Abstract
iTRAQ (isobaric Tags for Relative and Absolute Quantitation) is a technique that allows simultaneous quantitation of proteins in multiple samples. In this paper, we describe a Bayesian hierarchical model-based method to infer the relative protein expression levels and hence to identify differentially expressed proteins from iTRAQ data. Our model assumes that the measured peptide intensities are affected by both protein expression levels and peptide specific effects. The values of these two effects across experiments are modeled as random effects. The nonrandom missingness of peptide data is modeled with a logistic regression which relates the missingness probability for a peptide with the expression level of the protein that produces this peptide. We propose a Markov chain Monte Carlo method for the inference of model parameters, including the relative expression levels across samples. Our simulation results suggest that the estimates of relative protein expression levels based on the MCMC samples have smaller bias than those estimated from ANOVA models or fold changes. We apply our method to an iTRAQ dataset studying the roles of Caveolae for postnatal cardiovascular function.
Keywords: Bayesian hierarchical model, iTRAQ, Mixed-effects model, Nonignorable missing, Protein quantitation
1 Introduction
One main objective of proteomic research is to detect and quantify all proteins present in a biological sample. iTRAQ, a shotgun technique using Isobaric Tags for Relative and Absolute Quantitation, has become commonly used because of its improved quantitative reproducibility and higher quantification sensitivity [16] compared to other methods such as 2DE [9], ICAT [3], and DIGE [4, 10]. Using four or eight isobaric tags, iTRAQ can simultaneously analyze up to eight biological samples [2, 12]. Peptides digested from different samples of protein mixtures are labeled with different tags independently, mixed together, separated, and studied by MS (mass spectrometry) and MS/MS (tandem mass spectrometry). The resulting collection of mass spectra provides information on peptide identification and quantification, which can be utilized to identify and quantify relative protein expression levels.
We use the data from an iTRAQ experiment with four isobaric tags (114, 115, 116, and 117) as an example to illustrate the iTRAQ data format in Table 1. Each row represents a specific peptide identified from a software, such as MASCOT [11], which searches a protein sequence database to identify the peptide corresponding to a specific peak in the mass spectrum. The peptides thus identified are given in the second column. The peak areas for different samples labeled with different tags are shown in the last four columns, and their values can be used to calculate the relative abundance of a given peptide across samples. Each peptide may arise from different spectra and hence have multiple observations in an experiment. For example, the first three rows in Table 1 correspond to the same peptide across all the spectra. Missing peptides is a common phenomenon in iTRAQ data. That is, a peptide may be only observed in some of the samples, or some spectra, or some experiments. For example, the seventh row in Table 1 shows that peptide “DVDEIEAWISEK” is only observed in the samples labeled with 114 and 117. The fifth row indicates that in one spectrum, the intensities of the peptide “DLASVQALLR” are missing in all the samples. When multiple experiments are conducted, a peptide may be found to be missing in one experiment but observed in some other experiments (not shown in Table 1).
Table 1.
An example of the iTRAQ data when four isobaric tags are used. Each row corresponds to a specific peak in the mass spectrum. The first column gives the protein ID, the second column gives the peptide sequence that corresponds to this peak, and the last four columns give the peak areas for the four tags
| Protein accessions | Peptide sequence | Area114 | Area115 | Area116 | Area117 |
|---|---|---|---|---|---|
| IPI00798592.1 | ADVVESWIGEK | 22.03 | 29.88 | 29.08 | 36.89 |
| IPI00798592.1 | ADVVESWIGEK | 6.32 | 6.91 | 6.8 | 8.13 |
| IPI00798592.1 | ADVVESWIGEK | 5.3 | 3.84 | 3.66 | 10.26 |
| IPI00798592.1 | DLASVQALLR | 31.36 | 33.68 | 59.77 | 41.93 |
| IPI00798592.1 | DLASVQALLR | NA | NA | NA | NA |
| IPI00798592.1 | DLASVQALLR | 54.56 | 64.83 | 114.21 | 86.9 |
| IPI00798592.1 | DVDEIEAWISEK | NA | 7.11 | 13.6 | NA |
| IPI00798592.1 | DVDETIGWIK | 15.33 | 32.09 | 75.23 | 33.78 |
| IPI00798592.1 | … | … | … | … | … |
As seen above, the basic unit of iTRAQ data is the peptide. Each peptide has an associated intensity level. Several factors can affect the observed peptide intensities, i.e., the area columns in Table 1. The most obvious factor is the level of the protein in the sample that generates the peptide. Peptide specific features, such as ionization and fragmentation efficiency, affect the intensity levels for different peptides derived from the same protein. This is easily seen in Table 1, where all peptides are derived from the same protein. In addition, other factors such as sample preparation and experimental variation also contribute to the variabilities in the observed iTRAQ data. Hill et al. [5] illustrate in detail the possible sources of variations in iTRAQ data.
Another commonly encountered issue in iTRAQ data analysis is data missingness. Due to the nature of the technology, overlap in protein and peptide identifications between replicate experiments is less than ideal, and certain peptides are only observed for some samples in some spectra, leading to a large amount of missing data. Table 2 gives the number of proteins and peptides that are identified in only one, only two, or all three experiments when iTRAQ is performed three times on the same biological sample. It can be seen that only about 1/3 of the proteins we identified in all three experiments, whereas only about 1/4 of the peptides produced by these proteins we observed in all experiments. Liu et al. [6] and Wang et al. [15] suggested that the probability that a protein is missing is not random, but rather related to its abundance. Less abundant peptides are harder to detect due to the data-dependent acquisition of the analysis process, hence more likely to be missing. This is a nonignorable missing data problem. Ignoring the nonrandom missing pattern in statistical analysis may introduce significant bias in statistical inference and scientific conclusions.
Table 2.
Missing pattern in Caveolin-1 study. 8045 is the total number of peptides observed in the study for the 138 common proteins. Only about 1/3 of proteins and 1/4 of peptides are observed in all three experiments
| Number of experiments protein/peptide is present | ||||
|---|---|---|---|---|
| Counts | 1 | 2 | 3 | |
| proteins | 424 | 192 (45.3%) | 94 (22.2%) | 138 (32.5%) |
| peptides | 8045 | 4765 (59.2%) | 1156 (14.4%) | 2124 (26.4%) |
To identify differentially expressed proteins, one common approach is to calculate the ratio of the observed peptide intensities (the area columns in Table 1) between two samples and to compare the calculated ratios against prespecified upper and lower bounds. However, the criterion for threshold selection is subjective. For example, Seshi [14] considered iTRAQ ratios >5/4 or <4/5 as significant, whereas Salim et al. [13] used thresholds 1.20 and 0.83. These thresholds fail to consider the variability in data and are not statistically based. Oberg et al. [8] and Hill et al. [5] applied ANOVA models to incorporate the variability sources in inferring differentially expressed proteins. But they do not consider the nonrandom missingness, potentially biasing their results. In this paper, we introduce a novel approach to inferring the relative protein expression levels and hence to identify differentially expressed proteins. We model the measured peptide intensities as the results of both protein expression levels and peptide specific effects. For iTRAQ data from multiple experiments, we utilize a Bayesian hierarchical model in the sense that the model has an observation component that models the observed peptide intensities as random effects whose conditional distribution depends on the expected protein expression levels and peptide effects, and a second (hierarchical) component that defines the distributions of these expected values. If a sample is labeled with multiple tags in a single experiment, the variations across different tags are modeled as random effects. In this paper, we also describe a model for iTRAQ data from a single experiment. As for the nonrandom missingness, we use a logistic regression to model the missingness probability as a function of the protein expression level. Based on this model set-up, we infer differentially expressed proteins through posterior inferences.
The paper is organized as follows. Section 2 develops the hierarchical model and details the inferential procedure. Section 3 reports a simulation study comparing our method with ANOVA methods and ratio estimates, and studies the robustness of our method. Section 4 reports the analysis of a mouse caveolin-1 experiment, and discussion follows in Sect. 5. We describe the detailed MCMC scheme in Appendix A, and a model for iTRAQ data from a single experiment in Appendix B.
2 Model
We first describe the model for iTRAQ data from multiple experiments and estimate the relative expressions of proteins that are present in all experiments. We assume that the labeling effects have been removed by normalization methods such as quantile normalization [1]. Throughout the paper, we consider log-transformed peptide intensities and protein expressions. We assume that there are S (≥2) biological samples studied in K (≥2) experiments. Multiple isobaric tags may label the same sample in one experiment. We use Ls ≥ 1 to denote the number of tags labeling the sth sample. Then ∑s Ls = M is the number of isobaric tags used in one experiment, which is 4 when we use 4-plex isobaric reagents and M = 8 in the 8-plex version. Assume that there are I proteins in the sample and there are Ji peptides for the ith protein. For the lth label of the sth sample in the kth experiment, let ykijsln denote the observed intensity for the jth peptide of the ith protein from the nth spectrum. Note that j should be more appropriately denoted as j(i) to explicitly indicate that peptides are nested within proteins, and l should be denoted as l(s) to indicate the lth labeled tag of the sth sample. For notational simplification, we omit the parentheses. The measured intensity of a peptide depends on the protein expression level and the peptide effect. Let xkisl denote the expression level of the ith protein of the sth sample with the lth labeling tag in the kth experiment. Let zkij denote the peptide effect for the jth peptide of the ith protein in the kth experiment. We consider an additive model for ykijsln (k = 1, …, K; i = 1, …, I; j = 1, …, Ji; s = 1, …, S; l = 1, …, Ls; n = 1, …, Nkijsl):
| (1) |
which corresponds to a multiplicative model in the original scale. In (1), we assume independently, where denotes a Normal distribution with mean 0 and variance .
In addition to the additive model in (1), we also consider the multiplicative model ykijsln = xkisl × zkij + εkijsln on a small dataset with one protein and 11 peptides observed in three caveolin-1 experiments. The inferences from both models are quite close in terms of the magnitudes of the residual standard deviation (0.58 for the additive model vs. 0.60 for the multiplicative model) and the ratio of sum of squares of predicted values and sum of squares of original data R2 (0.73 for the additive model vs. 0.69 for the multiplicative model). The residuals vs. fitted values plots are also similar (not shown). This is also true when we apply both models to the data in the original scale. Since the multiplicative model in the logarithm scale and the additive model in the original scale do not greatly improve the inference (or even do worse), we use model (1) which is also easy to interpret.
Missing Data Mechanism
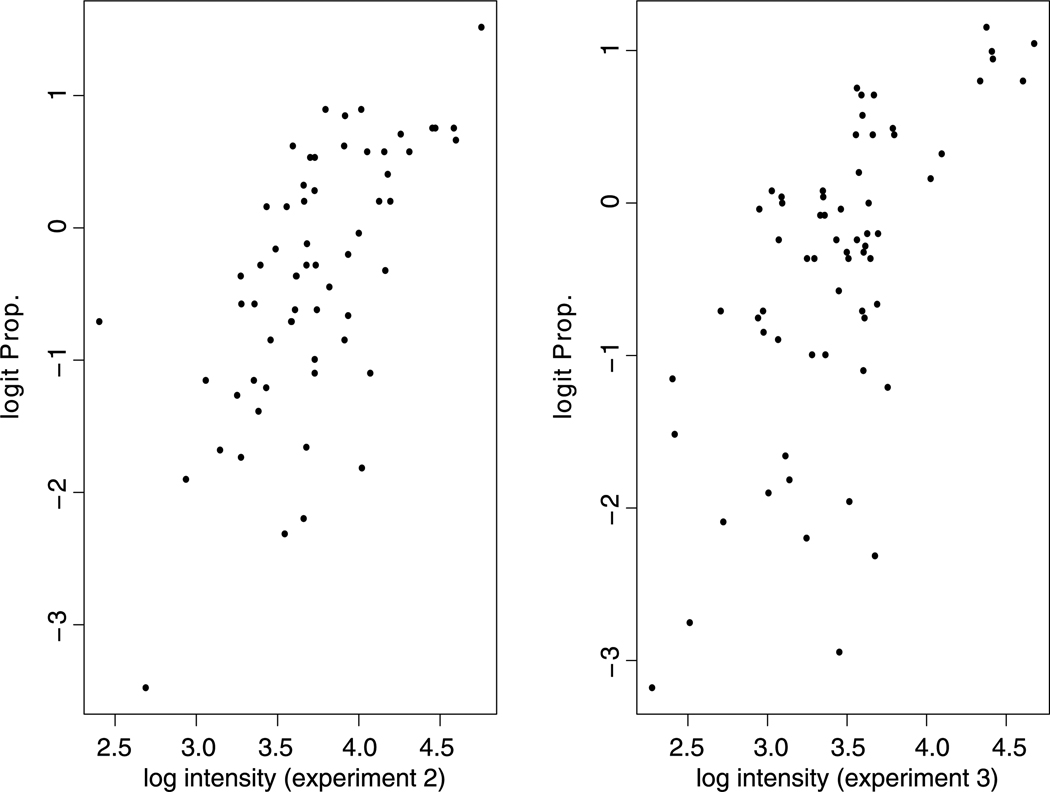
Peptide missingness presents a challenge even when we focus on proteins that are detected in all experiments. It is known that the probability of peptide missingness depends on the intensity of the peptide: lower intensity peptides are harder to detect. So there is a nonignorable missing data problem. To motivate a statistical model for missing peptide probability, we study the proportion of peptides observed in one experiment but missing in another experiment. As shown in Fig. 1, there is a negative correlation between missingness probability and peptide intensity. Furthermore, there is an approximate linear relationship between the peptide missingness probability and the observed intensity at the logit scale. Therefore, we model the missingness probability through a simple logistic regression,
| (2) |
where Ikijsln = 0 indicates that the jth peptide of the ith protein is missed in the kth experiment, the lth replicate of the sth sample and the nth spectrum. Formula (2) implies that the logit of the probability of peptide missingness is linearly dependent on its intensity. We expect b < 0, because peptides with lower intensities are more likely to be missing.
Fig. 1.
Relationship between the logit peptide missingness probability in one experiment and the observed peptide intensity in another experiment. X-axis: the log median intensity of 100 observed peptides binned based on their intensities in experiments 2 (left panel) and 3 (right panel). Y-axis: the logit proportion of the 100 peptides in each bin that are observed in the first experiment
Priors
Noting the hierarchical structure of the iTRAQ data and taking into account the variability across experiments and samples, we utilize a Bayesian hierarchical framework to model the data. We assume that xkisl and zkij are independently normally distributed across different experiments, i.e.,
| (3) |
| (4) |
where xisl and zij denote the protein and peptide effects averaged over multiple experiments, respectively. The protein expression levels in different replicates (labeled with different tags) of the same sample are also assumed to be normally distributed:
| (5) |
where xis denotes the expression level of the ith protein in the sth sample. Assumptions (3)–(5) lead to an equivalent form of (1):
| (6) |
where denote the random effects across experiments, and denotes the variation among multiple replicates of the same sample. Formula (6) is a mixed-effects model. To ensure the identifiability of the model, we restrict xi1 = 0. Then xis denotes the expression level of the ith protein in the sth sample relative to the first sample.
The second level of priors are normal distributions for xis and zij:
| (7) |
| (8) |
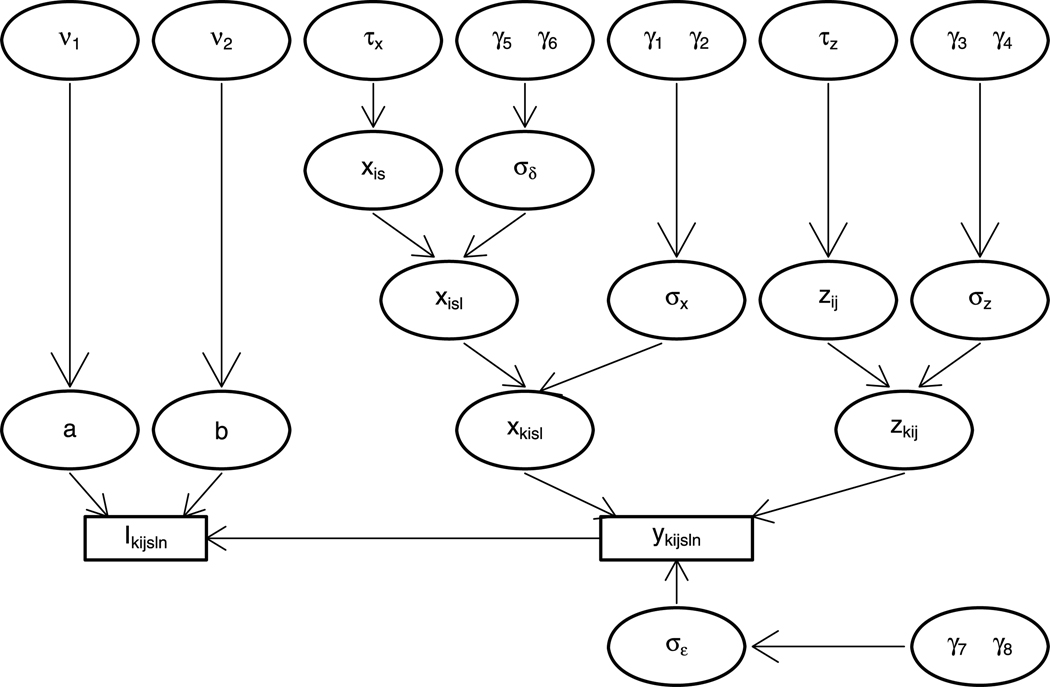
When we further assume hyperpriors for the hyperparameters, we finish the hierarchical model (Fig. 2) and can infer the posterior distributions of relevant parameters, xis, by MCMC simulations. Appendix A describes other hyperpriors and the MCMC updates in detail. Hence we can summarize the simulated posterior distributions with statistics such as posterior means, standard deviations and quantiles, and identify differentially expressed proteins.
Fig. 2.
The hierarchical structure of our model. The unknown parameters are in circles, and the observations are in rectangles
When a sample is labeled with a unique isobaric tag in an experiment, there is no replicate variation component within a sample. We note that it is easy to modify the model and the MCMC updates for statistical inference in this scenario. We will not discuss it further in this paper.
Single Experiment
When the iTRAQ data is from one experiment, we can similarly model the observed peptide intensities as the result of both protein expression levels and peptide effects, and model the nonrandom missingness through a logistic regression. We can further apply normal distributions as priors for protein expressions and peptide effects. The difference from the case of multiple experiments is that the experimental variability cannot be modeled. Appendix B describes this model and MCMC updates in more details.
Comparison to ANOVA Model
The most important difference between our Bayesian model and the ANOVA model proposed by Hill et al. [5] and Oberg et al. [8] is that we clearly model the nonignorable missingness in iTRAQ data. Oberg et al. [8] remarked at the end of their paper that using a censoring mechanism to fit the model would be a natural next step. Instead of censoring the data at an unknown threshold value, we model a higher probability of peptide missingness for lower peptide intensities. Our Bayesian model also differs from the ANOVA model in the sources of variations included in the model. In addition to the terms in our model, the ANOVA analysis also considers the labeling effect, the interaction between labeling and experimental effect, and variable peptide effects under different conditions (we talk about this in Discussion). The experimental effect and the replicative effect (when multiple tags label a sample) are considered constants for all proteins in the ANOVA model. In contrast, we model them as random effects that are specific to peptides and (or) proteins.
3 Simulation Study
We simulate data from a 4-plex version of iTRAQ on one protein containing ten peptides across three replicate experiments. Each sample is labeled with a distinct isobaric tag. In this case, there is no need to model the replicate effects specified by prior (5). We assume x = (0, −0.04, −0.48, −0.66) to be the true relative protein expression levels in log scale compared to the first sample. Under different parameter values for σx, σz, and σδ, we simulate data as follows: (1) sample , where zj ~ N(0, 1); here we dismiss subscripts i and l since there is only one protein and only one isobaric tag for a sample in an experiment; (2) sample , calculate the missing data probability P(Iksjn = 0), and determine the missing pattern Iksjn. We take a = −0.16 and b = −1.03 in the simulation, based on the posterior inference of a small subset of a real data.
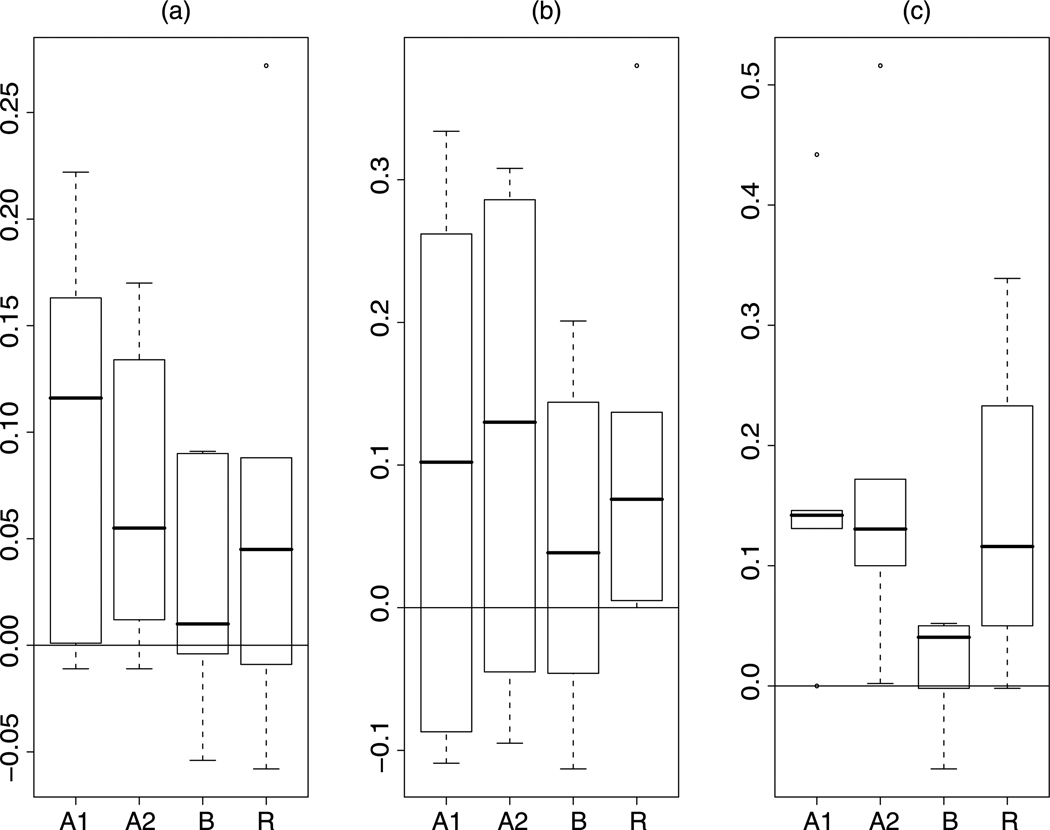
We analyze the simulated data with our Bayesian method and infer the relative protein expression levels through the MCMC samples. For comparison, we also analyze the data with the ANOVA model proposed by Hill et al. [5] and Oberg et al. [8], and calculate the means of the log ratios of peptide intensities. For each parameter setting, we simulate ten data sets and summarize the results from one data set in Table 3. The Bayesian method and the ANOVA analysis provide measures of the uncertainties of estimates. We either obtain the 95% credible intervals of the posterior distributions or the 95% confidence intervals for the estimates from the ANOVA analysis. When performing the ANOVA analysis, we consider two models. “ANOVA 1” includes the sample effect, peptide effect, experimental effect, and the interaction of sample effect and peptide effect. “ANOVA 2” removes the interaction term from “ANOVA 1.” From Table 3 we observe that all but one credible interval cover the true values when using our Bayesian method to analyze the data. But about 1/3 of the confidence intervals from ANOVA analysis fail to cover the true values, including the case where Bayesian analysis fails (estimate x3 for the simulated data when ). Comparing the estimates to the true values, we find that our Bayesian estimates have smaller bias than those from ANOVA analysis. Figure 3 draws the boxplots of the biases of the estimates using different methods for all six parameter settings. It is clear that the Bayesian method leads to the smallest bias. The better coverage and smaller bias of the Bayesian method are consistently observed in the analyses of the other nine simulated data sets. In the 60 analysis (10 data sets for each parameter setting), the 95% credible intervals from our Bayesian method fail to cover the true values 3% of the time, but the 95% confidence intervals from the ANOVA method fail in 1/3 of the cases. The means of the biases for estimates of x from the Bayesian analysis are at least 1/2 smaller than those from the ANOVA method. The lengths of the credible intervals and confidence intervals are specific to a data set or the parameter setting. Neither is consistently smaller than the other.
Table 3.
Comparison of the inference results for the relative protein expression levels under different settings of variations. The true values are x2 = −0.04, x3 = −0.48, and x4 = −0.66. “Bayesian” refers to our method. “ANOVA 1” considers the sample effect, peptide effect, experimental effect, and the interaction of sample and peptide effect. “ANOVA 2” removes the interaction term from the analysis of “ANOVA 1.” “Log-ratio” calculates the means of peptide log-ratios
| Bayesian | ANOVA 1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| x̂2 | x̂3 | x̂4 | x̂2 | x̂3 | x̂4 | ||||||
| 10−4 | 10−4 | 10−4 | −0.037 (−0.06, −0.01) | −0.480 (−0.50, −0.46) | −0.662 (−0.69, −0.64) | −0.051 (−0.053, −0.048) | −0.484 (−0.487, −0.481) | −0.660 (−0.663, −0.657) | |||
| 10−4 | 1 | 1 | −0.044 (−0.23, 0.14) | −0.526 (−0.73, −0.33) | −0.613 (−0.80, −0.43) | 0.100 (−0.13, 0.33) | −0.567 (−0.78, −0.35) | −0.529 (−0.75, −0.30) | |||
| 10−2 | 0.1 | 1.5 | −0.094 (−0.28, 0.09) | −0.593 (−0.79, −0.40) | −0.729 (−0.93, −0.52) | 0.052 (−0.12, 0.22) | −0.589 (−0.77, −0.41) | −0.516 (−0.71, −0.32) | |||
| 10−2 | 1 | 1.5 | 0.051 (−0.13, 0.23) | −0.279 (−0.46, −0.10) | −0.610 (−0.80, −0.42) | 0.123 (−0.05, 0.30) | −0.218 (−0.40, −0.03) | −0.514 (−0.70, −0.33) | |||
| 10−2 | 10 | 1.5 | −0.023 (−0.24, 0.19) | −0.336 (−0.55, −0.12) | −0.608 (−0.83, −0.39) | −0.039 (−0.31, 0.23) | −0.272 (−0.54, −0.01) | −0.218 (−0.52, 0.08) | |||
| 10−2 | 50 | 1.5 | 0.050 (−0.15, 0.25) | −0.403 (−0.60, −0.20) | −0.626 (−0.83, −0.42) | 0.182 (−0.07, 0.43) | −0.146 (−0.40, 0.11) | −0.520 (−0.77, −0.27) | |||
| ANOVA 2 | Log-ratio | ||||||||||
| 10−4 | 10−4 | 10−4 | −0.051 (−0.053, −0.049) | −0.484 (−0.486, −0.482) | −0.658 (−0.661, −0.656) | −0.036 | −0.480 | −0.662 | |||
| 10−4 | 1 | 1 | 0.005 (−0.17, 0.18) | −0.575 (−0.75, −0.40) | −0.560 (−0.74, −0.38) | −0.098 | −0.475 | −0.533 | |||
| 10−2 | 0.1 | 1.5 | 0.025 (−0.14, 0.19) | −0.525 (−0.70, −0.35) | −0.506 (−0.68, −0.33) | 0.046 | −0.429 | −0.427 | |||
| 10−2 | 1 | 1.5 | 0.094 (−0.08, 0.27) | −0.216 (−0.39, −0.04) | −0.488 (−0.67, −0.31) | −0.049 | −0.343 | −0.555 | |||
| 10−2 | 10 | 1.5 | −0.028 (−0.28, 0.22) | −0.194 (−0.45, 0.06) | −0.144 (−0.41, 0.12) | 0.232 | −0.100 | −0.321 | |||
| 10−2 | 50 | 1.5 | 0.130 (−0.10, 0.36) | −0.172 (−0.41, 0.07) | −0.553 (−0.79, −0.32) | 0.048 | −0.379 | −0.610 | |||
Fig. 3.
Boxplots of the biases of the estimates from Table 3 using different methods (A1: ANOVA 1; A2: ANOVA 2; B: Bayesian; R: log-ratio). a x̂2; b x̂3; c x̂4
In the above results, we simulated data according to our model, which may favor our approach. To study the robustness of our approach, we also consider a different missing mechanism. For each experiment, we first simulate whether each peptide is present from a Bernoulli distribution with probability p, which determines the potential frequency rj of the presence of peptide j in K = 3 experiments (rj = 0, 1, 2, or 3). Given rj, we sample the peptide effect zj | rj ~ logGamma (lrj, shrj, scrj) for rj > 0. The density function of a log-gamma distribution with shape a > 0, scale b > 0, and location c is
| (9) |
Peptides with rj = 0 are missed. Then we simulate the variabilities across experiments: . Finally we follow the second step in the previous study to simulate yksjn and Iksjn. This mechanism differs from our model in two ways: (1) the distribution for zj differs; and (2) the missing data mechanism differs since the simulation of possible presences of peptides from the Bernoulli distribution will also cause peptides missed. The resulting peptide frequency may be less than rj. We study how our method performs for the data simulated under this missing mechanism. We consider different values for the success probability in the Binomial distribution (0.9 and 0.2). For each case, we simulate ten data sets and analyze them with our method. Table 4 gives the means and standard deviations of the ten estimations. We find that the means are close to the true values and the inference is not sensitive to the new mechanism of peptide missing. Compared to the results obtained from the ANOVA analysis which contains the main effects of protein, peptide and experiments, the estimates from our Bayesian analysis are closer to the true values and have less variability (except x4 for Bi(3,0.2)) in the estimations.
Table 4.
Sensitivity of inference to a different missing mechanism and random number of observations. The true values are x2 = −0.04, x3 = −0.48, and x4 = −0.66. Values in parentheses are standard deviations of the estimates. The ANOVA model involves the sample effect, peptide effect, and experimental effect
| Bayesian | ANOVA 1 | |||||
|---|---|---|---|---|---|---|
| x̂2 | x̂3 | x̂4 | x̂2 | x̂3 | x̂4 | |
| Bi(3,0.9) | −0.027 (0.064) | −0.486 (0.069) | −0.666 (0.091) | −0.058 (0.150) | −0.442 (0.073) | −0.587 (0.181) |
| Bi(3,0.2) | −0.055 (0.056) | −0.500 (0.069) | −0.651 (0.048) | −0.054 (0.086) | −0.512 (0.079) | −0.630 (0.033) |
| Poisson | −0.036 (0.101) | −0.473 (0.136) | −0.634 (0.131) | −0.036 (0.108) | −0.469 (0.182) | −0.619 (0.212) |
In previous simulations, we fix the number of observations for each peptide as the same. When a peptide is not observed in an experiment, we assume that only one spectrum is missing and impute the values for all samples in only one observation. To study the effect of varying number of observations for different peptides on our inference, we randomly sample these numbers from a Poisson distribution. The rate of the distribution is randomly picked from a set of values. We also apply the missing mechanism described in the previous paragraph with p = 0.5. Under this scheme, we simulate ten data sets and analyze them with our method and the ANOVA model. From the calculated means and standard deviations in Table 4 we see that the distribution of the number of observations does not have great effect on the inference, and the estimates from our method have less variability than those obtained from ANOVA analysis.
4 Case Study
We apply our method to an iTRAQ dataset which aims to identify proteins affected by caveolin-1. Caveolin-1 is essential to the formation of caveolae, while the functional perturbations in the caveolae and the caveolae coat proteins may cause a wide range of diseases from cancer to a rare form of muscular dystrophy. Recent studies from mice suggest that they may be important for postnatal cardiovascular function [7]. Comparing the protein profiles from wild-type (WT) mice and knock-out Cav-1 (KO) mice using iTRAQ, we can explore the physiological and pathophysiological roles of caveolins for postnatal cardiovascular function. Samples from three KO mice and three WT mice were labeled with iTRAQ reagents as shown in Table 5. Among the 424 proteins identified in the study, a total of 138 common proteins were identified in all three comparisons of the WT/KO mice from iTRAQ analysis (Table 2). Focusing on the 4765 peptides of these 138 common proteins, we found that 2124 of them were observed in all three experiments.
Table 5.
Experimental design of the caveolin-1 study
| tag | ||||
|---|---|---|---|---|
| Experimental run order | 114 | 115 | 116 | 117 |
| 1 | WT | WT | KO | KO |
| 2 | WT | WT | KO | KO |
| 3 | WT | WT | KO | KO |
We first perform quantile normalization with each protein in the two replicates of each sample. Then we do two iterations of quantile normalization on each pair of samples to remove the systematic bias in the data. Applying our method to the log transformed value of the normalized data, we conduct 101000 iterations of MCMC updates and take the first 6000 as burn-in. The simulation takes 138 hours on the caveolin data with 4765 peptides and 200684 observations. Sampling every tenth iteration, we get 9500 samples, based on which we infer the posterior distributions of protein expression levels.
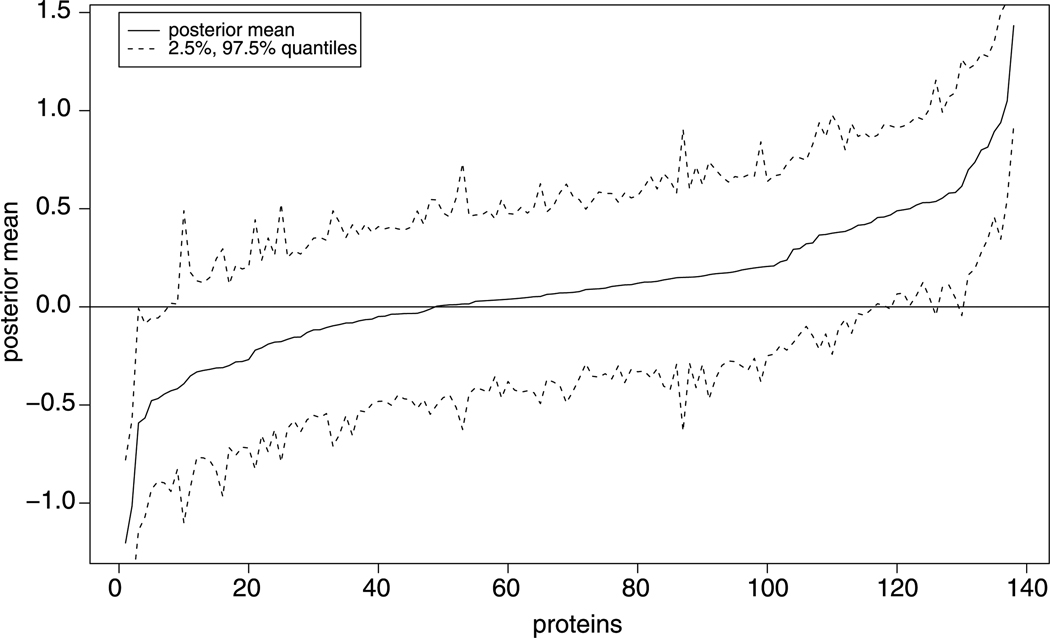
We illustrate the inferred posterior means of the relative protein expression levels in Fig. 4. We also depict the upper and lower 2.5% posterior quantiles in the figure. From these posterior inferences, we can further identify differentially expressed proteins. For example, if we require the 2.5% quantile above zero or the 97.5% quantile below zero, there are 19 up-regulated and 7 down-regulated proteins. We summarize the posterior inferences of other parameters in Table 6. For this normalized data, the randomness of peptide effects across experiments contributes the most significant source of variation (313.141). The replicate variation within a sample is almost ignorable (0.002). We infer the slope parameter in (2) to be negative (−0.217), implying that peptides with lower intensities are more susceptible to be missing.
Fig. 4.
Posterior means of the proteins sorted in increasing order
Table 6.
Posterior means and standard deviations of other parameters
| a | b | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mean | 1.351 | 0.141 | 0.002 | 313.141 | −0.442 | −0.217 | ||||
| sd | 0.004 | 0.008 | 0.002 | 23.160 | 0.021 | 0.005 |
To make a comparison with other methods, we also apply the ANOVA method to the data. Since there are 138 identified proteins and 4765 identified peptides, it is difficult to estimate all of the parameters in the ANOVA model simultaneously using current software and computers. Applying the stagewise regression idea in Oberg et al. [8], we first estimate the effects of experiments, proteins, and peptides (the first two groups of model (1) in Oberg et al. [8]), and then we take the residuals as responses for estimating the effects of samples, interactions between samples and proteins, peptides. The sample-related parameters are estimated for each protein individually, assuming that each protein has a different variance parameter, rather than a global variance parameter. Regarding the proteins as differentially expressed where the 95% confidence intervals do not cover zero, we find 60 up-regulated and 26 down-regulated proteins. They contain all the differentially expressed proteins inferred from our Bayesian model. Focusing on the proteins that are only found by ANOVA, we study their missing patterns and compare the estimates from both methods. We find that for 35 of the 41 (= 60 − 19) up-regulated and 15 of the 19 (= 26 − 7) down-regulated proteins, the differences of estimates of expression levels from both models may be due to missingness. Another reason that ANOVA identifies more proteins is likely due to the fact that protein-by-protein estimation leads to smaller variances than the global variance under our Bayesian approach. So the credible intervals from Bayesian analysis have wider, and more appropriate, ranges than the confidence intervals from ANOVA model.
5 Discussion
We have developed a novel Bayesian model to analyze iTRAQ data from multiple experiments or a single experiment. In our model, the observed peptide intensities are influenced by both the protein expression levels and the peptide effects. For data from multiple experiments, these two effects across experiments are modeled as random effects. If a sample is labeled with multiple isobaric tags, our model also allows random effects across replicates. We explicitly model the nonignorable missingness for peptides, which is a common phenomenon in iTRAQ data. The logit probability of peptide missingness is assumed to be linearly dependent on its intensity. We implement an MCMC approach to simulate the posterior distributions of relative protein expression levels. The MCMC samples provide both estimates of the expressions and measure of uncertainty for the estimates. Compared to the estimates from the ANOVA analysis and the simple log ratio calculation, we find that the estimates from the MCMC samples greatly reduces the bias due to missing data.
In our model, we assume that the logit of the missingness probability is linearly dependent on the peptide intensity ykijsln (2), and the later depends on the protein expression levels, peptide effects, and several variation terms (6). For a particular peptide j, in addition to the variation (εkijsln) across multiple spectra in an experiment, experimental variations are modeled at both the protein and peptide levels. A small peptide effect specific to a particular experiment (zkij) may cause the missingness of the peptide in that experiment (k). When both protein effect and peptide effect are large in an experiment k, this peptide will be observed in experiment k, but an extremely small value of εkijsln can lead to the missingness of this peptide in spectrum n of experiment k. So this model explains the peptide missingness both at the experiment level and at the spectrum level.
We have performed simulation studies to check whether our analysis is sensitive to this assumption of missingness. We simulate data sets from different missing mechanisms and analyze them with our Bayesian method. The estimated values are close to the true values and have smaller bias than the results from the ANOVA analysis. Furthermore, we also check how variable number of spectra for peptides affects our analysis, since when a peptide is missing in an experiment, we impute the values in only one MS spectrum. It is found that when we sample the number of MS spectra for a peptide from a Poisson distribution, our analysis leads to estimates close to the true values. This implies that our method is robust to these model violations.
Labeling effect is an issue that is not directly addressed in our model. The ability of peptides’ linkage to isobaric reagents may vary, implying peptide-tag specific labeling effect. Modeling all such labeling effects increases the number of model parameters dramatically. If we treat the labeling effects as constants for all peptides, this amounts to adding a constant specific to each tag in model (1). Due to the limitation of the data in caveolin study, the labeling effect is confounded with signals. In this paper, we first perform normalization to remove the labeling effect and systematic bias, and then apply our method to infer the relative protein expressions. In practical studies, we suggest to randomize the isobaric tags applied to samples when multiple experiments are conducted.
The fast convergence requirement is a challenge to our Bayesian approach. For a larger scale study, more MCMC iterations and hence longer time are needed to ensure the convergence. Although the Bayesian method is slower than the ANOVA method, the latter cannot fit all the involved parameters simultaneously using current software and computers. Oberg et al. [8] suggest to use the stagewise regression and then to infer the sample effects based on protein-by-protein estimation. But to get correct answers from the stagewise approach, it is necessary that the portions of the linear model design matrix corresponding to the multiple stages be orthogonal, which is not necessarily true.
In this study, we assume that all of the peptide-based observations accurately reflect the intact proteins. As a result, we ignore the possibility of homologous genes resulting in two or more proteins that share identical and nonidentical peptides as well as the possibility of post-transcriptional modifications. In addition to ignoring labeling effects, we do not include the interactions between peptide effects and sample conditions comparing to the ANOVA model. This corresponds to the assumption that certain proteins will have differential expressions under different conditions, but that any change in protein expression will affect all of the peptides for that protein equally. We expect this to be the common case, except for certain biological conditions: for example, a post-translational modification that involved a peptide substitution [8]. Despite these limitations, our method explicitly models the nonrandom missingness of iTRAQ data and provides a great improvement in estimating the relative expressions of proteins.
Acknowledgements
The work was supported in part by NIH grants HV28286, DA018343, GM59507 and NSF grant DMS 0714817. The work was also supported in part by “Yale University Biomedical High Performance Computing Center” and NIH grant RR19895, which funded the instrumentation.
Appendix A: MCMC Updates
We assume inverse gamma distributions as priors for the hyperparameters of variance: , where γ1 and γ2 denote the shape and scale parameters of a gamma distribution, respectively. We assume a ~ N(0, ν2) and b ~ N(0, ν2). The joint distribution of the model is
| (10) |
where MVN(․ | μ, Σ) denotes a multivariate normal distribution with mean vector μ and covariance matrix Σ, invGamma(․) denotes an inverse gamma distribution, and p(Ikijsl | ykijsl, a, b) can be determined by formula (2). The full conditional distributions for involved parameters are given below.
- Protein and peptide effects: xkisl, zkij, xisl, xis, and zij.
(11) (12) (13) (14)
When we take τx = τz = ∞, i.e., noninformative prior for xis and zij, .(15) -
Missing value ykijsln. Let μkijsl = xkisl + zkij; then
(16) Note that f(ykijsln | …) is log-concave. We can use Adaptive Rejection Sampling (ARS) method.
- Parameters in the logistic model for missing mechanism: a and b. Since
is log-concave, we can use ARS.(17) - Variances σε, σx, and σz.
(18) (19) (20) (21)
Appendix B: iTRAQ Data from One Experiment
We illustrate the model when each sample is labeled differently, or we treat the samples with distinct isobaric tags as different samples. It is easy to modify this model to take the replicates of samples into account. For the mth marker (or sample) and the ith protein, let yijmn denote the log value of the nth measured intensity for the jth peptide, and let xmi denote the (log) protein expression level in the experiment. Let zij be the peptide effect for the jth peptide of the ith protein. We consider the additive model for yijmn (m = 1, …, M; i = 1, …, I; j = 1, …, Ji; n = 1, …, Nijm) and missing mechanism:
| (22) |
| (23) |
and restrict xi1 = 0. We take normal distributions as priors for xmi and zij:
| (24) |
| (25) |
Priors for other parameters are the same as those in Sect. 2. Then the joint distribution of the model is
| (26) |
The full conditional distributions for missing yijmn, a and b are the same as those in multiple experiments. For xmi, zij, and σε, their full conditional distributions are given below:
| (27) |
| (28) |
| (29) |
Contributor Information
Ruiyan Luo, Email: ruiyan.luo@yale.edu, Department of Epidemiology and Public Health, Yale University School of Medicine, New Haven, CT 06520, USA.
Christopher M. Colangelo, W.M. Keck Foundation, Biotechnology Resource Laboratory, Yale University School of Medicine, New Haven, CT 06511, USA
William C. Sessa, Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06510, USA
Hongyu Zhao, Department of Epidemiology and Public Health, Yale University School of Medicine, New Haven, CT 06520, USA.
References
- 1.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 2.Choe L, D’Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 4.Hamdan M, Righetti PG. Modern strategies for protein quantification in proteome analysis: advantages and limitations. Mass Spectrom Rev. 2002;21:287–302. doi: 10.1002/mas.10032. [DOI] [PubMed] [Google Scholar]
- 5.Hill EG, Schwacke JH, Comte-Walters S, Slate EH, Oberg AL, Eckel-Passow JE, Therneau TM, Schey KL. A statistical model for iTRAQ data analysis. J Proteome Res. 2008;7:3091–3101. doi: 10.1021/pr070520u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 7.Marx J. Caveolae: a once-elusive structure gets some respect. Science. 2001;294:1862–1865. doi: 10.1126/science.294.5548.1862. [DOI] [PubMed] [Google Scholar]
- 8.Oberg A, Mahoney D, Eckel-Passow J, Malone C, Wolfinger R, Hill E, Cooper L, Onuma O, Spiro C, Therneau T, Bergen H. Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J Proteome Res. 2008;7:225–233. doi: 10.1021/pr700734f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4012. [PMC free article] [PubMed] [Google Scholar]
- 10.Patton WF. Detection technologies in proteome analysis. J Chromatogr B, Anal Technol Biomed Life Sci. 2002;771:3–31. doi: 10.1016/s1570-0232(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 11.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Salim K, Kehoe L, Minkoff MS, Bilsland JG, Munoz-Sanjuan I, Guest PC. Identification of differentiating neural progenitor cell markers using shotgun isobaric tagging mass spectrometry. Stem Cells Dev. 2006;15:461–470. doi: 10.1089/scd.2006.15.461. [DOI] [PubMed] [Google Scholar]
- 14.Seshi B. An integrated approach to mapping the proteome of the human bone marrow stromal cell. Proteomics. 2006;6:5169–5182. doi: 10.1002/pmic.200600209. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Tang H, Zhang H, Whiteaker J, Paulovich AG, Mcintosh M. Normalization regarding non-random missing values in high-throughput mass spectrometry data. Pac Symp Biocomput. 2006;11:315–326. [PubMed] [Google Scholar]
- 16.Wu WW, Wang G, Baek SJ, Shen R-F. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D Gel- or LC-MALDI TOF/TOF. J Proteome Res. 2006;5:651–658. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]