Abstract
Advanced transcriptome analyses have revealed the existence of various RNA species. In our previous study, a large number of non-protein-coding RNAs including antisense transcripts were identified using an Arabidopsis tiling array. Most of the antisense transcripts exhibited co-expression with sense transcripts during stress treatments or seed imbibition. Here, we report that antisense transcripts exhibit differential expression patterns to sense transcripts in distinct developmental tissues. In addition, RNA ligase-mediated RACE analysis identified the existence of 5′-capped and -uncapped antisense transcripts. These observations provide additional insight into antisense transcripts.
Key words: antisense transcript, non-protein-coding RNA, tiling array, Arabidopsis
Natural antisense transcripts exist widely in a variety of organisms, from animals to plants. Many of them are categorized as non-protein-coding RNA (ncRNAs). Generally, it is thought that natural antisense transcripts negatively regulate gene expression on the opposite strand. In addition, artificial overexpression of a complementary sequence against a protein-coding gene causes a decrease in its transcript levels. However, recent large-scale transcriptome analysis revealed that the expressions of endogenous sense and antisense transcripts were positively correlated in both mammals and plants.1–6 Conversely, inverse correlation of sense and antisense transcript expression has also been reported in other genome-wide studies.7,8 Therefore, it is likely that the expression of antisense transcripts is controlled in a more complex manner than has been thought. In this article, we focus on the developmental expression pattern and 5′-terminal structure of antisense transcripts that are expressed in Arabidopsis seeds.
Differential Expression Patterns of Sense and Antisense Transcripts in Distinct Tissues
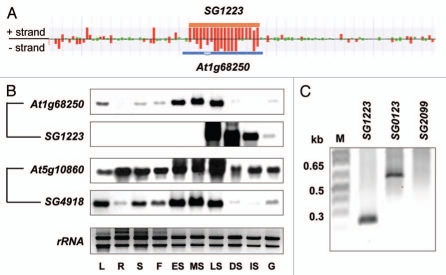
Transcriptome analysis using a genome tiling array covering the entire genomic sequence from both sense and antisense strands is an excellent techniques for finding natural antisense transcripts.3,9 For example, an antisense transcript (SG1223) highly expressed during seed imbibition was detected on the antisense strand to At1g68250, which encodes an unknown protein (Fig. 1A). In our previous report, we found that the expression patterns At1g68250 and SG1223 were similar during seed imbibition.5 We used RNA blot analysis to determine whether co-expression of sense and antisense transcripts was also observed in distinct developmental tissues. SG1223 expression was mainly observed from late maturation siliques to early germination stages (Fig. 1B). In contrast, At1g68250 was predominantly expressed during early to late maturation siliques, and its expression pattern was different from SG1223. This different expression pattern was also observed for the pair At5g10860/SG4918. Relative expression levels of At5g10860, which encodes a cystathionine beta-synthase, did not correlate with those of SG4918 in leaves and roots (Fig. 1B). These results indicated that expression of sense and antisense transcripts might be subject to complex control in a development- and environment-dependent manner.
Figure 1.
Expression patterns and 5′-terminal structures of antisense transcripts. (A) Detection of antisense transcripts by tiling arrays. SG1223 was detected as an antisense transcript against At1g68250. Red bars indicate a signal intensity >400 (in endosperm at 24 h after seed imbibition). (B) RNA blot analysis of sense and antisense transcripts in various tissues. Pairs of At1g68250/SG1223 and At5g10860/SG4918 are sense/antisense transcripts. Total RNAs were prepared from L, leaves; R, roots; S, stems; F, flowers; ES, early siliques; MS, middle siliques; LS, late siliques; DS, dry seeds; IS, 24-h-imbibed seeds and G, 48-h-imbibed seeds. A total of 40 µg RNA was loaded into each lane. Expression was detected using strand-specific RNA probes. Ethidium bromide-stained rRNA is shown as a loading control. (C) Detection of capped antisense transcripts by 5′-RACE PCR. RNA was subjected to dephosphorylation and decapping reactions and an RNA adapter was ligated to the 5′ end of the RNA. After an RT reaction by using an oligo(dT) primer, PCR was performed using adapter and antisense-specific primers.
Capped and Uncapped Antisense Transcripts
Most of the protein-coding mRNAs have a cap structure and a poly(A) tail, which have important roles in RNA stability, RNA processing and protein synthesis. In our study, a large number of novel ncRNAs, including antisense transcripts, probably had poly(A) tails, because we used the oligo(dT) primer for first-strand cDNA synthesis. Indeed, several stress- or abscisic acid-inducible antisense transcripts contained short poly(A) stretches at the 3′-terminus of the transcript.3 Interestingly, antisense transcripts for RD29A and CYP707A1 did not have a cap structure at their 5′-terminus.3 To reveal whether antisense transcripts identified in seeds also do not have a cap structure, we performed 5′-RACE PCR using a GeneRacer™ Kit, which is an RNA ligase-mediated RACE method. A 5′-RACE product was detected for SG1223 and SG0123, but not for SG2099, indicating that there are capped and uncapped products among the antisense transcripts (Fig. 1C). In fact, SG1223 is isolated as a full-length cDNA (RAFL09-44-D21). The functions of the fully overlapping natural antisense transcripts are unknown in most cases, and our knowledge of these is still limited in plants. Recently, however, it was reported that the cold-induced FLC antisense transcripts, COOLAIR, is involved in FLC silencing via chromatin modification during vernalization.10 Interestingly, COOLAIR has both a cap and a poly (A) tail, and is categorized as a long ncRNA (>100 nucleotide). On the other hand, many long ncRNAs tend to be degraded by the nonsense-mediated mRNA decay pathway, suggesting that the majority of ncRNAs are less stable than protein-coding RNA.11 Therefore, many ncRNAs might not have been isolated yet as full-length cDNAs.
Comprehensive transcriptome analysis via large-scale RNA sequencing and high-density tiling array identified a large number of ncRNAs. Next-generation sequencing technology will probably uncover more ncRNAs than expected. However, it is the most important to identify functional ncRNAs involved in plant development and environmental stress responses. For example, artificial microRNA technology against specific strands will be useful to examine the functions of antisense transcripts. In addition to antisense transcripts, it is reported that several long intergenic ncRNAs are involved in differentiation and stress response in Arabidopsis.12 Hence, together with small RNAs, further research on long ncRNAs will be necessary to understand plant growth, development and stress responses, as well as their effect on protein-coding genes.
Acknowledgements
This work was supported by the Special Postdoctoral Researcher's Program from RIKEN and the research fellowship from the Japan Society for the Promotion of Science for Young Scientists to (M.O.), and a grant from RIKEN Plant Science Center to (M.S.).
References
- 1.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 2.Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, et al. Complex loci in human and mouse genomes. Plos Genet. 2006;2:564–577. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 4.Oeder S, Mages J, Flicek P, Lang R. Uncovering information on expression of natural antisense transcripts in Affymetrix MOE430 datasets. BMC Genomics. 2007;8:200. doi: 10.1186/1471-2164-8-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamoto M, Tatematsu K, Matsui A, Morosawa T, Ishida J, Tanaka M, et al. Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J. 2010;62:39–51. doi: 10.1111/j.1365-313X.2010.04135.x. [DOI] [PubMed] [Google Scholar]
- 6.Grinchuk OV, Jenjaroenpun P, Orlov YL, Zhou JT, Kuznetsov VA. Integrative analysis of the human cis-antisense gene pairs, miRNAs and their transcription regulation patterns. Nucleic Acids Res. 2010;38:534–547. doi: 10.1093/nar/gkp954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen JJ, Sun M, Hurst LD, Carmichael GG, Rowley JD. Human antisense genes have unusually short introns: evidence for selection for rapid transcription. Trends Genet. 2005;21:203–207. doi: 10.1016/j.tig.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Jin H, Vacic V, Girke T, Lonardi S, Zhu JK. Small RNAs and the regulation of cis-natural antisense transcripts in Arabidopsis. BMC Mol Biol. 2008;9:6. doi: 10.1186/1471-2199-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada K, Lim J, Dale JM, Chen HM, Shinn P, Palm CJ, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 10.Swiezewski S, Liu FQ, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 11.Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, et al. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:2453–2458. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben Amor B, Wirth S, Merchan F, Laporte P, d'Aubenton-Carafa Y, Hirsch J, et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009;19:57–69. doi: 10.1101/gr.080275.108. [DOI] [PMC free article] [PubMed] [Google Scholar]