Abstract
N-Acylethanolamines (NAEs) are lipid metabolites derived from the hydrolysis of the membrane phospholipid N-acylphosphatidylethanolamine (NAPE). Recent work in Arabidopsis thaliana seedlings showed that combined treatments of NAE 12:0 and ABA inhibited seedling growth synergistically, suggesting low levels of NAE could potentiate the action of ABA. Here we examined the interplay between compound concentrations, growth inhibition and mutant genotypes with impaired sensitivities to these regulators. NAE 12:0 and ABA both induced dose-dependent increases in transcript levels of ABI3, and two ABI3 responsive genes, AtHVA22B and RD29B. Interestingly, even in the absence of growth inhibition, RD29B transcripts were elevated by ABA but not NAE treatment outside the sensitive window for ABA/NAE treatment, indicating some differences in the regulation of growth and the modulation of gene expression by these two compounds. Also noteworthy, the growth of ABA insensitive mutant (abi 3-1) seedlings was inhibited at higher concentrations of NAE 12:0 but not ABA, suggesting that NAE may act to inhibit early seedling establishment by both ABI3-dependent and ABI3-independent pathways. Collectively our results reinforce the concept that NAE12:0 interacts with ABA signaling in seedling establishment, but also points to a complexity in this interaction that modulates the sensitivity of young seedlings to phytohormone-mediated growth arrest.
Key words: Arabidopsis thaliana, N-Acylethanolamine, abscisic acid, ABI3
Introduction
N-Acylethanolamines (NAEs) are fatty acid amides that are derived from an N-acylated phosphatidylethanolamine precursor, a minor membrane lipid constituent of plant and animal cells. NAEs were first reported as constituents of soy lecithin and peanut meal in the 1950s.1 The occurrence and metabolism of NAEs is conserved among the eukaryotic organisms,2,3 however, the physiological functions of these lipids have been investigated mostly in vertebrates. In animal systems NAEs have been determined to have roles as regulators of important physiological processes such as embryo development, cell proliferation, immune responses and apoptosis.4
There is accumulating evidence that plants also use NAEs to regulate important physiological processes. This is supported by the identification of NAEs in a variety of plant tissues and the fact that NAE levels vary due to environmental and growth conditions.3,5 NAEs in plants, like animals, have potent biological activities at low concentrations, such as activation of defense gene expression by NAE 14:0.3,6 In other work, Arabidopsis seedlings treated with micromolar concentrations of N-lauroylethanolamine (NAE 12:0) show marked developmental abnormalities in roots, and these effects were specific for short/medium chain acylethanolamides.7 Endogenous levels of NAEs drop dramatically from the micromolar levels typically found in seeds to nanomolar levels during imbibition and germination in a variety of plant species, and remain low during subsequent seedling growth.8 This rapid depletion of NAEs suggests that for normal seedling establishment to occur, a system for NAE metabolism is required.
Key to understanding the functional roles of these lipids in all organisms is to identify the mechanisms that regulate their accumulation. NAEs in animal systems appear to be modulated by their degradation to free fatty acids and ethanolamine by fatty acid amide hydrolase (FAAH), a member of the amidase super-family of proteins.9 Thus, FAAH modulation of NAE levels has become an important focus for understanding the mechanistic action of the “endocannabinoid” signaling system in vertebrates.10
Bioinformatic approaches and heterologous protein expression led to the molecular identification of an amidohydrolase from Arabidopsis thaliana that hydrolyzed a wide range of acylethanolamides, including those in desiccated seeds. Research in the last decade has made it apparent that NAE metabolism occurs in plants by pathways analogous to those in vertebrates and invertebrates,5,11 pointing to the possibility that these lipids may be part of an evolutionarily conserved mechanism for the regulation of physiology in multicellular organisms. With the evidence of conserved enzymatic machinery in plants for the formation and degradation of NAEs, and the potent biological effects caused by altered exogenous NAE levels, it is now important to begin to address the question of how altered FAAH expression would affect NAE signaling in plants.
When FAAH is overexpressed, seeds are theoretically able to remove NAEs more rapidly and the seedlings grow more rapidly.12 These seedlings are also able to grow in high levels of exogenous NAE.12,13 When AtFAAH is not expressed (by T-DNA disruption), seeds cannot hydrolyze exogenous NAE as rapidly, and seedling growth is severely inhibited in the presence of added NAE 12:0.12 It is clear from these studies that FAAH expression influences NAE metabolism in Arabidopsis seedlings.
The reduction in early seedling growth mediated by exogenous NAE is similar to that caused by the plant hormone abscisic acid (ABA).13 ABA functions as a repressor of germination and aids in modulating seedling growth.14–16 Activation of the ABA signaling pathway leads to either immediate cellular changes such as the release of intracellular calcium, nitric oxide, sphingolipids and increases in reactive oxygen species17–19 or to changes in gene expression,14,15,20 especially ABA-responsive genes activated by ABA-responsive elements (ABREs).21 Addition of ABA to germinated seeds inhibits seedling growth, leading to reduced root length and smaller overall seedling size.15 The growth responses of seedlings toward ABA are roughly similar to those toward NAE 12:0, and the levels of both of these negative growth regulators are reduced during the course of normal seedling growth.13,15 It is therefore possible that NAE metabolism may interact with ABA signaling to produce changes in seedling growth and this hypothesis was put forward and supported recently by several lines experimental evidence.22 However, the relationship between ABA signaling and NAE metabolism is made complicated by the recent observation that the activity of FAAH toward NAE is independent of the hypersensitivity of FAAH overexpressors to ABA.22,23
In this manuscript we build on our previous findings that seedling development in Arabidopsis is in part mediated by converging ABA and NAE signaling pathways.13 Here we show that an intact ABA signaling pathway is required for the synergistic inhibition of seedling growth, yet higher levels of applied NAE can inhibit seedling growth in plants with an interrupted ABA signaling pathway whereas ABA cannot. In addition, the NAE/ABA window of sensitivity is not affected by altering FAAH levels; however, NAE can elevate ABA responsive gene expression within this window but may not affect expression of ABA regulated genes outside of it (i.e., RD29B). These results contribute to the previous evidence that NAE functions via ABA-dependent and ABA-independent mechanisms.
Results
Altered FAAH metabolism affects the sensitivity of Arabidopsis thaliana seedlings to NAE 12:0 and ABA and ABA-regulated transcript abundance.
Similar to our previous observations,7,12 NAE 12:0 induced a dose dependent reduction in growth of wild-type Arabidopsis seedlings (Fig. 1A). In addition, the response of Arabidopsis seedlings to exogenous NAE 12:0 was modified by altering the expression of the NAE hydrolyzing enzyme FAAH (Fig. 1A).12 Just as there was a dose dependent increase in growth inhibition in the presence of exogenous NAE 12:0, there was a corresponding increase of ABI3 expression (Fig. 1B). Other ABA-regulated genes also displayed differentially regulated expression associated with altered FAAH metabolism, where increased sensitivity to NAE (faah) is accompanied by elevated ABI3 transcript levels, and decreased sensitivity to NAE (FAAH OE) is accompanied by lower transcript levels of ABI3 (Fig. 1B).
Figure 1.
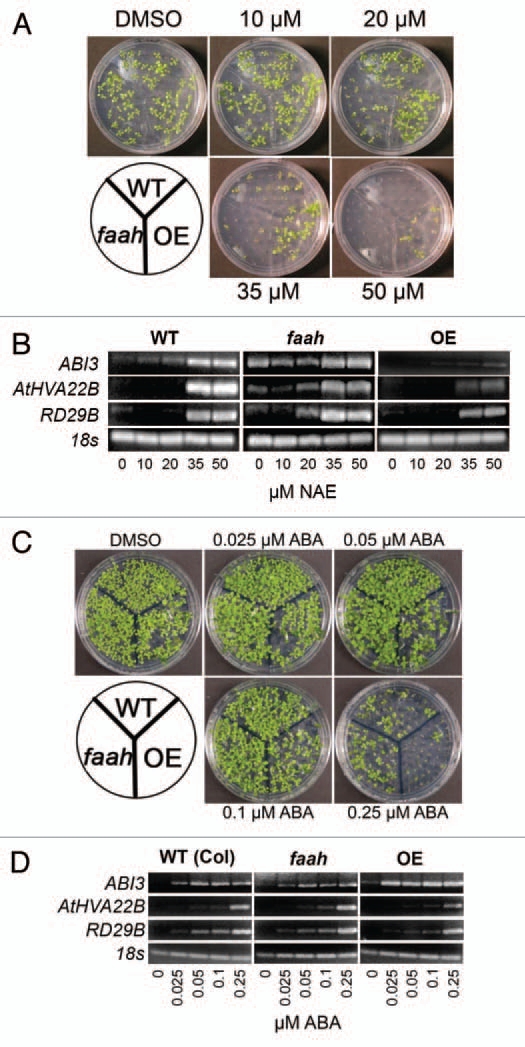
NAE and ABA induce dose dependent inhibition of seedling growth and modulate transcript levels of ABI3 and ABI3 responsive genes. (A) Representative images of 10-day-old seedlings at increasing concentrations of NAE 12:0 exhibit the tolerance of the FAAH overexpressor and the sensitivity of the FAAH knockout. (B) Agarose gel analysis of RT-PCR using gene specific primers to analyze gene expression in 10-day-old seedlings (treated with increasing concentrations of NAE 12:0). ABI3 was run for 40 cycles, AtHVA22B and RD29B for 22 cycles and 18s for 25 cycles. (C) Representative images of 10-day-old seedlings at increasing concentrations of (+) ABA exhibit the tolerance of the FAAH knockout and the sensitivity of the FAAH overexpressor. (D) Agarose gel analysis of RT-PCR using gene specific primers to analyze gene expression in 10-day-old seedlings (treated with increasing concentrations of (+) ABA). ABI3 was amplified for 40 cycles, AtHVA22B and RD29B for 22 cycles and 18s for 25 cycles.
Like NAE, exogenous ABA caused a dose dependent reduction of seedling growth (Fig. 1C). Furthermore, FAAH overexpressors demonstrated a hypersensitive response to ABA (Fig. 1C), which was again consistent with our previous findings.13,23 FAAH knockouts on the other hand appeared to show a slight tolerance to ABA (Fig. 1C). As with NAE, the seedlings with the highest sensitivity to ABA induced growth inhibition also had the highest level of ABI3 transcripts (Fig. 1D).
ABI3 encodes a transcription factor that is a key regulator of the seed to seedling transition.16 In previous work we showed that ABI3 expression in seedlings is correlated with the extent of seedling growth inhibition (i.e., seedlings with the strongest growth inhibition showed the highest ABI3 transcript levels13). Here we expanded our previous conclusions by monitoring transcript levels of ABI3 in wild-type and FAAH altered seedlings using a range of NAE 12:0 concentrations. We found that faah knockouts, which displayed the greatest growth inhibition to NAE 12:0, showed the highest ABI3 transcript levels even at NAE 12:0 treatments that did not visibly affect seedling growth (e.g., 10 µM; Fig. 1B). With FAAH overexpressors, induction of ABI3 transcripts by NAE 12:0 was minimal even at high concentrations (e.g., >40 µM; Fig. 1B). Without FAAH to metabolize NAE, very low levels of NAE (10 µM or lower) induced ABI3 and associated gene expression. This demonstrates that FAAH may be a pivotal regulator of NAE responsive gene expression.
In addition to ABI3, AtHVA22B and RD29B transcript levels were elevated by NAE 12:0 in a previous microarray study.13 Because these two genes have been shown to be regulated in part by ABI3,21 we examined their transcript levels in 7-day-old seedlings of WT, FAAH OE and faah seedlings grown in a range of NAE 12:0 concentrations (Fig. 1B). Both AtHVA22B and RD29B, showed a pattern of increased expression similar to ABI3 (Fig. 1B).
Because NAE and ABA both inhibited seedling growth in a dose dependent manner (Fig. 1A and C) and the fact that both metabolites are depleted during seed germination,13 the effects of ABA treatment on the same genes above were characterized by semi-quantitative real time RT-PCR. Like NAE-treated seedlings, ABA application enhanced the expression of ABI3 transcript (Fig. 1D). Furthermore, levels of ABI3 transcripts were lower in faah seedlings and higher in FAAH OE seedlings compared to WT at all levels of ABA treatment (Fig. 1D).
Combined NAE and ABA act to synergistically reduce arabidopsis seedling growth and modulate ABA-regulated transcript abundance.
In our previous work, we found that the combined application of ABA and NAE 12:0 resulted in a drastic reduction in seedling growth compared to when the two metabolites were applied separately (Fig. 2A).13 To determine whether the effect of both compounds is additive or synergistic, we performed calculations of overall growth after seedlings were treated. Using the relative difference in wild-type seedling fresh weight between the solvent control and each single treatment, and in the combined NAE and ABA treatment, the type of interaction between NAE and ABA was calculated.24 The total tissue fresh weight of wild-type seedlings in 20 µM NAE was 78% of the solvent-only controls and was 35% of controls in 0.1 µM ABA. If the combination of NAE and ABA was completely additive, the expected percent of control in the combined treatment would be ((35% × 78%)/100) = 27.3%. The actual measured percent of control in the combined treatment of 20 µM NAE and 0.1 µM ABA was 17%, indicative of treatment synergism. In addition, the pattern of growth inhibition was similar to that found in seedlings grown in exogenous NAE 12:0, in which the faah seedlings were most sensitive to the combined treatment (Fig. 2A).
Figure 2.
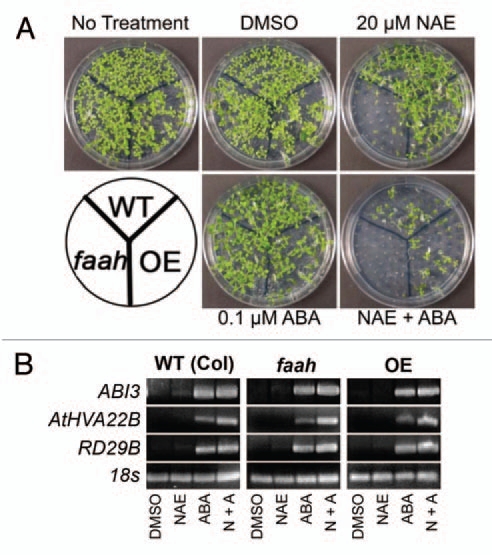
Low levels of NAE and ABA synergistically inhibit seedling growth and modulate transcript levels of ABI3 and ABI3 responsive genes. (A) Representative images of 10-day-old seedlings grown in a combination of NAE 12:0 and (+) ABA exhibit the tolerance of the FAAH overexpressor and the sensitivity of the FAAH knockout. (B) Agarose gel analysis of RT-PCR using gene specific primers to analyze gene expression in 10-day-old seedlings (treated with 20 µM NAE, 0.1 µM ABA or NAE + ABA). ABI3 was amplified for 40 cycles, AtHVA22B and RD29B for 22 cycles and 18s for 25 cycles.
The effects of the combined treatments on gene transcript levels also were characterized (Fig. 2B). Transcript levels of all three genes examined were increased in the combined treatment in the three genotypes (Fig. 2B). ABI3 levels in all three genotypes were similar in both ABA treatment and in the combined treatment. In wild-type seedlings, the levels of RD29B in the combined treatment also were similar to RD29B levels in ABA treatment alone. The levels of AtHVA22B in the combined treatment were similar to the ABA treatment alone in WT seedlings. In faah seedlings, AtHVA22B and RD29B levels were much higher in the combined treatment than in ABA alone. In the OE seedlings, levels of AtHVA22B were much higher in the combined treatment than in either treatment alone, but the levels of RD29B were similar in the combined treatment compared to the ABA treatment alone. Overall, the expression of ABI3, AtHVA22B and RD29B were not much different in seedlings grown in ABA alone versus those treated with both NAE and ABA, and seedling growth was dramatically reduced in the combined treatments. This suggests that reductions in seedling growth may not be entirely attributed to the expression of ABI3 and ABI3-regulated genes.
NAE and ABA synergistic growth arrest occurs only with an intact ABA signaling pathway.
We previously found that seedlings of various ABA signaling mutants are tolerant to the growth inhibition effects of NAE 12:0 and ABA.13 Here we further investigated the synergistic effects of combined NAE 12:0 and ABA treatment on the abi3 mutant because of the strong correlation of ABI3 transcript levels to NAE 12:0 or ABA-induced growth inhibition. As previously shown,13 a dramatic reduction of seedling growth due to combined NAE 12:0 and ABA treatment was observed in WT (Ler) background (Figure 3A). On the other hand, the ABA signaling mutant abi3-1 showed tolerance to the combined treatments (Fig. 3A). Growth measurements confirmed the resistance of abi3-1 and the necessity of a functioning ABA signaling pathway for a combined synergistic effect at these concentrations of NAE/ABA (Fig. 3B). In addition, NAE alone was able to partially inhibit seedling growth in the abi3-1. Growth inhibition of abi3-1 with NAE alone was very similar to the inhibition of the NAE/ABA combination, and both of these treatments inhibited abi3-1 more than ABA alone.
Figure 3.
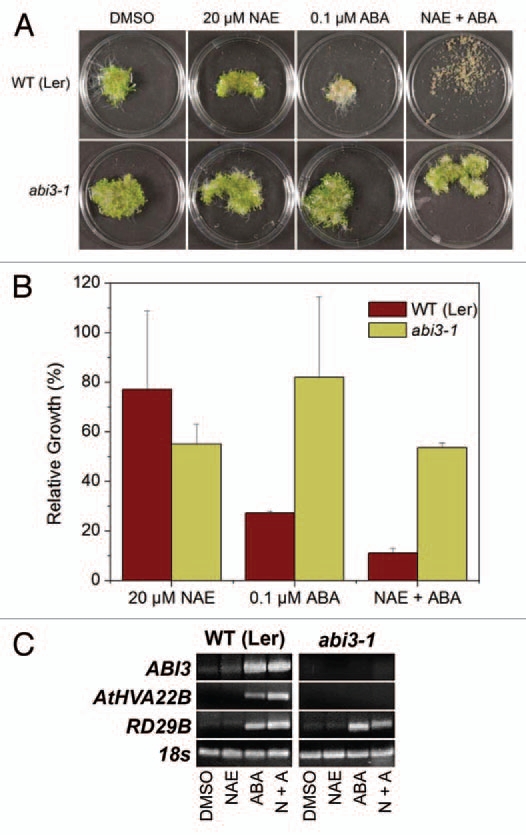
An intact ABA signaling pathway is required for synergistic growth arrest, but not for modulating transcript levels of RD29B. (A) Representative images of 10-day-old seedlings grown in NAE 12:0, ABA or NAE +ABA exhibit the tolerance of abi3-1 seedlings to synergistic growth arrest. (B) Growth of wild-type (Ler) and ABA signaling mutant (abi3-1) seedlings grown in 20 µM NAE 12:0, 0.1 µM ABA or NAE and ABA measured after 10 days of treatment. Values of change in fresh weight compared to DMSO control are the mean and SD of triplicate experiments. (C) Agarose gel analysis of RT-PCR using gene specific primers to analyze gene expression in 10-d-old seedlings (treated with 20 µM NAE, 0.1 µM ABA or a combination of both NAE + ABA). ABI3 was amplified for 40 cycles, AtHVA22B and RD29B for 22 cycles and 18s for 25 cycles.
The effects of combined treatments on ABA-dependent gene transcripts in ABA-signaling mutants were characterized (Fig. 3C). The abi3-1 mutant had no ABI3 transcript, as expected (Fig. 3C). It also showed no induction of the ABA-regulated gene AtHVA22B. However, RD29B did show an increase in transcript when treated with ABA alone and in the combined treatment (Fig. 3C), consistent with reports suggesting that this gene can be modulated independent of ABI3.21
Growth arrest and associated gene expression are induced only within a strict time period after germination.
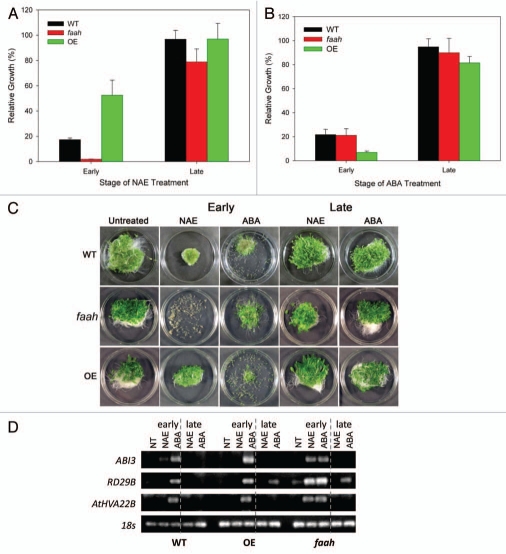
Arabidopsis possesses a defined sensitivity window to ABA during seed germination and seedling growth.13,16 Arabidopsis also has been found to possess a similar window of sensitivity to NAE 12:0.13 To determine if changes in the endogenous capacity for NAE metabolism would change this window of sensitivity to NAE/ABA, we investigated growth and gene expression in the early and late stages of seedling development of Arabidopsis with altered NAE metabolism. Growth of wild-type and FAAH-altered 14-day-old seedlings in response to exogenous NAE 12:0 and ABA was quantified by gain in fresh weight (Fig. 4A and B). Growth was reduced by both treatments in the early stage of seedling development (2 days); the FAAH OE showed a tolerance to the NAE treatment, while the faah displayed an increased sensitivity (Fig. 4A and C). The ABA treatment in the early stage reduced growth equally in the WT and faah mutants, but the FAAH OE revealed a heightened sensitivity to ABA (Fig. 4B and C). By contrast, growth was not substantially reduced by treatment with NAE or ABA in the later stage of seedling development (10 days) (Fig. 4A–C) in any genotype. These results indicate that the developmental sensitivity for either ABA or NAE was most pronounced in young seedlings and this sensitivity was not appreciably affected by altering FAAH expression.
Figure 4.
ABA and NAE 12:0 inhibit seedling growth and modulate gene expression within the NAE /ABA window of sensitivity (up to 6 days). (A and B) Growth of wild-type and FAAH -altered 14-day-old seedlings in response to exogenous NAE 12:0 (35 µM) and ABA (0.25 µM). Treatment at early stage of growth is at day 2 and treatment at late stage is on day 10. Values for gain in fresh weight are the mean and SE of triplicate experiments. (C) Images of 14-day-old wild-type and FAAH -altered seedlings in response to exogenous NAE 12:0 (35 µM), ABA (0.25 µM) and NT (not treated). (D) Agarose gel analysis of semi-quantitative RT-PCR using gene specific primers to analyze gene expression in 14-day-old seedlings (treated at early or late stage of seedling development with 35 µM NAE 12:0 or 0.25 µM ABA).
Gene expression levels of ABI3, AtHVA22B and RD29B were examined under this developmental context for these genotypes. The elevated levels of ABI3 transcripts were inversely associated with growth arrest (Fig. 4D). The 14-day-old faah seedlings treated at 2 days with NAE 12:0 display the least growth and highest levels of gene expression for all three genes, consistent with their hypersensitivity to NAE. By contrast, the 14-day-old FAAH OE treated with NAE at 2 days had much more growth and no transcripts were detected for any of the three ABA-responsive genes, consistent with their their ability to grow in NAE. NAE-treated wild-type seedlings (treated at 2 days) showed some growth, with intermediate levels of ABI3 transcripts. For seedlings treated with ABA, the FAAH OE showed the greatest reduction in growth and the highest ABI3 transcript levels. Treatments outside the phytohormone sensitive window did not show changes in ABI3 or AtHVA22B transcripts (or changes in growth); but the expression of RD29B appeared to be responsive to ABA, but not NAE at the late stages when seedling growth was not arrested. Yet RD29B is elevated by both NAE and ABA within the window of sensitivity, which suggests that NAE may not regulate ABA responsive genes outside of this window. Nonetheless, the association of elevated ABI3 transcript levels with seedling growth arrest by NAE or ABA was consistently observed, continuing to support a role for ABI3 in this interaction.
NAE induced growth arrest can occur independent of an intact ABA signaling pathway.
The abi3-1 mutant is insensitive to ABA, and recently ABA insensitive mutants were shown to be somewhat tolerant to NAE in early development.13 Growth data confirmed the ABA insensitivity of the abi3-1 mutants when grown in ABA from an early timepoint (starting at two days) (Fig. 5A and B). The Landsberg erecta (Ler) ecotype showed growth arrest when treated at the early stage of development, similar to that previously observed for Col(0). However, Ler showed greater inhibition of growth when treated at the late stage of seedling development with ABA than Col(0) (Figs. 4B and 5B), and may reflect slight differences in developmental timing of seedling establishment in these ecotypes. Nevertheless, treatment with NAE at the early stage of development resulted in growth arrest of both wild-type (Ler) and abi3-1 (Fig. 5A and B), while treatment at the late stage showed no inhibition of growth in either wild-types or abi3-1 (Fig. 5B). Wild-type seedlings treated with ABA or NAE showed a corresponding increase in transcript levels of the three surveyed genes (ABI3, AtHVA22B and RD29B), which were associated with growth arrest (Fig. 5C). The insensitivity of the abi3-1 seedlings to ABA treatment at the early stage of development was in stark contrast to its sensitivity when treated with NAE (Fig. 5A and B). As expected, there were no detectable ABI3 transcripts in the abi3-1 seedlings, and since the ABI3 transcription factor controls the expression of AtHVA22B, there were no transcripts for it either (Fig. 5C). The expression of RD29B seemed to be somewhat independent of ABI3, as seen before. However, most importantly, abi3-1 mutant seedlings showed growth arrest when treated with 35 µM NAE at the early stage of seedling growth (albeit at higher concentration than in Fig. 3), indicating that NAE induced growth arrest, at least partially, can be modulated outside the ABA signaling pathway controlled by ABI3.
Figure 5.
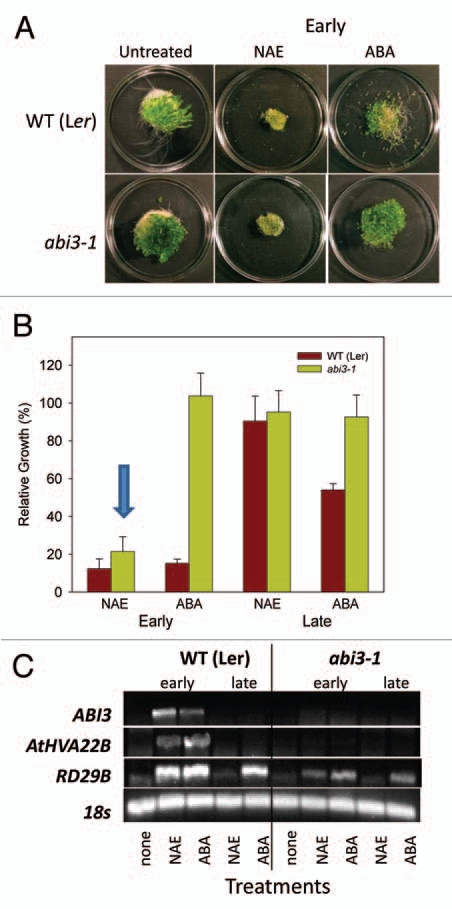
NAE 12:0, but not ABA, induces growth inhibition in an ABA insensitive mutant (abi3-1). (A) Treatment of wild-type (Landsberg erecta) and abi3-1 with NAE 12:0 (35 µM) or ABA (0.25 µM) at early stage of seedling development. (B) Inhibition of growth plotted as gain in fresh weight is shown for WT (Ler) and the abi3-1 mutant treated with NAE 12:0 (35 µM) or ABA (0.25 µM). Values for gain in fresh weight are the mean and SE of triplicate experiments. The arrow highlights the reduced growth of abi3-1 seedlings in NAE 12:0. (C) Agarose gel analysis of semiquantitative RT-PCR using gene specific primers to analyze gene expression in 14-day-old seedlings (treated at early or late stage of seedling development with 35 µM NAE 12:0 or 0.25 µM ABA).
Discussion
Previous studies have shown that Arabidopsis thaliana seeds germinated and grown in N-lauroylethanolamine (NAE 12:0) display a severe reduction in growth.7,25 However, the mechanism of this NAE-induced growth reduction remains largely unknown, although an interaction with ABA signaling has been suggested.13 The evidence presented here continues to support the hypothesis that NAE-induced growth arrest occurs primarily through an ABA/ABI3-regulated pathway, although an ABI3-independent mechanism may also be in place. Specifically, new information reported here includes the following: (1) Detailed concentration experiments link the amounts of ABA, NAE12:0 and combinations of these growth regulators to seedling growth reduction and elevated transcript levels for ABI3, HVA22B and RD29B (Figs. 1–4). (2) An intact ABA signaling pathway is required for synergistic growth arrest, but not for modulating transcript levels of RD29B (Fig. 3). (3) Higher concentrations of NAE 12:0, but not ABA, induced growth inhibition in an ABA insensitive mutant (abi3-1), indicating that NAE12:0 does not entirely overlap with ABA, and NAE can act outside the ABI3 pathway (Fig. 5). (4) ABA, but NAE12:0, can elevate transcript levels of RD29B outside of the secondary dormancy window, indicating that while there is considerable overlap between ABA and NAE12:0 in terms of early seedling growth arrest, this suggests that they also likely participate in distinct genetic regulation pathways (Figs. 4 and 5).
Reduction in seedling growth could be predicted at the molecular level by observing the levels of ABI3 transcripts in the seedlings. When seedlings developed normally, the levels of ABI3 transcript were low, when seedling growth was reduced, the ABI3 transcript levels were elevated (Figs. 1 and 4). While this inverse association between ABI3 transcript levels and growth had been suggested previously in reference 13, the evidence presented here extends these observations to genotypes with altered sensitivity to NAE or ABA, synergistic growth regulation by NAE and ABA, and developmental sensitivity to growth arrest. In addition, ABA-regulated genes (AtHVA22B and RD29B) were examined in these contexts as well to gain insights into the extent of ABA-regulation in seedling responses to NAE. While it is at present unclear whether high levels of ABI3 inhibited seedling growth or whether inhibiting seedling growth resulted in higher ABI3 levels, there is a consistent inverse relationship between ABI3 transcript levels and seedling growth. Moreover, there appears to be a genetic requirement for an intact ABA signaling pathway in the arrest of seedling growth by NAE and ABA. When the ABA signaling pathway was disrupted, as in the abi3-1 mutant, there was no synergistic reduction in growth by NAE and ABA (Fig. 3A and B). Similarly, previous experiments with other ABA insensitive mutants (abi1, abi2, abi4 and abi5) indicated a partial requirement of an intact ABA signaling pathway for NAE action.13
ABA-induced growth arrest and desiccation tolerance occur only within a narrow window of early post-germinative growth, and this is dependent upon ABI3 and an intact ABA signaling pathway.16 This arrest of seedling growth is proposed to function as a stress defense mechanism for seedlings in the activation of a secondary dormancy program. NAE 12:0 was found to induce growth arrest in wildtype seedlings within a similar narrow window during early seedling development.13 The data gathered here shows that the sensitivity window toward ABA and NAE is intact in the FAAH altered mutants, but they do have differing sensitivities to NAE and ABA (Fig. 4A–C). Interestingly, while the FAAH OE displays a tolerance toward NAE, it has heightened sensitivity to ABA (Fig. 4B and C) and the transcript levels of ABI3 are inversely associated with this difference in growth. We believe that when seedlings are exposed to elevated levels of NAE or ABA, within the window of sensitivity, a delay of the normal development of seedlings is induced by maintaining the expression, or resumption of expression, of genes that are normally associated with the ungerminated, desiccation-tolerant state. Either NAE or ABA alone, or low levels of both can induce this growth arrest presumably through ABI3-mediated transcriptional or post-transcriptional regulation.
Overexpression of FAAH with an inactive catalytic region resulted in plants with increased sensitivity to ABA but without tolerance to NAE.23 Therefore, the overexpression of FAAH, with the associated increased tolerance to NAE due to a higher capacity to metabolize NAE 12:0, does not contribute to the sensitivity of these seedlings to ABA. These results emphasize that FAAH is a regulator of NAE responses by its catalytic activity and functions in ABA responses through other mechanisms, perhaps via protein-protein interactions.
Although the ABI3-mediated signaling pathway appears to play a major role in NAE-induced growth arrest at lower levels of NAE, NAE 12:0 at higher concentrations induced seedling growth arrest through an ABA-independent mechanism since growth of abi3-1 seedlings was inhibited at higher concentrations of NAE (Fig. 5A and B). If NAE 12:0 was capable of inducing growth arrest only through the ABA signaling pathway, the abi3-1 seedlings would be expected to grow normally in exogenous NAE. Instead, abi3-1 seedlings showed a large reduction in growth (Fig. 5A and B). The observed growth reduction in abi3-1 seedlings treated with NAE also was associated with elevated levels of the ABA-regulated gene RD29B (Fig. 5C), despite the absence of the ABI3 transcription factor. RD29B transcript was induced by both NAE and ABA without ABI3, but once again only within the developmental window. Outside this temporal window, only ABA induced elevated transcripts of RD29B; also, RD29B transcript levels were much lower than wildtype suggesting that the ABI3 transcription factor contributes, at least in part, to RD29B transcript levels. RD29B is a general drought response gene that is strongly regulated by the ABI3 transcription factor,21 but can be activated independent of ABA through MAPK mechanisms.26 The abi3-1 growth inhibition suggests that NAE is capable of inhibiting seedling growth by mechanisms independent of the normal ABA response pathway, and the activation of RD29B expression may be part of this pathway.
In summary, NAE can induce growth arrest through both ABA signaling pathway-dependent and -independent mechanisms. The strong reduction in growth in seedlings treated with both NAE 12:0 and ABA suggests that NAEs, which are relatively minor lipids in Arabidopsis, may play a more significant role in early seedling development than currently appreciated. In addition, this interaction with an important phytohormone, ABA, illustrates the complex connections between components in plant development which still need to be identified and characterized. Overall, identifying the mechanism of NAE-induced growth arrest could help shed light on other general mechanisms of regulating seedling growth in the early stages of development.
Materials and Methods
Plant materials and growth assays.
The ABI mutant abi3-1 (CS24) and their corresponding parental ecotypes Landsberg erecta (Ler-0) and Columbia (Col) were obtained from the Arabidopsis Biological Resource Center (ABRC). The faah T-DNA insertion mutant (Salk_095108) and the 35S::AtFAAH overexpressor lines were generated as described previously in reference 12. For growth experiments, seeds were surface-sterilized with 95% ethanol followed by 30% bleach with 0.1% Tween-20 for 3 minutes each and rinsed several times with sterile, deionized water. Seeds were stratified for 3 days at 4°C in the dark and grown in either liquid nutrient media (0.5x Murashige and Skoog salts containing 1% sucrose) or solid nutrient media (0.5x MS salts with 1% sucrose and 0.8% phytagel) as previously described in reference 12. Germination and growth occurred in a controlled environment room with a 16 h light (∼60 µmolm−2s−1), 8 h dark cycle at 20 to 22°C. Liquid cultured seedlings were incubated with shaking (∼75 rpm) and growth was quantified as fresh weight accumulation after collection with a Buchner funnel and rinsing three times with deionized water to remove exogenous NAE and ABA from the seedlings. ABA and/or NAEs were added from concentrated stocks, dissolved in DMSO, to the appropriate final concentrations after media was allowed to cool to 50°C. Untreated controls contained equivalent final concentrations of DMSO solvent alone, ranging from 0.01 to 0.05%. Concentrations of exogenous ABA (Calbiochem-Novabiochem Corp., catalogue #100111) were calculated based on the active cis-isomer, although the ABA used includes an equimolar concentration of the inactive trans-isomer. N-lauroylethanolamine (NAE 12:0) was synthesized as previously described in reference 27. NAE and ABA were both added to media at only one time point in each experiment and were not continuously replaced. To determine if the combined effect of NAE and ABA was additive, synergistic or antagonistic, the expected response from a combination of NAE and ABA was calculated.24 Growth parameters for each treatment were converted to percent of control values. The expected values for an additive response is calculated by the equation Expected % of control = (% of control in treatment A × % of control in treatment B)/100. If the actual percent of control for the combined treatment is lower than the expected value, the combination is synergistic; if higher, the combination is antagonistic.
Semi-quantitative RT-PCR.
Total RNA was isolated from seed and seedling samples using the Qiagen RNeasy Plant Mini Kit (Qiagen, catalogue #74904). RNA was quantified and evaluated for purity by UV spectroscopy and agarose gel electrophoresis.28 Changes in mRNA transcripts were analyzed by semi-quantitative RT-PCR performed with a Smart Cycler II (Cepheid) instrument using a real-time one-step assay system (Takara Bio). The following gene-specific primer pairs were used: ABI3 (At3g24650) (F) 5′-GAG CTG GCT CAG CTT CTG CTA TG-3′ and (R) 5′-AGG CCA AAA CCT GTA GCG CAT GTT C-3′, AtHVA22B (At5g62490) (F) 5′-CAT CGC TGG ACC TGC ATT AC-3′ and (R) 5′-GGA TAT AAT GGG ATC CAT TCG AGG-3′, and RD29B (At5g62490) (F) 5′-CAT AAA GGT GGA GAA GCT GGA GTA-3′ and (R) 5′-CCT CCA AAT CTT GCC GGA GAA TTC-3′. All primers were designed to span one intron to distinguish cDNA amplification from genomic DNA contamination. Relative transcript levels in all samples were normalized using 18S rRNA as a constitutively expressed internal control, with primers (F) 5′-TCC TAG TAA GCG CGA GTC ATC A-3′ and (R) 5′-CGA ACA CTT CAC CGG ATC AT-3′.29 Semi-quantitative RT-PCR reactions were performed in duplicate with 0.2 µg of total RNA and 0.5 µL of 10 µM gene-specific primers in each 25 µL reaction. The reaction mix was subjected to the following RT-PCR conditions: 42°C for 15 min, one cycle; 95°C for 2 minutes, one cycle; 94°C for 10 s, 58°C for 25 s, 72°C for 20 s. The number of cycles and annealing temperature were experimentally determined for each set of gene-specific primers. RT-PCR products were examined by gel electrophoresis.
Acknowledgements
This work was supported by a grant from the United States Department of Energy, Office of Basic Energy Sciences (BES, grant number DE-FG02-05ER15647).
Abbreviations
- NAE
N-Acylethanolamine
- NAE 12:0
N-lauroylethanolamine
- ABA
abscisic acid
- FAAH
fatty acid amide hydrolase
- ABI
ABA insensitive
References
- 1.Kuehl FA, Jacob TA, Ganley OH, Ormond RE, Meisinger MAP. The identification of N-(2-hydroxyelthyl)-pal, itamide as a naturallly occurring anti-inflammatory agent. J Am Chem Soc. 1957;79:5577–5578. [Google Scholar]
- 2.Schmid HH, Schmid PC, Natarajan V. The N-acylation-phosphodiesterase pathway and cell signalling. Chem Phys Lipids. 1996;80:133–142. doi: 10.1016/0009-3084(96)02554-6. [DOI] [PubMed] [Google Scholar]
- 3.Chapman KD. Occurrence, metabolism and prospective functions of N-acylethanolamines in plants. Prog Lipid Res. 2004;43:302–327. doi: 10.1016/j.plipres.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 4.De Petrocellis L, Cascio MG, Di Marzo V. The endocannabinoid system: a general view and latest additions. Br J Pharmacol. 2004;141:765–774. doi: 10.1038/sj.bjp.0705666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman KD. Emerging physiological roles for N-acylphosphatidylethanolamine metabolism in plants: signal transduction and membrane protection. Chem Phys Lipids. 2000;108:221–229. doi: 10.1016/s0009-3084(00)00198-5. [DOI] [PubMed] [Google Scholar]
- 6.Kilaru A, Blancaflor FB, Venables BJ, Tripathy S, Mysore KS, Chapman KD. The N-acylethanolamine-mediated regulatory pathway in plants. Chem Biodivers. 2007;4:1933–1955. doi: 10.1002/cbdv.200790161. [DOI] [PubMed] [Google Scholar]
- 7.Blancaflor EB, Hou G, Chapman KD. Elevated levels of N-lauroylethanolamine, an endogenous constituent of desiccated seeds, disrupt normal root development in Arabidopsis thaliana seedlings. Planta. 2003;217:206–217. doi: 10.1007/s00425-003-0985-8. [DOI] [PubMed] [Google Scholar]
- 8.Chapman KD, Venables B, Markovic R, Blair R, Jr, Bettinger C. N-Acylethanolamines in seeds. Quantification of molecular species and their degradation upon imbibition. Plant Physiol. 1999;120:1157–1164. doi: 10.1104/pp.120.4.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 10.Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, et al. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci USA. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrestha R, Noordermeer MA, van der Stelt M, Veldink GA, Chapman KD. N-Acylethanolamines are metabolized by lipoxygenase and amidohydrolase in competing pathways during cottonseed imbibition. Plant Physiol. 2002;130:391–401. doi: 10.1104/pp.004689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YS, Shrestha R, Kilaru A, Wiant W, Venables BJ, Chapman KD, et al. Manipulation of Arabidopsis fatty acid amide hydrolase expression modifies plant growth and sensitivity to N-acylethanolamines. Proc Natl Acad Sci USA. 2006;103:12197–12202. doi: 10.1073/pnas.0603571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teaster ND, Motes CM, Tang Y, Wiant WC, Cotter MQ, Wang YS, et al. N-Acylethanolamine metabolism interacts with abscisic acid signaling in Arabidopsis thaliana seedlings. Plant Cell. 2007;19:2454–2469. doi: 10.1105/tpc.106.048702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Physiol Plant Mol Biol. 2009;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 15.Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:15. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- 17.Bright J, Desikan R, Hancock JT, Weir IS, Neill SJ. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006;45:113–122. doi: 10.1111/j.1365-313X.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- 18.Chai MF, Wei PC, Chen QJ, An R, Chen J, Yang S, et al. NADK3, a novel cytoplasmic source of NADPH, is required under conditions of oxidative stress and modulates abscisic acid responses in Arabidopsis. Plant J. 2006;47:665–674. doi: 10.1111/j.1365-313X.2006.02816.x. [DOI] [PubMed] [Google Scholar]
- 19.Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature (London) 2003;423:651–654. doi: 10.1038/nature01643. [DOI] [PubMed] [Google Scholar]
- 20.Rock CD. Tansley Review No. 120. Pathways to abscisic acid-regulated gene expression. New Phytol. 2000;148:357–396. doi: 10.1046/j.1469-8137.2000.00769.x. [DOI] [PubMed] [Google Scholar]
- 21.Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, et al. Transcriptional regulation of ABI3-and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos and seedlings of Arabidopsis. Plant Mol Biol. 2006;60:51–68. doi: 10.1007/s11103-005-2418-5. [DOI] [PubMed] [Google Scholar]
- 22.Kim SC, Chapman KD, Blancaflor EB. Fatty acid amide lipid mediators in plants. Plant Science (Oxford) 2010;178:411–419. [Google Scholar]
- 23.Kim SC, Kang L, Nagaraj S, Blancaflor EB, Mysore KS, Chapman KD. Mutations in Arabidopsis fatty acid amide hydrolase reveal that catalytic activity influences growth but not sensitivity to abscisic acid or pathogens. J Biol Chem. 2009;284:34065–34074. doi: 10.1074/jbc.M109.059022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colby SR. Calculating synergistic and antagonistic responses of herbicide combinations. Weeds. 1967;15:20–22. [Google Scholar]
- 25.Motes CM, Pechter P, Yoo CM, Wang YS, Chapman KD, Blancaflor EB. Differential effects of two phospholipase D inhibitors, 1-butanol and N-acylethanolamine, on in vivo cytoskeletal organization and Arabidopsis seedling growth. Protoplasma. 2005;226:109. doi: 10.1007/s00709-005-0124-4. [DOI] [PubMed] [Google Scholar]
- 26.Hua ZM, Yang X, Fromm ME. Activation of the NaCl- and drought-induced RD29A and RD29B promoters by constitutively active Arabidopsis MAPKK or MAPK proteins. Plant Cell Environ. 2006;29:1761–1770. doi: 10.1111/j.1365-3040.2006.01552.x. [DOI] [PubMed] [Google Scholar]
- 27.Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 28.Mehra M. RNA isolation from cells and tissues. Wiley-Liss, Inc., Wiley-Liss, Ltd; 1996. [Google Scholar]
- 29.Rider S, Jr, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J. Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 2003;35:33–43. doi: 10.1046/j.1365-313x.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]