Abstract
Satellite cells (SCs) are the main source of adult skeletal muscle stem cells responsible for muscle growth and regeneration. By interpreting extracellular cues, developmental regulators control quiescence, proliferation, and differentiation of SCs by influencing coordinate gene expression. The scope of this review is limited to the description and discussion of protein complexes that introduce and decode heritable histone and chromatin modifications and how these modifications are relevant for SC biology.
1. Introduction
Epigenetics can be defined as the ensemble of heritable changes in gene function that occur without modifications of primary DNA sequence (Bird, 2007; Russo et al., 1996) to include DNA methylation and chromatin structural changes introduced by histone modifications and nucleosome positioning. During specification, proliferation, and differentiation of skeletal muscle cells (myogenesis), the epigenetic marks deposited by chromatin-modifying enzymes at specific loci determine whether different subsets of genes will be repressed or activated, effectively controlling the fate of muscle progenitors and the transition between each developmental phase. Therefore, the balance between cell self-renewal and differentiation is ultimately regulated by coordination of stage-specific transcriptional machineries through epigenetic mechanisms. Many excellent and comprehensive reviews on satellite cells (SCs) have been published (Buckingham, 2007; Charge and Rudnicki, 2004; Dhawan and Rando, 2005; Kang and Krauss, 2010) and the reader is referred to them.
2. Satellite Cells
SCs represent the primary source of multipotent stem cells responsible for postnatal skeletal muscle growth and regeneration (Charge and Rudnicki, 2004; Partridge, 2004). They originate from Pax3/7-expressing muscle progenitors in the embryonic dermomyotome (Gros et al., 2005; Relaix et al., 2005). In the mouse, SCs expressing Pax7 appear at approximately embryonic day 16.5 (E16.5) in the developing limb muscle (Kassar-Duchossoy et al., 2005) and acquire their characteristic position under the basal lamina of mature muscle fibers (Mauro, 1961). SCs comprise about 30% of total muscle nuclei in neonatal mice and 2–7% of muscle nuclei in the adult mice. They express a distinct profile of surface markers and transcription factors (TFs; Charge and Rudnicki, 2004). Recent studies have revealed that SCs are a heterogeneous population (Beauchamp et al., 2000; Collins et al., 2005; Kuang et al., 2007; Sherwood et al., 2004) composed of two groups of cells: noncommitted stem cells which maintain self-renewal ability and committed myogenic progenitors which undergo lineage-specific differentiation (Kuang et al., 2007). Because of these two distinguished characteristics, SCs qualify as a bona fide tissue-specific adult stem cell. Under normal circumstances, adult SCs are in a quiescent state. In response to environmental cues, such as mechanical stress, muscle injury, or degenerative muscle diseases, SCs exit quiescence, enter cell cycle, and actively proliferate (activated state). Following activation, a subset of SC returns to its niche under the basal lamina to replenish the stem cell reservoir. Majority of SCs activate expression of the myogenic factor MyoD and give rise to committed myoblasts, which undergo rapid proliferation and subsequently exit from the cell cycle to terminally differentiate and fuse to form new muscle fibers or repair damaged muscle (Dhawan and Rando, 2005). The identification of mesangioblasts and other non-SC muscle resident and circulating progenitors expand the repertoire of cells potentially participating to muscle growth and regeneration (Benchaouir et al., 2007; Corbel et al., 2003; Cossu and Bianco, 2003; Dellavalle et al., 2007; Joe et al., 2010; Mitchell et al., 2010; Sampaolesi et al., 2003, 2006; reviewed in Tedesco et al., 2010).
3. Repressing Muscle Gene Expression: Skeletal Muscle Developmental Regulators and Polycomb Proteins in Embryonic Stem (ES) and Nonmuscle Cells
Totipotent ES cells retain the ability of self-renewing and generating all adult cell types. These two properties are mutually exclusive: an individual ES cell either renews or differentiates into a given cell lineage. Understanding the molecular mechanisms that regulate this dichotomic decision is not only of scientific interest to developmental biologists but also holds great promise for regenerative medicine. Under defined culture conditions, ES cells can be indefinitely propagated in an undifferentiated state. ES cell differentiation into specialized cell lineages occurs in culture or during development when appropriate signals are provided and correctly interpreted. For these processes to be properly executed, transcription and translation of developmental regulators (DRs) need to be tightly controlled. DRs are TFs or related molecules that coordinate temporal and spatial expression of battery of genes involved in the specification and maintenance of a given cell state. DRs favoring ES self-renewal (such as Oct4, Nanog, and Sox2) are expressed in undifferentiated ES cells but extinguished upon lineage commitment and differentiation (Chambers et al., 2003, 2007; Masui et al., 2007; Nichols et al., 1998). Conversely, cell type-specific DRs, such as MyoD (Davis et al., 1987), are repressed in undifferentiated ES cells and their expression is activated once a given cell fate is specified. Polycomb group (PcG) proteins are pivotal regulators of DRs expression in ES cells (Bernstein et al., 2006; Boyer et al., 2006; Lee et al., 2006). PcG proteins are assembled in multiple Polycomb Repressive Complexes (PRCs) that exert their regulatory functions by introducing epigenetic modifications to chromatin structure conducive to transcriptional repression (Simon and Kingston, 2009). PRC2—and related PRC3 and PRC4—contains the core components Ezh2, Suz12, and Eed. Ezh2 is the PRC2 catalytic component with methyltransferase activity directed at lysine 27 of histone H3 (H3K27; Cao et al., 2002; Czermin et al., 2002; Kirmizis et al., 2004; Kuzmichev et al., 2002; Muller et al., 2002), with Suz12 being required for the enzymatic activity (Cao and Zhang, 2004; Pasini et al., 2004). Eed specifically recognizes and binds to trimethylated repressive marks, including H3K27me3. This event leads to the allosteric activation of the methyltransferase activity of PRC2, resulting in further propagation of the H3K27me3 mark to extended chromatin blocks and possibly transmission and retention of this histone modification through DNA replication (Margueron et al., 2009). H3K27me3 serves, in most cases, as a docking site for PRC1 recruitment, which promotes further chromatin condensation to ensure gene silencing (Francis et al., 2004).
The paired domain- and homeobox-containing TFs Pax3 and Pax7 and the myogenic determinant factor MyoD are nodal DRs expressed in quiescent and activated SCs (Kassar-Duchossoy et al., 2005; Megeney et al., 1996; Relaix et al., 2005; Rudnicki et al., 1993; Seale et al., 2000). Myf6 (also referred as to MRF4) is transcribed during SC differentiation. In ES cells, Pax3, Pax7, MyoD, and MRF4 are not expressed and their respective loci are occupied by Suz12 and nucleosomes with H3K27me3 repressive mark (Lee et al., 2006). However, in addition to the repressive H3K27me3 mark, the regulatory regions of Pax3, Pax7, MyoD, and MRF4 also contain the H3K4me3 mark (Zhao et al., 2007), a histone modification associated with transcriptional activation (Barski et al., 2007). The coexistence of negative and positive histone marks (bivalent domains; Bernstein et al., 2006) would serve the purpose of silencing DRs in ES cells while keeping them poised for immediate activation once signals are received and decoded to initiate cell lineage specific gene expression. Consistent with this proposed function, bivalent domains are resolved in monovalent regions retaining only H3K4me3 when they experience gene activation or H3K27me3 when remaining repressed. The Pax3, Pax7, MyoD, and myogenin—another pivotal regulator of SC differentiation—loci engage Suz12, are H3K27me3 marked, and repressed also in human embryonic fibroblasts (Bracken et al., 2006) and numerous nonepidermal “master” DRs, including MyoD, are marked by H3K27me3 and transcriptionally repressed in committed embryonic basal epidermal progenitors (Ezhkova et al., 2009). Pax3 and Pax7 are also occupied by Suz12 and marked with H3K27me3 in F9 embryonic carcinoma cells (Squazzo et al., 2006). Thus, DRs may need to be continuously repressed not only in ES cells but also in cells that have entered a cell lineage-specific fate, which is incompatible with expression of the repressed DRs. Alternatively, H3K27me3 may be a remnant mark introduced by PcG in ES cells and self-propagated (Hansen et al., 2008) in specified cells. The presence of PRC2 at some DRs is indicative of an active and continuously repressive role exerted by PcG in specified cell lineages. ES cells in which either Suz12 or Eed has been genetically ablated (Boyer et al., 2006; Lee et al., 2006) or Ezh2 has been reduced by miR-214 overexpression (Juan et al., 2009) display an aberrant expression of certain cell lineage-specific DRs, including Pax7, Gata1-6, and Sox17. Neither MyoD nor MRF4 appear to be expressed in Suz12- or Eed-null ES cells nor in ES cells with miR-214-mediated reduced Ezh2, indicating that derepres-sion does not necessarily coincides with transcriptional activation, at least for certain loci. Conditional Ezh2 inactivation in committed embryonic basal epidermal progenitors is not sufficient to activate expression of nonepider-mal PcG targets (Ezhkova et al., 2009). Interestingly, mouse embryonic fibroblasts (MEFs) derived from mice carrying an hypomorphic allele and expressing ~25% of the normal complement for the TF YY1—a protein known to participate in PcG-mediated repression (Atchison et al., 2003; Satijn et al., 2001; Srinivasan and Atchison, 2004; Woo et al., 2010; Yue et al., 2009)—display increased expression of several muscle-specific transcripts (actins, myosins, troponins), which are physiologically absent in MEFs (Affar el et al., 2006).
4. Sculpting Chromatin for Transcription in Skeletal Muscle Cells
Chromatin accessibility to enzymes and TFs characterizes different cell types (Weintraub and Groudine, 1976) and different developmental stages within the same cell lineage (Groudine and Weintraub, 1981), including skeletal muscle cells (Carmon et al., 1982). This is a reflection of different conformations that nucleosomes adopt as a consequence of histone and DNA modifications and positioning introduced by specific enzymatic complexes. These enzymatic protein complexes are recruited at discrete DNA regions via interaction with proteins (TFs) that recognize sequence-specific DNA modules. Chromatin-modifying complexes are transferred to stalled RNA polymerases or travel in association with elongating RNA polymerases. Noncoding RNAs can also recruit enzymatic complexes at defined chromatin regions (Mohammad et al., 2008; Rinn et al., 2007; Zhao et al., 2008).
4.1. Pax3/Pax7 and alternative SC fates
As already mentioned, Pax3 and Pax7 are important regulators of SC biology (Oustanina et al., 2004; Relaix et al., 2005; Seale et al., 2000). They both regulate expression of numerous genes including the myogenic determinant factor Myf5 (Maroto et al., 1997; Tajbakhsh et al., 1997). In C2C12 cell and satellite-derived primary myoblasts, Pax7 interacts with and engages the Wdr5-Ash2-MLL2 histone methyltransferase at the Myf5 locus, promoting H3K4me3 and gene activation (McKinnell et al., 2008). Pax3 activates Myf5 through the intermediate induction of Dmrt2, a TF that interacts with one of the Myf5 enhancers (Sato et al., 2010). Differences were noted in transcriptional potency, with Pax3 being a poor activator compared to Pax7 (McKinnell et al., 2008; Relaix et al., 2003), perhaps reflective of recruitment of distinct regulatory protein complexes. An important question relates to the inability of Pax7 to activate Myf5 in SCs returning to their niche. In these cells (Pax7+/Myf5−; Kuang et al., 2007), the Myf5 locus may retain H3K27me3 introduced by PcG in ES cells (epigenetic memory; Lee et al., 2006) or be occupied by histones, such as histone linker H1, or histone variants that may preclude Pax7 chromatin access. Alternatively, the Pax7 protein may be modified to avoid association with regulatory protein complexes required to activate Myf5 or such regulatory complexes may be absent in the self-renewing daughter cell. The hypothesized H3K27me3-marked Myf5 alleles may segregate with Pax7+/Myf5− cells and bear methylated DNA. Indeed, Ezh2 directly controls DNA methylation by recruiting DNA methyltransferase proteins (Vire et al., 2006). Preferential asymmetric segregation of histone- and/or DNA-methylated Myf5 alleles in the self-renewing cell maybe achieved by DNA cosegregation with the immortal DNA strand (Conboy et al., 2007; Shinin et al., 2006).
4.2. Myogenic bHLH protein binding modalities
MyoD, Myf5, myogenin, and MRF4 heterodimerize with ubiquitously expressed bHLH E-proteins and recognize the DNA core consensus sequence CANNTG (the E-box; reviewed in Tapscott, 2005). Chromatin immunoprecipitation (ChIP) combined with either microarray hybridization or high-throughput sequencing and gene expression profiling (Bergstrom et al., 2002; Blais et al., 2005; Cao et al., 2006, 2010) has allowed identification of myogenic bHLH targets on a genome-wide scale. As anticipated, MyoD and myogenin bind to regulatory regions of genes whose transcription is regulated during skeletal muscle differentiation (Blais et al., 2005; Cao et al., 2006). Unexpectedly, MyoD binding is evident at several sites before differentiation (Blais et al., 2005) and majority of MyoD binding occurs at either introns or intergenic regions, coinciding with local histone acetylation (Cao et al., 2010). One interpretation of these findings is that, in addition to regulating gene expression, MyoD may have a role in the epigenetic reprogramming of skeletal muscle cells (Cao et al., 2010). Importantly, genome-wide MyoD binding distribution in the C2C12 cell line, primary muscle cells, and fibroblasts expressing MyoD are extremely similar (Cao et al., 2010), a reassuring finding further confirming coincidence of regulatory transcriptional modalities in different cellular model of skeletal muscle differentiation.
4.3. Marking chromatin for repression
The composition of the transcripitonal machinery (protein complexes) and the chromatin asset (histone and DNA) at muscle-specific genes reflects the different states of the cell. In the vicinity of transcriptional start sites (TSS) of genes not expressed in undifferentiated myoblasts, core histones H3 and H4 are mainly deacetylated and methylated at specific lysines (H3K9,14,18 deAc, H4K5,8,16 deAc, H3K9me2, and H3K27me3) by histone deaceylases (HDACs; Fulco et al., 2003; Lu et al., 2000b; Mal et al., 2001; Ohkawa et al., 2006; Puri et al., 2001) and methyltransferases (HMTs; Suv39h1/KMT1A and PcG; Caretti et al., 2004; Mal, 2006). These enzymes have developed the ability to function as cooperative modules (Margueron and Reinberg, 2010). This is a property deriving by mutually exclusive histone modifications. Acetylation and methylation cannot occur on the same lysine residue and deacetylation is often a prerequisite for subsequent methylation. Thus, HDACs are often found in protein complexes containing HMTs. Class I, II, and III HDACs, SUv39h1/KMT1A, the chromodomain-containing methyl-binding protein HP1, and PcG Ezh2 associate in different protein complexes (Caretti et al., 2004; Kuzmichev et al., 2005; van der Vlag and Otte, 1999; Vaquero et al., 2007; Vaute et al., 2002) and are recruited, along with the PcG-related TF YY1, at chromatin regulatory regions of inactive muscle genes (Fig. 3.1A; Caretti et al., 2004; Fulco et al., 2003; Mal, 2006; Ohkawa et al., 2006; Wang et al., 2007; Zhang et al., 2002). Initially identified as histone deacetylases and methyltransferases, HDACs, and HMTs were subsequently found to act also on nonhistone proteins, including TFs such as MyoD, MEF2, and p53 (Chuikov et al., 2004; Fulco et al., 2003; Huang et al., 2006, 2010; Mal et al., 2001; Puri et al., 2001; Zhao et al., 2005).
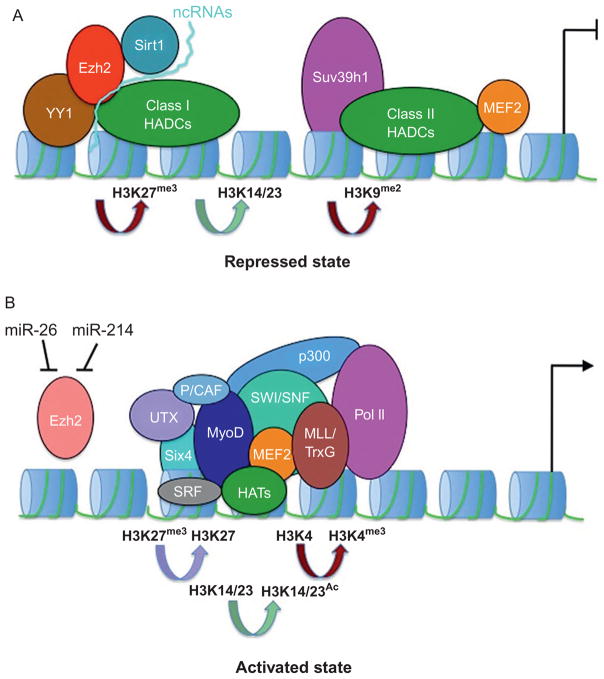
Figure 3.1.
(A) Repressive histone marks are introduced by protein complexes composed by class I, II, and III (SIRT1) deacetylases and histone methyltransferases (Suv39h1 and Polycomb Ezh2). Ezh2 may be recruited by the Pleiohomeotic (PHO)-related YY1 protein or noncoding RNAs (ncRNAs). Lysines 14–23 of histone H3 are deacetylated and lysine 27 (Ezh2) and lysine 9 (Suv39h1) are methylated. (B) Repressors are replaced by a new set of protein complexes at transcribed genes. Via interaction with the transcriptional activators MyoD, MEF2, SRF, and Six4, acetyltransferases (HATs, p300, P/CAF) and nucleosome remodeling machines (SWI/SNF) promote lysine acetylation (H3K14-23AC) and nucleosome remodeling, respectively. Demethylation of lysine 27 is achieved by transcriptional downregulation and microRNA-mediated (miR-26 and miR-214) repression of Ezh2. In addition, the lysine 27-specific UTX actively removes methyl groups from lysine 27. The Trithorax-related mixed lineage leukemia (MLL) proteins promote methylation of histone H3 lysine 4, a mark associated with polymerase II (PolII) recruitment and gene activation.
Specific histone isoforms participate in shaping repressive chromatin structure. Through specific interaction with the homeoprotein Msx1, histone H1b is deposited at the core enhancer region of the MyoD locus within a repressive chromatin region characterized by H3K9 methylation, decreased H3K9 and 14 acetylation, and reduced H3Ser10 phosphorylation (Lee et al., 2004). H1b depletion abrogated the ability of Msx1 to inhibit muscle cell differentiation, directly linking the repressive function of Msx1 to histone H1b.
4.4. SIRT1: A multifaceted regulator
SIRT1 is an NAD+-dependent protein deacetylase (Imai et al., 2000) expressed in quiescent and activated SCs (J. G. Ryall, A. Pasut, M. A. Rudnicki, and V. S., unpublished results). Reducing SIRT1 in the C2C12 cell line or in primary myoblasts results in their premature differentiation (Fulco et al., 2003) and, analogously, primary myoblasts derived from heterozygous SIRT1+/− exhibit early differentiation and are resistant to antidifferentiative cues, such as glucose restriction (Fulco et al., 2008). Overexpression experiments have revealed that SIRT1 increases SC proliferation (Rathbone et al., 2008). It is likely that the effects of SIRT1 on muscle cell differentiation results from its deacetylase activity on different substrates, including histones, acetyltransferases, and transcriptional regulators (Brunet et al., 2004; Fulco et al., 2003; Motta et al., 2004). Indeed, SIRT1 deacetylates MyoD and P/CAF (Fulco et al., 2003), an acetyltrans-ferase that activates MyoD (Dilworth et al., 2004; Kuninger et al., 2006; Sartorelli et al., 1999). MEF2D, an important TF that collaborates with MyoD to activate genes critical for muscle differentiation, is also deacety-lated by SIRT1 (Zhao et al., 2005). In addition to controlling cell differentiation, SIRT1, and related sirtuins, may have other functions in SCs that remain to be explored. For instance, SIRT1 interacts with and mediates transcriptional repression induced by hairy/Hes1 and HEY2 (Bianchi-Frias et al., 2004; Rosenberg and Parkhurst, 2002; Takata and Ishikawa, 2003), two transcriptional repressors whose activation is promoted by the Notch pathway. Given the central role played by the Notch pathway in regulating SC activation and cell fate determination in postnatal myogenesis (Brack et al., 2008; Conboy and Rando, 2002), asymmetric cell division (Conboy et al., 2007; Kuang et al., 2007; Shinin et al., 2006), restoring the regenerative potential of aged muscle (Conboy et al., 2003; Luo et al., 2005), and the ability of SIRT1 to control SC proliferation and differentiation (Fulco et al., 2003; Rathbone et al., 2008), to retard degenerative processes and increase lifespan across species (reviewed in Imai and Guarente, 2010), it will be of great interest to determine whether the Notch and SIRT1 pathways crosstalk. Another process that may be potentially influenced by SIRT1 or other sirtuins is the cell response to oxidative stress. It is intriguing that quiescent SCs have developed strategies that protect them from xenobiotics, geno-toxics, and oxidative stress (Pallafacchina et al., 2010), likely to maintain genome integrity. Since SIRT1 is activated by and protects from the deleterious effects induced by oxidative stress, DNA damage, and hypoxia (Brunet et al., 2004; Dioum et al., 2009; Motta et al., 2004; Oberdoerffer et al., 2008), it may contribute to SC resistance to environmetal insults. In principle, SIRT1 transcription needs not to be regulated in quiescent versus activated SCs, as its enzymatic activity can be modulated by the redox state (Fulco et al., 2003).
4.5. Turning on transcription: Erasing and writing
Reception and decodification of prodifferentiative signals induce modification of the chromatin structure conducive to gene activation. Repressive marks need to be erased and substituted with histone modifications and nucleosome restructuring compatible with RNA polymerase II (PolII) engagement and elongation. A pivotal step in this process is the recruitment of histone acetyltransferases (HATs), ATP-dependent chromatin remodeling complexes, methyltransferases, demethylases, and specific subunits of the basal transcriptional machinery.
H3K27me3 is reduced at specific muscle regulatory regions upon gene activation (Caretti et al., 2004). Several independent, but functionally coherent, mechanisms contribute to this phenomenon (Fig. 3.1B). H3K27me3 deposition is curtailed as Ezh2 transcription is reduced (Caretti et al., 2004) and the residual Ezh2 transcripts are targeted for translational inhibition by the microRNA miR-26 and miR-214 (Juan et al., 2009; Wong and Tellam, 2008). Active H3K27 demethylation is concomitantly brought about by Six4-mediated recruitment of the UTX demethylase (Seenundun et al., 2010). The presence of the H3K4 methyl-transferases mixed lineage leukemia (MLL) proteins within the UTX complex provides a parsimonious and elegant mechanism whereby a repressive mark is erased and one with activation property is introduced by the same protein complex (Fig. 3.1B; Rampalli et al., 2007). Jmjd1, a specific H3K9me2 demethylase (Yamane et al., 2006) may be involved in erasing H3K9me2 from muscle regulatory regions. During the differentiation process, class I HDACs are disengaged from MyoD and possibly other transcriptional regulators and redistributed to alternative protein complexes via cell cycle-regulated events involving pRb hypophosphorylation (Puri et al., 2001) and calcium-mediated activation of the calmodulin-dependent protein kinase (CaMK) stimulates MEF2 activity by dissociating it from class II HDACs (Lu et al., 2000a). Pharmacological HDACs inhibition promotes, in a temporal-specific manner, muscle gene expression (Iezzi et al., 2002, 2004), favoring a functional and morphological amelioration in two different animal models of muscle dystrophies (Minetti et al., 2006; reviewed in (Mozzetta et al., 2009). Removal of HDACs from transcribed regions has been classically seen as a prerequisite for gene activation and HATs and HDACs cooccupancy mutually exclusive. However, in genome-wide mapping studies, HDACs have been detected with HATs at active genes with acetylated histones and their recruitment imputed to phosphorylated PolII, providing the basis for dynamic cycles of acetylation/deacetylation required to reset chromatin after PolII elongation (Wang et al., 2009). These findings are consistent with the observation that SIRT1 and PCAF cooc-cupy transcribed chromatin regions (Fulco et al., 2003). However, it cannot be excluded that, at activated genes, SIRT1 deacetylase activity may be tempered by unfavorable [NAD+]/[NADH] ratio (Sartorelli and Caretti, 2005). The HATs p300, PCAF, and related GNC5 are engaged at muscle-activated genes by different TFs—including myogenic bHLH, MEF2 factors, SRF, six proteins, and ubiquitous transcriptional regulators (reviewed in Guasconi and Puri, 2009; Perdiguero et al., 2009). Association of MyoD with the HATs p300 and PCAF (Puri et al., 1997b) is promoted by AKT1 and 2 kinases via direct phosphorylation of p300 (Serra et al., 2007). Several TFs regulating muscle gene expression are acetylated by the same HATs they recruit on the chromatin (Avantaggiati et al., 1997; Duquet et al., 2006; Gu et al., 1997; Lill et al., 1997; Polesskaya et al., 2000; Sartorelli et al., 1999; Simone et al., 2004). In some instances, that is, p53 and MyoD, acetylation increases DNA binding affinity (Gu et al., 1997; Sartorelli et al., 1999), providing a positive feedback mechanism to increase HATs recruitment and consequent histone acetylation.
4.6. Moving obstacles
Nucleosomes containing epigenetic marks compatible with transcriptional activation need to be accordingly redistributed as to avoid occluding access of regulatory DNA regions to TFs. Nucleosome positioning can be achieved by ATP-dependent chromatin remodeling complexes or by TFs in the absence of additional factors (reviewed in Radman-Livaja and Rando, 2010). Binding of a TF near the entry/exit point of a nucleosome modifies the chromatin structure making accessible other sites located toward the center of the nucleosome (Polach and Widom, 1996). With a related mechanism, an NFkB-p65 subunit mutant without activation domains continues to regulate transcription of a subset of target genes by permitting access to secondary TFs (van Essen et al., 2009). Pbx1/Meis1 is a “pioneer” TF capable of penetrating repressive chromatin. While the detailed mechanisms endowing Pbx1/Meis with such ability are not known, it may be that its binding sites are preferentially situated in nucleo-some free-regions, as in the case of the IFN-b promoter, or that Pbx1/Meis1 may directly interact with and displace histones, as it occurs for HNF3 (Sagerstrom, 2004). Pbx1/Meis1 is constitutively bound to muscle regulatory regions before transcriptional activation and in the absence of MyoD. Pbx1/Meis interacts with MyoD (Knoepfler et al., 1999) and can position it and tether it even at imperfect MyoD binding sites (Berkes et al., 2004). Interaction of Pbx1/Meis1 with MyoD occurs via two domains required for initiation of chromatin remodeling (Bergstrom and Tapscott, 2001; Gerber et al., 1997). These studies provide a mechanistic explanation of how MyoD can penetrate repressive chromatin to initiate remodeling. Pbx1/Meis1 may need to be somehow modified or interact with transcriptional repressors to repel MyoD and avoid premature gene activation. Alternatively, regulated interaction of Pbx1/Meis1 with SWI/SNF may permit productive MyoD recruitment (de la Serna et al., 2005). Recruitment of the ATP-dependent SWI/SNF BRM and brahma-like 1 (BRG-1) chromatin remodeling complex through interaction with MyoD (de la Serna et al., 2001; reviewed in de la Serna et al., 2006) is regulated via direct phosphorylation of the SWI/SNF BAF60c subunit mediated by the MAPK p38 (mitogen-activated kinases; Simone et al., 2004; reviewed in Albini and Puri, 2010) and required for binding of MyoD, myogenin, and MEF2 (de la Serna et al., 2005) and sustained gene expression (Ohkawa et al., 2007). At the myogenin promoter, MyoD induces histone hyperacetylation before and independently of BRG-1 activity (de la Serna et al., 2005), suggesting the formation of a temporally regulated transcriptional protein complex where MyoD-mediated HATs recruitment (Eckner et al., 1996; Puri et al., 1997a, b; Sartorelli et al., 1997; Yuan et al., 1996) precedes BRG-1-dependent chromatin remodeling. BRG-1-meditaed remodeling at different muscle loci is temporally regulated by sequential activities of the arginine methyltransferases PRMT5 and CARM1/PRMT4 (Dacwag et al., 2009; Mallappa et al., 2010). An interesting relation between SWI/SNF and PcG proteins was uncovered when brahma was identified as a suppressor of PcG mutants in homeotic gene expression, suggesting that SWI/SNF may regulate gene expression by antagonizing repressive chromatin structure organized by PcG proteins (Tamkun et al., 1992). A dedicated role of BAF60c to myogenesis is suggested by its preferential expression in developing heart and somites and has been confirmed by its genetic ablation, which results in cardiac and skeletal muscle defects (Lickert et al., 2004). Intriguingly, and consistent with its chromatin remodeling ability, BRG-1 has been found to facilitate iPS formation (Singhal et al., 2010).
4.7. RNA helicases: Connecting histone acetylation, chromatin remodeling, and microRNA maturation
The highly related RNA helicases p68 and p72 (p68/p72) and the long noncoding RNA (lncRNA) steroid receptor activator (SRA) associate with MyoD and coregulate its transcriptional activity (Caretti et al., 2006). They also serve as coregulators of other TFs, including p53, estrogen, and androgen receptors. Their mechanistic role is not fully understood as the RNA helicase activity seems dispensable to regulate transcription (Bates et al., 2005; Caretti et al., 2006). However, several other features endow p68/p72 with regulatory properties. P68 interacts with HDAC1, repressing transcription in a promoter-specific manner (Wilson et al., 2004). In other biological circumstances, p68 can interact with and is acetylated by p300. Acetylation increases p68/p72 stability, thus stimulating its ability to coac-tivate estrogen receptor-mediated transcription (Mooney et al., 2010). Sumoylation may constitute a regulatory switch as it enhances p68 transcriptional repression by favoring its association with HDAC1 (Fig. 3.2A; Jacobs et al., 2007). In differentiated skeletal muscle cells, p68/p72 were detected on chromatin regulatory regions in association with the lncRNA SRA and MyoD (Fig. 3.2B). Reducing p68 level by RNAi in either C2C12 cell line or satellite-cell-derived primary muscle cells hampered differentiation, transcription of selected muscle genes, and chromatin engagement of BRG-1, TATA-binding protein (TBP), and PolII without affecting either MyoD or p300 recruitment (Caretti et al., 2006). Genes whose expression was affected by p68 require BRG-1 (de la Serna et al., 2005). More recently, the estrogen receptor ERα has been demonstrated to form a protein complex with p300, p68/p72, and MyoD to regulate BRCA2 transcription (Jin et al., 2008). The HMG-related small chromatin protein p8 also associates and is recruited at muscle-specific genes with p300, p68, and MyoD (Fig. 3.2B). p8 knockdown compromises chromatin recruitment of p300, p68, and MyoD, thus impairing muscle transcription (Sambasivan et al., 2009). In addition to a direct role on the formation and stabilization of chromatin-modifying protein complexes, p68/p72 influence also other processes related to gene expression. P68/p72 are part of the nuclear RNase III Drosha complex, which cleaves primary transcripts of miRNA genes (pri-miRNAs) into hairpin precursor intermediates (pre-miRNAs), further processed to mature miRNAs by the cytosolic RNase III Dicer. Association of p68/72 with the TFs p53 or SMAD regulates the processing of primary miRNAs into precurors miRNAs (Davis et al., 2008; Suzuki et al., 2009). Gene disruption of either p68 or p72 results in early lethality, with specific defects in maturation of selected miRNAs. P72 deletion negatively impacts maturation of miR-214 (Fukuda et al., 2007), a micro-RNA regulating Ezh2 availability and involved in muscle cell differentiation (Juan et al., 2009). Thus, p72 may regulate muscle gene expression by two independent mechanisms, by coactivating MyoD-dependent transcription and favoring miR-214 processing (Juan and Sartorelli, 2010).
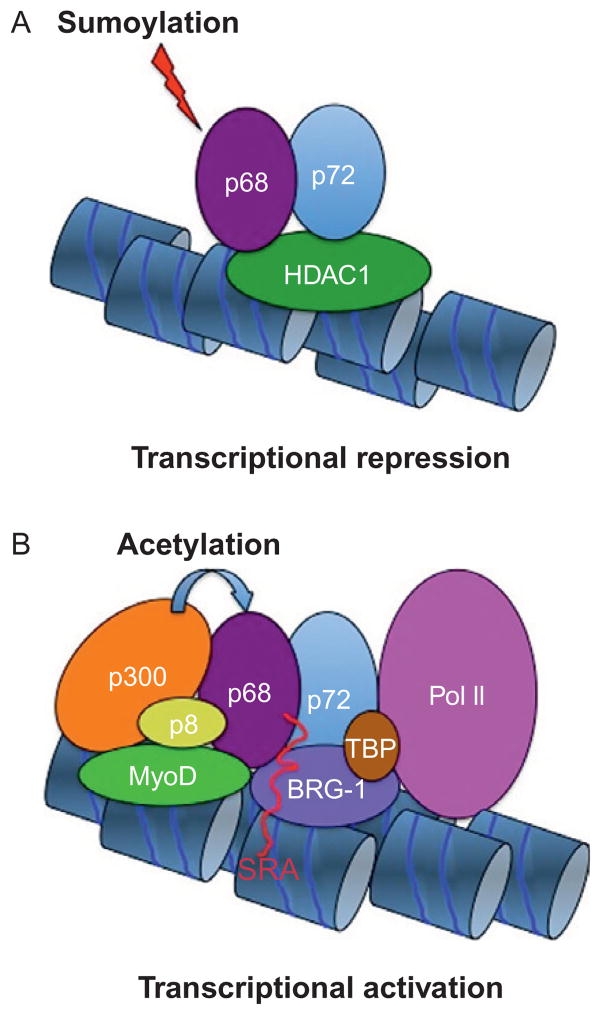
Figure 3.2.
(A) Sumoylation of the RNA helicase p68 (forming heterodimers with the RNA helicase p72) can favor repression by promoting p68 interaction with the histone deacetylase HDAC1. (B) At transcribed genes, the RNA helicases p68/p72, the noncoding RNA SRA, form a transcriptional complex with the HAT p300, the high-mobility group-related p8, MyoD, the nucleosome remodeling SWI/SNF complex, the TATA-binding protein (TBP), and PolII. In addition to histones, p300 also acetylates p68.
4.8. Switching basal players: TRF3 and TAF3
It might have been naively predicted that a family of tissue-specific TFs (activators), such as the myogenic bHLH, would suffice to ensure cell-lineage restricted gene expression, with the constant core basal transcriptional machinery serving many masters. It turned out that activators and core machinery members display mutual preferences (D’Alessio et al., 2009). Upon muscle cell differentiation, TBP and several TAFs—expressed in undifferentiated cells—are degraded and replaced by TRF3 and TAF3. Indeed, MyoD can directly target TAF3, and such interaction support in vitro transcription. Moreover, TRF3/TAF3 are recruited at the myo-genin promoter and interference with their accumulation prevents myotube formation (Deato and Tjian, 2007; Deato et al., 2008). As MyoD is tran-scriptionally active in undifferentiated myoblasts, these findings imply that MyoD can communicate and direct transcription with two alternative core basal machineries. As reported for SWI/SNF (Tamkun et al., 1992), also tissue-specific TAFs counteract PcG proteins to promote terminal differentiation (Chen et al., 2005).
4.9. Conveying signals to the nucleus by the MAPK p38
In skeletal muscle cells, extracellular cues mediated by morphogen gradients, ligand–receptor, and soluble growth factor–receptor interactions are decoded and ultimately conveyed to the genome by chromatin-modifying complexes to regulate transcriptional output (reviewed in Guasconi and Puri, 2009). Protein modifications mediated by kinases and phosphates rapidly and effectively mediate cellular responses to environmental signals and internal processes by regulating protein interactions, enzymes activity, and protein localization (Pawson, 2007).
Within this context, signaling mediated by the MAPK p38 has received a great deal of experimental attention. The important role exerted by p38a in regulating the satellite quiescent state (Jones et al., 2005) is evidenced by the analysis of p38a mutant mice, which shows delayed-cell-cycle exit and altered expression of cell cycle regulators in cultured myoblasts (Perdiguero et al., 2007). Activation of p38, initiated by the transmembrane Ig-fibro-nectin—type III repeat CDO protein—promotes formation of MyoD-E47 dimers, which in turn activate CDO transcription (Cole et al., 2004; Takaesu et al., 2006). E47 is a direct target of p38-mediated phosphorylation promoting MyoD-E47 heterodimer formation (Lluis et al., 2005). Different members of the MAPK p38 family exert opposing effects on muscle gene expression. While p38a/b favor muscle transcription by promoting MyoD-E47 heterodimer formation (Lluis et al., 2005) and MEF2D binding (Penn et al., 2004), engagement of the SWI/SNF chromatin remodeling complex via phosphorylation of the BAF60c subunit (Simone et al., 2004), and recruitment of the Ash2L-containing MLL protein complex through MEF2D phosphorylation (Rampalli et al., 2007), p38g antagonizes muscle gene expression by promoting recruitment of the H3K9 methyltransferase Suv39h1/KMT1 at the myogenin promoter through direct phosphoryla-tion of Ser199 and Ser200 of MyoD (Gillespie et al., 2009).
An unexpected role of p38a in linking tumor necrosis factor (TNF) signaling to PcG proteins during muscle regeneration has been recently uncovered. Inflammatory cells populating sites of damaged muscles are an essential component of the regenerative phase, characterized by SC expansion and differentiation. Locally released cytokines, including interleukin 1,4,6, and TNFa promote muscle regeneration (Charge and Rudnicki, 2004; Dhawan and Rando, 2005; Horsley et al., 2003; Serrano et al., 2008). TNFa regulates muscle regeneration by activating MAPK p38 (Chen et al., 2007). In proliferating, undifferentiated muscle cells, where p38a signaling is ineffective (Wu et al., 2000), the PRC2 complex prevents unscheduled gene expression by repressing transcription of muscle-specific myosins and muscle creatine kinase genes (Caretti et al., 2004). When proliferating SCs undergo differentiation, PRC2 is released from muscle structural genes—where is replaced by an activation complex (Caretti et al., 2004)—and relocated to Pax7 regulatory regions to repress gene expression (Palacios et al., 2010). Such PRC2 redistribution from muscle structural genes to Pax7 is regulated by p38a, which directly phosphorylates the human PRC2 catalytic subunit Ezh2 at Thr372 (corresponding to mouse Ezh2 Thr367), influencing recruitment of the Polycomb-related transcriptional repressor YY1 (Palacios et al., 2010). Indeed, p38a, Ezh2, YY1, and H3K27me3 are codetected at the Pax7 regulatory regions of differentiating primary myoblasts. Genetically or pharmacologically interfering with p38 or PRC2, results in continued Pax7 expression and expansion of SCs, which retain their ability to differentiate once p38 or PRC2 blockade is removed (Palacios et al., 2010). While chromatin engagement of MAPKs p38 has been previously documented (Chow and Davis, 2006; de Nadal and Posas, 2010; Pokholok et al., 2006), the contribution of p38α to redistribute PcG proteins at specific genomic loci in response to inflammatory signals introduces an additional layer of regulatory refinement.
5. Conclusions
As a result of the concerted action of acetyltransferases and deacety-lases, methyltransferases and demethylases, kinases and phosphatases, epige-netic modifications are dynamic and reversible. This property of the epigenome, associated with the availability and development of new small molecules with the ability of regulating the activities of specific chromatin-modifying protein complexes, allows, in principle, a regulatable manipulation of cell fate decisions.
In the context of muscle stem cell biology, the combined investigation of molecules and mechanisms regulating the epigenome and small molecule screening may prove useful toward the development of effective therapies for degenerative diseases.
Acknowledgments
The authors are supported by the Intramural Research Program of the National Institute of Arthritis, Musculoskeletal, and Skin Diseases of the National Institutes of Health.
References
- Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini S, Puri PL. SWI/SNF complexes, chromatin remodeling and skeletal myogenesis: It’s time to exchange! Exp Cell Res. 2010;316:3073–3080. doi: 10.1016/j.yexcr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, Gregory DJ, Lane DP, Perkins ND, Fuller-Pace FV. The DEAD box protein p68: A novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 2005;24:543–553. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, Buckingham ME, Partridge TA, Zammit PS. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchaouir R, Meregalli M, Farini A, D’Antona G, Belicchi M, Goyenvalle A, Battistelli M, Bresolin N, Bottinelli R, Garcia L, Torrente Y. Restoration of human dystrophin following transplantation of exon-skipping-engineered DMD patient stem cells into dystrophic mice. Cell Stem Cell. 2007;1:646–657. doi: 10.1016/j.stem.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol Cell Biol. 2001;21:2404–2412. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brack AS, Conboy IM, Conboy MJ, Shen J, Rando TA. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell. 2008;2:50–59. doi: 10.1016/j.stem.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C R Biol. 2007;330:530–533. doi: 10.1016/j.crvi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ. Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Carmon Y, Czosnek H, Nudel U, Shani M, Yaffe D. DNAase I sensitivity of genes expressed during myogenesis. Nucleic Acids Res. 1982;10:3085–3098. doi: 10.1093/nar/10.10.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- Chen X, Hiller M, Sancak Y, Fuller MT. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science. 2005;310:869–872. doi: 10.1126/science.1118101. [DOI] [PubMed] [Google Scholar]
- Chen SE, Jin B, Li YP. TNF-alpha regulates myogenesis and muscle regeneration by activating p38 MAPK. Am J Physiol Cell Physiol. 2007;292:C1660–C1671. doi: 10.1152/ajpcell.00486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CW, Davis RJ. Proteins kinases: Chromatin-associated enzymes? Cell. 2006;127:887–890. doi: 10.1016/j.cell.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Cole F, Zhang W, Geyra A, Kang JS, Krauss RS. Positive regulation of myogenic bHLH factors and skeletal muscle development by the cell surface receptor CDO. Dev Cell. 2004;7:843–854. doi: 10.1016/j.devcel.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- Conboy MJ, Karasov AO, Rando TA. High incidence of non-random template strand segregation and asymmetric fate determination in dividing stem cells and their progeny. PLoS Biol. 2007;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbel SY, Lee A, Yi L, Duenas J, Brazelton TR, Blau HM, Rossi FM. Contribution of hematopoietic stem cells to skeletal muscle. Nat Med. 2003;9:1528–1532. doi: 10.1038/nm959. [DOI] [PubMed] [Google Scholar]
- Cossu G, Bianco P. Mesoangioblasts–vascular progenitors for extravascular mesodermal tissues. Curr Opin Genet Dev. 2003;13:537–542. doi: 10.1016/j.gde.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dacwag CS, Bedford MT, Sif S, Imbalzano AN. Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol Cell Biol. 2009;29:1909–1921. doi: 10.1128/MCB.00742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio JA, Wright KJ, Tjian R. Shifting players and paradigms in cell-specific transcription. Mol Cell. 2009;36:924–931. doi: 10.1016/j.molcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25:3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: Lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Posas F. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 2010;29:4–13. doi: 10.1038/emboj.2009.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellavalle A, Sampaolesi M, Tonlorenzi R, Tagliafico E, Sacchetti B, Perani L, Innocenzi A, Galvez BG, Messina G, Morosetti R, Li S, Belicchi M, et al. Pericytes of human skeletal muscle are myogenic precursors distinct from satellite cells. Nat Cell Biol. 2007;9:255–267. doi: 10.1038/ncb1542. [DOI] [PubMed] [Google Scholar]
- Dhawan J, Rando TA. Stem cells in postnatal myogenesis: Molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 2005;15:666–673. doi: 10.1016/j.tcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc Natl Acad Sci USA. 2004;101:11593–11598. doi: 10.1073/pnas.0404192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- Duquet A, Polesskaya A, Cuvellier S, Ait-Si-Ali S, Hery P, Pritchard LL, Gerard M, Harel-Bellan A. Acetylation is important for MyoD function in adult mice. EMBO Rep. 2006;7:1140–1146. doi: 10.1038/sj.embor.7400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, Akimoto C, Yamamoto Y, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: A mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Gillespie MA, Le Grand F, Scime A, Kuang S, von Maltzahn J, Seale V, Cuenda A, Ranish JA, Rudnicki MA. p38-{gamma}-dependent gene silencing restricts entry into the myogenic differentiation program. J Cell Biol. 2009;187:991–1005. doi: 10.1083/jcb.200907037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Manceau M, Thome V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- Groudine M, Weintraub H. Activation of globin genes during chicken development. Cell. 1981;24:393–401. doi: 10.1016/0092-8674(81)90329-9. [DOI] [PubMed] [Google Scholar]
- Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- Guasconi V, Puri PL. Chromatin: The interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iezzi S, Cossu G, Nervi C, Sartorelli V, Puri PL. Stage-specific modulation of skeletal myogenesis by inhibitors of nuclear deacetylases. Proc Natl Acad Sci USA. 2002;99:7757–7762. doi: 10.1073/pnas.112218599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iezzi S, Di Padova M, Serra C, Caretti G, Simone C, Maklan E, Minetti G, Zhao P, Hoffman EP, Puri PL, Sartorelli V. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Dev Cell. 2004;6:673–684. doi: 10.1016/s1534-5807(04)00107-8. [DOI] [PubMed] [Google Scholar]
- Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: Implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jacobs AM, Nicol SM, Hislop RG, Jaffray EG, Hay RT, Fuller-Pace FV. SUMO modification of the DEAD box protein p68 modulates its transcriptional activity and promotes its interaction with HDAC1. Oncogene. 2007;26:5866–5876. doi: 10.1038/sj.onc.1210387. [DOI] [PubMed] [Google Scholar]
- Jin W, Chen Y, Di GH, Miron P, Hou YF, Gao H, Shao ZM. Estrogen receptor (ER) beta or p53 attenuates ERalpha-mediated transcriptional activation on the BRCA2 promoter. J Biol Chem. 2008;283:29671–29680. doi: 10.1074/jbc.M802785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NC, Tyner KJ, Nibarger L, Stanley HM, Cornelison DD, Fedorov YV, Olwin BB. The p38alpha/beta MAPK functions as a molecular switch to activate the quiescent satellite cell. J Cell Biol. 2005;169:105–116. doi: 10.1083/jcb.200408066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Sartorelli V. MicroRNA-214 and polycomb group proteins: A regulatory circuit controlling differentiation and cell fate decisions. Cell Cycle. 2010;9:1445–1446. doi: 10.4161/cc.9.8.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Krauss RS. Muscle stem cells in developmental and regenerative myogenesis. Curr Opin Clin Nutr Metab Care. 2010;13:243–248. doi: 10.1097/MCO.0b013e328336ea98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoepfler PS, Bergstrom DA, Uetsuki T, Dac-Korytko I, Sun YH, Wright WE, Tapscott SJ, Kamps MP. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis1/Prep1. Nucleic Acids Res. 1999;27:3752–3761. doi: 10.1093/nar/27.18.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuninger D, Wright A, Rotwein P. Muscle cell survival mediated by the transcriptional coactivators p300 and PCAF displays different requirements for acetyltransferase activity. Am J Physiol Cell Physiol. 2006;291:C699–C709. doi: 10.1152/ajpcell.00056.2006. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Lluis F, Ballestar E, Suelves M, Esteller M, Munoz-Canoves P. E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J. 2005;24:974–984. doi: 10.1038/sj.emboj.7600528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci USA. 2000a;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000b;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- Luo D, Renault VM, Rando TA. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin Cell Dev Biol. 2005;16:612–622. doi: 10.1016/j.semcdb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: Inhibition of the myogenic program. EMBO J. 2001;20:1739–1753. doi: 10.1093/emboj/20.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallappa C, Nasipak BT, Etheridge L, Androphy EJ, Jones SN, Sagerstrom CG, Ohkawa Y, Imbalzano AN. Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function. Mol Cell Biol. 2010;30:3176–3186. doi: 10.1128/MCB.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroto M, Reshef R, Münsterberg A, Koester S, Goulding M, Lassar A. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89:139–148. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Minetti GC, Colussi C, Adami R, Serra C, Mozzetta C, Parente V, Fortuni S, Straino S, Sampaolesi M, Di Padova M, Illi B, Gallinari P, et al. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat Med. 2006;12:1147–1150. doi: 10.1038/nm1479. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Pannerec A, Cadot B, Parlakian A, Besson V, Gomes ER, Marazzi G, Sassoon DA. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12:257–266. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P, Kanduri C. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney SM, Goel A, D’Assoro AB, Salisbury JL, Janknecht R. Pleiotropic effects of p300-mediated acetylation on p68 and p72 RNA helicase. J Biol Chem. 2010;285:30443–30452. doi: 10.1074/jbc.M110.143792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Mozzetta C, Minetti G, Puri PL. Regenerative pharmacology in the treatment of genetic diseases: The paradigm of muscular dystrophy. Int J Biochem Cell Biol. 2009;41:701–710. doi: 10.1016/j.biocel.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y, Yoshimura S, Higashi C, Marfella CG, Dacwag CS, Tachibana T, Imbalzano AN. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. J Biol Chem. 2007;282:6564–6570. doi: 10.1074/jbc.M608898200. [DOI] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, Marquez VE, Valente S, Mai A, Forcales SV, Sartorelli V, Puri PL. TNF/p38alpha/Polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallafacchina G, Francois S, Regnault B, Czarny B, Dive V, Cumano A, Montarras D, Buckingham M. An adult tissue-specific stem cell in its niche: A gene profiling analysis of in vivo quiescent and activated muscle satellite cells. Stem Cell Res. 2010;4:77–91. doi: 10.1016/j.scr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Partridge T. Reenthronement of the muscle satellite cell. Cell. 2004;119:447–448. doi: 10.1016/j.cell.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T. Dynamic control of signaling by modular adaptor proteins. Curr Opin Cell Biol. 2007;19:112–116. doi: 10.1016/j.ceb.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev. 2004;18:2348–2353. doi: 10.1101/gad.1234304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E, Ruiz-Bonilla V, Gresh L, Hui L, Ballestar E, Sousa-Victor P, Baeza-Raja B, Jardi M, Bosch-Comas A, Esteller M, Caelles C, Serrano AL, et al. Genetic analysis of p38 MAP kinases in myogenesis: Fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J. 2007;26:1245–1256. doi: 10.1038/sj.emboj.7601587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E, Sousa-Victor P, Ballestar E, Munoz-Canoves P. Epigenetic regulation of myogenesis. Epigenetics. 2009;4:541–550. doi: 10.4161/epi.4.8.10258. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Duquet A, Naguibneva I, Weise C, Vervisch A, Bengal E, Hucho F, Robin P, Harel-Bellan A. CREB-binding protein/p300 activates MyoD by acetylation. J Biol Chem. 2000;275:34359–34364. doi: 10.1074/jbc.M003815200. [DOI] [PubMed] [Google Scholar]
- Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997a;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997b;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE, Giordano A, Kedes L, Wang JY, Sartorelli V. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell. 2001;8:885–897. doi: 10.1016/s1097-2765(01)00373-2. [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. Nucleosome positioning: How is it established and why does it matter? Dev Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, Dilworth FJ. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol. 2007;14:1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbone CR, Booth FW, Lees SJ. Sirt1 increases skeletal muscle precursor cell proliferation. Eur J Cell Biol. 2008;88:35–44. doi: 10.1016/j.ejcb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Polimeni M, Rocancourt D, Ponzetto C, Schafer BW, Buckingham M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes Dev. 2003;17:2950–2965. doi: 10.1101/gad.281203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MI, Parkhurst SM. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell. 2002;109:447–458. doi: 10.1016/s0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Russo VEA, Martinssen RA, Riggs AD, editors. Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press; Woodbury: 1996. [Google Scholar]
- Sagerstrom CG. PbX marks the spot. Dev Cell. 2004;6:737–738. doi: 10.1016/j.devcel.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Sambasivan R, Cheedipudi S, Pasupuleti N, Saleh A, Pavlath GK, Dhawan J. The small chromatin-binding protein p8 coordinates the association of anti-proliferative and pro-myogenic proteins at the myogenin promoter. J Cell Sci. 2009;122:3481–3491. doi: 10.1242/jcs.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaolesi M, Torrente Y, Innocenzi A, Tonlorenzi R, D’Antona G, Pellegrino MA, Barresi R, Bresolin N, De Angelis MG, Campbell KP, Bottinelli R, Cossu G. Cell therapy of alpha-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 2003;301:487–492. doi: 10.1126/science.1082254. [DOI] [PubMed] [Google Scholar]
- Sampaolesi M, Blot S, D’Antona G, Granger N, Tonlorenzi R, Innocenzi A, Mognol P, Thibaud JL, Galvez BG, Barthelemy I, Perani L, Mantero S, et al. Mesoangioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: Direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Rocancourt D, Marques L, Thorsteinsdottir S, Buckingham M. A Pax3/Dmrt2/Myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet. 2010;6:e1000897. doi: 10.1371/journal.pgen.1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, Jones DR, Du K, Jhala US, Simone C, Puri PL. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol Cell. 2007;28:200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano AL, Baeza-Raja B, Perdiguero E, Jardi M, Munoz-Canoves P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008;7:33–44. doi: 10.1016/j.cmet.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Sherwood RI, Christensen JL, Weissman IL, Wagers AJ. Determinants of skeletal muscle contributions from circulating cells, bone marrow cells, and hemato-poietic stem cells. Stem Cells. 2004;22:1292–1304. doi: 10.1634/stemcells.2004-0090. [DOI] [PubMed] [Google Scholar]
- Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI–SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L, Atchison ML. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004;18:2596–2601. doi: 10.1101/gad.1228204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- Takaesu G, Kang JS, Bae GU, Yi MJ, Lee CM, Reddy EP, Krauss RS. Activation of p38alpha/beta MAPK in myogenesis via binding of the scaffold protein JLP to the cell surface protein Cdo. J Cell Biol. 2006;175:383–388. doi: 10.1083/jcb.200608031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata T, Ishikawa F. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem Biophys Res Commun. 2003;301:250–257. doi: 10.1016/s0006-291x(02)03020-6. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: A regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Tedesco FS, Dellavalle A, Diaz-Manera J, Messina G, Cossu G. Repairing skeletal muscle: Regenerative potential of skeletal muscle stem cells. J Clin Invest. 2010;120:11–19. doi: 10.1172/JCI40373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- van Essen D, Engist B, Natoli G, Saccani S. Two modes of transcriptional activation at native promoters by NF-kappaB p65. PLoS Biol. 2009;7:e73. doi: 10.1371/journal.pbio.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wang H, Hertlein E, Bakkar N, Sun H, Acharyya S, Wang J, Carathers M, Davuluri R, Guttridge DC. NF-kappaB regulation of YY1 inhibits skeletal myogenesis through transcriptional silencing of myofibrillar genes. Mol Cell Biol. 2007;27:4374–4387. doi: 10.1128/MCB.02020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wilson BJ, Bates GJ, Nicol SM, Gregory DJ, Perkins ND, Fuller-Pace FV. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CF, Tellam RL. microRNA-26a targets the histone methyltransfer-ase enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR, Karin M, Wang JY, Puri PL. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol Cell Biol. 2000;20:3951–3964. doi: 10.1128/mcb.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- Yue R, Kang J, Zhao C, Hu W, Tang Y, Liu X, Pei G. Beta-arrestin1 regulates zebrafish hematopoiesis through binding to YY1 and relieving polycomb group repression. Cell. 2009;139:535–546. doi: 10.1016/j.cell.2009.08.038. [DOI] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: Potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]