Abstract
The formation of root hairs is a unique developmental process that requires the concerted action of a multitude of proteins. Root hair development is controlled by intrinsic programs, but fine-tuning of these programs occurs in response to environmental signals, dictating the shape and function of epidermal cells. In particular, low availability of soil-immobile mineral nutrients such as phosphate (Pi) affects the density and length of root hairs, resulting in an increased absorptive surface area. We recently reported on a time-course transcriptional profiling study aimed at identifying gene networks that signal Pi deficiency and mediate adaptation to Pi shortage. Using root-specific coexpression analysis of early Pi-responsive genes, we identified a subset of novel loci crucial for the development of root hairs under Pi-deficient conditions. Remodeling of cell wall structures may be associated with the TOR (Target of Rapamycin) pathway, a highly conserved central regulator of growth and development in eukaryotic cells that senses nutrient availability.
Key words: phosphate deficiency, coexpression networks, cell walls, TOR pathway, root hairs
Plant cell walls are dynamic structures that play important roles in defense responses, in intercellular communication and in determining cell shape. Growth and differentiation requires controlled loosening and remodeling of polysaccharide networks, resulting in complex restructuring during development and in response to environmental signals. For example, under Pi-deficient conditions longitudinal elongation of epidermal cells is reduced while tip growth is enhanced in cells that develop into root hairs.
In a previous transcriptional profiling study on roots that have been subjected to short-term deficiency treatment, we found that several genes encoding hydrolytic enzymes that mediate the rearrangement of polysaccharides such as pectate lyases, polygalacturonases and pectin methylesterases were downregulated early after transfer of the plants to media deprived of Pi. Pectins, in adition to hemicelluloses, are the third major component of plant cell walls and contribute to the mechanical stability of the walls. Three arabinogalactan proteins (AGPs) genes were found to be coexpressed with this group of genes encoding hydrolytic enzymes and also showed decreased transcript abundance. AGPs are a diverse class of macromolecules belonging to the hydroxyproline-rich glycoprotein superfamily. AGPs were shown to be involved in growth, signaling and development; although, for most of the AGPs a precise role in these processes remains to be determined.1,2 AGPs are transiently attached to the plasma membrane by a GPI-anchor and released into the periplasm after cleavage of the anchor by a phospholipase.
AGPs can be trapped by pectins. Alterations in pectin composition lead to changes in the mechanical properties, but also to changes in the interactions with other cell wall components. The downregulation of pectate lyases during Pi deficiency increases cell wall porosity.3 Thus, Pi-deficient plants may possess a more loosely cross-linked pectic matrix, supporting the migration of AGPs. In our study, homozygous agp14 mutants showed altered root hair elongation under both control and Pi-deficient conditions, indicating that AGP14 is a crucial component of the machinery involved in rearranging cell walls during root hair formation.
The TOR Pathway Connects Stress, Growth and Cell Wall Remodeling
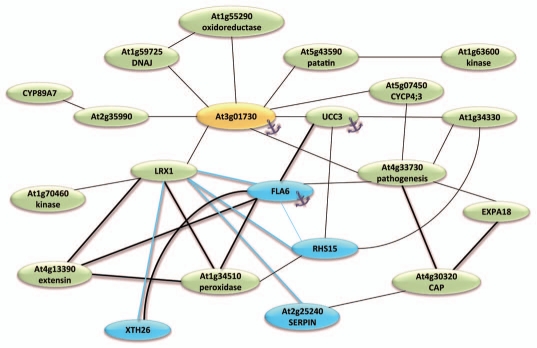
Little is known about the processes that regulate cell wall remodeling. One of the few components identified is the chimeric LRR-extensin 1 (LRX1). LRX1 is predominantly expressed in root hairs, probably functioning in the establishment of the extracellular matrix as a signaling intermediate.4,5 lrx1 mutants form aberrant root hairs that often swell or branch.4 Under Pi deficient conditions, lrx1 mutant plants form fewer root hairs than wild-type roots, but the number of root hairs does not deviate from the wild-type when grown on Pi-replete media. This indicates that the functional LRX1 protein is required for the response to Pi starvation.6 LRX1 is closely coexpressed with XTH26, FLA6, RHS15, the serine-type endopeptidase SERPIN and the peroxidase gene At1g34510 (Fig. 1). All genes showed reduced expression in Pi-deficient per1 plants, a mutant that has been isolated in a genetic screen for plants that display inhibited root hair elongation phenotype specifically upon Pi deficiency.7 A suppressor mutant of lrx1, rol1 (repressor of lrx1) was shown to code for a protein involved in the biosynthesis of rhamnose, a major component of pectin.8 Interestingly, one gene from our newly indentified loci in Pi deficiency-induced root hair formation, At3g01730, was overexpressed in rol1 plants. The root morphology of rol1 plants resemble that of Pi-deficient plants with regard to reduced length of primary roots, trichoblasts and root hairs, providing a further connection of LRX1 with Pi deficiency signaling.
Figure 1.
Coexpression network around At3g0170. Blue nodes represent genes that are less expressed in Pi-deficient per1 roots when compared with the wild-type. Anchor symbols denote genes encoding GPI-anchored proteins. The gene network was determined with ATTED-II version 5.5 (atted.jp/).
Another suppressor of the lrx1 phenotype named rol5 showed decreased cell elongation associated with reduced LM5 immunolabeling, indicative of reduced pectin and increased LM2 labeling, indicative of altered AGP levels. A functional homolog of ROL5 in yeast, Ncs6p, affects the Target of Rapamycin (TOR) pathway, which was shown to be involved in cell wall integrity sensing in yeast.9 In yeast and mammals, TOR is thought to link the energy status of the cell and nutrient availability to translation efficiency and growth. The TOR pathway also exists in plants;10 mutations in the central component, the Ser/Thr kinase TOR, are lethal, but only some components of the pathway have been identified in plants so far. Downregulation of a putative upstream component of TOR in Arabidopsis, the translationally controlled tumor protein (TCTP), compromises pollen tube growth and root hair development,11 implying a potential role for TOR in tip growth. The TOR pathway is thus a prime candidate for adapting growth related cell wall restructuring processes with cell division rates and energy status that are altered by Pi deficiency.
Final Remarks
Integration of external signals into developmental programs requires the concerted action of an array of proteins involved in sensing, responding and adapting to the prevailing environmental conditions. The case of cell wall remodeling during growth under Pi deficient conditions vividly shows the complexity of these processes. A plethora of genes is differentially expressed upon Pi starvation many of which have functional redundancies. Coexpression analysis is a valuable tool to gain a more integrative view of these processes. Other regulatory levels however, both up-stream and down-steam of transcription, contribute to the phenotype and need to be addressed experimentally to complete the picture.
References
- 1.Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annu Rev Plant Biol. 2007;58:137–161. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- 2.Ellis M, Egelund J, Schultz CJ, Bacic A. Arabinogalactan-proteins: Key regulators at the cell surface? Plant Physiol. 2010;153:403–419. doi: 10.1104/pp.110.156000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron-Epel O, Hernandez D, Jiang LW, Meiners S, Schindler M. Dynamic continuity of cytoplasmic and membrane compartments between plant cells. J Cell Biol. 1988;106:715–721. doi: 10.1083/jcb.106.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumberger N, Ringli C, Keller B. The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 2001;15:1128–1139. doi: 10.1101/gad.200201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ringli C. Monitoring the outside: cell wall-sensing mechanisms. Plant Physiol. 2010;153:1445–1452. doi: 10.1104/pp.110.154518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller M, Schmidt W. Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol. 2004;134:409–419. doi: 10.1104/pp.103.029066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li WF, Perry PJ, Prafulla NN, Schmidt W. Ubiquitin-specific protease 14 (UBP14) is involved in root responses to phosphate deficiency in Arabidopsis. Mol Plant. 2010;3:212–223. doi: 10.1093/mp/ssp086. [DOI] [PubMed] [Google Scholar]
- 8.Diet A, Link B, Seifert GJ, Schellenberg B, Wagner U, Pauly M, et al. The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell. 2006;18:1630–1641. doi: 10.1105/tpc.105.038653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahfouz MM, Kim S, Delauney AJ, Verma DP. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkowitz O, Jost R, Pollmann S, Masle J. Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell. 2008;20:3430–3447. doi: 10.1105/tpc.108.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]