Abstract
In plants, as in animals, recent work has established that many developmental and defense response pathways are regulated by E3 ubiquitin ligases which control the level or the activity of key proteins through ubiquitination. Nodule formation is a tightly regulated process that integrates specific signal exchange and the coordinated activation of developmental mechanisms to synchronize bacterial infection and organ development. In the last decade, the characterization of several E3 ubiquitin ligase with roles during nodulation has been reported. These are mainly RING-finger and U-Box proteins involved either in nodule organogenesis or in the infection process. In this review, we summarize the knowledge in this field and conclude that the major challenge will be the identification of the regulation and targets of these E3 ubiquitin ligases.
Key words: symbiosis, legume, signaling, post-translational modifications
E3 ubiquitin ligases (E3s) in eukaryotes are involved in protein ubiquitination, a post-translational regulatory process essential for growth and interaction with the environment. E3s work in association with the E1 and E2 ubiquitin conjugating proteins and catalyse the transfer of ubiquitin to the protein target. E3s select the targets and thus control the specificity of the ubiquitination. Although ubiquitination is often a signal for degradation by the proteasome,1 it has become clear that this modification can lead to a variety of outcomes such as modification of protein activity, creation of docking sites or internalization of cell surface proteins.2 On the basis of their subunit composition and their mechanisms of action, E3s can be divided into four groups: the Skp-Cullin-F-box (SCF) complex, the Anaphase Promoting Complex (APC), the Homology to E6-associated protein Carboxyl Terminus (HECT) domain and the Really Interesting New Gene (RING)/U-box domain (U-box domain is a degenerate version of the RING-finger domain).3 The first two groups define multimeric E3s in which target specificity is determined by the F-box subunit in SCF complex and two adaptors CDC20 or Cdh1 in APC. The other groups constitute monomeric E3s and they are named by their E2 interacting domain (HECT, RING and U-Box), whereas target specificity is determined largely by other associated domains.
In plants, the E3s are encoded by a very large number of genes, for example more than 1,200 genes in Arabidopsis.4 Plant E3s belong to multigenic families. The F-box protein family has particularly experienced an expansion in plants in comparison to other eukaryotic species,5 and this is also true to a lesser extent for the U-box protein family.6 This high number of genes relative to other eukaryotes underlines the importance of E3s in regulating plant processes. E3s have been identified as key regulators in plants of hormone signaling, photomorphogenesis, cell cycling and plant-microbe interactions.7
The legume-rhizobia symbiosis is initiated by a signal exchange between the partners which leads to the production of nodules on the roots of the plants, in which the bacteria fix dinitrogen. Root nodulation requires the dual activation of nodule organogenesis and infection processes in the root cells.8 Both processes depend on the perception of Rhizobial Nod factor (NFs), lipochitooligosaccharadic signals which are perceived by symbiotic receptors in the plant roots. The characterization of receptor like kinase (RLKs) genes and their mutant phenotypes indicates that they play multiple roles in the perception of NFs and their transduction via calcium mediated responses and transcriptional regulation. Among these RLK, LYK3 is involved in the earliest steps of symbiotic interactions such as NFs recognition and the infection signaling networks that conduct to the invasion of rhizobia into root hair cells through infections threads (ITs).9 Extensive research in legume models, such as Medicago truncatula and Lotus japonicus has identified roles of E3s in nodulation. The only example of the involvement of a multimeric E3 complex in nodulation is the requirement of the APC ubiquitin ligase activator, Ccs52A for DNA endoreduplication during cell differentiation in nodules.10 However, several reports have described roles of monomeric E3s in nodulation. In this review we summarize this work, focusing on the identification, the developmental roles and the biochemical function of the E3s.
Identification and Structure of E3s Involved in Nodulation
Forward genetic approaches have identified several putative E3s involved in nodulation. The first gene encoding a putative E3, described to play such a role, was ASTRAY/LjBZF in L. japonicus, identified from a hypernodulating and photomorphogenic mutant. Because this latter phenotype was similar to several characteristic responses of the Arabidopsis mutant Hy5 (LONG HYPOCOTYL5), a homolog of Hy5 was identified from L. japonicus and complementation experiments confirmed the role of this gene in both the symbiotic and non symbiotic phenotypes described in astray.11,12 ASTRAY/LjBZF encodes a protein of 321 amino acids containing, from the N-terminus, a RING-finger domain involved in protein-protein interactions, an acidic region and a typical bZIP (basic leucine zipper) domain involved in DNA binding (Fig. 1).12 It is worth noting that ASTRAY is a divergent Lotus homolog of Arabidopsis HY5 because the RING-finger domain and the acidic region are not present in HY5. It seems that the structural organization of ASTRAY is specifically found in legumes. It would be interesting to examine whether the RING-finger domain is important to the nodulation phenotype.
Figure 1.
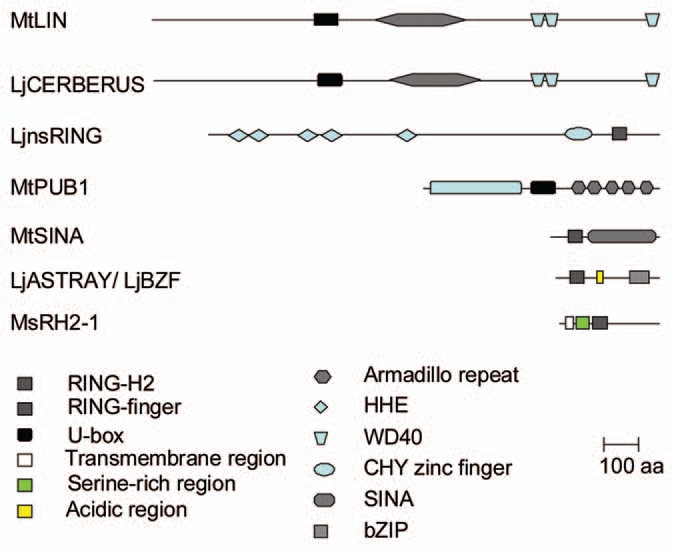
Schematic representation of the structure of the E3 ubiquitin ligases identified to play a role in nodulation.
The MsRH2-1 gene was identified during the analysis of non nodulating alfalfa mutants as a unique polymorphic PCR fragment between the Nod−/Nod+ germplasms of alfalfa. A 295 amino acid protein contains from the N-terminal end one putative transmembrane domain, followed by a serine-rich region and a RING-H2 domain (a version of RING-finger domain in which one cysteine is replaced by a histidine) (Fig. 1).13
Lin (lumpy infection) and cerberus were firstly described as mutants defective in the early infection processes in M. truncatula and L. japonicus, respectively and because of their very similar phenotypes they were predicted to be orthologous genes.14,15 This was confirmed by positional cloning which showed that LIN shares 86% sequence identity with CERBERUS. LIN and CERBERUS encode proteins of 1,488 and 1,477 amino acids, respectively, which contain from the N-terminal end about 500 amino acids without any homology with known domains, followed by a U-box domain, then a large region with an ARMADILLO (ARM) repeat and at least three WD-40 repeats all presumably involved in protein-protein interactions (Fig. 1).16,17
Three others E3s have been shown to play symbiotic roles by reverse genetic approaches. The LjnsRING gene was studied due to its specific expression in early stages of nodulation in L. japonicus.18 Nodule-specific genes are good candidates to play crucial roles during symbiosis. A1236 amino acid protein was predicted from LjnsRING cDNA. From the N-terminus, LjnsRING has five HHE domains with unknown function, a CHY zinc-finger motif and a RING-H2 domain, both probably involved in protein-protein interactions (Fig. 1).19
Studies in Arabidopsis revealed that SINAT5, a RING-finger domain containing protein related to the Drosophila SEVEN IN ABSENTIA protein, involved in photoreceptor differentiation, was an E3 that affected lateral root formation.20 Because of the similarities between lateral root and nodule formation, the role of SINA proteins was investigated in nodule formation of M. truncatula. Six M. truncatula genes were identified encoding proteins (304 to 333 amino acids) with a RING-finger domain at the N-terminus part followed by a large conserved SINA domain involved in protein-protein interactions (Fig. 1).20
Recently, Plant U-box protein 1 of M. truncatula (MtPUB1) was identified as an interacting partner of the LYK3 RLK using a yeast two-hybrid screening.21 MtPUB1 encodes a protein of 694 amino acids containing from the N-terminal part the large U-box N-terminal Domain (UND) with unknown function (which is found in a subset of Arabidopsis PUB proteins22) followed by the U-box domain and at least five ARM repeats (Fig. 1).21
Functional Characterization
In addition to the phenotypical analysis of mutants, several functional characterizations were carried out by RNAi knockdown and/or overexpression strategies using the hairy root transformation system, available in L. japonicus and M. truncatula. The phenotypes suggest a role of ASTRAY/LjBZF, MsRH2-1, LjnsRING and SINA proteins in nodulation, although other root and shoot phenotypes were described showing that these proteins either play a more general role in plant development or have additional roles.12,13,19,20
LjBZF/ASTRAY is a divergent Lotus homolog of Arabidopsis HY5, a transcription factor that activates photomorphogenesis responses and root development in Arabidopsis.23 Defects in light and gravity responses in astray mutant were very similar to those of hy5. However astray did not show increase of lateral root initiation like hy5, but an increase of nodule initiation. In astray, nodule primordia appeared earlier than in wild type plants and approximately two-fold more nodules were formed compared to the wild type, on a wider zone but with normal density. In contrast to other hypernodulating mutants, astray showed normal sensitivity to general inhibitors of nodulation such as ethylene and nitrate, thus astray has been categorized as a light-insensitive mutant.12
The overexpression of MsRH2-1 in alfalfa led to dramatic alterations in plant growth. The size of the plants was reduced and exhibited weaker shoot branching and lateral root development than the control. In addition, leaves started their development normally but the cells did not expand. The nature of the changes in plant architecture and morphology suggests disruption of auxin-related signaling pathways. MsRH2-1 was weakly expressed in all organs tested. During lateral root and nodule development MsRH2-1 was initially expressed at the base of the primordia, and later in the regions specific to the differentiating of the vascular bundles. Overexpression of MsRH2-1 resulted in a very low number of nodules. Auxin is required for both initiation and elaboration of the vasculature, two stages common to both lateral root and nodule development.24,25 Therefore, it is difficult to conclude whether MsRH2-1 is directly involved in the specific development of nodules or if its role is a consequence of its involvement in more general plant growth and development processes.
Although the basal expression of the LjnsRING gene was very low in all organs, the shoot and root growth of transgenic RNAi plants was strongly retarded. Transgenic hairy roots were impaired in the development of lateral roots leading to many short or aborted lateral roots. Unsuccessful experiments of regeneration of stable transgenic lines led the authors to suggest that LjnsRING causes a serious imbalance of phytohormones. In addition, in RNAi transgenic plants, significant inhibition of nodule formation was observed likely due to strong inhibition of IT formation. However, how LjnsRING affects the formation of ITs in RNAi plants was not elucidated. In addition, expression of LjnsRING was shown to be upregulated in roots 4 to 7 days post-inoculation by rhizobium and is highly nodule-specific but because of the early infection phenotype and its consequence on nodule formation, it is difficult to evaluate the role of LjnsRING in nodule functioning. Thus, LjnsRING, seems to be involved in general processes of growth and development but seems also to have an additional role in nodulation.
It has been previously demonstrated that Arabidopsis SINAT5 attenuates auxin signals in roots by ubiquitination of the NAC1 (NO APICAL MERISTEM/CUP-SHAPED COTYLEDON 1) transcription factor to promote its degradation.26 SINAT5 also controls floral transition by regulating flowering-related proteins through its E3 activity.27,28 In M. truncatula, the six SINA genes were all expressed in roots and nodules. A significant increase in transcript level was observed in inoculated roots only for MtSINA4, 6 days post inoculation.20 Because SINA proteins often act as dimers, pairwise yeast two-hybrid analysis was used to study oligomerization and showed that each of the MtSINA proteins is capable of interacting with the others and also with the Arabidopsis SINAT5 protein. The authors used ectopic expression of Arabidopsis proteins to study the role of MtSINA in nodulation. Both SINAT5 and SINAT5DN, a dominant-negative form of this protein affected in E3 activity, were used.26,29 In this way, only a global role of the SINA family in Medicago could be investigated. Expression of SINAT5DN modified the size of leaves and the number of lateral roots of M. truncatula transgenic plants similarly to its effect in Arabidopsis transgenic lines.20 With regard to nodulation, ectopic expression of SINAT5DN affected nodule initiation which was delayed but not inhibited. The high number of infection events observed and the lower number of functional nodules obtained suggest an interference with infection control rather than nodule primordium development. More precisely, observations showed that ITs were formed but infection did not progress well. ITs were deformed, broader than those of the WT with a dense matrix containing few bacteria and an irregular cell wall structure. Some ITs reached the nodule and released bacteria but the symbiosomes were enlarged with large spaces between the bacteroid and the symbiosome membrane, leading to small Fix− nodules which senesced earlier.20 This work clearly identified a role of MtSINA proteins during nodulation, but it is not clear if some members of the MtSINA protein family have pleiotropic roles including nodulation or if some members have a specific role in symbiosis.
For the E3s containing a U-box domain, specific roles in nodulation processes have been reported. The lin and cerberus mutants formed small white bumps following rhizobia inoculation but, although rhizobia were able to colonize curled root hair tips, infections never progressed beyond the root hair cells. Gain of function mutations (autoactive versions) in Calcium Calmodulin-dependant protein Kinase (CCaMK) confer spontaneous nodulation in the absence of rhizobial infection. The use of these mutants revealed that LIN and CERBERUS were not necessary for nodule organogenesis.16,17 Thus, the arrest of nodule development in lin or cerberus roots is an indirect consequence of the arrest of the rhizobial infection process and additional data implicate CERBERUS specifically in the IT pathway.30
The role of MtPUB1 was characterized by RNAi knockdown and overexpression approaches. Overexpression of MtPUB1 led to a delay in nodulation which was apparent 7 days after infection but not at 14 days post-infection. No difference in the number of nodules was observed in RNAi knockdown plants after infection with the wild-type S. meliloti strain. M. truncatula plants inoculated with mutated S. meliloti strains producing modified NFs showed a marked reduction in infection and nodulation.9,31 In contrast, nodulation experiments with the same S. meliloti strains producing modified NFs show a very strong increase in the number of nodules produced in RNAi-MtPUB1 plants.21 MtPUB1 is a partner of the LYK3 receptor. LYK3 is primarily involved in infection, as shown by the characterization of the weak lyk3-4 (hcl-4) allele, in which nodulation by the wild-type rhizobia is very poor because of the abnormal infection process: bacteria remain as microcolonies in the root hair curl or ITs are rapidly blocked, forming saclike structures in the epidermal cells.32 A dramatic increase in the number of normal ITs and the number of nodules was obtained in RNAi-MtPUB1 lyk3-4 plants. Thus, MtPUB1 plays a negative role in nodulation, affecting primarily the initiation and growth of ITs, in relation to the activity of the LYK3 receptor.
Biochemical Characterization
The proteins described in these studies are classified in the enzymatic class of E3s by the presence of sequences homologous to highly conserved domains identified in biochemically characterized E3s. Only, MtPUB1 has, to date, been shown to possess in vitro ubiquitination activity as demonstrated by its auto-ubiquitination,21 while experiments to demonstrate the E3 activity of LjnsRING or CERBERUS were unsuccessful.17,19 In addition, MtPUB1 was phosphorylated by the LYK3 kinase domain in vitro but no ubiquitination or alteration of LYK3 stability was found, following interaction with MtPUB1.21
Conclusions
In conclusion, all the monomeric E3s currently identified with roles in nodulation contain a RING-finger or a U-Box domain. These E3s are involved at different steps of nodulation, some in nodule development and others in the infection process. Because some are also involved in general developmental processes for example, photomorphogenesis or hormone regulation, it is difficult to decipher a specific effect from a general one. However, LIN, CERBERUS and MtPUB1 have clear specific roles in rhizobial infection. SINA and LjnsRING have roles in developmental processes outside of nodulation and their roles in the highly specific process of infection are ambiguous. Surprisingly, these five proteins all exhibit a clear negative role on the early steps of the infection process.
Because of functional redundancies due to the large expansion of several E3 families in plants, which is associated with crucial role in many general processes of plant development, it leaves the possibility that additional E3s are involved in nodulation but not yet identified. Reverse genetics, transcriptional or proteomic approaches could be useful to identified new E3s involved in symbiosis.
More biochemical analysis on E3s identified is necessary to characterize their putative E3 ubiquitin ligase activity. In addition, the regulation of the E3 activity by phosphorylation or self-ubiquitination for example, has also to be determined to understand how these proteins play their roles. In the case of MtPUB1, although auto-ubiquitination and phosphorylation by LYK3 have been determined in vitro, it will be very interesting to correlate these post-translational modifications to a specific signaling event in infection and/or nodulation. Thus, additional experiments are required to evaluate the implication of MtPUB1 E3 activity and its regulation on the phenotype observed.
Finally, the targets of all the symbiotic E3s still remain to be identified. LYK3 did not appear to be a target of MtPUB1 ubiquitin ligase activity in vitro. Recently, using a yeast two-hybrid screen, an exocyst complex component has been found as a target of a related PUB protein, ARC1, involved in self-incompatibility in Brassica napus.33 By analogy, such a component might be the target of MtPUB1. ARC1 was also previously identified by yeast two-hybrid screening as a partner of the S-locus receptor kinase (SRK1) which phosphorylates ARC1.34 Thus, yeast two-hybrid screening seems a method of choice to identify targets of symbiotic E3s.
References
- 1.Hershko A, Ciechanover A. Mechanisms of intracellular protein breakdown. Annu Rev Biochem. 1982;51:335–364. doi: 10.1146/annurev.bi.51.070182.002003. [DOI] [PubMed] [Google Scholar]
- 2.Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Koonin EV. The U box is a modified RING finger: a common domain in ubiquitination. Current Biology. 2000;10:132–134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 4.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 5.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson C. A new gun in town: the U box is a ubiquitin ligase domain. Sci STKE. 2002;2002:1–4. doi: 10.1126/stke.2002.116.pe4. [DOI] [PubMed] [Google Scholar]
- 7.Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot. 2009;60:1109–1121. doi: 10.1093/jxb/ern369. [DOI] [PubMed] [Google Scholar]
- 8.Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 9.Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 10.Vinardell JM, Fedorova E, Cebolla A, Kevei Z, Horvath G, Kelemen Z, et al. Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell. 2003;15:2093–2105. doi: 10.1105/tpc.014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura R, Ohmori M, Fujita H, Kawaguchi M. A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc Natl Acad Sci USA. 2002;99:15206–15210. doi: 10.1073/pnas.222302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura R, Ohmori M, Kawaguchi M. The novel symbiotic phenotype of enhanced-nodulating mutant of Lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol. 2002;43:853–859. doi: 10.1093/pcp/pcf098. [DOI] [PubMed] [Google Scholar]
- 13.Karlowski WM, Hirsch AM. The overexpression of an alfalfa RING-H2 gene induces pleiotropic effects on plant growth and development. Plant Mol Biol. 2003;52:121–133. doi: 10.1023/a:1023916701669. [DOI] [PubMed] [Google Scholar]
- 14.Kuppusamy KT, Endre G, Prabhu R, Penmetsa RV, Veereshlingam H, Cook DR, et al. LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol. 2004;136:3682–3691. doi: 10.1104/pp.104.045575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombardo F, Heckmann AB, Miwa H, Perry JA, Yano K, Hayashi M, et al. Identification of symbiotically defective mutants of Lotus japonicus affected in infection thread growth. Mol Plant Microbe Interact. 2006;19:1444–1450. doi: 10.1094/MPMI-19-1444. [DOI] [PubMed] [Google Scholar]
- 16.Kiss E, Olah B, Kalo P, Morales M, Heckmann AB, Borbola A, et al. LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol. 2009;151:1239–1249. doi: 10.1104/pp.109.143933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yano K, Shibata S, Chen WL, Sato S, Kaneko T, Jurkiewicz A, et al. CERBERUS, a novel U-box protein containing WD-40 repeats, is required for formation of the infection thread and nodule development in the legume-Rhizobium symbiosis. Plant J. 2009;60:168–180. doi: 10.1111/j.1365-313X.2009.03943.x. [DOI] [PubMed] [Google Scholar]
- 18.Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, et al. Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res. 2004;11:263–274. doi: 10.1093/dnares/11.4.263. [DOI] [PubMed] [Google Scholar]
- 19.Shimomura K, Nomura M, Tajima S, Kouchi H. LjnsRING, a novel RING Finger protein, is required for symbiotic interactions between Mesorhizobium loti and Lotus japonicus. Plant Cell Physiol. 2006;47:1572–1581. doi: 10.1093/pcp/pcl022. [DOI] [PubMed] [Google Scholar]
- 20.Den Herder G, De Keyser A, De Rycke R, Rombauts S, Van de Velde W, Clemente MR, et al. Seven in Absentia proteins affect plant growth and nodulation in Medicago truncatula. Plant Physiol. 2008;148:369–382. doi: 10.1104/pp.108.119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, et al. The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell. 2010;10:3474–3488. doi: 10.1105/tpc.110.075861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004;134:59–66. doi: 10.1104/pp.103.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathesius U, Schlaman HR, Spaink HP, Of Sautter C, Rolfe BG, Djordjevic MA. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998;14:23–34. doi: 10.1046/j.1365-313X.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- 25.de Billy F, Grosjean C, May S, Bennett M, Cullimore JV. Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol Plant Microbe Interact. 2001;14:267–277. doi: 10.1094/MPMI.2001.14.3.267. [DOI] [PubMed] [Google Scholar]
- 26.Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature. 2002;19:167–170. doi: 10.1038/nature00998. [DOI] [PubMed] [Google Scholar]
- 27.Park BS, Sang WG, Yeu SY, Choi YD, Paek NC, Kim MC, et al. Post-translational regulation of FLC is mediated by an E3 ubiquitin ligase activity of SINAT5 in Arabidopsis. Plant Sci. 2007;173:269–275. [Google Scholar]
- 28.Park BS, Eo HJ, Jang IC, Kang HG, Song JT, Seo HS. Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem Biophys Res Commun. 2010;398:242–246. doi: 10.1016/j.bbrc.2010.06.067. [DOI] [PubMed] [Google Scholar]
- 29.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, et al. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun. 2010;1:1–12. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, et al. Rhízobíum meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, et al. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR. Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell. 2009;9:2655–2671. doi: 10.1105/tpc.109.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR. Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA. 1998;95:382–387. doi: 10.1073/pnas.95.1.382. [DOI] [PMC free article] [PubMed] [Google Scholar]


