Abstract
Bienertia sinuspersici is one of only three higher land plant species known to perform C4 photosynthesis without Kranz anatomy through partitioning of photosynthetic functions between dimorphic chloroplasts in a single photosynthetic cell. We recently reported the successful separation of the two chloroplast types and biochemical and functional analyses revealed differences in protein composition and specialization of photosynthetic functions. In Kranz type C4 species, spatial (or cell-specific) control of transcription of nuclear genes contributes to development of dimorphic chloroplasts, but obviously this cannot be involved in formation of dimorphic chloroplasts within individual photosynthetic cells. Therefore, we address here the question of how nuclear encoded proteins could be selectively targeted to plastids within a cell to form two types of chloroplasts. We discuss current knowledge of chloroplast differentiation in single cell C4 species and present three hypothetical mechanisms for how this could occur.
Key words: chloroplast import, chloroplast proteases, chloroplast targeting, Kranz C4 photosynthesis, mRNA targeting, plastid development, single cell C4 photosynthesis
Higher plants typically contain different plastid types with diverse morphology and function in different tissues. Amyloplasts, chromoplasts, leucoplasts and chloroplasts, to name a few, all originate and develop from the same proplastids during ontogenesis. Plastids are considered semi-autonomous organelles having their own genome and being capable of replication independent from their host cell cycle. Nevertheless, the vast majority of proteins needed for functional plastids are encoded by the nuclear genome and must be imported into the plastids. How proplastids differentiate into the final functional plastids is not yet fully understood, but there is evidence that plastid fate is determined at least in part by the differentiation of the host cell type.1 Therefore, different tissues transcribe genes for different sets of proteins which, in turn, are imported in the respective plastid type for proper function.
C4 plants are an interesting system to study plastid development, since protochloroplasts differentiate into two morphologically and functionally different chloroplast types in mesophyll (M) and bundle sheath (BS) cells to support the C4 carbon concentrating mechanism. In this system the C4 cycle delivers CO2 to Rubisco for assimilation in the C3 cycle. All the enzymes and metabolite transporters for the process are nuclear encoded, with the exception of the large subunit of Rubisco where the gene is chloroplastic. In the C4 plant maize, it has been shown in a series of large scale proteomics experiments that a vast number of nuclear encoded proteins accumulate preferentially either in M or BS chloroplasts.2–4 Considerable effort is being made to understand the nature of this selective protein accumulation. It is a useful system for studying regulation of cellular differentiation in plants. Discovery of mechanisms controlling the process may provide the knowledge needed for engineering the C4 carbon concentrating mechanisms into major C3 crops to improve yield, and water and nitrogen use efficiency.5,6
For most C4 photosynthetic genes, cell-specific regulation of transcript abundance of chloroplast targeted proteins is considered to have a significant role in determining chloroplast differentiation. Yet, no common motif or master regulator has been found, and regulation of individual genes does not appear to be conserved across different species. For example, it has been clearly established that posttranscriptional and posttranslational mechanisms are involved in M and BS specific chloroplast differentiation.7–9
Bienertia sinuspersici is a recently discovered species with a unique form of C4 photosynthesis. In this single cell C4 species (SCC4), the carbon concentrating mechanism does not depend on cooperation between M and BS cells, as in Kranz type C4 species. Rather, it possesses a unique chlorenchyma with two functional and biochemical different chloroplast types within photosynthetic cells (Fig. 1A). Here, the so called peripheral chloroplasts (P-CP) are spatially separated by a large vacuole from chloroplasts clustered in a central compartment (C-CP). This structural arrangement allows for enrichment of CO2 in the Rubisco containing C-CP, ultimately repressing photorespiration, similar to the mechanism in Kranz type C4 plants.10 During leaf development very young leaves transition from a C3 default mode with one type of chloroplast to form two cytoplasmic domains with two types of chloroplasts which then complete differentiation for C4 function.11
Figure 1.
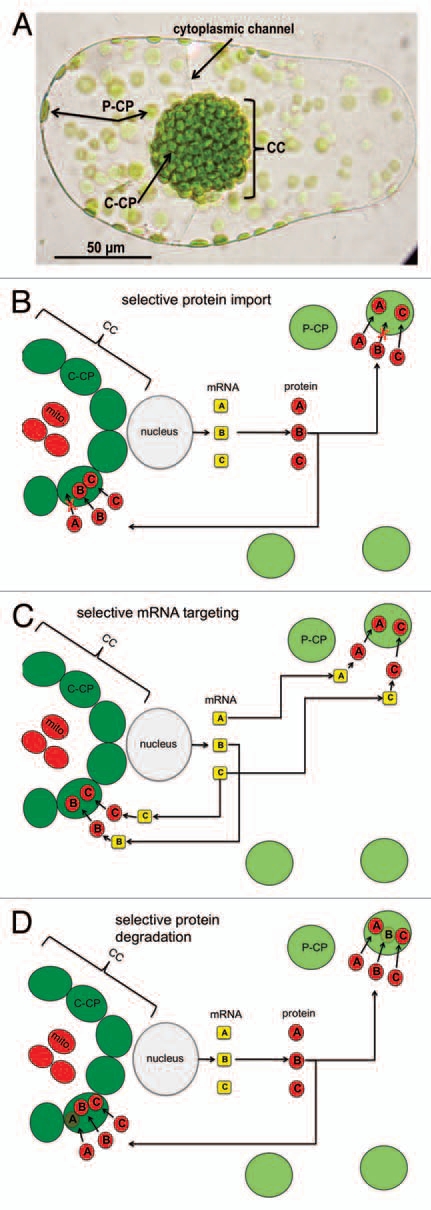
Hypothetical mechanisms of chloroplast differentiation in SCC4 species. Part (A) Micrograph of a mature B. sinuspersici chlorenchyma cell showing peripheral chloroplasts (P-CP), the central cytoplasmic compartment (CC) and the chloroplasts of the central cytoplasmic compartment (C-CP). The two compartments are connected via cytoplasmic channels. (B–D) Models for selective accumulation of proteins making functionally different chloroplasts. Protein A specifically localizes to the P-CP, whereas B localizes only to the C-CP, protein C localizes to both chloroplast types. Capital letters in yellow boxes correspond to mRNAs and letters in red boxes to proteins. mito, mitochondria.
We recently developed a protocol for isolation and purification of intact dimorphic chloroplasts from B. sinuspersici. Biochemical and functional characterization showed these chloroplasts operate analogous to M and BS chloroplasts in Kranz type C4 species with respect to carbon fixation.12 Several proteins were found to accumulate selectively in the C-CP [e.g., the nuclear encoded small subunit of Rubisco (RBCS) and the chloroplast encoded large subunit of Rubisco (RBCL)] and in the P-CP [e.g., pyruvate, Pi dikinase (PPDK) and the putative pyruvate transporter (PyT1)]. In addition, preliminary results from two dimensional differential gel electrophoreses (2D-DIGE), as well as from quantitative shotgun proteomic analysis of the two chloroplast types revealed that large numbers of proteins accumulate preferentially in the C-CP or the P-CP, similar to the situation previously reported for the M and BS chloroplasts of maize (Offermann S, Doroshenk K, Okita T, Friso R, Wijk Kv and Edwards G, unpublished data).
Obviously in SCC4 species, control of protein composition and therefore functional specialization of the two chloroplast types can not be achieved by cell-specific transcriptional control, as is observed in Kranz type C4 species. Consequently, in the following section we discuss alternative molecular mechanisms, which may be operating in SCC4 species to allow for development of two distinctive chloroplast types within a single photosynthetic cell.
Hypothesis 1: Selective Protein Import
The first hypothesis is plastid centric, i.e., that chloroplasts have the ability to selectively import the required proteins for differential chloroplast function (Fig. 1B). In this model, certain proteins (e.g., RBCS, required for Rubisco and C3 cycle function) are imported only into the C-CP. In contrast, some others which are required to support the carboxylation phase of the C4 cycle (e.g., PPDK) are imported only into the P-CP. Proteins which are required to support certain functions in both chloroplast types (e.g., photosystems) are not subject to selective import. This model requires a specific recognition process between the preprotein and the chloroplast import apparatus. Preproteins must therefore encode information about their different destinations either in the transit peptide or in an internal targeting site, and the chloroplast import apparatus must decode this information upon import. Evidence suggests that chloroplasts are, in principle, capable of forming selective TIC-TOC complexes, as has been shown for the differential import pathways of highly expressed photosynthetic proteins and low abundance housekeeping proteins through specialized TIC-TOC complexes.13–15 Recently, it has been shown that, in Arabidopsis, multiple sequence motifs in the transit peptide of RBCS contribute independently to selective import through specialized TIC-TOC complexes, and there is emerging evidence that redox regulation might be involved in regulating the formation of specific TIC-TOC complexes as well.16,17 In this context, it is tempting to speculate that preproteins sharing the same chloroplast type destination in SCC4 species would likely share a common motif in their transit peptide, or elsewhere in the protein structure, which would allow for an efficient recognition system. We are currently working towards sequencing the B. sinuspersici transcriptome, which in combination with our proteomics approach, should enable us to address the question of whether proteins selectively targeted to one chloroplast type share a common motif.
Hypothesis 2: Selective mRNA Targeting
In an alternative hypothesis, the required partitioning of proteins between the two chloroplast types could be accomplished through selective mRNA targeting rather than selective import of proteins into the chloroplasts. In this model, nuclear encoded mRNA accumulates on ribosomes in close proximity to the respective chloroplast types. After translation, the preproteins are imported and spatial separation of the two chloroplast types by the vacuole ensures the correct localization (Fig. 1C). Mechanistically, such selective mRNA localization can be achieved either by directional transport involving the cytoskeleton, selective mRNA trapping, or by degradation of mRNAs within distinct regions of the cell, and it has been reported that the signals directing these processes are mostly, but not exclusively, found within the 3′UTR of the mRNA.18
mRNA targeting has been shown to be involved in plant processes which require localized intracellular protein accumulation. In rice for example, mRNAs for different seed storage proteins are targeted to specific subdomains of the ER.19,20 Although it is generally assumed that nuclear encoded mRNAs for chloroplast targeted proteins are translated on randomly distributed cytosolic ribosomes,21,22 recent evidence suggests that targeting of RBCS and LHCII in algae involves localized translation of their mRNAs.23 Similarly, it has been shown that certain nuclear encoded mRNAs which code for mitochondrial proteins in potato are selectively targeted to the mitochondrial surface.24 In an extreme case, it has even been reported that a nuclear encoded mRNA directly enters the chloroplasts, thereby bypassing the canonical chloroplast protein import pathway completely.25 In a recent study, Lung et al. reported that the B. sinuspersici RBCS transit peptide is insufficient for correct localization of the protein exclusively to the C-CP when fused to GFP in a transient protoplast transformation system.26 We therefore speculate that important localization signals found either in the coding region of the protein and/or in the coding region and UTR of the corresponding mRNA that were not analyzed in that study might be important for correct localization of proteins in the SCC4 systems. The latter condition was observed for selective targeting of the rice prolamine RNA where cis-localization signals are distributed to the coding sequence and 3′UTR.19
Hypothesis 3: Selective Protein Degradation
Finally, a third hypothesis is that differentiation requires neither selective protein import nor differential localization of certain C4 photosynthesis transcripts, but rather a selective protein degradation mechanism operating within the two different chloroplast types. In this model, proteases specifically degrade certain proteins which are not required or which interfere with proper function of the respective chloroplast type (Fig. 1D). Although this pathway is certainly the most speculative, since very little is known about substrate specificity of chloroplast proteases,27 recent evidence suggests a highly specific protein degradation mechanism might be operating in the M chloroplasts of Kranz type C4 species. Here, the accumulation of RBCL has been shown to be partly under transcriptional control, but the observed differences were not sufficient to explain the exclusive accumulation of RBCL in BS chloroplasts. Since RBCL synthesis was detected in purified M cells, it was proposed that the RBCL protein must be specifically degraded in this chloroplast type.28 If chloroplast proteases are indeed involved in regulation of protein composition in the two chloroplast types of SCC4 species, an open question would certainly be how the proteases, or the factors referring their specificity, accumulate differentially in the two chloroplast types in the first place.
It is possible that two or more of the aforementioned mechanisms work in concert to achieve differentiation of chloroplasts with specialized functions in SCC4 species. This could occur by differential import of certain protein into the chloroplasts by selective expression of TIC-TOC complexes in hypothesis 1, differential targeting of mRNAs for functional proteins in hypothesis 2, and by differential expression of proteases in chloroplasts which would selectively degrade certain proteins in hypothesis 3.
In summary, there is little known in plant biology about development of functional domains and their integration into complex functions, as occurs in single cell C4 photosynthesis. This is an exquisite example of complex structural and functional polarity in a single cell. The exceptional development of dimorphic chloroplasts in a single cell provides opportunities for studying the mechanism of selectively expressing proteins from nuclear encoding genes in chloroplasts which are polarized to different cellular compartments. Future research could lead to the development of new models of plant cell organization, and structural and biochemical differentiation.
Acknowledgments
Supported by the National Science Foundation (grant no. IBN-0641232).
References
- 1.Lopez-Juez E. Plastid biogenesis, between light and shadows. J Exp Bot. 2007;58:11–26. doi: 10.1093/jxb/erl196. [DOI] [PubMed] [Google Scholar]
- 2.Friso G, Majeran W, Huang MS, Sun Q, van Wijk KJ. Reconstruction of metabolic pathways, protein expression and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: Large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 2010;152:1219–1250. doi: 10.1104/pp.109.152694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Majeran W, Cai Y, Sun Q, van Wijk KJ. Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell. 2005;17:3111–3140. doi: 10.1105/tpc.105.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majeran W, Zybailov B, Ytterberg AJ, Dunsmore J, Sun Q, van Wijk KJ. Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol Cell Proteomics. 2008;7:1609–1638. doi: 10.1074/mcp.M800016-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hibberd JM, Sheehy JE, Langdale JA. Using C4 photosynthesis to increase the yield of rice—rationale and feasibility. Curr Opin Plant Biol. 2008;11:228–231. doi: 10.1016/j.pbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Zhu XG, Long SP, Ort DR. Improving photosynthetic efficiency for greater yield. Annu Rev Plant Biol. 2010;61:235–261. doi: 10.1146/annurev-arplant-042809-112206. [DOI] [PubMed] [Google Scholar]
- 7.Berry JO, Patel M, Zielinski A. C4 gene expression in mesophyll and bundle sheath cells. In: Raghavendra AS, Sage RF,, editors. C4 Photosynthesis and related CO2 concentrating mechanisms. Advances in photosynthesis and respiration. Dordrecht, Netherlands: Springer; 2011. pp. 221–250. [Google Scholar]
- 8.Hibberd JM, Covshoff S. The regulation of gene expression required for C4 photosynthesis. Annu Rev Plant Biol. 2010;61:181–207. doi: 10.1146/annurev-arplant-042809-112238. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Peterson RB, Brutnell TP. Regulatory mechanisms underlying C4 photosynthesis. New Phytol. 2011;190:9–20. doi: 10.1111/j.1469-8137.2011.03649.x. [DOI] [PubMed] [Google Scholar]
- 10.Smith ME, Koteyeva NK, Voznesenskaya EV, Okita TW, Edwards GE. Photosynthetic features of non-Kranz type C4 versus Kranz type C4 and C3 species in subfamily Suaedoideae (Chenopodiaceae) Funct Plant Biol. 2009;36:770–782. doi: 10.1071/FP09120. [DOI] [PubMed] [Google Scholar]
- 11.Voznesenskaya EV, Koteyeva NK, Chuong SDX, Akhani H, Edwards GE, Franceschi VR. Differentiation of cellular and biochemical features of the single-cell C4 syndrome during leaf development in Bienertia cycloptera (Chenopodiaceae) Am J Bot. 2005;92:1784–1795. doi: 10.3732/ajb.92.11.1784. [DOI] [PubMed] [Google Scholar]
- 12.Offermann S, Okita TW, Edwards GE. Resolving the compartmentation and function of C4 photosynthesis in the single-cell C4 species Bienertia sinuspersici. Plant Physiol. 2011;155:1612–1628. doi: 10.1104/pp.110.170381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanova Y, Smith MD, Chen KH, Schnell DJ. Members of the Toc159 import receptor family represent distinct pathways for protein targeting to plastids. Mol Biol Cell. 2004;15:3379–3392. doi: 10.1091/mbc.E03-12-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubis S, Patel R, Combe J, Bedard J, Kovacheva S, Lilley K, et al. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell. 2004;16:2059–2077. doi: 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith MD, Rounds CM, Wang F, Chen KH, Afitlhile M, Schnell DJ. AtToc159 is a selective transit peptide receptor for the import of nucleus-encoded chloroplast proteins. J Cell Biol. 2004;165:323–334. doi: 10.1083/jcb.200311074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee DW, Lee S, Oh YJ, Hwang I. Multiple sequence motifs in the Rubisco small subunit transit peptide independently contribute to Toc159-dependent import of proteins into chloroplasts. Plant Physiol. 2009;151:129–141. doi: 10.1104/pp.109.140673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stengel A, Benz JP, Buchanan BB, Soll J, Bolter B. Preprotein import into chloroplasts via the Toc and Tic complexes is regulated by redox signals in Pisum sativum. Mol Plant. 2009;2:1181–1197. doi: 10.1093/mp/ssp043. [DOI] [PubMed] [Google Scholar]
- 18.Kloc M, Zearfoss NR, Etkin LD. Mechanisms of sub-cellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- 19.Choi SB, Wang CL, Muench DG, Ozawa K, Franceschi VR, Wu YJ, et al. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407:765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- 20.Doroshenk KA, Crofts AJ, Washida H, Satoh-Cruz M, Crofts N, Sugino A, et al. Characterization of the rice glup4 mutant suggests a role for the small GTPase Rab5 in the biosynthesis of carbon and nitrogen storage reserves in developing endosperm. Breeding Sci. 2010;60:556–567. [Google Scholar]
- 21.Carde JP, Joyard J, Douce R. Electron-microscopic studies of envelope membranes from spinach plastids. Biol Cell. 1982;44:315. [Google Scholar]
- 22.Chua NH, Schmidt GW. Transport of proteins into mitochondria and chloroplasts. J Cell Biol. 1979;81:461–483. doi: 10.1083/jcb.81.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uniacke J, Zerges W. Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc Nat Acad Sci USA. 2009;106:1439–1444. doi: 10.1073/pnas.0811268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaud M, Marechal-Drouard L, Duchene AM. RNA trafficking in plant cells: Targeting of cytosolic mRNAs to the mitochondrial surface. Plant Mol Biol. 2010;73:697–704. doi: 10.1007/s11103-010-9650-3. [DOI] [PubMed] [Google Scholar]
- 25.Nicolai M, Duprat A, Sormani R, Rodriguez C, Roncato MA, Rolland N, et al. Higher plant chloroplasts import the mRNA coding for the eucaryotic translation initiation factor 4E. Febs Lett. 2007;581:3921–3926. doi: 10.1016/j.febslet.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 26.Lung SC, Yanagisawa M, Chuong SD. Protoplast isolation and transient gene expression in the single-cell C4 species, Bienertia sinuspersici. Plant Cell Rep. 2011;30:473–484. doi: 10.1007/s00299-010-0953-2. [DOI] [PubMed] [Google Scholar]
- 27.Kato Y, Sakamoto W. New Insights into the types and function of proteases in plastids. Int Rev Cell Mol Biol. 2010;280:185–218. doi: 10.1016/S1937-6448(10)80004-8. [DOI] [PubMed] [Google Scholar]
- 28.Oswald A, Streubel M, Ljungberg U, Hermans J, Eskins K, Westhoff P. Differential biogenesis of photosystem-II in mesophyll and bundle-sheath cells of malic enzyme NADP+-type-C4 plants—a comparative protein and RNA analysis. Eur J Biochem. 1990;190:185–194. doi: 10.1111/j.1432-1033.1990.tb15563.x. [DOI] [PubMed] [Google Scholar]


