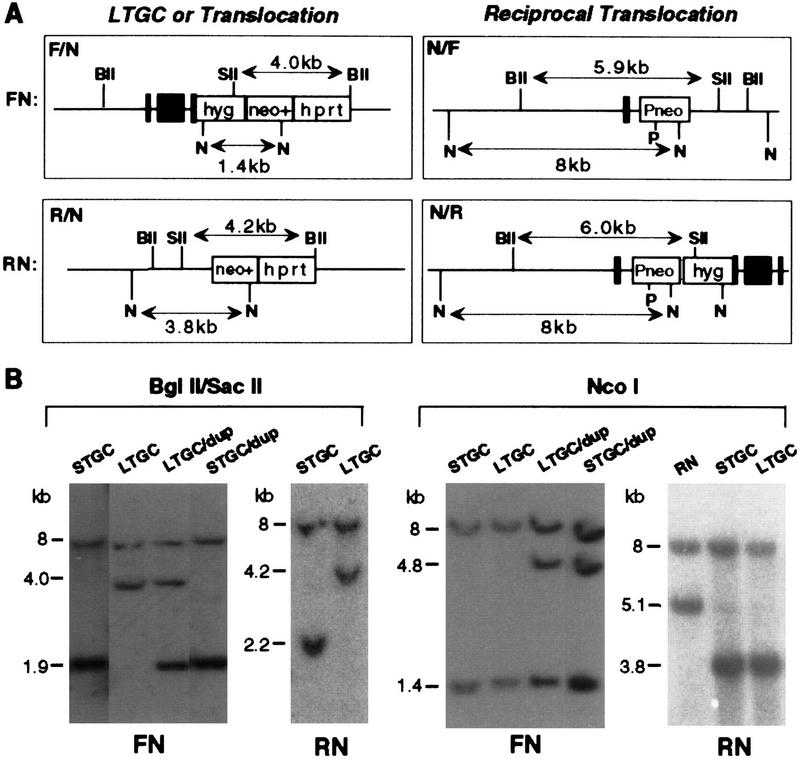
Figure 4.
Rare interchromosomal recombination products. A small number of the interchromosomal recombination products arose from LTGC events. (A) Restriction maps of the LTGC and, for comparison, translocation alleles. The LTGC clones have conversion tracts that extend at least as far as the BglII site at the beginning of the hprt gene, forming hybrid F/N and R/N alleles. The BglII–SacII fragments of these alleles are 4.0 and 4.2 kb, respectively. Reciprocal translocations will give F/N and R/N alleles that are identical with the LTGC alleles in this region of the chromosome, along with hybrid N/F and N/R alleles. (B) Southern blot analysis of LTGC recombinants and recombinants with duplication (dup) alleles. The duplication alleles are the same size as the parental F allele. DNA was probed with a 5′ neo gene fragment.
Gene structures of the LTGC clones are shown in Figure 4. Whereas both BglII–SacII fragments of the STGC clones were of parental size, the LTGC clones had one parental- and one novel-sized fragment. The parental-sized fragment was from the N allele, which remained unchanged at 8 kb, as in the STGC clones (Fig. 4). However, the F and R alleles were altered in this digest, exhibiting bands of 4.0 and 4.2 kb, respectively. These sizes are consistent with the 5′ portion of the fragment being derived from the F or R allele and the 3′ portion of the fragment being derived from the N allele to create hybrid F/N and R/N alleles, respectively. For two of the three FN LTGC clones and the one RN LTGC clone, only two bands are seen, corresponding to the parental N allele and the hybrid allele. In the remaining FN LTGC clone, termed LTGC/dup, there is one additional band that is the size of the F allele, indicating that a duplication of the F allele had occurred (see below). That the LTGC events were bona fide recombination events was verified by the NcoI digest in which the 1.4-kb (FN) and 3.8-kb (RN) recombination products are seen (Fig. 4). As with the BglII–SacII digest, the N allele in the LTGC clones remained unchanged from the parental cell line. PacI digestion confirmed that the PacI site in the N allele was retained in these clones (data not shown).
The reciprocal hybrid alleles, N/F and N/R, were not seen in any of the four LTGC clones (Fig. 4). The LTGC alleles could represent alleles in which conversion of the F (or R) allele extended 3′ of the neo gene into the N allele but then rejoined the F (or R) allele, presumably with the junction occurring at a position of heterology. Alternatively, it was possible that the hybrid F/N or R/N alleles in the LTGC clones were derived from nonreciprocal translocations or reciprocal translocations during G2 with the reciprocal neo− alleles segregating from the neo+ alleles at mitosis.
To distinguish between LTGCs and translocations, we used fluorescence in situ hybridization (FISH) of metaphase chromosome spreads. Chromosomes 14 and 17 were identified with chromosome-specific paints labeled with FITC and rhodamine, respectively. Using this technique, no chromosomal aberrations were apparent (Fig. 5; Table 2). Two intact chromosome 14s and two intact chromosome 17s were identified in each of the three LTGC clones examined. (The fourth LTGC clone was not available for analysis.) There was no evidence for translocations in any of the clones, as neither of the red chromosome 17s was joined to green chromosome 14 sequences. Translocations would be apparent, as both the pim-1 and Rb loci are located in the middle third of their respective chromosomes (see legend to Fig. 1). Alternatively, if the gene conversion tracts were extensive, we would see a green or yellow patch (green + red) within one of the red chromosome 17s. However, we do not observe this, which suggests that the conversion tract did not extend into a large portion of the chromosome. Furthermore, in the LTGC/dup clone no additional copy of chromosome 17 was observed (Fig. 5C). Taken together, these results indicate that none of the LTGC events resulted in gross chromosomal aberrations.
Figure 5.
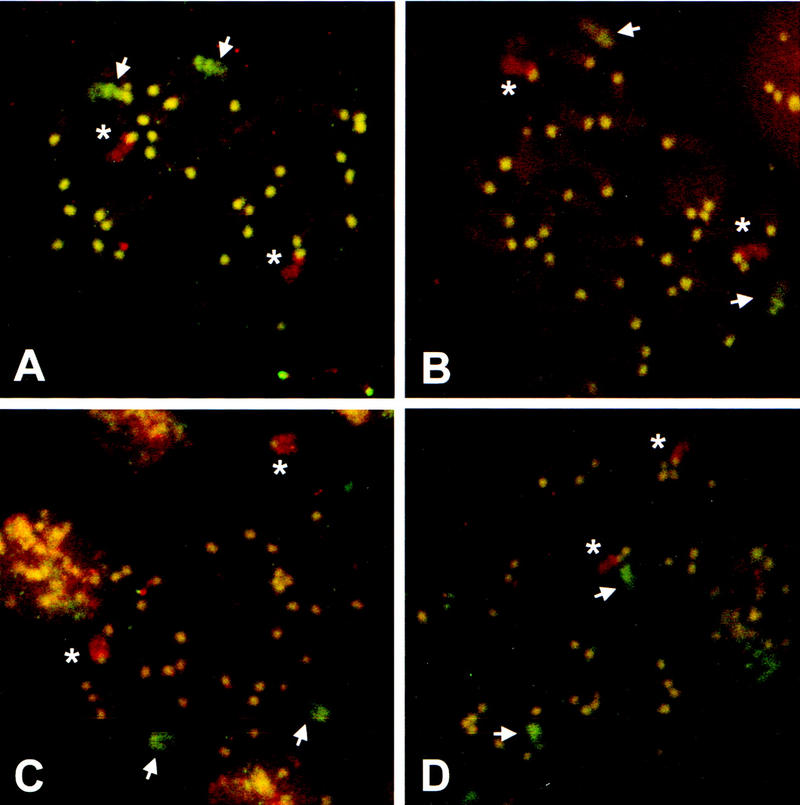
FISH analysis. Chromosome spreads were hybridized to mouse chromosome 17 and 14 probes that were rhodamine and FITC labeled, respectively. A representative spread is shown for each of the control E14TG2a ES cells (A) and cell lines with LTGC events (B–D). The LTGC panels are an FN LTGC cell line (B), the FN LTGC/dup cell line (C), and the RN LTGC cell line (D). Asterisks mark the chromosome 17 pair; arrows mark the chromosome 14 pair. The yellow dots are the centromeres that cross-hybridize to the probes. In each of the spreads, a normal chromosome count of 40 was verified.
Duplications of the targeted allele at the pim-1 locus
A fraction of clones were found to have additional copies of the F or R allele. This was evident upon NcoI digestion, as three bands were apparent—the N allele, the recombinant NcoI+ F (or R) allele, and an NcoI− F (or R) allele (Fig. 4B; data not shown). These clones fell into two classes. One class consisted of 7 of 32 G418 R clones (6 STGC and 1 LTGC) that were derived from the parental FN cell line 4-15. Southern blot analysis of the pim-1 locus in six of these clones (see Fig. 2) demonstrated that both pim-1 loci in these clones contained the F targeting construct (data not shown). Consistent with this, only two chromosome 17s were observed with FISH analysis, at least for the LTGC clone that was checked (Fig. 5C). It is possible that sometime during growth of the parental 4-15 clone in hygromycin-selective medium, enrichment for a cell with two F allele chromosomes occurred. The fate of the I-SceI site was examined in the second F allele in these seven clones. In four clones, the I-SceI site remained intact. In the other three clones, the I-SceI site was lost but was not converted to an NcoI site (data not shown), apparently as a result of the ends being rejoined by a nonhomologous repair pathway.
The other class consisted of one clone from an RN cell line. Unlike the subpopulation of FN 4-15 clones with neo at both pim-1 alleles, this clone remained heterozygous at the pim-1 locus having one wild-type pim-1 allele and one targeted allele. The extent of the duplication of chromosome 17 sequences is unknown, as is whether it was pre-existing in the population or arose by an unusual mechanism during the STGC. The I-SceI site in the additional R sequences remained intact in this clone (data not shown).
Interchromosomal vs. allelic recombination
We have demonstrated previously that a broken chromosome can use its allelic partner as a homologous repair template (Moynahan and Jasin 1997). Because homologs share extensive homology, we wanted to determine whether DSBs are repaired at a substantially lower frequency from the limited amount of homology available on a nonhomologous chromosome. To examine allelic recombination, both alleles of the Rb locus on chromosome 14 were marked with the neo gene substrates, creating the KN cell line. In this cell line, the N allele contains the Pneo gene and the K allele contains the Sneo gene (Fig. 1C). The Sneo gene is similar to S2neo but differs in that nonhomologous, as well as homologous, repair can restore neo gene function. Previous studies have demonstrated that approximately one-third of G418R clones with such a system arise from allelic recombination accompanied by loss of heterozygosity downstream of the neo gene (see Moynahan and Jasin 1997). The other events are either nonhomologous repair events or allelic recombination involving STGC, which are not distinguishable in this system. We directly compared homologous DSB repair in the FN and KN cell lines. Similar to what was seen above, we found that DSB-induced interchromosomal recombination in the FN parental line occurred at a frequency of 2 × 10−6 to 3.5 × 10−6 (data not shown). Allelic recombination in the KN line was deduced to occur at a frequency of 9 × 10−6. (Although the frequency discounts nonhomologous events from the analysis, some allelic events may also be excluded; however, we expect the total frequency of allelic events to be no more than 1.8 × 10−5; Moynahan and Jasin 1997). Thus, DSB-induced allelic recombination and DSB-induced recombination between nonhomologous chromosomes are both observed but with interchromosomal recombination occurring at a somewhat lower frequency.
Discussion
In this paper we report that a single DSB can induce recombination between loci on nonhomologous chromosomes. Recombination is induced at least three orders of magnitude, from <1 × 10−9 to >1 × 10−6. This large induction is similar to that seen with allelic recombination, gene targeting, and direct repeat recombination (Table 1; Moynahan and Jasin 1997; Donoho et al. 1998; Liang et al. 1998). However, for both gene targeting and direct repeat recombination, the absolute frequency of induced recombination is much higher, reaching levels of 1 × 10−3 or more. Thus, recombination between heterologs is substantially induced by a DSB, but the absolute level is ∼100-fold lower than other types of recombination where the repair template is presumably at a higher local concentration to the break site. Most (98%) of the interchromosomal recombination events are gene conversions in which a relatively short region around the chromosome break site is converted (Table 2). The remaining events involve longer tracts of gene conversion encompassing 2.5 kb or more, which extend into a region that is not homologous between the two chromosomes.
Interestingly, none of the events resulted in chromosomal translocations, whether reciprocal or not. We would have predicted that if a number of single crossovers had occurred, then one orientation, either FN or RN, would produce a dicentric and an acentric product (see Fig. 1). This potentially lethal event would have been evident by an overall reduced number of recombinants from one parental configuration and a significantly greater proportion of translocation events from the second parental configuration. In contrast, we observed very similar recombination frequencies and very similar types of recombinants from both FN and RN parental cells with no evidence for translocations from either. Although dicentric chromosomes should be inviable, isodicentric chromosomes can be stabilized by inactivation of one centromere in some cases, and have been postulated to occur in some intrachromosomal recombination events (Godwin and Liskay 1994). However, FISH analysis of the LTGC events observed here did not indicate any abnormal chromosome structures or extra centromeres (see Fig. 5).
Gene conversions associated with crossovers are frequent in meiotic recombination, but the association of these two repair events is less clear in somatic cells. In mitotic yeast cells, analysis of plasmid integration had originally demonstrated that crossovers were as frequent as noncrossover events in the conversion of double-stranded gaps (Orr-Weaver and Szostak 1983). However, the majority of spontaneous intrachromosomal recombination events seen in both yeast and mammalian systems are gene conversions (Jackson and Fink 1981; Bollag and Liskay 1991). When crossovers are observed, they are predominantly associated with LTGCs (Aguilera and Klein 1989; Godwin and Liskay 1994). Thus, the lack of crossovers observed here may be a reflection of the predominance of STGC events in which recombination did not extend to distances long enough to permit crossover events. As with spontaneous intrachromosomal recombination, spontaneous allelic and interchromosomal events in yeast are also predominantly noncrossover gene conversions, yet in all three types of recombination, crossovers still account for 5%–20% of events (Harris et al. 1993 and references therein). The absence of crossovers in our assay system suggests that crossovers are more strongly suppressed (≤1%) in mammalian cells than in yeast. This strong bias against crossing-over is also seen in Drosophila during P-element-induced recombination (Gloor and Lankenau 1998), suggesting that organisms containing large numbers of repetitive elements may have developed a stronger crossover suppression to maintain integrity of genomes.
Homologous recombination and genome integrity
Homologous recombination recently has been demonstrated to be one of two major DSB repair pathways in mammalian cells—the other being nonhomologous repair—and in one system, homology-directed repair was found to account for up to 50% of the repair events (Liang et al. 1998). In the experiments presented here, we estimate that 0.1%–1% of the electroporated cells express I-SceI and undergo cleavage. With interchromosomal recombinational repair measured at 2 × 10−6 to 4 × 10−6, a large number of the repair events were therefore undetectable by neo gene selection. We expect that these repair events included religation of the I-SceI overhangs, nonhomologous repair in which the two chromosome ends were rejoined imprecisely, and homologous repair from the sister chromatid, providing that the I-SceI site on the sister chromatid was not cleaved simultaneously.
Homologous recombination as a major DSB repair pathway had presented a dilemma in terms of how complex mammalian genomes with large fractions of repetitive DNA are maintained without frequent genome rearrangements. This study and others implicate several levels of control in protecting mammalian genomes from rearrangement events. Repetitive elements have diverged from each other, and it is known that sequence divergence can suppress recombination (for mammalian cells studies, see, e.g, Waldman and Liskay 1987). In a related system to that presented here, it has been shown that 1.2% heterology can reduce DSB-promoted gene conversion in ES cells up to eightfold (Elliott et al. 1998). As demonstrated here, at least two other levels of control operate—a reduced frequency with which repeats on heterologous chromosomes (and presumably repeats at more distant locations on the same chromosome) recombine, and a suppression of crossover events when they do. Thus, in the case of two repetitive elements located on different chromosomes that are 1%–2% divergent, we would predict that a translocation event would be at least five orders of magnitude lower than recombination between identical sequences on the sister chromatid (10-fold reduction due to heterology and a 100-fold reduction for each ectopic location and crossover suppression). More significant divergence would likely add greater magnitude to this equation, as there appears to be a requirement for a minimal length of perfect homology for recombination (Waldman and Liskay 1988). These multiple levels of control can prevent massive genome rearrangements when a single genetic mutation occurs. For example, even though recombination between diverged sequences is substantially elevated in mismatch repair-deficient cell lines (Rayssiguier et al. 1989; de Wind et al. 1995) and between repeats on nonhomologous chromosomes (Bailis et al. 1990), massive rearrangements have not been reported in these mutants, even for cells with highly repetitive genomes (de Wind et al. 1995).
Replication-based recombination model
The profound bias toward noncrossover gene conversion may be due to a general preference for recombinational repair without crossing-over or to a specific restriction against crossing-over when nonhomologous chromosomes recombine. The DSB repair model put forth in yeast to explain meiotic recombination and plasmid integration predicts that crossing-over would frequently accompany gene conversion (Szostak et al. 1983), and recent evidence supports the basic outline of this model (Stahl 1996). However, alternative DSB repair models proposed for mitotic cells in which recombination is coupled to replication do not invoke an association of gene conversion with crossing-over (Gloor and Lankenau 1998; Paques et al. 1998). Occasional crossovers can be accommodated in these models, but they are a minority of events. In mammalian cells, crossovers have not as yet been detected in recombination events between homologs (Moynahan and Jasin 1997), and they appear to be only a fraction of repair events between introduced plasmids and genomic DNA (Jasin et al. 1985; Jasin and Berg 1988; C. Richardson, J. Winderbaum and M. Jasin, unpubl.). It is currently unknown if recombination between sister chromatids, predicted to be the most prevalent type of recombinational repair in mammalian cells (Liang et al. 1998), shows a similar bias toward noncrossover events. Nevertheless, a replication-based recombination model would explain the bias toward noncrossover events seen here for interchromosomal recombination.
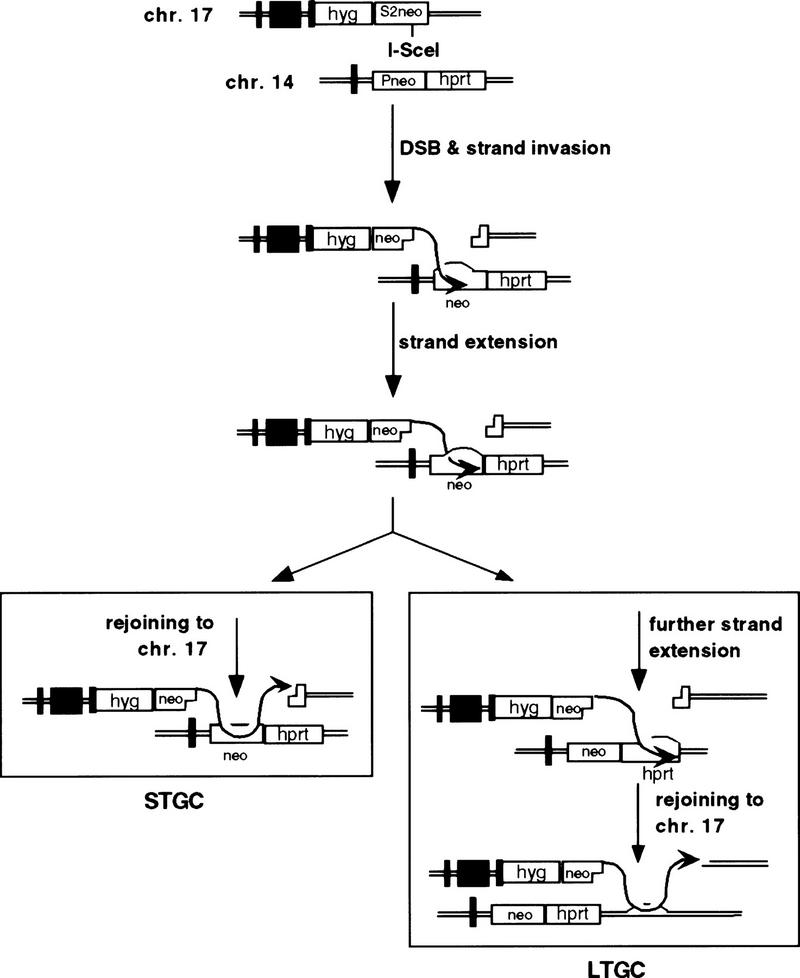
A replication-based recombination model, in addition to accounting for STGC, may also account for the small proportion of LTGC events. In this model, one of the broken ends from chromosome 17 would invade the homologous sequence on chromosome 14, forming a D-loop (Fig. 6). The invading end would prime DNA synthesis, which would extend a short distance past the correcting sequences at the NcoI site and then reanneal to the homologous sequences on chromosome 17. In the rare LTGC events, more extensive amounts of DNA synthesis would occur, resulting in migration of the D loop into nonhomologous sequences downstream of the neo gene. Eventually, the newly synthesized strand would rejoin chromosome 17, either in a region of fortuitous homology between the two chromosomes or in a region of little or no homology. In the latter case, the robust nonhomologous repair machinery for chromosomal break repair in mammalian cells would be utilized to rejoin the two chromosome 17 ends. A coupling of homologous recombination with nonhomologous repair has been observed during other forms of recombination (Bailis et al. 1992; Godwin and Liskay 1994), including DSB-promoted recombination (Rouet et al. 1994; Moynahan and Jasin 1997). The rarity of LTGC events observed in this study is consistent with the finding that gene conversion tracts are generally short (Taghian and Nickoloff 1997; Elliott et al. 1998). Longer tracts may be involved when homology is extensive, for example, when homologs recombine (Moynahan and Jasin 1997). In both the STGC and LTGC recombination events the uncleaved N allele remained unchanged, a feature that is also consistent with replication-based recombination models.
Figure 6.
Model for interchromosomal recombination. For both the STGC and LTGC events, a model in which one of the 3′ ends from chromosome 17 invades the homologous neo sequence on chromosome 14 is consistent with the lack of crossover events. For the STGC events the invasion can be from the 3′ strand on either the 5′ (as shown) or 3′ side of the break site. For the LTGC events the invasion is limited to the strand on the 5′ side of the break site, as shown, as events that result in neo+ genes are selected. Following invasion, the strand is extended by new DNA synthesis. In the case of the STGC events, the newly extended strand can rejoin chromosome 17 by reannealing to homologous neo sequences. In the case of the LTGC events, the amount of extension is longer and can rejoin chromosome 17 at a region of fortuitous homology or at a region of nonhomology.
Multiple lesions, aberrant repair, and the genetic control of recombination
In this study we examined the ability of a single precise DSB to induce recombination rather than a number of undefined lesions as produced by γ-irradiation or other global DNA-damaging agents. Investigations employing global DNA-damaging agents have demonstrated that genome rearrangements such as translocations can be induced, but by mechanisms other than conservative homologous recombination. Translocations apparently occur from interactions between multiple broken ends on nonhomologous chromosomes, each able to rejoin with another partner (Edwards et al. 1994; Phillips and Morgan 1994; Haber and Leung 1996; Wang et al. 1997). Details of the joined sequences in many cases look very much like the rejoining of two ends from a single chromosome break (Godwin et al. 1994; Phillips and Morgan 1994; Rouet et al. 1994; Liang et al. 1998). Whether nonhomologous repair is inherently more mutagenic than homologous repair is unclear, but it is interesting to note that bleomycin-induced translocations are only readily induced in growth-arrested cells (Wang et al. 1997) when precise homologous repair from a sister chromatid would not be possible. Similarly, it has also been shown in yeast that an increased frequency of translocations occurs when the homologous recombination machinery is disrupted (Chen et al. 1998).
Although normal cells apparently have mechanisms to suppress the occurrence of chromosomal rearrangements such as translocations, altered expression of genes involved in DNA repair (i.e., caretaker genes) may permit these events to occur. Similarly, altered expression of genes involved in DNA-damage sensing (i.e., gatekeeper genes) may permit cells that have undergone these events to survive by escaping apoptosis (see Levine 1997 and references therein). For example, the survival of damaged cells is thought to be responsible for the high tumor incidence in p53-deficient mice. The high frequency of mutations in oncogenes and cell cycle regulators seen in leukemic cells of patients also provides support to this observation (Lotem and Sachs 1993), although it often is difficult to assess which genetic change is responsible for the initial chromosomal alteration. Variations in the cellular environment may also be important for the survival and proliferation of cells containing large chromosomal alterations.
Although it is known that recombination facilitates segregation of homologous chromosomes during meiosis (Stahl 1996), its requirement during mitotic cell division is less clear. It is tantalizing to speculate that homologous recombination during mitotic divisions in mammalian cells is critical for the repair of naturally occurring lesions because Rad51 mutation results in early embryonic lethality. As in yeast and prokaryotes, homologous recombination is also likely to have an important role in the repair of lesions induced by DNA-damaging agents, as human X-ray repair cross complementing (XRCC) genes, which share homology to Rad51 (Liu et al. 1998), as well as the mouse Rad54 gene (Essers et al. 1997), are involved in the repair of damage generated by ionizing radiation and cross-linking agents. Other putative Rad51 homologs may also be involved in recombinational repair (Albala et al. 1997; Dosanjh et al. 1998; Pittman et al. 1998); however, direct evidence of the involvement of any Rad51 homolog has yet to be demonstrated. One gene that has a demonstrated effect on homology-directed translocation events in yeast is the rad9 cell cycle checkpoint gene (Fasullo et al. 1998). Mutation of rad9 results in a higher incidence of both spontaneous and damage-induced reciprocal translocations. With the defined genetic system presented here, it should now be possible to determine the components involved in interchromosomal recombination in mammalian cells and also unravel the safeguards that prevent the occurrence of translocation events.
Materials and methods
DNA and cell line constructions
The pim-1 allele was gene targeted by modifying a previously described gene-targeting vector (p59, te Riele et al. 1990). A XhoI–SalI fragment containing the S2neo gene (Smih et al. 1995) was inserted into the unique SalI site of p59 downstream of the hyg coding sequence and upstream of pim-1 exon 4. Plasmids with S2neo in the same orientation as hyg were designated as the F targeting vector. Those with the S2neo gene in the reverse orientation were further manipulated. A SacI–SacI fragment containing the polyadenylation signal of the bovine growth hormone was subcloned from the β-geo plasmid (Friedrich and Soriano 1991) into pUC19 so that a SalI–XhoI fragment could be isolated and inserted into the SalI site of the R vector. Plasmids were screened for those with the polyadenylation signal in the same orientation as hyg to complete construction of the R targeting vector. The targeting constructs were cleaved away from the plasmid backbone prior to electroporation.
ES cell lines E14TG2a (Hooper et al. 1987) and the N cell line (Moynahan and Jasin 1997) were grown in standard media supplemented with leukemia inhibitory factor (LIF) at 105 U/ml (GIBCO/Life Technologies). For gene targeting, 0.8 ml of cells at 2 × 107 cells/ml were electroporated at 250 V and 960 μF with 100 μg of the linearized pim-1 targeting fragment. Selection medium containing hygromycin (110 μg/ml) was added 48 hr after transfection. Targeted clones (FN and RN parental lines and the control F cell line) were identified by screening Southern blots of genomic DNA cleaved with HincII, using a genomic HincII–BstXI pim-1 fragment as a probe (te Riele et al. 1990). Verification of the N allele targeting was done by screening Southern blots of genomic DNA cleaved with PstI, using a genomic PstI–PvuII Rb fragment as a probe (te Riele et al. 1992).
The I-SceI expression vector was constructed by inserting a blunt-ended EcoRI–SalI fragment that contained the I-SceI coding sequence from pCMV3xnlsI–SceI (Donoho et al. 1998; C. Richardson and M. Jasin, unpubl.) into a blunted XhoI site of pCAGGS (Niwa et al. 1991). In this vector, termed pCBASce, the CMV enhancer and the chicken β-actin promoter drives I-SceI endonuclease expression on a spliced mRNA (P.J. Romanienko and M. Jasin, unpubl.). For control transfections, the previously described pneo-WT plasmid was used (Elliott et al. 1998).
DSB induction and Southern blot analysis
Electroporations were performed as described above with 0.8 ml of cells at 2 × 107 or 1 × 108 cells/ml. To examine DSB-induced recombination, 50 μg of the circular pCBASce expression vector was electroporated. For control gene targeting experiments, 40 μg of circular pneo-WT plasmid was used alone or coelectroporated with 50 μg of the circular pCBASce expression vector. One day after electroporation, a cell suspension from one plate was counted to determine the number of cells surviving electroporation. G418 was added to the remaining plates at 200 μg/ml. After 12 days of selection, G418R colonies were counted and individually expanded. Southern blots of genomic DNA from neo+ clones were probed with the 5′ neo gene XhoI–NarI fragment (Rouet et al. 1994).
FISH
Metaphase spreads from cell lines were prepared by standard procedures. A biotin-labeled chromosome 14 paint and a digoxigenin-labeled chromosome 17 paint (Oncor) were denatured at 72°C for 10 min and preannealed at 37°C for 2 hr prior to hybridization to the chromosome spreads for 16 hr, as per manufacturer’s instructions. A biotin–digoxigenin dual detection kit (Oncor) was used for fluorescent detection of chromosome paint signals and DAPI counterstaining of chromosomes.
Acknowledgments
We thank Hein te Riele (Amsterdam) for materials and the reviewers for their helpful comments on the manuscript. C.R. is a Leukemia Society of America Fellow (recipient of the Vrushalli Ranadive Fellowship). This work was supported by a grant from the National Science Foundation (MCB-9728333) to M.J.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL m-jasin@ski.mskcc.org; FAX (212) 717-3317.
References
- Albala JS, Thelen MP, Prange C, Fan W, Christensen M, Thompson LH, Lennon GG. Identification of a novel human RAD51 homolog, RAD51B. Genomics. 1997;46:476–479. doi: 10.1006/geno.1997.5062. [DOI] [PubMed] [Google Scholar]
- Aguilera A, Klein HL. Yeast intrachromosomal recombination: Long gene conversion tracts are preferentially associated with reciprocal exchange and require RAD1 and RAD3 gene products. Genetics. 1989;123:683–694. doi: 10.1093/genetics/123.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis AM, Arthur L, Rothstein R. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homologous genes by an excision repair dependent process. Genetics. 1990;126:535–547. doi: 10.1093/genetics/126.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis AM, Arthur L, Rothstein R. Genome rearrangement in top3 mutants of Saccharomyces cerevisiae requires a functional RAD1 excision repair gene. Mol Cell Biol. 1992;12:4988–4993. doi: 10.1128/mcb.12.11.4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MD, Read LR. Ectopic recombination within homologous immunologlobin μ gene constant regions in a mouse hybridoma cell line. Mol Cell Biol. 1992;12:4422–4432. doi: 10.1128/mcb.12.10.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag RJ, Liskay RM. Direct-repeat analysis of chromatid interactions during intrachromosomal recombination in mouse L cells. Mol Cell Biol. 1991;11:4839–4845. doi: 10.1128/mcb.11.10.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, Suh H, Qin XF, Besmer E, Kenter A, Rajewsky K, Nussenzweig MC. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Umezu K, Kolodner RD. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- Choulika A, Perrin A, Dujon B, Nicolas J-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1963–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleaux L, d’Auriol L, Gailbert F, Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M, Antonarakis SE. The nature and mechanisms of human gene mutation. In: Volgelstein B, Kinsler KW, editors. The genetic basis of human cancer. New York, NY: McGraw-Hill; 1998. pp. 65–94. [Google Scholar]
- Cuypers HT, Selton G, Quint W, Zijlstra M, Maandag ER, Boelens W, ven Wezenbeck P, Melief C, Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: Integration of proviruses in a distinct chromosomal region. Cell. 1984;37:141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosanjh MK, Collins DW, Fan W, Lennon GG, Albal JS, Shen Z, Schild D. Isolation and characterization of RAD51C, a new human member of the RAD51 family of related genes. Nucleic Acid Res. 1998;26:1179–1184. doi: 10.1093/nar/26.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AA, Moiseenko VV, Nikjoos H. Modelling of DNA breaks and the formation of chromosome aberrations. Int J Radiat Res. 1994;66:633–637. doi: 10.1080/09553009414551741. [DOI] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from doublestrand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Hendriks RW, Swagemakers SMA, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JHJ, Kanaar R. Diruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- Fasullo M, Bennett T, Ah Ching P, Koudelik J. The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations. Mol Cell Biol. 1998;18:1190–1200. doi: 10.1128/mcb.18.3.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: A genetic screen to identify and mutate developmental genes in mice. Genes & Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Lankenau DH. Gene conversion in mitotically dividing cells: A view from Drosophila. Trends Genet. 1998;14:43–46. doi: 10.1016/s0168-9525(97)01365-6. [DOI] [PubMed] [Google Scholar]
- Godwin AR, Liskay RM. The effects of insertions on mammalian intrachromsomal recombination. Genetics. 1994;136:607–617. doi: 10.1093/genetics/136.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin AR, Bollag RJ, Christie D-M, Liskay RM. Spontaneous and restriction enzyme-induced chromosomal recombination in mammalian cells. Proc Natl Acad Sci. 1994;91:12554–12558. doi: 10.1073/pnas.91.26.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Leung WY. Lack of chromosome territoriality in yeast: Promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S, Rudnicki KS, Haber JE. Gene conversions and crossing over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics. 1993;135:5–16. doi: 10.1093/genetics/135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- Jackson JA, Fink GR. Gene conversion between duplicated genetic elements in yeast. Nature. 1981;292:306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Jeggo PA. DNA double-strand break repair and V(D)J recombination: Involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- Jasin M, Berg P. Homologous integration in mammalian cells without target gene selction. Genes & Dev. 1988;2:1353–1363. doi: 10.1101/gad.2.11.1353. [DOI] [PubMed] [Google Scholar]
- Jasin M, deVilliers J, Weber F, Schaffner W. High frequency of homologous recombination in mammalian cells between endogenous and introduced SV40 genomes. Cell. 1985;43:695–703. doi: 10.1016/0092-8674(85)90242-9. [DOI] [PubMed] [Google Scholar]
- Jeffs AR, Benjes SM, Smith TL, Sowerby SJ, Morris C. The BCR gene recombines preferentially with Alu elements in complex BCR-ABL translocations of chronic myeloid leukemia. Hum Mol Gen. 1998;7:767–776. doi: 10.1093/hmg/7.5.767. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D-S, Hasty P. A mutation in mouse rad51 results in early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Narayana LS, Zhou Z-Q, Adamson AW, Sorensen KJ, Chen DJ, Jones NJ, Thompson LH. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- Lotem J, Sachs L. Regulation by bcl-2, c-myc, and p53 of susceptibility to induction of apoptosis by heat shock and cancer chemotherapy compounds in differentiation-competent and -defective myeloid leukemic cells. Cell Growth Differ. 1993;4:41–47. [PubMed] [Google Scholar]
- Morris C, Jeffs A, Smith T, McDonald M, Board P, Kennedy M, Fitzgerald P. BCR gene recombines with genomically distinct sites on band 11q13 in complex BCR-ABL translocations of chronic myeloid leukemia. Oncogene. 1996;12:677–685. [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. Proc Natl Acad Sci. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murti JR, Bumbulis M, Schimenti JC. Gene conversion between unlinked sequences in the germline of mice. Genetics. 1994;137:837–843. doi: 10.1093/genetics/137.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J-I. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW. Yeast recombination: The association between double-strand gap repair and crossing-over. Proc Natl Acad Sci. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Leung W, Haber J. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JW, Morgan MF. Illegitimate recombination induced by DNA double-strand breaks in a mammalian chromosome. Mol Cell Biol. 1994;14:5794–5803. doi: 10.1128/mcb.14.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Weinberg LR, Schimenti JC. Identification, characterization, and genetic mapping of Rad51d, a new mouse and human RAD51/Rec A related gene. Genomics. 1998;49:103–111. doi: 10.1006/geno.1998.5226. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CW. Alu: Structure, origin, evolution, significance, and function of one-tenth of human DNA. Prog Nucleic Acid Res. 1996;53:283–319. doi: 10.1016/s0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acid Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl F. Meiotic recombination in yeast: Coronation of the double-strand-break repair model. Cell. 1996;87:965–968. doi: 10.1016/s0092-8674(00)81791-2. [DOI] [PubMed] [Google Scholar]
- Stone JC, Crosby JL, Kozak CA, Schievella AR, Bernards R, Nadeau JH. The murine retinoblastoma homolog maps to chromosome 14 near Es-10. Genomics. 1989;5:70–75. doi: 10.1016/0888-7543(89)90088-8. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H, Maandag ER, Clarke A, Hooper M, Berns A. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature. 1990;348:649–651. doi: 10.1038/348649a0. [DOI] [PubMed] [Google Scholar]
- te Riele H, Maandag ER, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci. 1996;93:6236–6340. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman AS, Liskay RM. Differential effects of base-pair mismatch on intrachromsomal versus extrachromosomal recombination in mouse cells. Proc Natl Acad Sci. 1987;84:5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol Cell Biol. 1988;8:5350–5357. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhou R, Zou Y, Jackson-Cook C, Povirk L. Highly conservative reciprocal translocations formed by apparent joining of exchanged DNA double-strand break ends. Proc Natl Acad Sci. 1997;94:12018–12023. doi: 10.1073/pnas.94.22.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]