Abstract
Background
Understanding the diversity of interactions between RNA aptamers and nucleotide cofactors promises both to facilitate the design of new RNA enzymes that utilize these cofactors and to constrain models of RNA World evolution. In previous work, we isolated six pools of high affinity RNA aptamers to coenzyme A (CoA), the principle cofactor in biological acyltransfer reactions. Interpretation of the evolutionary significance of those results was made difficult by the fact that the affinity resin attachment strongly influenced the outcome of those selections. Here we describe the selection of four new pools isolated on a disulfide-linked CoA affinity matrix to minimize context-dependent recognition imposed by the attachment to the solid support.
Results
The four new aptamer libraries show no sequence or structural relation to a previously dominant CoA-binding species, even though they were isolated from the same initial random libraries. Recognition appears to be limited to the adenosine portion of the CoA – in particular the Höogsteen edge – for most isolates surveyed, even when a counter selection was employed to remove such RNAs. Two of the recovered isolates are eluted with intact CoA more efficiently than with AMP alone suggesting a possible pantotheine interaction. However, a detailed examination of recognition specificity revealed that the 3' phosphate of CoA, and not the pantotheine arm, determined recognition by these two isolates.
Conclusion
Most aptamers that have been targeted towards cofactors containing adenosine recognize only the adenosine portion of the cofactor. They do not distinguish substituents on the 5' carbon, even when those substituents have offered hydrogen bonding opportunities and the selection conditions discouraged adenosine recognition. Beyond hydrogen bonding, additional factors that guide the selection towards adenosine recognition include aromatic stacking interactions and relatively few rotational degrees of freedom. In the present work, a sterically accessible, disulfide-linked CoA affinity resin afforded the selection of a more diverse aptamer collection then previous work with a N6 linked CoA resin.
Keywords: CoA aptamers, Nucleotide cofactors, affinity chromatography matrix, SELEX, Acyltransferase ribozyme
Background
RNA aptamers offer unique opportunities for studying host-guest relations between macromolecules and their small molecule binding partners, while simultaneously providing starting points for the development of new RNA enzymes for important physiological reactions. Dozens of small-molecule targets have yielded aptamers with a wide range of specificities and affinities, including nucleotide phosphates and cofactors, amino acids, carbohydrates, antibiotics, and transition state analogs. Among nucleotide cofactors, the adenine moiety has proven to be a particularly potent target for eliciting aptamers. Most RNA [1] and ssDNA [2] aptamers selected to bind ATP or other adenosine cofactors bind their targets through the adenosine without discriminating among analogs with substituents at the 5' position. For example, selections targeted to nicotinamide adenine dinucleotide (NAD+) [3] and S-adenosyl methionine (SAM) [4] yielded the same RNA motif found in previous ATP selections [1,5]. This evolutionary convergence arises from three factors: i) similarities in the design of each experiment (all targets were immobilized through the C8 position of the base), ii) the solvent-exposed location of the C8 and 5' positions in the bound complex, as revealed in the solution structure of the aptamer [6,7] (allowing bulky substitution without affecting binding interactions), and iii) the relatively high frequency of this simple motif in populations of random sequence (at least one in 1011 [8]).
Coenzyme A (CoA) is the predominant cofactor in biological acyl-transfer reactions. The sulfhydryl at the end of its unique pantotheine arm allows for the formation of high energy thioester bonds and serves as the leaving group during acyltransfer reactions. Jadhav and Yarus isolated an RNA species that can form such thioesters from pre-activated acyl-adenylate substrates when the CoA portion is covalently tethered to the RNA chain [9]. No ribozymes have yet been described that generate or utilize thioesters involving free coenzyme A. CoA-binding RNA aptamers were previously isolated from two independent RNA libraries [8]. The dominant structural motif from one pool ("70A", for 70 random nucleotides, selected to bind CoA) binds ATP in solution with a dissociation constant, Kd, of approximately 1 μM. Binding is achieved primarily through the Hoögsteen face of the adenine, and is independent of the ribose and pantotheine arm of CoA. Another CoA-binding pool, designated "80A" [80 random nucleotides), was selected at the same time, using different primer-binding sequences than the 70A pool. For both the 70A and 80A pools, nearly all of the RNA was removed from the CoA affinity resin with AMP alone, suggesting that those CoA aptamers interact exclusively with the AMP portion of CoA, and probably only with the adenine or adenosine [8]. The CoA aptamers also bind ATP, but they differ markedly from the previous "ATP aptamers" in sequence, secondary structures, dependence on pH and metal ions, and the recognition specificity among adenosine-containing analogs [8].
The choice of attachment between a cofactor target and its solid support can heavily influence the outcome of a SELEX experiment. The previously identified ATP and CoA aptamers recognize ATP in solution with similar affinities, but neither binds to the affinity resin used to isolate the other [8]. During the original CoA selection, CoA was immobilized to the affinity matrix through an amide linkage to its exocyclic N6 (Figure 1B), while during the original ATP, NAD+ and SAM selections, the cofactors were immobilized through an alkyl chain to the C8 positions of the base (Figure 1C) [1,3,4]. We reasoned that these selections may have artificially eliminated desirable binding structures because of steric constraints imposed by the solid support.
Figure 1.
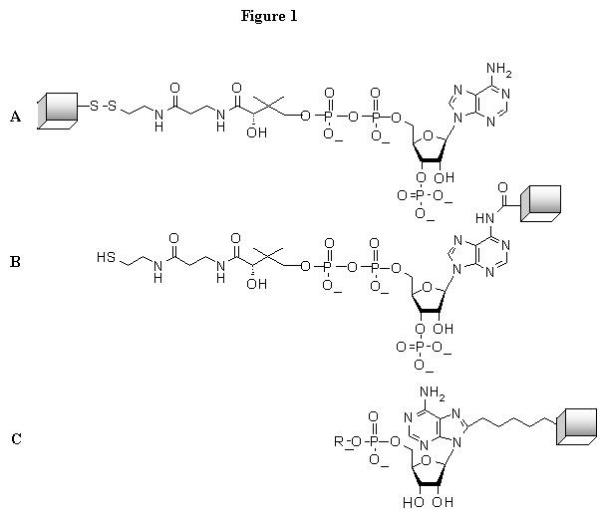
Affinity matrices used in isolation of aptamers to adenosine cofactors. A) CoA attached to sepharose solid support through its pantotheine arm (disufide-linked CoA resin); B) CoA attached to sepharose through its primary amine at N6 (Amide-linked CoA resin); C) Other adenosine cofactors attached through the C8 of adenine. ATP, R = PPi; AMP, R = H, NAD+, R = nicotinamide mononucleotide.
To obtain a population of CoA-binding aptamers that was less constrained by experimental design, a new affinity matrix was synthesized in which the CoA was immobilized through a disulfide linkage to the sulfhydryl on the pantotheine arm (Figure 1A). We reasoned that removing steric constraints on the adenosine could facilitate the isolation of a more diverse collection of CoA aptamers, and that this diversity could increase the probability of identifying CoA-dependent acyltransfer ribozymes in future experiments. This resin simultaneously offers the opportunity to seek aptamers that contact the pantotheine arm. Pantotheine carries numerous hydrogen bonding donors and acceptors that could potentially interact with RNA, and might thus be thought to be an attractive target for putative RNA aptamers. We find that while the requirements for recognition for most aptamers are fully satisfied with adenine alone, a few aptamers additionally require the 3' phosphate found in CoA, but none is shown unambiguously to make contacts with the pantotheine.
Results
Selection of SA and PSA pools
To evaluate the effect of affinity resin linkage on the selection of CoA aptamers and to obtain new libraries of CoA-binding RNA aptamers, CoA was immobilized on an activated thiopropyl sepharose matrix through a disulfide bond to its unique sulfur. The two RNA pools used for the selection, containing either 70 or 80 random nucleotides to allow distinction by size (total transcript sizes 118 and 134 nucleotides, respectively), carry different primer binding sequences to prevent excessive bias due to experimenter-imposed sequence constraints. After seven cycles of selection using affinity elution, 35% to 45% of the input RNA was retained on the resin and could be eluted with free CoA (Figure 2). The 70SA and 80SA pools (70 or 80 random nucleotides targeted to S-linked CoA) were cloned for sequencing following the seventh cycle of selection.
Figure 2.
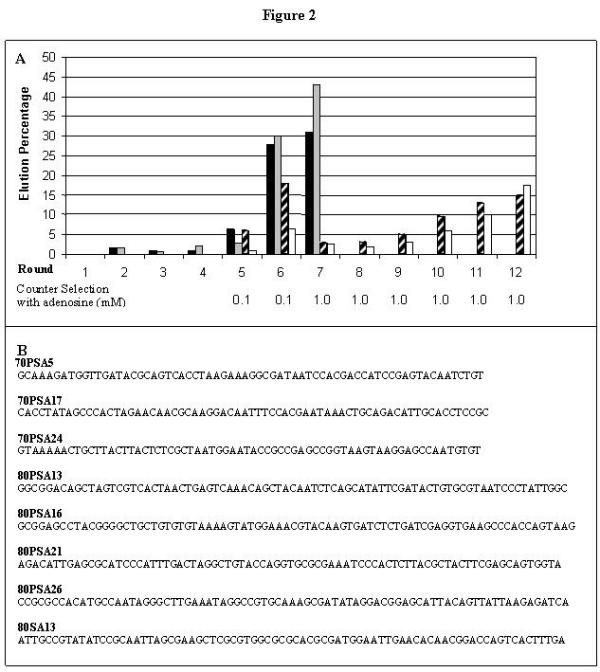
Enrichment of aptamers to CoA. A) Black and gray columns represent the enrichment of 70SA and 80SA pools, respectively. White and striped columns bars represent the enrichment of 70PSA and 80PSA pools, respectively. Adenosine concentrations used during counter-selections for PSA pools are given beneath the graph. B) Sequences of aptamers that are used in experiments described in figure 3 and 4. Only the initially random portions are shown. Not shown are the 5' and 3' primer-binding sequences (see methods). Complete sequences of all aptamers from these selections are given.
The 70SA and 80SA selections were split into two additional parallel selection pathways with increased stringency. During selection cycles 5 and 6, 0.1 mM adenosine counter-selection was included during the washing steps. This is expected to discourage adenosine-specific aptamers with fast dissociation rates, and simultaneously to encourage aptamers that recognize pantotheine. For rounds 7 through 12, counter-selection stringency was increased by raising the adenosine concentration to 1.0 mM. These two selected populations comprise the 70PSA and 80PSA sets (70 or 80 random nucleotides targeted to the Pantotheine arm of S-linked CoA). During the twelfth cycle of selection under these conditions, 14–18% of input RNA withstood the adenosine washes and was eluted with CoA (Figure 2A). The material eluting during round 12 was converted to dsDNA and cloned for sequencing.
A total of 96 individual isolates were sequenced from the 70SA, 80SA, 70PSA and 80PSA aptamer pools. The complete sequences of the eight of these aptamers are given in figure 2B; all sequences are available at http://aptos.chem.indiana.edu/dhburke/all_seqs.html. Forty-five sequences were each encountered only once, 20 as pairs of identical sequences, 15 as triples, 10 in two groups of five identical sequences, and 6 as a single group of identical sequences. No shared sequence motifs were evident within or between the populations, and no sequence was found in more than one of the four selections. Two sequences from the 80SA pool matched those of aptamers identified in the original 80A selection [8] (80SA5 = 80A#68; 80SA13 = 80A#41). No sequence suggested similarity with other 80A sequences, with the dominant 70A sequence [8], or other ATP aptamers identified previously [1-5]. An initial survey of binding activities using radiolabeled RNAs identified 18 isolates that bound and eluted from the CoA resin in the absence of any counterselection, with 20 to 75% of the input RNA eluting with CoA. A sample of some of these elution results are shown in Figure 3A (gray bars).
Figure 3.
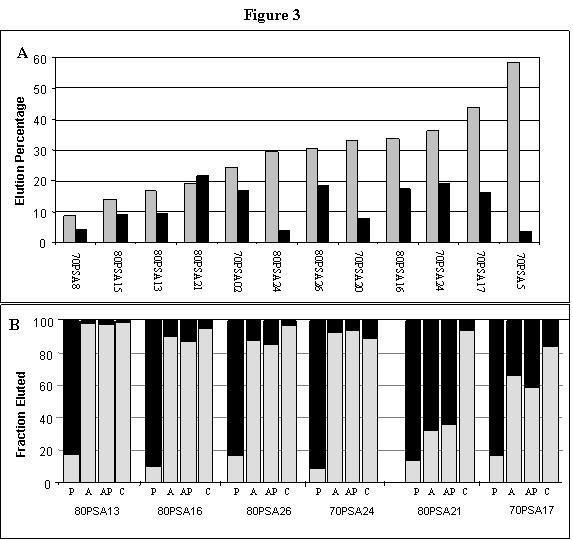
Elution specificities of selected aptamers. A) The indicated aptamers were loaded onto the disulfide-linked CoA affinity resin. The percentage of input RNA eluted with 5 mM CoA are plotted on the graph. Prior to the elution, non-specifically-bound RNAs were removed with 10 column volumes of binding buffer (gray bars) or with 10 volumes of 1 mM adenosine (black bars). B) The aptamers indicated below each set of columns were bound to the disulfide-linked CoA resin, then washed with five volumes each of binding buffer, then 1 mM analog (gray bars), and finally of 5 mM CoA (black bars). Analogs were either pantotheine alone (P), AMP alone (A), AMP and pantotheine together (AP), or CoA (C). Percent elution is normalized to total RNA eluted in five "analog" fractions plus five "CoA" washes for ease of comparison. Under the conditions used in this assay, all of the aptamers gave approximately 50% total elution.
Specificity of individual 70PSA and 80PSA aptamers
Two experiments were performed to determine the elution specificity for several aptamers from the two PSA populations. The first experiment evaluated the effect of the adenosine-wash on the elution percentages of twelve PSA aptamers. Radiolabeled RNAs were loaded onto disulfide-linked CoA affinity resins and assayed under the same conditions as those used during the PSA selection (washed with 10 column volumes of 1 mM adenosine and then eluted with 6 volumes of 5 mM CoA). In most cases, the adenosine wash substantially reduced the amount of RNA available for elution by CoA (Figure 3A, solid bars). Isolate 80PSA21 eluted equivalently with CoA irrespective of whether an adenosine wash was included, suggesting that this aptamer's interaction with adenosine and its interaction with intact CoA are not equivalent.
The second experiment employed a variation on a specificity assay that we have used previously [5,8,12] involving analog affinity elution. First, the lowest CoA concentrations that effectively eluted aptamers from the disulfide-linked resin was determined to be 1 mM. Then PSA aptamers were loaded onto the disulfide-linked CoA resin, and eluted with a 1 mM solution of structural analog of CoA (AMP, pantotheine, AMP + pantotheine, or CoA itself). Any RNA remaining after the analog elution was stripped from the resin by washing with 5 volumes of 5 mM CoA. Of the total eluted RNA, the fraction that is eluted by the analog is an indication of the extent to which the analog contains the elements recognized by the aptamer in the intact CoA. Most of the aptamers were efficiently eluted with 1 mM AMP, while pantotheine did not appreciably elute any of the RNAs. (Figure 3B). Elution was not significantly stimulated when both AMP and pantotheine were used together as analogs. Thus, most RNAs from the PSA populations require nothing beyond the AMP portion of CoA for recognition. The exceptions to this rule were 80PSA21 and to a lesser degree 70PSA17 for which complete elutions were only possible with intact CoA. Either these aptamers contact some portion of the pantotheine arm of CoA or some other difference between AMP and CoA is responsible for the observed specificity.
To differentiate between these two possibilities for the CoA-specific PSA aptamers, and to refine the specificity definitions of two SA aptamers that eluted with 1 mM AMP alone, radiolabeled RNA from isolates 80PSA21, 70PSA17, 70PSA5 and 80SA13 were immobilized on the CoA resin and challenged with 15 different CoA analogs. In each case, more than 90% of the total eluted RNA was removed from the resin by 1 mM CoA as the analog, and less then 10 percent was removed by elution with purine as the analog. Aptamers 70PSA5 and 80SA13 were also eluted efficiently with adenine alone. As adenine differs from purine only in containing an exocyclic N6 amine, this moiety is shown to be essential for the binding of these two aptamers to CoA. These results were further confirmed by challenging these RNAs with N6 methyl adenine, which removed less then 10 percent of the total eluted RNA. The N7 position is also important for recognition, as N7-deaza-adenosine failed to elute these two aptamers. Sugar modifications had no effect on the elution of these two aptamers, nor did N3 methylation of adenine. From these results we conclude that 70PSA5 and 80SA13 recognize CoA predominantly through the Höogsteen face of adenine.
Aptamers 80PSA21 and 70PSA17 gave strikingly different elution patterns from those of 70PSA5 and 80SA13, although they are similar to each other. In general for 70PSA17, the distinction between recognition and non-recognition of the analogs is less clear than for 80PSA21. For both aptamers, 3'-AMP and 3',5'-cyclic AMP removed most of the RNA from the resin. N7-deaza adenosine and N6-methyl adenosine failed to elute these RNAs from the CoA resin. Other analogs gave intermediate elution values. For 80PSA21, the partial elution by adenosine was notably greater than that of the other reagents. Recognition by both of these two CoA aptamers thus appears to be dominated by interactions with both the Höogsteen face of adenine and the 3' phosphate. Thus, for the aptamers that appeared in figure 3 to have a strong preference for intact CoA, it is actually the 3' phosphate that is essential for recognition.
Relative preferences of 70A and 70SA aptamers for disulfide-linked vs. amide-linked CoA resins
The 70A [8], 70SA, and 70PSA pools are all derived from the same initial 70N random pool [10]. The 70A and 70SA pools were selected under nearly identical conditions, albeit utilizing different CoA affinity resins. To evaluate the effects of each resin on the two sets of selected RNAs, representative RNAs from each pool were passed over both the disulfide-linked and amide-linked CoA resins. The 70SA pool showed a strong preference for the sterically accessible disulfide-linked CoA. Seventy percent of the total RNA eluted from the disulfide-linked CoA resin, while only twenty percent eluted from the amide-linked CoA resin. In contrast, aptamer CoA-min52, a 52-nucleotide core minimal aptamer representing the dominant motif from the original 70A pool [8], does not evidence a strong preference for either resin, yielding approximately 30% retention in each case (Table 1). Thus, steric inaccessibility of the target ligand may have limited the repertoire of previous selections, while the greater accessibility afforded by the disulfide-linked resin permitted selection of diverse population of high-affinity CoA aptamers.
Table 1.
Effect of CoA linkage on fitness for selection for disulfide-linked versus amide-linked CoA resins (compare structures in figure 1).
| RNA | Percent of input RNA eluted from... | |
| Amide linked CoA resin | Disulfide linked CoA resin | |
| 70CoA-min52 | 34% | 28% |
| 70SA selected pool | 20% | 70% |
Transcripts of aptamer 70CoAmin-52 and of the 70SA pool (500 pmole) were loaded onto each of the indicated CoA resins, washed with 10 column volumes of binding buffer and eluted with 5 mM CoA.
Discussion and Conclusion
Four new pools of RNA aptamers that recognize coenzyme A and various adenosine derivatives are described. The 70SA and 80SA pools were selected under standard conditions, although the use of a disulfide-linked CoA profoundly affected the SELEX outcome relative to previous selections. Selection of the 70PSA and 80PSA pools included counterselection against simple adenosine recognition. Previous selections for CoA aptamers have also included counterselections [8], but those utilized 3'AMP rather than adenosine as the counterselection agent. The great majority of aptamers in all four of the pools described in the present work recognize AMP as readily as they recognize CoA, judging from their abilities to bind the CoA affinity resin and to be eluted by adenosine or AMP rather than CoA. A small fraction of the 70PSA and 80PSA resist elution by adenosine, eluting efficiently only when provided with intact CoA. This specificity appears to result from a requirement for a phosphate at the 3' position. To our knowledge, these aptamers are the first to be characterized in which the presence of a phosphate on the nucleotide is required for maximal binding. Previously, aptamers to cAMP were selected [13]. These RNAs eluted from a cAMP affinity resin in the presence of adenosine or cAMP, but not AMP or other analogs having large groups on the 2', 3', or 5' positions. Thus, while they discriminated among various phosphorylated nucleotides, they did not require the phosphate for maximal binding.
The tyranny of adenosine
Recognition of adenosine – or of adenine alone – dominates all four pools from the current selections, as it has dominated previous selections for aptamers to ATP [1], NAD+ [3], S-adenosyl methionine [4], and CoA [8]. In principle, the 5' substituents in these other cofactors could have been recognized by aptamers – such as positive charges in SAM and NAD+, hydrogen bond-rich environments in the pantotheine arm of CoA and in the nicotinamide of NAD+, and the planar aromatic nicotinamide ring of NAD+. Nevertheless, the aptamers from those selections recognize only adenosine. Adenosine therefore must be a more attractive target for aptamer evolution than any of the other components in these cofactors. Adenosine offers many binding opportunities: i) hydrogen bonding to its Watson-Crick edge, to its major and minor groove edges, and to its ribose hydroxyls, ii) aromatic stacking interaction potential on each face of the base, and iii) relatively few rotational degrees of freedom. While some aptamers from previous selections have shown specificity for the Watson-Crick edge [1], the Höogsteen edge of adenine dominates the selections involving CoA, even when the resin attachment is far removed from the nucleoside moiety.
Adenosine has also been a major determinant of substrate recognition in some ribozymes selected to act on adenine-containing substrates. Illangasekare and Yarus described the first ribozymes capable of transferring an amino acid from an aminoacyl adenylate (Phe-AMP) onto their own 3' ends [14]. Free AMP is a competitive inhibitor of the reaction, but Phe is not [15], indicating that recognition of the Phe-AMP occurs exclusively through the AMP portion. Jenne and Famulok observed a similar inhibition by AMP in an independently isolated ribozyme for acyltransfer from biotin-AMP to an internal 2'OH of the RNA [16], again suggesting that substrate recognition occurs primarily through the adenosine.
The tyrannical influence of adenosine is not ubiquitous, however, as illustrated by several ribozyme and aptamer selections. First, Wiegand et al. selected ribozymes that catalyze synthesis of an amide bond. In these reactions, a primary amine tethered to the RNA condenses with biotinyl adenylate (bio-AMP), displacing the AMP [17]. Free AMP did not inhibit the reaction even when present at 500-fold molar excess over bio-AMP, and bio-UMP was accepted by the ribozyme as a substrate with nearly identical Km and kcat as compared to bio-AMP [17]. Both lines of evidence confirm that the adenosine portion of bio-AMP is not required for this binding interaction. Second, Li and Breaker identified self-phosphorylating deoxyribozymes that transfer the gamma phosphate from a mixture of all four NTPs onto their own 5' ends. The initial winners were mostly specific for GTP rather than for ATP [18]. Guanosine has one additional hydrogen bonding partner on its Watson-Crick face, which may reduce the structural and hence evolutionary requirements for making a G-binding pocket. This would increase the number of ribozyme sequences that utilize GTP relative to those that utilize ATP, thereby accounting for the observed preference. Third, a selection for aptamers to flavin adenine dinucleotide (FAD) produced aptamers with specificity only for the flavin moiety and no discernable adenosine requirement [12]. Apparently when the random pool had access to both the adenine and flavin rings, the larger flavin ring competed more effectively for attention from the RNA. Based on the specificities of aptamers selected to the various nucleotide cofactors, it is tempting to rank their relative attractiveness to incipient aptamer pools according to the number of aromatic rings they carry: flavin (3 rings) > adenine (2 rings) > nicotinamides (1 rings), with a further ranking based on hydrogen bonding potential: guanosine (3 hydrogen bonds on Watson-Crick face) > adenosine (2 bonds).
The presence of adenosine in so many of the common biological cofactors has been postulated to reflect an evolutionary origin for modern metabolism in an RNA World. The adenosine cofactors (such as ATP, CoA, FAD, NAD+, SAM, adenosyl cobalamine [coenzyme B12]) are used in many classes of chemical reactions, including acyl, methyl, and phosphoryl group transfer, redox reactions, and radical-mediated rearrangements. Other nucleotides are used in more restricted classes of reactions. Adenine, which is readily synthesized under abiotic conditions, has been proposed to have arisen in these cofactors as a shared handle with which enzymes (or ribozymes) could interact through interchangeable structural modules. Such modules are seen in protein enzymes in the form of the Walker A and B motifs for ATPases and in the Rossman fold for binding dinucleotide cofactors, although it should be pointed out that in those cases conservation is much stronger among residues that build the framework for cofactor binding than among the residues that contact the cofactor directly. The utility of cofactor-binding modules in the design and evolution of new enzymes – or the lack of such utility – has yet to be established.
Removing constraints imposed by resin attachment
The primary difference between the present 70SA and 80SA selections and the previous 70A and 80A selections for CoA aptamers is the nature of the covalent linkage of the CoA to the solid support. The original 70A pool was dominated by a single structural motif, represented by isolate CoA-min52, which binds ATP in solution with a Kd value around 1.0 μM [8]. Because the structure of CoA-min52 is partially derived from nucleotides in the 5' and 3' primer binding sites, its core motif is estimated to have been present at a frequency of 6 × 10-8 in the initial random 70N pool [8]. We therefore expected this motif to dominate the 70SA pool as well, yet it was not encountered among 25 isolates sequenced from the 70SA pool. This can be understood by examining the relative retention on the two affinity resins by each library.
When both RNAs are assayed on the amide-linked CoA resin, the CoAmin52 RNA displayed 1.7-fold greater elution than the selected 70SA pool (34% vs. 20%, Table 1). Thus, the greater initial abundance of the motif in CoAmin52 and its unique adaptation to the amide-linked resin both contributed to its dominance in the previous selection [8]. In contrast, 2.5-fold more RNA from the 70SA pool is eluted from the less-constrained disulfide-linked resin than the amount eluted when CoAmin52 RNA is applied to this resin (70% vs. 28%, Table 1). The amount of input RNA eluted from the affinity matrix – and therefore the fraction of the RNA that survives the selection cycle – defines adaptive fitness for this in vitro selection. The 2.5-fold difference in survival in each selection cycle is expected to have propagated throughout the seven rounds of the 70SA selection, producing a differential amplification of 2.57 = 610-fold for the 70SA pool vs. the CoAmin52 element over the course of the selection. Multiplying 25 (number of 70SA isolates sequenced without encountering the CoAmin52 element) times 6 × 10-8 (frequency of previously dominant motif in the shared starting 70N random pool) divided by 610 (inherent 2.57-fold advantage enjoyed by the isolates recovered at the end of 7 cycles of the 70SA selection) gives an estimate of the frequency of unique, proficient CoA-binding structures in the initial random library to be at least 2.5 × 10-9. The differential fitness therefore erases the numerical advantage of the CoA-min52 motif and explains the absence of this motif from the 70SA and PSA pools.
The motif in CoA-min52, then, represents the simplest structure capable of binding CoA in the context of the amide linkage, but not in the less-constrained context of the disulfide linkage. In the previous selection, other, more proficient CoA aptamers must have been unintentionally eliminated due to the steric constraints of the resin attachment. The disulfide-linked CoA affinity resin places the adenine base of CoA as far as possible from the immobilization matrix, and this appears to have greatly stimulated both the number of isolates obtained and their binding abilities.
The principle is likely to be general, that a broader spectrum of high-affinity ligands can be isolated using affinity matrices that are sterically less constrained than from those that are more constrained. By extension, our results suggest that steric inaccessibility may have also limited the repertoire of previous selections, such as those that have yielded the well-characterized "ATP aptamer" through the use of C8-linked adenosine nucleotide targets [1,3-5]. Linkages through the base or sugar preclude direct binding to these locations; therefore, fewer tertiary structures will be capable of binding to the target. This constraint likely contributed to the emergence of a single consensus sequence in each of these selections. In contrast, the greater accessibility afforded by the disulfide-linked resin used in this work permitted the selection of diverse high-affinity CoA aptamers. Sterically unconstrained adenosyl resins could be similarly constructed using dephosphoCoA or γ-thio-ATP, β-thio-ADP, etc. Peptides coupled to solid supports through their termini or through long, flexible linkers may also be superior SELEX targets as compared to those coupled through an interior position.
Methods
Affinity resin
Activated thiopropyl sepharose (2.46 g, approximately 50 micromoles of coupling sites) (Sigma) swelled to 12 ml upon hydration in 50 mM Bis-Tris buffer, pH 5.8. CoA was added to a final concentration of 15 mM (approximately 3-fold molar excess above resin capacity) and the slurry was incubated at room temperature for one hour on a rotating rack. Formation of the desired disulfide bond linking CoA to the resin was monitored spectroscopically in three ways: first, by observing the displacement of 2-thiopyridone from the resin into solution and monitoring absorbance at 345 nm; second, by observing the decrease in CoA signal at 260 nm in the supernatant following the coupling reaction; and finally, by washing the coupled resin extensively with buffer and then reducing a small aliquot with dithiothreitol to release the bound CoA (260 nm). CoA concentration in various resin preparations is calculated to be 3 to 7 mM.
Selection and affinity elution screens
The 70N and 80N initial random pools were the same as those we have used in previous RNA selections [4,5,8,10-12]. Selections and enzymatic manipulations of nucleic acids were performed as in previous selections for CoA aptamers [8] except where noted. In the first cycle of the selection, 5 nanomoles of gel-purified RNA (10–25 copies/sequence) was loaded in 1 mL binding buffer (50 mM Bis-Tris pH 6.4, 200 mM NaCl, 10 mM MgCl2) onto 250 μl bed volume of resin in 4 × 250 μl fractions, followed by 6 × 250 μl washes with 1× binding buffer and 6 × 250 μl elutions with 5 mM CoA in 1 × binding buffer supplemented with 5 mM additional MgCl2. Input RNA was reduced to 1 nanomole in the second round of selection and 0.1 nanomole for all subsequent rounds. All rounds after the first used 150 μl resin bed volume, 9 × 150 μl washes and 6 × 150 μl elutions. For analytical surveys shown in figure 3 and table 1, elutions were performed using 6 × 200 μl. The fraction of uniformly radiolabeled RNA eluting in each fraction was monitored by Cerenkov counting both during the selection and in post-selection screens for activity. Assays of the activity and specificity of individual aptamers were performed essentially as described previously [8], with modifications noted in the text.
Author's Contribution
DS gathered the data and performed most of the experimental analyses. JF did sequencing and the initial activity screening, DHB performed the selection. DS and DHB wrote the manuscript. All authors have read and approved the manuscript.
Figure 4.
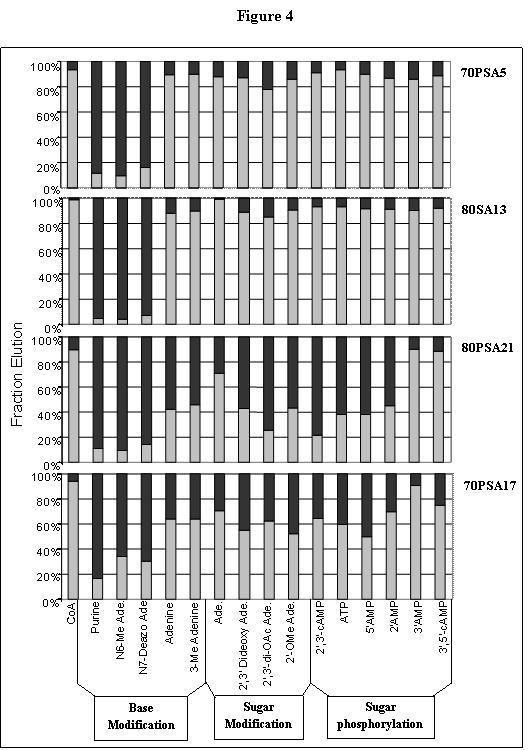
Refining the recognition specificity of four aptamers. Experimental design is as described in Figure 3. Analogs used in this assay are listed blow in the figure; Ade, adenosine; OAc, acetyl. The designation of each aptamer is indicated to the right of the corresponding panel.
Acknowledgments
Acknowledgement
We thank Amrish Malhi for technical assistance early in this work. Dr. David Nickens, Dr. Sanchita Hati, Daniel Held, and Steven Rhee offered critical comments on the manuscript. This material is based upon work supported by the National Science Foundation under Grant MCB 9896363 and by a Young Investigator Award from the Arnold and Mabel Beckman Foundation.
Contributor Information
Dayal Saran, Email: dsaran@indiana.edu.
Joseph Frank, Email: josephwfrank@hotmail.com.
Donald H Burke, Email: dhburke@indiana.edu.
References
- Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–3. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–65. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- Burgstaller P, Famulok M. Isolation of RNA aptamers for Biological Cofactors by In Vitro Selection. Angew Chem Int Ed Engl. 1994;33:1084–1087. doi: 10.1002/anie.199410841. [DOI] [Google Scholar]
- Burke DH, Gold L. RNA aptamers to the adenosine moiety of S-adenosyl methionine: structural inferences from variations on a theme and the reproducibility of SELEX. Nucleic Acids Res. 1997;25:2020–4. doi: 10.1093/nar/25.10.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lato SM, Ozerova ND, He K, Sergueeva Z, Shaw BR, Burke DH. Boron-containing aptamers to ATP. Nucleic Acids Res. 2002;30:1401–7. doi: 10.1093/nar/30.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann T, Suzuki E, Nakamura GK, Feigon J. Solution structure of an ATP-binding RNA aptamer reveals a novel fold. RNA. 1996;2:628–40. [PMC free article] [PubMed] [Google Scholar]
- Patel DJ. Structural analysis of nucleic acid aptamers. Curr Opin Chem Biol. 1997;1:32–46. doi: 10.1016/S1367-5931(97)80106-8. [DOI] [PubMed] [Google Scholar]
- Burke DH, Hoffman DC. A novel acidophilic RNA motif that recognizes coenzyme A. Biochemistry. 1998;37:4653–63. doi: 10.1021/bi972877p. [DOI] [PubMed] [Google Scholar]
- Jadhav VR, Yarus M. Acyl-CoAs from coenzyme ribozymes. Biochemistry. 2002;41:723–9. doi: 10.1021/bi011803h. [DOI] [PubMed] [Google Scholar]
- Burke DH, Scates L, Andrews K, L G. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J Mol Biol. 1996;264:650–66. doi: 10.1006/jmbi.1996.0667. [DOI] [PubMed] [Google Scholar]
- Burke DH, Hoffman DC, Brown A, Hansen M, Pardi A, Gold L. RNA aptamers to the peptidyl transferase inhibitor chloramphenicol. Chem Biol. 1997;4:833–43. doi: 10.1016/S1074-5521(97)90116-2. [DOI] [PubMed] [Google Scholar]
- Roychowdhury-Saha M, Lato SM, Shank ED, Burke DH. Flavin recognition by an RNA aptamer targeted toward FAD. Biochemistry. 2002;41:2492–9. doi: 10.1021/bi015719d. [DOI] [PubMed] [Google Scholar]
- Koizumi M, Breaker RR. Molecular recognition of cAMP by an RNA aptamer. Biochemisty. 2000;39:983–92. doi: 10.1021/bi000149n. [DOI] [PubMed] [Google Scholar]
- Illangasekare M, Sanchez G, Nickles T, Yarus M. Aminoacyl-RNA synthesis catalyzed by an RNA. Science. 1995;267:643–7. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]
- Illangasekare M, Yarus M. Small-molecule-substrate interactions with a self-aminoacylating ribozyme. J Mol Biol. 1997;268:631–9. doi: 10.1006/jmbi.1997.0988. [DOI] [PubMed] [Google Scholar]
- Jenne A, Famulok M. A novel ribozyme with ester transferase activity. Chem Biol. 1998;5:23–24. doi: 10.1016/S1074-5521(98)90084-9. [DOI] [PubMed] [Google Scholar]
- Wiegand T, Janssen R, Eaton B. Selection of RNA amide synthases. Chem Biol. 1997;4:675–83. doi: 10.1016/S1074-5521(97)90223-4. [DOI] [PubMed] [Google Scholar]
- Li Y, Breaker R. Phosphorylating DNA with DNA. Proc Natl Acad Sci U S A. 1999;96:2746–51. doi: 10.1073/pnas.96.6.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]


