Abstract
Background
Molecular experiments using multiplex strategies such as cDNA microarrays or proteomic approaches generate large datasets requiring biological interpretation. Text based data mining tools have recently been developed to query large biological datasets of this type of data. PubMatrix is a web-based tool that allows simple text based mining of the NCBI literature search service PubMed using any two lists of keywords terms, resulting in a frequency matrix of term co-occurrence.
Results
For example, a simple term selection procedure allows automatic pair-wise comparisons of approximately 1–100 search terms versus approximately 1–10 modifier terms, resulting in up to 1,000 pair wise comparisons. The matrix table of pair-wise comparisons can then be surveyed, queried individually, and archived. Lists of keywords can include any terms currently capable of being searched in PubMed. In the context of cDNA microarray studies, this may be used for the annotation of gene lists from clusters of genes that are expressed coordinately. An associated PubMatrix public archive provides previous searches using common useful lists of keyword terms.
Conclusions
In this way, lists of terms, such as gene names, or functional assignments can be assigned genetic, biological, or clinical relevance in a rapid flexible systematic fashion. http://pubmatrix.grc.nia.nih.gov/
Keywords: Literature mining, microarray, Proteomic, PubMed
Background
With the advent of high throughput genomic and proteomic approaches, the ability to generate data has outstripped the ability to assign biological relevance. Searching the MEDLINE literature database of greater than 14 million entries one-by-one makes establishing biological significance a daunting task. The basic PubMed search window contains a typical single search box for the input of simple keyword combinations. PubMed does allows complex searches using advanced search options, but this requires some knowledge of string search assembly, and an understanding of the PubMed Entrez programming utilities. These are still relatively obscure for many molecular biologists.
Literature mining approaches have been developed to place multiplex biological datasets into context relative to published medical literature. Computational tools such as PubGene [1], VxInsight [2], MedMiner [3], EASE [4], MeshMap[5], XPLORMED [11], AbXtract [12], and HAPI [13] are available which allow the user to query more complex gene name or keyword combinations, including multiplex proteomic or cDNA microarray results, versus literature citations in PubMed with defined types of output. These computational tools use different strategies in literature mining and in some cases produce statistical significance or graphical displays.
Similarly, literature based annotation projects such as the GO project [9] have devised a complex hierarchical annotation system using expert annotators based on a defined controlled vocabulary. This is important due to significant problems with nomenclature in molecular biology. Although quite useful, the GO approach does not allow for flexibility, variable interpretation, or individual investigator input in the hierarchies assigned to individual genes. In this report, we describe a simple, freely accessible, web based application for basic literature mining of PubMed that performs automatic multiplex Boolean queries. This allows the naive user to assign biological relevance and annotate gene lists through PubMed.
Implementation
PubMatrix is a CGI front-end application, which submits queries consisting of search and modifier terms against NCBI's PubMed database and presents the results as a matrix of document hits. Results are stored in a database for retrieval and are presented as hyperlinks to the user for rerunning individual queries of interest. The application runs on an Apache http server using the PERL programming language and a MySQL database for storing terms and results.
Results
PubMatrix is a simple intuitive multiplex comparison tool requiring no user understanding of algorithm design, perl, or scripting, and minimal instruction time. It allows automatic systematic searching of PubMed followed by a quick intuitive survey of results, and as such, dramatically reduces the investigator time needed to query PubMed as compared to a gene-by-gene approach. Moreover, it is systematic and objective. Unlike typical PubMed searches performed on an ad hoc basis, the 1st and the 1000th query are searched in an unbiased manner. This helps avoid becoming distracted with seemingly interesting comparisons early on in a traditional literature search session. It is flexible in the sense that any list of terms in almost any combination may be used. With PubMatrix, large lists of keywords can be compared and, when used with lists of gene names and function, may be used to analyze and annotate microarray gene lists and datasets. GO keywords may be used to annotate gene lists in a semi-automated fashion. Additionally, the ability to save searches in an archive allows sharing among collaborators and public accessibility of curated lists of useful search results and search terms.
Figure 1 shows the web interface for PubMatrix. Data entry begins with creating two lists of keyword terms, (figure 1) SEARCH TERMS and MODIFIER TERMS. These can be any simple words or strings of words. Typically official gene symbols (TP53, CTLA4, etc.) can be used for microarray or proteomic data. These lists can be pasted into the appropriate data entry boxes on the page or loaded as a text file through the browser. Essentially any search combination that can be loaded into the basic PubMed search window can be run in PubMatrix in matrix file format. This gives a broad search capability as compared to PubGene [1], which is limited to gene identifiers only. Terms in either list can contain PubMed field limits (i.e. author, title, abstract, journal, publication date, etc.) subject to PubMed/Entrez formatting specifications [10]. Compound terms in either list can be used as well. Like PubMed, success in using PubMatrix is dependent upon the quality of the input terms. If terms are general, you may end up with general results, if they are too specific, you may limit the search inappropriately. As in PubMed, PubMatrix makes no provision for nomenclature curation and provides no measure of statistical significance. General terms may be appropriate for some searches while specific terms may be useful for others. If the terms used are inappropriate or non-specific, the search will return inappropriate and low quality results; quisquiliarum ineo quisquiliarum egredior.
Figure 1.
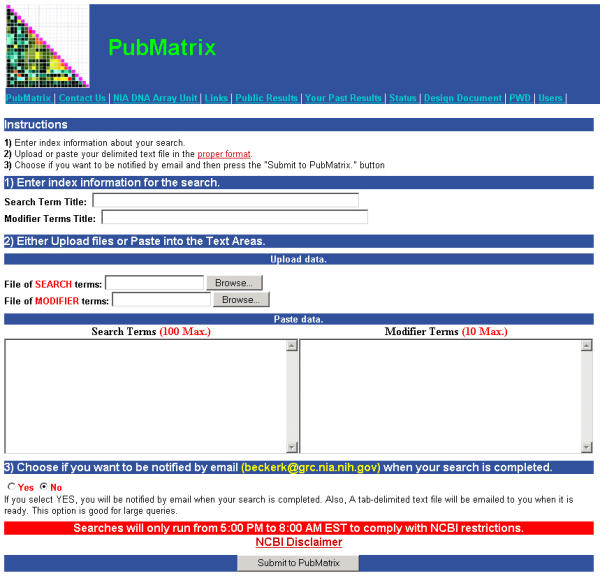
The PubMatrix Data Input Page http://pubmatrix.grc.nia.nih.gov/
There is currently a 100 SEARCH TERM and 10 MODIFIER TERM limit on the search capability due to NLM automatic script searching restrictions. The size and speed of PubMatrix is largely determined by NLM restrictions. In unrestricted tests, PubMatrix rapidly returned results using term lists of over 10,000 terms. The size limit of 100 × 10 will generally accommodate a gene list from a published summary table of microarray results or genes from a cluster produced from a typical clustering program. After data entry, the PubMatrix program then performs pairwise comparisons of each SEARCH term against each MODIFIER term using the Boolean operator "AND", capturing only the pairwise frequency counts found in PubMed. It does not capture the actual PubMed record. Relative frequencies are then presented in an html matrix table (Fig 2) of control terms versus search terms. This table can then be surveyed or sorted for positive hits. Clicking on a single frequency number in the matrix table then performs a de-novo individual query for that single pairwise combination in PubMed. In this way the user can drill down or focus on specific interesting combinations.
Figure 2.
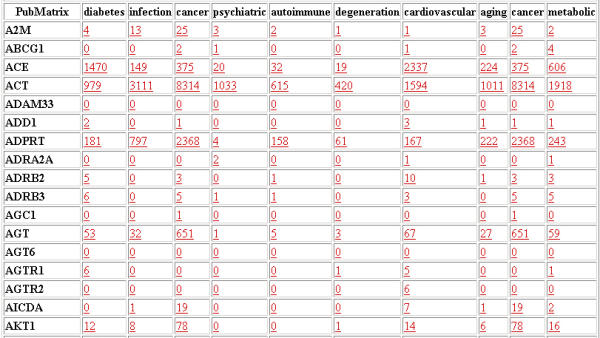
The report page from a simple search of gene names versus neural modifier terms. All reported numbers are hyperlinked and will initiate a de-novo search for that specific term combination.
Productive searches using broadly useful control term lists have been assembled into an associated PubMatrix public archive. These previously used search term lists may be commonly useful and may aid other investigators in subsequent searches. Examples include, official gene symbols, functional descriptors, cytogenetic bands, or polymorphic markers. Likewise, non-molecular term lists have been used and archived which may be broadly useful, such as commonly prescribed drugs, common diseases, and meeting speakers, among others. For example, prior to a meeting, an attendee can rapidly search all the meeting speakers names against important keywords found in the meeting program to provide a one-page summary of speakers scientific interests. Archived term lists can be copied to initiate new searches with new modifier term combinations. Table 1 gives examples of categorical lists of terms used in PubMatrix. Searches using these term lists can be found in the PubMatrix: Public Results page.
Table 1.
Examples of categorical search lists
| Category | Examples |
| Official Gene Symbols | APOB, ACE, BDNF, CD45, ... |
| Polymorphic markers | D1S478, D6S470, D13S193, ... |
| DNA sites | AAATTT, CAGCAG, TTTTTT, ... |
| Chromosomal bands | 1ter*, 1p36*, 1p35*....Xq27*, Xter* |
| Countries | sweden, canad*, mexic*, finland, ... |
| Common Prescription drugs | acetaminophen, acyclovir, albuterol, alprazolam, ... |
| Common diseases | atopic dermatitis, asthma, crohn's, Celiac, Graves',... |
| Date of Publication | 1973 [dp], 1974 [dp]......2000 [dp], 2001 [dp], 2002 [dp] |
| Meeting Speakers | Weiss A, Pierce SK, Kupfer A,... |
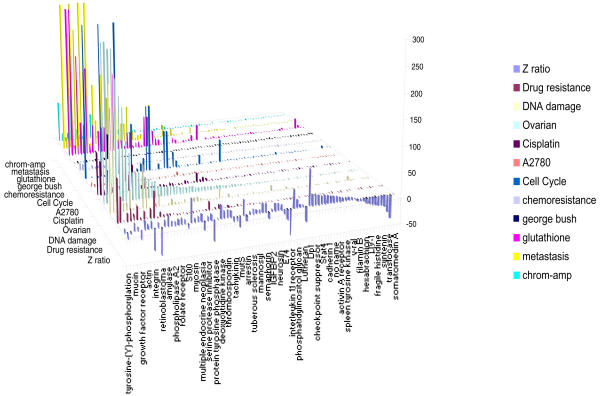
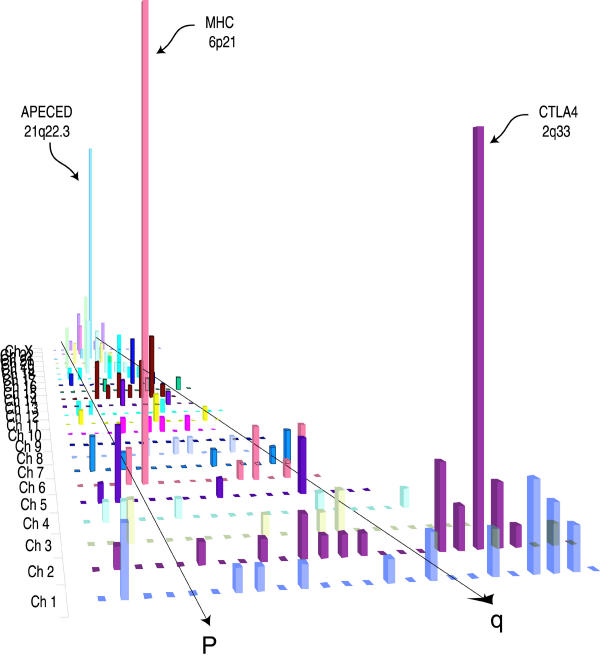
PubMatrix tables can be combined or processed further to provide visual displays or summaries of complete experiments or large datasets in the context of medical literature. Figure 3 shows a display of cDNA microarray results of an ovarian cancer cell line treated with the drug cisplatin. The literature results are displayed in the context of gene expression ratios (Z-ratios). Similarly, Figure 4 shows a comprehensive term list of human cytogeneic bands covering the human genome (1p36*, 1p35*, 1p34*...Xter*) versus the single term "Autoimmune". When displayed by chromosome, major autoimmune loci are identified. These include the major histocompatibility complex, chromosome 2q (CTLA4), as well as APECED on 21q. Figures 3 and 4 were displayed in MS EXCEL using the 3-D chart view option. Moreover, PubMatrix tables can be pasted into spreadsheet programs and imported into other types of data visualization programs such as CLUSTER/TREEVIEW [11] or SPOTFIRE [12] for graphic display and manipulation.
Figure 3.
Gene expression results (Z-ratio) of cisplatin treatment of an ovarian tumor cell line versus keywords relevant to cisplatin resistance. This graph was constructed in MS EXCEL directly from a PubMatrix search result table using the 3D chart view option after adding gene expression values (Z-ratios).
Figure 4.
Visual display of chromosomal-band term list versus the term. "autoimmune". Search terms were 313 sequential human chromosomal bands (1pter, 1p36, 1p35, 1p34, etc....Xq26, Xq27, Xq28, Xqter) versus the single modifier term "autoimmune". This graph was constructed in MS EXCEL using 3D chart view option after separating individual chromosome results into individual columns.
Conclusions
PubMatrix allows a simple systematic approach to query the medical literature in PubMed with comparative keyword lists. It performs simple automatic queries and greatly reduces analysis time. In this way, increasingly large datasets generated by high-throughput multiplex assays such as proteomic or microarray assays can be mined, archived, displayed, and annotated for biological and disease relevance.
Availability and requirements
PubMatrix is available for free use at this URL: http://pubmatrix.grc.nia.nih.gov/
Authors' contributions
KGB and TJB conceived the approach and participated in early design and testing. DAH, GD, RAL, and CC participated in software design and testing. JE participated in web design, database development, and algorithm modification. All authors read and approved the final manuscript
Acknowledgments
Acknowledgements
The authors would like to thank E. Hovig and D. Wheeler for helpful discussions, and C. Sherman-Baust and P. Morin for sharing of data.
Contributor Information
Kevin G Becker, Email: beckerk@grc.nia.nih.gov.
Douglas A Hosack, Email: DHosack@niaid.nih.gov.
Glynn Dennis, Jr, Email: GDennis@niaid.nih.gov.
Richard A Lempicki, Email: RLempicki@niaid.nih.gov.
Tiffani J Bright, Email: bright1@umbc.edu.
Chris Cheadle, Email: Cheadlec@mail.nih.gov.
Jim Engel, Email: engelj@grc.nia.nih.gov.
References
- Jenssen TK, Laegreid A, Komorowski J, Hovig E. A literature network of human genes for high-throughput analysis of gene expression. Nat Genet. 2001;28:21–28. doi: 10.1038/88213. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Tanabe L, Scherf U, Smith LH, Lee JK, Hunter L, Weinstein JN. MedMiner: an Internet text-mining tool for biomedical information, with application to gene expression profiling. Biotechniques. 1999;27:1210–4. doi: 10.2144/99276bc03. 1216–1217. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biology. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan P. MeSHmap: a text mining tool for MEDLINE. Proc AMIA Symp. 2001:642–646. [PMC free article] [PubMed] [Google Scholar]
- Perez-Iratxeta C, Perez AJ, Bork P, Andrade MA. Update on XplorMed: A web server for exploring scientific literature. Nucleic Acids Res. 2003;31:3866–3868. doi: 10.1093/nar/gkg538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Valencia A. Automatic annotation for biological sequences by extraction of keywords from MEDLINE abstracts. Development of a prototype system. Proc Int Conf Intell Syst Mol Biol. 1997;5:25–32. [PubMed] [Google Scholar]
- Masys DR, Welsh JB, Fink JL, Gribskov M, Klacansky I, Corbeil J. Use of keyword hierarchies to interpret gene expression patterns. Bioinformatics. 2001;7:319–26. doi: 10.1093/bioinformatics/17.4.319. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entrez Programming Utilities http://eutils.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher B. Decision analytics software solutions for proteomics analysis. J Mol Graph Model. 2000;18:79–82. [PubMed] [Google Scholar]