Abstract
Microsomal PGE synthase 1 (mPGES-1) is the terminal enzyme in the induced state of prostaglandin E2 (PGE2) synthesis and constitutes a therapeutic target for rheumatoid arthritis (RA) treatment. We examined the role of the prostaglandin E synthase (PTGES) gene polymorphism in susceptibility to and severity of RA and related variations in the gene to its function. The PTGES gene polymorphism was analyzed in 3081 RA patients and 1900 controls from two study populations: Swedish Epidemiological Investigation of Rheumatoid Arthritis (EIRA) and the Leiden Early Arthritis Clinic (Leiden EAC). Baseline disease activity score (DAS28) was employed as a disease severity measure. mPGES-1 expression was analyzed in synovial tissue from RA patients with known genotypes using immunohistochemistry. In the Swedish study population, among women a significant association with risk for RA was observed for PTGES single-nucleotide polymorphisms (SNPs) in univariate analysis and for the distinct haplotype. These results were substantiated by meta-analysis of data from EIRA and Leiden EAC studies with overall OR 1.31 (95% confidence interval 1.11–1.56). Several PTGES SNPs were associated with earlier onset of disease or with higher DAS28 in women with RA. Patients with the genotype associated with higher DAS28 exhibited significantly higher mPGES-1 expression in synovial tissue. Our data reveal a possible influence of PTGES polymorphism on the pathogenesis of RA and on disease severity through upregulation of mPGES-1 at the sites of inflammation. Genetically predisposed individuals may develop earlier and more active disease owing to this mechanism.
Keywords: mPGES-1, gene polymorphism, rheumatoid arthritis, gender
INTRODUCTION
Rheumatoid arthritis (RA) is a chronic inflammatory disease characterized by persistent synovial inflammation resulting in joint destruction and severe disability. It is one of the most common complex autoimmune diseases, with up to 1% prevalence, and a higher prevalence, activity and earlier disease onset in women. Genetic factors significantly contribute to the development of RA and its severity.1 The major genetic risk factor for RA is the shared epitope alleles of the HLA-DRB1 gene locus.2, 3 Recently several common genetic variants have been discovered outside HLA-DRB1 locus, identifying important biological pathways predisposing to RA (for a review, see Raychaudhuri4).
Prostaglandin E2 (PGE2) is one of the key mediators of inflammation, pain and joint destruction in RA, and its production is an important target of anti-inflammatory drugs. Hence PGE2 biosynthetic pathway is of considerable interest to gene association and pharmacogenetic studies.5, 6 PGE2 is produced by conversion of arachidonic acid to PGH2 by cyclooxygenases (COX-1/COX-2) and subsequent conversion of PGH2 to PGE2 by terminal PGE synthases. Microsomal PGE synthase 1 (mPGES-1) is the terminal enzyme in the induced PGE2 production at the sites of inflammation7, 8 and has an important role in the pathogenesis of inflammatory arthritis.9, 10 In humans, mPGES-1 expression is induced by proinflammatory cytokines in RA synovial fibroblasts and synovial fluid monocytes11, 12 and markedly up-regulated in RA synovial tissue.13 In mice with experimental arthritis, genetic deletion of mPGES-1 results in significantly reduced disease incidence, severity and pain.9, 10 mPGES-1 is recognized as an alternative target for anti-inflammatory treatment with improved selectivity and safety compared with COX-1/COX-2.8
The human mPGES-1 is encoded by prostaglandin E synthase (PTGES) gene, which is localized at chromosome 9q34.3, spans 14.8 kb and contains three exons. According to NCBI single-nucleotide polymorphisms (SNPs) database, there are 249 SNPs in the PTGES gene region, including four coding SNPs (two synonymous and two non-synonymous SNPs). Whereas none of the studied variants in PTGES gene had a significant effect on the risk of lymphoma,14 non-fatal myocardian infarction or ischemic stroke,15 and breast cancer susceptibility,16 in a large-scale association study one SNP in the 5′ region of the PTGES gene was associated with hypertension in women in the Japanese population.17 SNPs in the PTGES gene could modify enzyme expression levels or activity and alter inducible PGE2 biosynthesis. Consequently, PTGES gene polymorphisms might contribute to inter-individual differences in susceptibility to or severity of RA, as well as clinical response to future mPGES-1 inhibitors. Hence, PTGES genetics may have clinical importance from disease risk assessment to choice of personalized treatment in RA.
Here, we studied the association between PTGES gene polymorphisms and risk and severity of RA. In addition, we examined whether PTGES gene polymorphism could contribute to altered mPGES-1 expression.
MATERIALS AND METHODS
Patients
In this study, two independent cohorts have been included: Swedish Epidemiological Investigation of Rheumatoid Arthritis (EIRA) and Leiden Early Arthritis Clinic (Leiden EAC). The first cohort consisted of 2012 RA patients and 1033 controls matched by age, sex and residential area from the EIRA, a population-based case–control study in Sweden (Table 1). A case was defined as a person who was diagnosed for the first time with RA based on the diagnostic criteria of the American College of Rheumatology.18 Data regarding age of disease onset were available for all patients, and data regarding baseline disease activity score (DAS28) for 887 (44%) RA patients. Further characteristics of the EIRA study are described elsewhere.19
Table 1. Clinical characteristics of EIRA and Leiden EAC studies.
| Characteristics | EIRA | Leiden EAC |
|---|---|---|
| No. of patients | 2012 | 1069 |
| No. of controls | 1033 | 867 |
| Country of origin | Sweden | The Netherlands |
| Female sex, patients (%) | 70.4 | 64 |
| Female sex, controls (%) | 74.6 | 21.9 |
| Age of onset, years (mean±SD) | 51.7±12.7 | 56.4±15.4 |
| Disease duration, years | <1 | <2 |
| ACPA positive (%) | (1265/2012) 62.8 | (375/625a) 60 |
| HLA-DRB1 SE (patients) (%) | (1486/2012) 73.8 | (590/842b) 70.1 |
At the replication stage, the second cohort consisted of 1069 Dutch Caucasian individuals with RA who fulfilled the ACR classification criteria for RA and 867 controls, who were unrelated Dutch Caucasians with no history of RA from EAC (Table 1). More details about the EAC study population have been described elsewhere.20 Approval was granted by the regional ethics committees and all participants gave their informed consent to participate in the study.
SNP genotyping
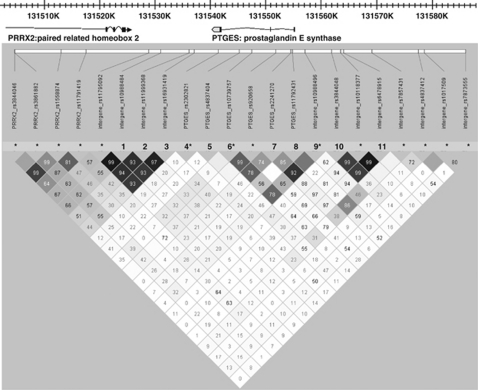
In all, 11 SNPs were selected across the PTGES gene locus and flanking regions (Figure 1). The selection was based on the minor allele frequency being higher than 10% according to NCBI for all SNPs except for rs11792431 (minor allele frequency 2.5%). In the replication phase, we genotyped three SNPs in PTGES locus, which showed significant association with RA risk. Genotyping was performed using TaqMan SNP genotyping assays (Applied Biosystems, Europe BV, The Netherlands). The average genotyping call rate was 96.6%, varying from 95 to 98% for different SNPs.
Figure 1.
Linkage disequilibrium map for the PTGES locus (r2). The SNPs marked with numbers were analyzed in the EIRA case–control study. The SNPs marked with * were analyzed using data from our previous genome-wide association scan.
In addition, we included in the analysis data from our previous genome-wide association scan21 for 14 SNPs in the PTGES locus and inter-gene regions with neighboring PRRX2 and TOR1B genes (Figure 1). Genotyping was performed on the Illumina HumanHap300 (version 1.0) array (Illumina, San Diego, CA, USA).21
Immunohistochemical analysis
Synovial tissue biopsy specimens from 24 RA patients from EIRA carrying different PTGES genotypes were obtained during arthroscopy. Synovial tissue samples were snap frozen and kept at −70°C until sectioned. Staining of cryostat sections with rabbit polyclonal anti-human mPGES-1 antiserum was performed as previously described.13 Irrelevant rabbit IgG was used as a negative control. Stained tissue sections were examined using a Polyvar II microscope (Reichert-Jung, Vienna, Austria) and photographed with a digital Leica camera 300F (Leica, Cambridge, UK). The positive staining was assessed quantitatively using computer-assisted image analysis and expressed as the percentage of the total area of counterstained tissue.
Statistical analysis
Genotype and allele frequencies in the patient and the control groups were compared using the χ2-test and/or Fisher's exact test when appropriate. Haplotype frequencies were estimated using HaploView 4.0 (http://www.broad.mit.edu/mpg/haploview/index.php), and the permutation test was set to 1 000 000 permutations for the single markers and for the haplotypes, with P<0.05 for significance of empirical P-value. An odds ratio (OR) with 95% confidence interval (CI) was calculated for the association between different genetic factors and risk of RA. Evaluation of linkage disequilibrium and deviations from Hardy–Weinberg equilibrium were performed using HaploView. The distribution of genotypes for all studied SNPs within the RA group and within the control group was in agreement with Hardy–Weinberg equilibrium (P>0.05). Before combining for meta-analysis, the EIRA and Leiden EAC data were tested for the homogeneity in the ORs, using a Pearson χ2 goodness-of-fit test as described elsewhere.22 In the meta-analysis of haplotype-frequency data, the RA cases versus controls were compared in EIRA and Leiden EAC studies using Mantel–Haenszel analysis assuming random effects for the ORs.22
In RA patients stratified by sex, the associations between PTGES polymorphisms and DAS28 and age at the first diagnosis of RA, as well as the results of functional study, were analyzed using Mann–Whitney's U-test with Bonferroni corrections for multiple comparisons. P-values <0.05 were considered statistically significant.
RESULTS
PTGES polymorphisms and risk of developing RA
In the discovery phase, we analyzed allelic and haplotype frequencies of 11 SNPs in the PTGES gene in the entire Swedish cohort, as well as in men and women separately. An association between the SNP rs16931419 and risk of RA was observed in the allelic model (women and men combined), but the difference was not significant after permutation test (Table 2). Among women three SNPs, rs10988484, rs11999368 and rs16931419, from a single linkage disequilibrium block in the gene locus downstream the PTGES gene showed higher minor allele frequencies in female RA patients compared with those in female controls (Table 2). The association of the SNP rs16931419 with the risk of RA was still apparent after permutation test (OR 1.38; 95% CI 1.14–1.67, empirical P-value=0.0082). Among men, a trend toward an association between the SNP rs3844048 and risk of RA was observed, but this association was not statistically significant after permutation test (Table 2).
Table 2. Minor allele frequencies of SNPs at PTGES locus in the Swedish and the Dutch populations.
| No. | SNP | Position | MA | MAF, cases | MAF, controls | OR (95% CI) | P | P* | |
|---|---|---|---|---|---|---|---|---|---|
| The Swedish population | |||||||||
| 1 | rs10988484 | 3′ near gene | T | All | 0.154 | 0.138 | 1.14 (0.98–1.33) | 0.0966 | NS |
| F | 0.160 | 0.129 | 1.28 (1.07–1.54) | 0.0076 | 0.0591 | ||||
| M | 0.141 | 0.163 | 0.84 (0.63–1.12) | 0.23 | NS | ||||
| 2 | rs11999368 | 3′ near gene | T | All | 0.148 | 0.134 | 1.12 (0.96–1.31) | 0.15 | NS |
| F | 0.152 | 0.124 | 1.26 (1.05–1.52) | 0.0138 | 0.1035 | ||||
| M | 0.138 | 0.162 | 0.83 (0.62–1.10) | 0.19 | NS | ||||
| 3 | rs16931419 | 3′ near gene | C | All | 0.149 | 0.127 | 1.2 (1.02–1.41) | 0.02 | NS |
| F | 0.153 | 0.116 | 1.38 (1.14–1.67) | 0.0010 | 0.0082 | ||||
| M | 0.139 | 0.160 | 0.84 (0.63–1.14) | 0.26 | NS | ||||
| 4 | rs2302821 | 3′-UNR | C | All | 0.091 | 0.091 | 1.0 (0.83–1.21) | 0.95 | NS |
| F | 0.093 | 1.0 | 0.93 (0.75–1.15) | 0.49 | NS | ||||
| M | 0.087 | 0.065 | 1.36 (0.91–2.04) | 0.13 | NS | ||||
| 5 | rs4837404 | Intron 2 | G | All | 0.333 | 0.325 | 1.04 (0.92–1.16) | 0.54 | NS |
| F | 0.334 | 0.323 | 1.05 (0.92–1.20) | 0.44 | NS | ||||
| M | 0.330 | 0.331 | 0.99 (0.80–1.24) | 0.93 | NS | ||||
| 6 | rs10739757 | Intron 2 | C | All | 0.095 | 0.101 | 0.93 (0.78–1.12) | 0.44 | NS |
| F | 0.092 | 0.102 | 0.89 (0.72–1.1) | 0.27 | NS | ||||
| M | 0.103 | 0.099 | 1.01 (0.91–1.12) | 0.82 | NS | ||||
| 7 | rs2241270 | Intron 1 | T | All | 0.096 | 0.097 | 1.00 (0.94–1.06) | 0.93 | NS |
| F | 0.096 | 0.096 | 1.00 (0.83–1.23) | 0.95 | NS | ||||
| M | 0.098 | 0.100 | 0.99 (0.70–1.39) | 0.93 | NS | ||||
| 8 | rs11792431 | Exon 1 | A | All | 0.056 | 0.054 | 1.04 (0.82–1.32) | 0.74 | NS |
| F | 0.053 | 0.052 | 1.00 (0.76–1.33) | 0.94 | NS | ||||
| M | 0.063 | 0.058 | 1.09 (0.71–1.69) | 0.69 | NS | ||||
| 9 | rs10988496 | 5′ near gene | G | All | 0.104 | 0.104 | 1.00 (0.84–1.20) | 0.96 | NS |
| F | 0.101 | 0.103 | 0.97 (0.79–1.20) | 0.80 | NS | ||||
| M | 0.101 | 0.107 | 1.00 (0.75–1.45) | 0.81 | NS | ||||
| 10 | rs3844048 | 5′ near gene | G | All | 0.301 | 0.308 | 0.97 (0.86–1.09) | 0.58 | NS |
| F | 0.303 | 0.291 | 1.05 (0.92–1.22) | 0.40 | NS | ||||
| M | 0.295 | 0.358 | 0.75 (0.60–0.93) | 0.0114 | NS | ||||
| 11 | rs6478915 | 5′ near gene | G | All | 0.250 | 0.247 | 1.01 (0.89–1.15) | 0.82 | NS |
| F | 0.252 | 0.232 | 1.11 (0.96–1.29) | 0.16 | NS | ||||
| M | 0.244 | 0.289 | 0.79 (0.63–1.01) | 0.05 | NS | ||||
| The Dutch population | |||||||||
| 1 | rs10988484 | 3′ near gene | T | All | 0.152 | 0.152 | 1.00 (0.83–1.19) | 0.95 | NS |
| F | 0.151 | 0.129 | 1.20 (0.85–1.70) | 0.28 | NS | ||||
| M | 0.152 | 0.159 | 0.95 (0.74–1.22) | 0.72 | NS | ||||
| 2 | rs11999368 | 3′ near gene | T | All | 0.145 | 0.141 | 1.03 (0.86–1.24) | 0.74 | NS |
| F | 0.138 | 0.124 | 1.14 (0.82–1.62) | 0.46 | NS | ||||
| M | 0.157 | 0.146 | 1.08 (0.85–1.39) | 0.49 | NS | ||||
| 3 | rs16931419 | 3′ near gene | C | All | 0.139 | 0.138 | 1.01 (0.84–1.22) | 0.93 | NS |
| F | 0.131 | 0.116 | 1.16 (0.80–1.67) | 0.42 | NS | ||||
| M | 0.153 | 0.145 | 1.06 (0.83–1.36) | 0.59 | NS | ||||
Abbreviations: MAF, minor allele frequency; NS, not significant; F, females; M, males.
*P-value after 1 000 000 permutations.
To exclude possible correlation with neighboring genes owing to extended linkage disequilibrium, we expanded the analysis by including 14 SNPs in the PTGES locus and intergenic areas with the PRPX2 or the TOR1B genes using data from our previous genome-wide association scan21 (Figure 1). There was no evidence for an association between SNPs in proximity to the PRPX2 gene or the TOR1B gene and susceptibility to RA (data are not shown).
Further, we examined the frequency of haplotypes (only with the frequency above 0.01) based on the three SNPs significantly associated with the risk of RA. The frequency of the haplotype TTC (susceptible haplotype) was significantly higher in women with RA than in women without RA (OR 1.34; 95% CI 1.11–1.63, empirical P-value=0.0077) (Table 3). The frequency of the opposite haplotype CCT (protective haplotype) was significantly lower in women with RA compared with controls (OR 0.78; 95% CI 0.66–0.93, empirical P-value=0.0172). We did not find significant associations between these haplotypes and RA risk among men (Table 3).
Table 3. Haplotype frequencies of PTGES variations in female patients with RA and female controls from the Swedish and the Dutch populations.
| Haplotype | HF | HF, cases | HF, controls | OR (95% CI) | P | P* | |
|---|---|---|---|---|---|---|---|
| The Swedish population | |||||||
| CCT | All | 0.843 | 0.837 | 0.854 | 0.88 (0.76–1.02) | 0.09 | NS |
| F | 0.842 | 0.831 | 0.862 | 0.78 (0.66–0.93) | 0.0066 | 0.0172 | |
| M | 0.844 | 0.851 | 0.828 | 1.19 (0.90–1.57) | 0.22 | NS | |
| TTC | All | 0.130 | 0.137 | 0.117 | 1.19 (1.01–1.40) | 0.03 | NS |
| F | 0.128 | 0.139 | 0.107 | 1.34 (1.11–1.63) | 0.0030 | 0.0077 | |
| M | 0.136 | 0.131 | 0.146 | 0.88 (0.65–1.18) | 0.39 | NS | |
| TCT | All | 0.011 | 0.011 | 0.011 | 0.93 (0.56–1.57) | 0.75 | NS |
| F | 0.012 | 0.012 | 0.013 | 0.97 (0.55–1.71) | 0.86 | NS | |
| M | 0.005 | 0.004 | 0.007 | 0.54 (0.16–1.8) | 0.30 | NS | |
| The Dutch populations | |||||||
| CCT | All | 0.835 | 0.836 | 0.834 | 1.01 (0.85–1.20) | 0.83 | NS |
| F | 0.844 | 0.841 | 0.855 | 0.89 (0.65–1.24) | 0.50 | NS | |
| M | 0.827 | 0.826 | 0.827 | 0.99 (0.78–1.25) | 0.97 | NS | |
| TTC | All | 0.126 | 0.128 | 0.125 | 1.01 (0.84–1.23) | 0.82 | NS |
| F | 0.12 | 0.125 | 0.105 | 1.21 (0.84–1.75) | 0.29 | NS | |
| M | 0.131 | 0.132 | 0.131 | 1.00 (0.77–1.30) | 0.97 | NS | |
| TCT | All | 0.02 | 0.018 | 0.025 | 0.16 (0.46–1.13) | 0.13 | NS |
| F | 0.02 | 0.019 | 0.021 | 0.90 (0.40–2.00) | 0.80 | NS | |
| M | 0.02 | 0.015 | 0.025 | 0.61 (0.31–1.19) | 0.14 | NS | |
Abbreviations: NS, not significant; HF, haplotype frequency; F, females; M, males.
Haplotypes are based on PTGES SNPs rs10988484, rs11999368 and rs16931419 and are considered only with the frequency above 0.01. *P-value after 1 000 000 permutations.
In the replication phase, we analyzed the association of the three SNPs, rs10988484, rs11999368 and rs16931419, with RA susceptibility in the Dutch cohort. Similarly to the Swedish cohort, these SNPs showed higher minor allelic frequencies in female patients with RA compared with the frequencies in female controls, though the difference was not statistically significant (Table 2). In the same way, the frequency of the susceptible haplotype TTC was higher in women with RA than in women without RA, though the difference was not significant (Table 3). The allelic frequencies among female controls in the two populations were identical, whereas the haplotype frequencies were highly similar.
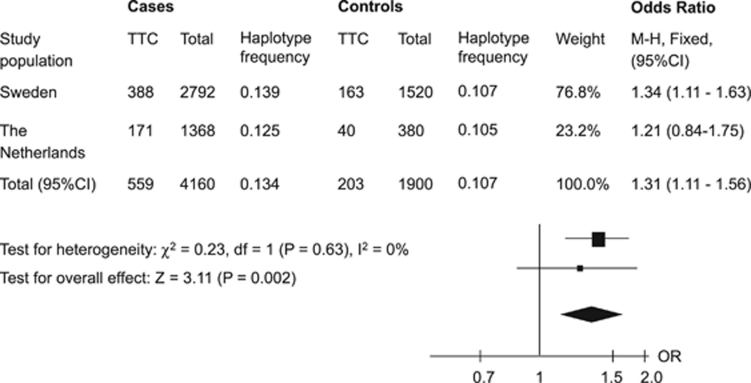
Genetic variations with low or moderate effects on disease susceptibility may not be evident in an individual study owing to low statistical power. Thus, we performed a meta-analysis combining our results from the Swedish and the Dutch cohorts to verify the significance of PTGES polymorphisms for susceptibility to RA. Test for the heterogeneity between EIRA and EAC showed no significant differences (P>0.05). The overall statistical power in the combined analysis was >80% (in two-tailed test) to detect associations with OR≥1.3, given the prevalence of studied genetic factors. The meta-analysis demonstrated a statistically significant association between the susceptible haplotype TTC and risk of developing RA in women with an overall OR of 1.31 (95% CI 1.11–1.56) and P=0.002 (Figure 2).
Figure 2.
The association of haplotype TTC with susceptibility to rheumatoid arthritis in women in two study populations and the meta-analysis. The squares represent odds ratios (ORs) and lines represent confidence intervals (95% CI).
PTGES polymorphisms and age of onset and activity of RA
RA is a heterogeneous syndrome with relatively broad variations in disease onset and activity. Therefore, we tested the correlation between these parameters and PTGES genetic markers in patients from EIRA. Baseline DAS28 and mPGES-1 expression in RA synovial tissue were used as disease activity measures.
There was no association of RA risk haplotype TTC with baseline DAS28 scores or age of disease onset in women or men. The minor allele of the SNP rs10739757 corresponded to 3 years earlier onset of RA in female patients (median 51, range 19–74 vs median 54, range 16–82, P=0.0065) (Table 4).
Table 4. Association of PTGES genotypes with disease onset and activity in female patients with RA.
| Disease parameters | SNP | Genotype | Median (range) | Counts | P-value |
|---|---|---|---|---|---|
| Disease onset, years | rs10739757 | C | 51 (19–74) | 246 | 0.0065 |
| TT | 54 (16–82) | 1136 | |||
| DAS28, score | rs4837404 | AA | 5.57 (1.74–8.89) | 266 | 0.0080 |
| G | 5.28 (1.57–8.29) | 321 | |||
| DAS28, score | rs2302821 | C | 5.57 (2.39–8.89) | 114 | 0.0321 |
| AA | 5.31 (1.57–8.37) | 491 | |||
| DAS28, score | rs11792431 | A | 5.63 (3.82–8.89) | 63 | 0.0369 |
| GG | 5.32 (1.57–8.66) | 536 |
Abbreviation: DAS28, disease activity score.
Furthermore, we observed that the homozygous major allele in the SNP rs4837404 was associated with higher DAS28 scores in women with RA (median 5.57, range 1.74–8.89, vs median 5.28, range 1.57–8.29, P=0.0080), but not in men. In addition, the minor allele in the SNP rs2302821 showed a trend toward association with higher DAS28 scores in women with RA (median 5.57, range 2.39–8.89 vs median 5.31, range 1.57–8.37, P=0.0321). Also, the female RA patients carrying the minor allele of the SNP rs11792431 showed a trend toward higher DAS28 scores (median 5.63, range 3.82–8.89 vs median 5.32, range 1.57–8.66, P=0.0369) (Table 4).
All mentioned variations related to disease course were in low linkage disequilibrium (r2≤0.34) with SNPs included in the susceptibility haplotype TTC.
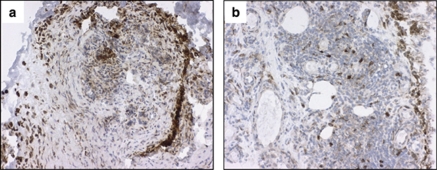
Using immunohistochemical analysis, we evaluated the expression of mPGES-1 in RA synovial tissue in relation to PTGES genotypes. There was no difference in mPGES-1 expression in relation to RA risk haplotype TTC. mPGES-1 expression was significantly higher in RA synovial tissue from patients (mean±SD age, 52.8±14.4 years) carrying the minor allele C in SNP rs2302821 compared with the patients (mean±SD age, 54.4±14.1 years) who did not carry that allele (median 12.9% of the stained area, range 8.1–23.6 vs median 7.1%, range 0.9–10.5, P<0.001, n=9 and 15, respectively) (Figure 3). No differences were observed in mPGES-1 expression in relation to gender or treatment of patients with NSAIDs, MTX or prednisolone.
Figure 3.
The expression of mPGES-1 in synovial tissue of RA patients in relation to PTGES genotypes. Photographs illustrating brown immunoperoxidase staining of mPGES-1-positive cells in representative synovial tissue sections from RA patients. (a) Sample carrying the minor allele C in SNP rs2302821; (b) sample that did not carry that allele, counterstained with hematoxylin (original magnification × 250).
Discussion
The present study demonstrates the association of variations in the PTGES gene, encoding mPGES-1, with susceptibility to RA in women. The data are consistent for single SNPs and haplotypes in the large study from Sweden. Moreover, the finding was substantiated by a meta-analysis of data from the EIRA and Leiden EAC studies. We also found that the PTGES genotypes are likely to be related to earlier disease onset and disease severity, as they were associated with higher baseline disease activity scores and related to higher expression of mPGES-1 in synovial tissue biopsies from RA patients.
PGE2 is actively involved in the regulation of immune response, inflammation and pain, and polymorphisms in the genes in the PGE2 biosynthetic pathway are important risk factors for cardiovascular, neoplastic and inflammatory diseases.6, 23 The role of gene polymorphisms in the PGE2 pathway in relation to RA risk and severity is less investigated. Two case–control studies have revealed a possible protective influence of COX-2 −765C allele on the susceptibility to RA and disease activity.24, 25 To our knowledge, the present study is the first one to examine an association between PTGES polymorphisms and the risk of developing RA.
We performed an association study in two independent populations. In a large sample from Sweden, we found the association between three SNPs from the same recombination block, which spread approximately within 9 kb downstream to PTGES, and RA risk in women. This significant association was apparent for the SNP rs16931419 (3 kb downstream to PTGES) after 1 000 000 permutations. Moreover, the TTC haplotype based on the SNPs from this recombination block demonstrated a significant association with RA risk in women in the Swedish population.
We expanded the analysis by including 14 SNPs in the PTGES locus and intergenic areas with the PRPX2 or the TOR1B genes to exclude possible association with neighboring genes. No evidence for the association between the SNPs in proximity to the PRPX2 and the TOR1B genes and RA risk was seen in our study population.
In order to replicate our finding, we analyzed the association between three PTGES SNPs included in the haplotype and risk of RA in a Dutch cohort. Though the respective case–control frequencies had a similar pattern in RA females from Leiden EAC as in EIRA, there was no significant association between single SNPs or haplotype and susceptibility to RA in the Dutch cohort. The power of the replication study was though limited by the small size of the female control group. To increase the power of the study, we performed a meta-analysis of the data from EIRA and Leiden EAC studies and confirmed the association between TTC haplotype and increased risk to RA in women. This finding substantiates the evidence that PTGES polymorphisms might be important for susceptibility to RA in women.
The revealed genetic association was consistent for women but not detected for men. The higher prevalence of RA in women suggests that genetic risk of RA might vary with sex. Indeed, there are a number of associations that show genetic effects in only one sex, or reciprocal effects on risk of RA in males and females.26, 27 Thus, there is strong association of SNP rs2900180 in the TRAF-C5 locus with RA without obvious evidence of an effect in males.27 Also, SNP rs11761231 has been reported in a large study to be associated with RA in females, but has no effect on disease status in males.26 On the other hand, several SNPs in NCF4 (rs729749), NCF2 (rs789181) and RAC2 (rs1476002) genes were found to be mildly associated with RA only in men.28
Moreover, the association of COX-2 polymorphisms with disease risk and response to NSAID varies between sexes,29, 30 most likely as a result of sexual dimorphism in PGE2 metabolism. Systemic biosynthesis of PGE2 differs markedly between males and females in human, as in mice.31, 32 In rodents, variations in COX-2 and mPGES-1 expression at basal and pathological conditions33, 34 and different patterns of EP receptor expression35 have been reported in females and males. Thus, sex-dependent association of PTGES polymorphisms with RA susceptibility may reflect different response to this genetic trigger in females and males owing to differences in PGE2 metabolism. However, the absence of an association between PTGES polymorphism and RA risk in men might also reflect the small number of individuals and low study power, which needs further validation.
The SNPs associated with RA are in a single linkage disequilibrium block and located downstream to the 3′ untranslated region (3′UTR) of the PTGES gene. 3′UTR is involved in post-transcriptional gene regulation via mRNA stability, the mechanism for which is revealed in the regulation of mouse Ptges gene expression.36 These SNPs may be regulatory or in linkage disequilibrium with other regulatory polymorphisms in 3′UTR that could similarly modulate the human mPGES-1 expression and predispose individuals to RA.
In our study, we found that gene polymorphisms in PTGES might have not only a disease-predisposing impact but also a disease-modifying effect, as two SNPs within PTGES gene were related to early disease onset or baseline DAS28 scores. Revealing the relations between genetic variations and certain functions or phenotypes is currently the most challenging task for the genetics of complex diseases. We investigated whether gene variations associated with disease activity could relate to mPGES-1 expression. We previously demonstrated the expression of mPGES-1 in synovial biopsies as a reliable parameter of inflammatory activity in RA.12 Our data from the current study indicate that mPGES-1 expression seems to be dependent on the genotype, which was also reflected by genetic association with the clinical measure of baseline DAS28. Individuals with the minor allele C in PTGES rs2302821 that showed a trend toward association with higher DAS28 had significantly higher mPGES-1 expression in RA synovial tissue, which represents additional phenotypic replication for this association. Although the total patient number in our experiment is small, it represents a unique collection of tissue samples from RA patients with distinct genotypes.
In conclusion, we provide the first evidence that PTGES gene polymorphisms are associated with susceptibility to RA in women. In addition, PTGES polymorphisms were related to earlier disease onset, higher baseline disease activity scores and might contribute to higher expression of mPGES-1 in synovial tissue biopsies from RA patients. These data suggest the importance of PTGES gene variants in the assessment of RA risk and severity.
Acknowledgments
We thank the RA patients and controls for participating in the study, and Eva Jemseby and Gul-Britt Almgren for invaluable contributions to the collection of data and maintenance of the EIRA database. This study is supported by the Swedish Research Council, Swedish Rheumatism Association, King Gustaf V 80 years Foundation, Swedish Society of Medicine, Karolinska Institutet Foundation, European Commission Sixth Framework Program Autocure and European Commission Seventh Framework Program Masterswitch.
There is potential duality of interest since Dr M Korotkova was recently partially employed by Actar AB.
References
- Klareskog L, Alfredsson L, Rantapaa-Dahlqvist S, Berglin E, Stolt P, Padyukov L. What precedes development of rheumatoid arthritis. Ann Rheum Dis. 2004;63 (Suppl 2:ii28–ii31. doi: 10.1136/ard.2004.028225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XJ, Olsen NJ, Pincus T, Stastny P. HLA-DR alleles with naturally occurring amino acid substitutions and risk for development of rheumatoid arthritis. Arthritis Rheum. 1990;33:939–946. doi: 10.1002/art.1780330704. [DOI] [PubMed] [Google Scholar]
- Moreno I, Valenzuela A, Garcia A, Yelamos J, Sanchez B, Hernanz W. Association of the shared epitope with radiological severity of rheumatoid arthritis. J Rheumatol. 1996;23:6–9. [PubMed] [Google Scholar]
- Raychaudhuri S. Recent advances in the genetics of rheumatoid arthritis. Curr Opin Rheumatol. 2010;22:109–118. doi: 10.1097/BOR.0b013e328336474d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halushka MK, Halushka PV. Toward individualized analgesic therapy: functional cyclooxygenase 1 and 2 haplotypes. Clin Pharmacol Ther. 2006;79:404–406. doi: 10.1016/j.clpt.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Skarke C, Schuss P, Kirchhof A, Doehring A, Geisslinger G, Lotsch J. Pyrosequencing of polymorphisms in the COX-2 gene (PTGS2) with reported clinical relevance. Pharmacogenomics. 2007;8:1643–1660. doi: 10.2217/14622416.8.12.1643. [DOI] [PubMed] [Google Scholar]
- Jakobsson PJ, Thoren S, Morgenstern R, Samuelsson B. Identification of human prostaglandin E synthase: a microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc Natl Acad Sci USA. 1999;96:7220–7225. doi: 10.1073/pnas.96.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson B, Morgenstern R, Jakobsson P-J. Membrane prostaglandin E synthase-1: a novel therapeutic target. Pharmacol Rev. 2007;59:207–224. doi: 10.1124/pr.59.3.1. [DOI] [PubMed] [Google Scholar]
- Kamei D, Yamakawa K, Takegoshi Y, et al. Reduced pain hypersensitivity and inflammation in mice lacking microsomal prostaglandin E synthase-1. J Biol Chem. 2004;279:33684–33695. doi: 10.1074/jbc.M400199200. [DOI] [PubMed] [Google Scholar]
- Trebino C, Stock J, Gibbons C, et al. Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proc Natl Acad Sci USA. 2003;100:9044–9049. doi: 10.1073/pnas.1332766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stichtenoth DO, Thoren S, Bian H, Peters-Golden M, Jakobsson PJ, Crofford LJ. Microsomal prostaglandin E synthase is regulated by proinflammatory cytokines and glucocorticoids in primary rheumatoid synovial cells. J Immunol. 2001;167:469–474. doi: 10.4049/jimmunol.167.1.469. [DOI] [PubMed] [Google Scholar]
- Korotkova M, Westman M, Gheorghe KR, et al. Effects of antirheumatic treatments on the prostaglandin E2 biosynthetic pathway. Arthritis Rheum. 2005;52:3439–3447. doi: 10.1002/art.21390. [DOI] [PubMed] [Google Scholar]
- Westman M, Korotkova M, af Klint E, et al. Expression of microsomal prostaglandin E synthase in rheumatoid arthritis synovium. Arthritis Rheum. 2004;50:1774–1780. doi: 10.1002/art.20286. [DOI] [PubMed] [Google Scholar]
- Hoeft B, Becker N, Deeg E, Beckmann L, Nieters A. Joint effect between regular use of non-steroidal anti-inflammatory drugs, variants in inflammatory genes and risk of lymphoma. Cancer Causes Control. 2008;19:163–173. doi: 10.1007/s10552-007-9082-9. [DOI] [PubMed] [Google Scholar]
- Lemaitre RN, Rice K, Marciante K, et al. Variation in eicosanoid genes, non-fatal myocardial infarction and ischemic stroke. Atherosclerosis. 2009;204:e58–e63. doi: 10.1016/j.atherosclerosis.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham JE, Harrington P, Driver KE, et al. Common polymorphisms in the prostaglandin pathway genes and their association with breast cancer susceptibility and survival. Clin Cancer Res. 2009;15:2181–2191. doi: 10.1158/1078-0432.CCR-08-0716. [DOI] [PubMed] [Google Scholar]
- Iwai N, Tago N, Yasui N, et al. Genetic analysis of 22 candidate genes for hypertension in the Japanese population. J Hypertens. 2004;22:1119–1126. doi: 10.1097/00004872-200406000-00012. [DOI] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Klareskog L, Stolt P, Lundberg K, et al. A new model for an etiology of rheumatoid arthritis. Smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- van Aken J, van Bilsen JH, Allaart CF, Huizinga TW, Breedveld FC. The Leiden Early Arthritis Clinic. Clin Exp Rheumatol. 2003;21 (Suppl 31:S100–S105. [PubMed] [Google Scholar]
- Plenge RM, Seielstad M, Padyukov L, et al. TRAF1–C5 as a risk locus for rheumatoid arthritis — a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- Helmersson J, Arnlov J, Axelsson T, Basu S. A polymorphism in the cyclooxygenase 1 gene is associated with decreased inflammatory prostaglandin F2alpha formation and lower risk of cardiovascular disease. Prostaglandins Leukot Essent Fatty Acids. 2009;80:51–56. doi: 10.1016/j.plefa.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Lee KH, Kim HS, El-Sohemy A, Cornelis MC, Uhm WS, Bae SC. Cyclooxygenase-2 genotype and rheumatoid arthritis. J Rheumatol. 2006;33:1231–1234. [PubMed] [Google Scholar]
- Yun HR, Lee SO, Choi EJ, Shin HD, Jun JB, Bae SC. Cyclooxygenase-2 polymorphisms and risk of rheumatoid arthritis in Koreans. J Rheumatol. 2008;35:763–769. [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of 14 000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang JJ, Morris AP. Assessment of sex-specific effects in a genome-wide association study of rheumatoid arthritis. BMC Proc. 2009;3 (Suppl 7:S90. doi: 10.1186/1753-6561-3-s7-s90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson LM, Lindqvist AK, Kallberg H, et al. A case-control study of rheumatoid arthritis identifies an associated single nucleotide polymorphism in the NCF4 gene, supporting a role for the NADPH-oxidase complex in autoimmunity. Arthritis Res Ther. 2007;9:R98. doi: 10.1186/ar2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay R, Jain M, Kumar S, Ghoshal UC, Mittal B. Functional polymorphisms of cyclooxygenase-2 (COX-2) gene and risk for esophageal squamous cell carcinoma. Mutat Res. 2009;663:52–59. doi: 10.1016/j.mrfmmm.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Vogel U, Segel S, Dethlefsen C, et al. Associations between COX-2 polymorphisms, blood cholesterol and risk of acute coronary syndrome. Atherosclerosis. 2010;1:155–162. doi: 10.1016/j.atherosclerosis.2009.08.036. [DOI] [PubMed] [Google Scholar]
- Murphey LJ, Williams MK, Sanchez SC, et al. Quantification of the major urinary metabolite of PGE2 by a liquid chromatographic/mass spectrometric assay: determination of cyclooxygenase-specific PGE2 synthesis in healthy humans and those with lung cancer. Anal Biochem. 2004;334:266–275. doi: 10.1016/j.ab.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wang M, Yu Y, Lawson J, Funk CD, Fitzgerald GA. Cyclooxygenases, microsomal prostaglandin E synthase-1, and cardiovascular function. J Clin Invest. 2006;116:1391–1399. doi: 10.1172/JCI27540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JC, Sasser JM, Pollock DM, Pollock JS. Sexual dimorphism in renal production of prostanoids in spontaneously hypertensive rats. Hypertension. 2005;45:406–411. doi: 10.1161/01.HYP.0000156879.83448.93. [DOI] [PubMed] [Google Scholar]
- Francois H, Coffman TM. Prostanoids and blood pressure: which way is up. J Clin Invest. 2004;114:757–759. doi: 10.1172/JCI22929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audoly LP, Tilley SL, Goulet J, et al. Identification of specific EP receptors responsible for the hemodynamic effects of PGE2. Am J Physiol. 1999;277:H924–H930. doi: 10.1152/ajpheart.1999.277.3.H924. [DOI] [PubMed] [Google Scholar]
- Degousee N, Angoulvant D, Fazel S, et al. c-Jun N-terminal kinase-mediated stabilization of microsomal prostaglandin E2 synthase-1 mRNA regulates delayed microsomal prostaglandin E2 synthase-1 expression and prostaglandin E2 biosynthesis by cardiomyocytes. J Biol Chem. 2006;281:16443–16452. doi: 10.1074/jbc.M602815200. [DOI] [PubMed] [Google Scholar]