Abstract
Saliva, a biofluid historically well-studied biochemically and physiologically, has entered the post-genomic ‘omics’ era, where its proteomic, genomic, and microbiome constituents have been comprehensively deciphered. The translational path of these salivary constituents has begun toward a variety of personalized individual medical applications, including early detection of cancer. Salivary diagnostics is a late-comer, but it is catching up where dedicated resources, like the Salivaomics Knowledge Base (SKB), now have taken center stage in the dissemination of the diagnostic potentials of salivary biomarkers and other translational and clinical utilities.
Keywords: salivary biochemistry and physiology, proteome, biomarkers, early detection, genomics, microbiome
Saliva: The Biofluid
Saliva has a critical role in maintaining the health and function of the upper gastrointestinal tract (Mandel, 1989; Amerongen and Veerman, 2002). The secretion of saliva by the major and minor salivary glands is tightly regulated through neurotransmitter stimulation in what is classically considered to be a two-step process (Thaysen et al., 1954; Baum, 1993; Catalán et al., 2009). The first step involves secretion of an isotonic primary fluid by the acinar cells. The second step, occurring in the duct system, involves the re-absorption of most Na+ and Cl− found in the primary fluid, along with the secretion of K+ and HCO3−. However, in humans, these 2 steps appear to be more complicated than is typically considered (Baum et al., 1984). The ion fluxes leading to saliva formation are mediated through multiple ion transporters and channels (Catalán et al., 2009). Human acinar cells are water-permeable, a result of the presence of at least 2 water channels, aquaporin 3 and 5, in their plasma membranes (Gresz et al., 2001). Conversely, duct cells are considered to be water-impermeable and contain no known facilitated water permeability pathway. The final saliva that enters the mouth is markedly hypotonic.
Most protein secreted into saliva originates in acinar cells (~80%), with ~20% secreted by duct cells. A small number of proteins/protein families constitute the overwhelming majority of salivary proteins. These include amylase, mucins, histatins, proline-rich proteins, statherin, and cystatin (Oppenheim et al., 2007). However, >1000 polypeptides have been identified in saliva (Denny et al., 2008; Siqueira et al., 2008; Yan et al., 2009), most in trace amounts. Importantly, when the total mass of secreted constituents is considered, very little material enters a healthy gland’s saliva from extra-glandular sources, e.g., the serum.
The 3 paired major salivary glands (parotid, submandibular, sublingual) produce generally similar, but nonetheless unique, secretions. As a result of multiple processes, whole saliva, the fluid in the mouth, is not the simple sum of these individual gland secretions. For example, proteins found in gland saliva can adhere to mucosa and the dentition, be subjected to proteolysis or de-glycosylation, as well as aggregate into complexes (Helmerhorst and Oppenheim, 2007). Additionally, whole saliva contains food debris and bacterial products, as well as leaking serum constituents from any diseased tissue present. Thus, depending on the purpose, there are several types of saliva that can be collected: individual secretions from the major and minor salivary glands, as well as whole saliva (Sreebny and Vissink, 2010).
Additionally, saliva can be collected under unstimulated (resting) or stimulated conditions, with several means of stimulating secretion. Stimulation, e.g., by 2% citrate, the chewing of wax, or sucking on a lemon drop, does not necessarily lead to equivalent secretions (Dawes and Jenkins, 1964; Dawes, 1966). A further important consideration is the time of collection, since there are significant circadian rhythms recognized for both salivary flow rate and composition (Dawes, 1972). These and other variables mean that the collection of saliva samples for use as a diagnostic fluid must be performed purposefully, and be carefully described in subsequent reports, to allow for meaningful interpretation of the analytical information obtained (Dawes, 1993). Indeed, even the use of standardized collections of gland saliva in healthy adults can lead to quite variable results (Fox et al., 1987; Ship et al., 1991; Wu et al., 1993).
Given all of these concerns and caveats, it is reasonable to ask if examining salivary constituents presents more variability than would be found in the analysis of serum. The answer is clearly ‘no’; however, there has been markedly more study of serum composition, and considerably more is known about the normal ranges of and reasons for variability in most serum constituents. Thus, it seems fair to say that the current value of using saliva as a diagnostic fluid depends on the intended purpose. If the purpose is to monitor a patient’s general health status, then saliva is not now a suitable diagnostic fluid. If, however, the purpose is to ask very specific questions unrelated to normalcy, with ‘yes’ vs. ‘no’ answers desirable—e.g., Is there evidence of an illicit drug or an existing pathological process present?—then using saliva as a diagnostic fluid may be quite suitable.
The Saliva Proteome
Different approaches are required to compile a comprehensive catalogue of proteins, due to the variability of the sources and the composition of human saliva. Mass spectrometry (MS)-based methods are of greatest utility since they are unbiased, requiring no prior knowledge of protein composition. Several groups have reported data that they generated by applying these types of approaches to the analysis of saliva, with different groups concentrating on different aspects of the saliva proteome, resulting in the identification of various numbers of proteins. Many of these investigators used samples of whole saliva, which, in addition, to secretions from the parotid and the submandibular/sublingual (SM/SL) glands, contains the protein products of the cells that comprise the oral cavity as well as contributions from micro-organisms (Wilmarth et al., 2004; Vitorino et al., 2004; Huang, 2004; Hu et al., 2005; Guo et al., 2006). Other investigators have focused on salivary-gland-specific secretions. For example, 2D SDS-PAGE separation followed by MS identification was used to characterize parotid, SM, SL, and/or SM/SL saliva collected as ductal secretions (Hu et al., 2004; Hardt et al., 2005; Walz et al., 2006). Other investigators focused on pellicle components, the subset of proteins that adhere to the tooth surface (Yao et al., 2003; Vitorino et al., 2006). The National Institute of Dental and Craniofacial Research (NIDCR) funded an effort to compile a catalogue of human salivary proteins that involved three teams of investigators—The Scripps Research Institute/University of Rochester, The University of California, Los Angeles/University of Southern California and The University of California, San Francisco— with the vision that this catalogue will serve as a valuable resource in the investigation of proteins with unknown functions in saliva, as well as an initial step in the development of saliva-based diagnostic tests (Denny et al., 2008).
Functional Characterization of Salivary Proteins
The investigators searched parotid and SM/SL proteins against the gene ontology database (GO) to achieve a functional overview of the salivary proteins. GO definitions were used for the analysis of the distribution of salivary proteins across 3 different categories. The parotid and SM/SL proteins showed generally similar distribution across the GO categories. The highest proportion of both the parotid and SM/SL proteins is extracellular, which is consistent with the secretory role of salivary glands. In addition, organelle, cytoplasm, cytoskeleton, plasma membrane, protein complex, and other cellular components such as nuclear proteins occupy the rest of the categories almost entirely. These cellular proteins are found mostly in low abundance compared with well-characterized secretory salivary proteins. The source of these proteins is probably exosomal (Gonzalez-Begne et al., 2009; and see below) or from shed glandular epithelial cells. GO molecular function distribution showed that parotid and SM/SL proteins have the highest distributions in proteins with binding activity, as well as proteins involved in catalytic function. A high proportion of the proteins with structural molecular activity was also found. GO distribution of the biological process revealed that high proportions of parotid and SM/SL proteins are involved in metabolic processes and regulation of biological processes.
Gene ontology annotation of the proteins integrated from the results of all three research groups revealed interesting features of salivary proteins (Denny et al., 2008). Among many of the enzymes identified, proteins associated with oxidoreductase activity were overrepresented. Similarly, proteins with anti-oxidation activity were also overrepresented. In contrast, proteins with transferase activity were underrepresented. Proteins with hydrolase activity remained unchanged; however, a specific hydrolase, which acts on acid anhydrides, was found to be enriched. Among these hydrolases, carbonic anhydrase VI is a well-studied and abundant salivary hydrolase. Not surprisingly, among the proteins with binding activity, antigen- and lipid-binding proteins were enriched, while nucleic-acid-binding proteins were underrepresented. Finally, it has been well-documented that salivary fluid contains many proteins with enzyme-inhibiting, especially protease-inhibiting, functions, to serve as a preventive mechanism for oral pathogen invasion (Ruzindana-Umunyana and Weber, 2001; Jespersgaard et al., 2002). Consistently, the category of proteins as enzyme inhibitors and protease inhibitors is enriched. Overall, the identification of functional versatile protein molecules in saliva indicates that this body fluid may carry more complex functions than previously thought.
Hypothetical Proteins and Their Functional Implications
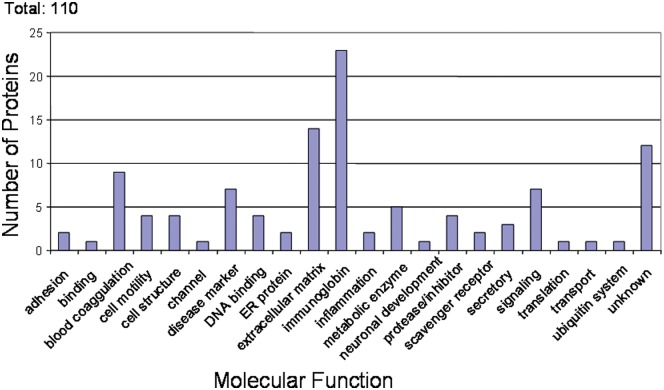
Many of the salivary proteins reported by Denny et al. (2008) were hypothetical proteins that have not been annotated. To provide insight, investigators have postulated the functions of the proteins based on their sequence similarities to the proteins with known function through a PSI-blast search engine (Fig. 1) (Altschul et al., 1997). A large group of these hypothetical proteins could be matched to the immunoglobulins. Immunoglobulins were among the most abundant proteins identified, which is consistent with previously reported studies (Gomez et al., 1991). Together with the identification of defensin peptides by Denny et al. (2008), and in previous reports (Abiko et al., 2003; Brogden et al., 2003), this suggests the importance of saliva as a defense mechanism against oral pathogens. Extracellular matrix proteins such as mucins were also abundant; these proteins might serve to maintain saliva’s lubricating function. Several blood coagulation factors were found, suggesting the overlap of proteins between serum and saliva. Several proteins with known linkage to multiple system disorders were mapped. These included: moesin, a protein with a role in lymph node metastasis of oral squamous cell carcinoma (Kobayashi et al., 2004; Belbin et al., 2005); dystrophin, gene product of Duchenne muscular dystrophy locus (Hoffman and Kunkel, 1989); and DMBT1, whose expression is frequently suppressed in human lung cancer (Takeshita et al., 1999; Wu et al., 1999). The biological significance of these proteins in saliva is not clear; nevertheless, these results suggest that salivary proteins could serve as a useful data resource for further search of disease biomarkers.
Figure 1.
Molecular function of hypothetical proteins. The functions of the proteins were postulated based on their sequence similarities to the proteins with known function through PSI-blast search engine.
The hypothetical salivary proteins were also searched against protein pathway databases BioCarta (Ogata et al., 1998; Mlecnik et al., 2005). As expected, these salivary proteins are involved in a number of metabolic pathways, including amino-acid-related metabolism, carbohydrate metabolism, energy metabolism, and glycan biosynthesis and metabolism. Interestingly, several salivary proteins were found in a few systemic diseases, such as amyloid beta A4 protein precursor (Alzheimer’s), DJ-1 (Parkinson’s diseases), and colon cancer secreted protein-2 (colorectal cancer). The biological roles of these proteins in saliva await further characterization.
The identification of the proteins present in human saliva provides an inventory that allows for further analysis for variations that may be associated with disease or changes associated with age. However, understanding how normal saliva changes with normal factors such as aging will be key to the development of saliva-based markers of disease. Ambatipudi et al. (2009) performed a comprehensive proteomic profile of pooled saliva collected from the parotid glands of healthy females, divided into two age groups of 20-30 and 55-65 years old, respectively. A pre-fractionation of the proteins by hydrophobic charge interaction chromatography was used to reduce complexity of the proteins and to deplete high- from low-abundant proteins. Proteins were then identified by multidimensional protein identification technology (MudPIT). In total, 532 proteins were identified in the two age groups, with 266 identified exclusively in one age group, while another 266 proteins were found in both groups.
The majority of the proteins identified in the two age groups belonged to the defense and immune response categories. Of note, several defense-related proteins (e.g., lysozyme, lactoferrin, and histatin-1) were significantly more abundant in the older females as determined by G-test. This study supports the use of high-throughput proteomics as a robust discovery tool. Such results provide a foundation for future studies to identify specific salivary proteins which may be linked to age-related diseases specific to women.
State Of The Science In Cancer Biomarkers
Numerous startling discoveries on the carcinogenic process have occurred in the past two decades. Several pathways and networks of molecular circuitries have revealed the connectivity of processes leading to cancer. It is the accumulation of these molecular changes that has been implicated in disease progression. Since the enactment of the 1971 National Cancer Act, significant progress has been made in both the understanding and treatment of cancer, and from 1992 to 1998, cancer death rates declined modestly (1.4%) in both men and women (Weir et al., 2003). However, cancer remains the second leading cause of death in the United States. For many cancers, successful prevention depends on the accurate evaluation of risk, and successful treatment depends on early detection. For example, the five-year survival for colorectal cancer is 91% if it is detected while still localized, 66% if detected with lymph node involvement, and only 9% if it has metastasized to distant sites (Eyre et al., 2002). Although the primary tumor can usually be controlled by local therapy, most cancer deaths are caused by metastatic disease. The goal of early detection and screening is therefore the diagnosis and treatment of cancer before it spreads beyond the organ of origin, perhaps even in its pre-invasive state. Unfortunately, available early detection and screening techniques pick up many tumors at a relatively late stage in their natural history. As a result, decrements in mortality with the current available detection modalities are likely to be modest. New technologies coming from the field of molecular and cellular biology are able to identify genetic as well as antigenic changes during the early stages of malignant progression. Some of these changes show promise as biomarkers for pre-neoplastic development or for early malignant transformation. Consequently, many oncologists and cancer biologists are working to develop methods to detect cancers at their early stages of development.
At the present time, some biomarkers, such as CA 125 and PSA, along with imaging technologies have remained the mainstay of the diagnostic realm. For example, spiral computerized tomography (CT) and x-rays are used for lung cancer detection; however, the latter detects smaller lesions than do x-rays. Both spiral CT and x-rays will detect small lesions (Swensen et al., 2002). The challenge is how to differentiate between a deadly tumor and a scar or other non-cancerous lesions. Today, tools that detect changes at the molecular level take the observer out of the picture, providing a much clearer answer about whether cancer gene markers or proteins produced by cancer genes are present. Research has shown that some molecular markers are present in sputum three or more years prior to the diagnosis of lung cancer (Palmisano et al., 2000).
However, progress in the application of biomarker-based detection and diagnosis is currently impeded by some practical hurdles. Studies on the application of biomarkers for earlier cancer detection or even for risk assessment have been fragmented and not well-organized. While studies conducted by individual investigators have been useful in advancing our understanding of carcinogenesis, there has been a lack of research emphasis on the continuum of pre-clinical tumor development, early evaluation of new techniques, and their clinical application. In many of these reported studies, the investigators have not been able to explore fully the biological implications or to test systematically the clinical application of these molecular markers. The lack of availability of high-quality matched specimens from normal, suspicious, pre-neoplastic, and multistage neoplastic lesions along with demographic and follow-up data has been another major impediment in progress toward molecular-based diagnosis. As a consequence, much work in this area has been fragmented into numerous small and disconnected studies without complete evaluation. Usually, the results of these studies cannot even be generalized to the population as a whole.
Biomarkers
Since the definition of ‘biomarker’ varies, and there is no definition that adequately encompasses all aspects of biomarker applications, the National Cancer Institute’s (NCI) Early Detection Research Network (EDRN) uses our own definition for describing a biomarker. Biomarkers are defined as cellular, biochemical, molecular, or genetic alterations by which a normal, abnormal, or simply biologic process can be recognized or monitored. Biomarkers are measurable in biological media, such as in tissues, cells, or fluids. A biomarker may be a molecule secreted by a malignancy itself, or it can be a specific response of the body to the presence of cancer (Wagner et al., 2004). For example, alterations in gene sequence or expression and in protein structure and function can be used to detect cancer, determine prognosis, and monitor disease progression and therapeutic response. Ideally, these changes should be quantifiable in serum, plasma, saliva, sputum, urine, and other bodily fluids for potential non-invasive methods of detection.
State of the Biomarkers in Cancer Diagnostics
Only a few biomarkers have made it to clinical applications, despite the fact that hundreds of thousands of published papers claim to have discovered biomarker(s) for potential utility in clinical application (Ransohoff, 2005; also see review by Ludwig and Weinstein, 2005). Why is progress so marginal in this field? Let us examine this issue by understanding the requirements that a potential biomarker must meet before being considered for clinical application. It is unlikely that a single molecular biomarker will be able to distinguish disease from non-disease states because of the high degrees of heterogeneity across larger populations. To illustrate the complexity, first consider a biomarker test that may have high sensitivity but low specificity. In this case, most individuals with disease will test positive, but the low specificity rating indicates that many people without the disease will also test positive. Biomarkers with this performance profile certainly would not be desirable, since unnecessary treatment might be administered to patients who are free of disease. Conversely, a biomarker with low sensitivity and high specificity will help ensure that an individual is disease-free, but will often fail to detect the genuine cases of disease. At times, this type of performance might have limited clinical value, particularly in helping to rule out negative cases where additional tests were performed on a patient, yielding uncertainties in the prediction of disease. Clearly, a biomarker possessing the qualities of both high sensitivity and specificity is desired for the accurate diagnosis of disease. It is likely that many diagnostic tests of the future will encompass analysis of panels of such discriminatory molecules.
There have been several excellent articles on these issues (Feng et al., 2004; Ransohoff, 2004, 2005), and therefore they are not covered in this article. Just one important note: Most initial observations are derived from limited numbers of cases and controls which later form the basis for a larger case-control design in subsequent validation studies. However, case-control studies will require independent replication and eventual confirmation in large prospective cohort studies. Also, initial studies are not usually hypothesis-driven and therefore are more prone to bias. In non-hypothesis-driven studies, there are many unknowns: The identity of the biomarker is not known, and the size of the panel is likely to be undefined, e.g., the protein profile pattern of many peaks without the identification of an individual peak. Because of the lack of such information, investigators will not be able to anticipate the source of potential bias. To make this point clear, let us take an example of a defined panel in which PSA is one of the biomarkers. Investigators would be able to anticipate that patients with prostate-related diseases, other than prostate cancer, will likely have elevated PSA. To avoid the identification of markers that are diagnostic for prostate-related disease, investigators may include a separate control group in their study design. Sample collection, storage conditions, and duration of storage and processing are other sources for biases. “Wired-in bias” refers to the differences in biomarker measurements between case and control specimens that have nothing to do with case or control biologically, but are due to the differences in case and control specimen selection, collection, processing, and storage. Such a “wired-in-bias” is different from the biases that are due to inappropriate data analyses or interpretations. The latter could be eliminated by appropriate analyses and careful interpretations, while the former could not, hence the name “wired-in-bias”. “wired-in-bias” could be eliminated only by tight study design and adherence to protocol. For case-control study design, there is almost no way to eliminate “wired-in-bias” due to the settings in which cases and controls are selected, collected, processed, and stored. For cohort pre-clinical specimen, “wired-in bias” is basically eliminated because at the time of specimen collection, one does not know which is the case or control, and the protocol is usually followed. Therefore, case and control specimens were collected under the same setting. In summary, good quality samples from well-designed cohorts or trials should be made available to investigators for initial discovery work.
In light of these hurdles in discovery and evaluation of biomarkers, the NCI’s Early Detection Research Network (EDRN; www.cancer.gov/edrn) has developed and implemented systematic, comprehensive guidelines to develop, evaluate, and validate biomarkers. This five-phase approach establishes both a standard and a road map for successfully translating research on biomarker applications from the laboratory to the bedside (Fig. 2).
Figure 2.
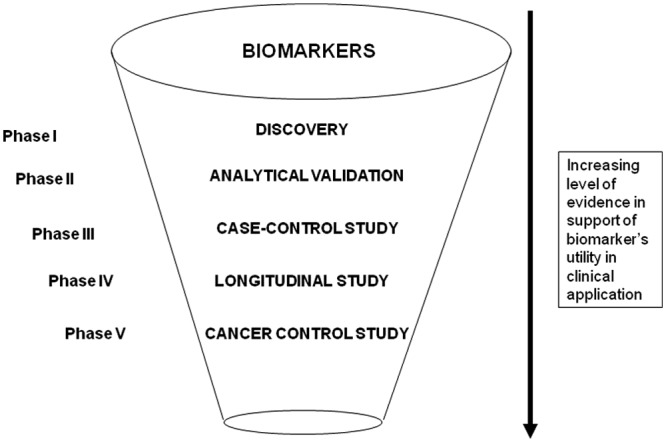
Funneling of potential biomarkers for clinical validation.
Phase 1 includes exploratory study to identify potentially useful biomarkers. In Phase 2, biomarkers are studied to determine their capacity for distinguishing between cases with cancer and those without. Phase 2 is called the validation phase. Repositories of longitudinally collected clinical specimens from research cohorts are used in Phase 3 to determine the capacity of a biomarker to detect pre-clinical disease. Prospective screening studies are considered part of the next phase, Phase 4. Finally, large-scale population studies that evaluate not just the role of the biomarker for detection of cancer but also the overall impact of screening on the population comprise Phase 5. Researchers have welcomed the five-phase structure that EDRN proposes for biomarker development, because it provides an orderly series of studies that build upon each other to yield an efficient and thorough approach to biomarker development. The key aspects of the study design for each of the phases have been discussed (Pepe et al., 2001, 2008), generating many interesting thoughts on how to take the phase of biomarkers forward in aiding study designs for biomarker validation. Pepe and colleagues proposed a prospective-specimen-collection, retrospective-blinded-evaluation (PRoBE) design in which biologic specimens are collected prospectively from a cohort that represents the target population of interest for clinical application of the biomarker. Specimens and clinical data are collected in the absence of knowledge about patient outcome. After outcome status is ascertained, case patients with the outcome and control individuals without it are selected randomly from the cohort, and their specimens are assayed for the biomarker in a fashion that is blinded to case–control status. Although every biomarker study has its own special considerations, as does every randomized clinical trial, the proposed PRoBE guidelines elucidate the key design issues. The guidelines describe the design issues in relation to the clinical context, biomarker performance criteria, the biomarker test, and study size. The principles can be applied to studies of biomarkers intended for use in disease diagnosis, screening, or prognosis. Common biases that pervade the biomarker research literature would be eliminated if these rigorous standards were adopted. An example of this approach is illustrated below.
Let us consider a case in which we would like to design a study to validate biomarkers that may be specific to benign or invasive cancer among women with suspicious breast lesions. Our goal is to reduce unnecessary biopsies while ensuring that women with invasive cancer are biopsied. So the priority is to maintain detection in almost all women with invasive cancer. The PRoBE design stipulates the minimally acceptable marker that would be able to detect at least 98% of the cases (TPR = 98%; TPR is defined as a proportion of cases detected). Since currently all women with suspicious lesions are biopsied, our goal is to reduce biopsy by 25% (FPR = 75%; FPR is defined as a proportion of controls falsely detected or 1−specificity). Considering anticipated performance of TPR 98% and FPR of 50%, the study size would be 300 invasive cancer cases and 100 benign non-proliferative controls. This is the design EDRN has adopted to test biomarkers. Samples are collected at centers where women are undergoing diagnostic biopsies. Samples, including blood, are collected prospectively prior to biopsy, with some exclusion criteria: women under 18 years, having a history of cancer, pregnant, or nursing. Demographics and medical and family history are obtained. Following a pathology report, cases and controls are selected randomly for reference sets to be used for testing potential biomarkers.
Future Research Trends in Biomarkers
The future for molecular screening requires a close collaboration between imaging and biomarker-based investigations. While biomarkers will have the ability to provide quantifiable characteristics, imaging will provide temporal and special features of the disease space and amplify biomarker visualization in real time. Smart reagents and probes are needed to exploit the power of imaging, and biomarkers could provide some needed probes. A comprehensive database of biomarkers needs to be developed, integrating genomics, proteomics, metabolomics, glycomics, and imaging features of a broad spectrum of disease states. As technologies continue to mature, reference reagents and samples must be made available to investigators to provide standards for their exploratory studies and allow for consistencies in cross-validation among studies. However, standards should not be imposed in initial discovery processes that could impede innovations.
Salivaomics Knowledge Base (SKB)
The holy grail of diagnostics is non-invasiveness. Saliva fulfills this goal. However, while salivary diagnostics is recognized for oral diseases, its clinical utility and scientific credibility for systemic diseases are unsubstantiated. The clinical and scientific credentialing of saliva for systemic disease detection will present a ground-breaking technology that is impactful and sustainable, and will transform molecular diagnostics globally.
However, to provide credibility in these endeavors, the critical hypothesis that needs to be tested is “Can Salivary Biomarkers Detect Systemic Diseases?” There are two essential gaps that must be bridged. First is a translational and clinical gap that salivary biomarkers can detect systemic diseases. To achieve this goal, a definitive Food and Drug Administration(FDA)-level clinical salivary biomarker development and validation study for systemic disease detection must be performed. Should the salivary biomarkers survive this FDA-level clinical study, the biomarkers would have achieved a pivotal validation status that can be advanced for FDA regulatory approval and diagnostic product development. The second gap is mechanistic. How can a distal disease like pancreatic or breast cancer communicate/signal salivary glands and contribute to salivary biomarker signature for cancer detection? To address this issue, one needs to perform rigorous mechanistic studies using animal models to test the hypothesis that, upon development of a systemic disease, salivary biomarkers are consistently altered and a unique detection panel can be derived. This two-pronged approach will provide the definitive translational and clinical data as well as mechanistic rationale for systemic diseases reflecting through salivary biomarkers. We have recently made inroads using this dual approach to investigate the scientific and clinical frontiers of salivary diagnostics.
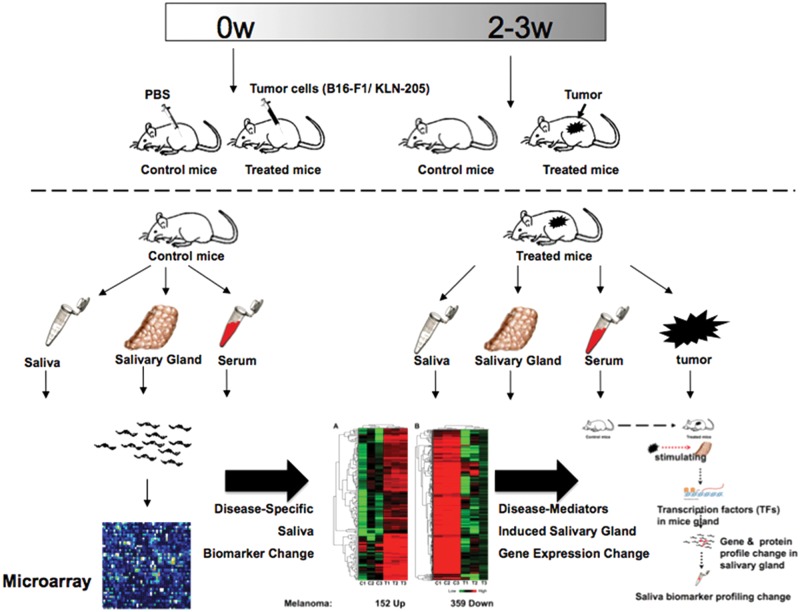
In a study by Gao et al. (2009), mouse models of melanoma and non-small-cell lung cancer were used to compare the transcriptome biomarker profiles of tumor-bearing mice with those of control mice (Fig. 3). Microarray analysis showed that salivary transcriptomes were significantly altered in tumor-bearing mice vs. controls. Significant overlapping among transcriptomes of mouse tumors, serum, salivary glands, and saliva suggests that salivary biomarkers have multiple origins. Furthermore, the expression of two groups of significantly altered transcription factors (TFs)—Runx1, Mlxipl, Trim30 and Egr1, Tbx1, Nr1d1—was identified in salivary gland tissue of melanoma-bearing mice and can potentially be responsible for 82.6% of the up-regulated gene expression and 62.5% of the down-regulated gene expression, respectively, in the saliva of melanoma-bearing mice. We also showed that the ectopic production of nerve growth factor (NGF) in the melanoma tumor tissue as a tumor-released mediator can induce expression of the TF Egr-1 in the salivary gland. Analysis of these data, taken together, supports the conclusion that, upon systemic disease development, significant changes can occur in the salivary biomarker profile. Although the origins of the disease-induced salivary biomarkers may be both systemic and local, stimulation of salivary glands by mediators released from remote tumors plays an important role in regulating the salivary surrogate biomarker profiles.
Figure 3.
Mice (either C57BL/6 mice or DBA/2 mice) were randomly divided into 2 groups as follows: control group (control mice) and tumor group (tumor mice) (15 animals per group). PBS was injected into control mice, while mice in the tumor group were injected with tumor cells. Tumor establishment took ~ 3 weeks. Saliva, salivary gland, serum, and tumor tissues were collected from each mouse. Five mice each were pooled into one group and processed to profile their transcriptome by the expression microarrays (Gao et al., 2009).
These studies provide early insights into the mechanistic foundation of salivary diagnostics for systemic disease detection. While many more additional studies need to be conducted to map out the detailed mechanisms and signaling connectivities, what is clear from these studies is that when systemic diseases developed, robust and validatable salivary biomarker changes ensued. A recent discovery is that salivary biomarkers (proteomic and transcriptomic) are contained in exosomes, which are small-membrane vesicles (Palanisamy, 2010; Sharma, 2010), 40-100 nm in diameter, corresponding to the internal vesicles present in multivesicular endosomes (MVEs), and have been known to be key for intercellular communication elsewhere in the immune system (Chairoungdua et al., 2010). The revelation that the salivary transcriptome and proteome are contained in exosomes not only explained their unusual stability in saliva but also provided a mechanism whereby these nano-vesicles can mediate a signal transduction/communication axis to shuttle disease-specific exosomal contents (biomarkers, molecular constituents of mRNA, miRNA, and proto-oncogenic and pro-angiogenic proteins) to anatomical targets, including salivary glands. This systemic communicative capability of exosomes has been recently revealed, since they have been shown to be released from many different cell types to be important modulators of the immune system as well as oncogenesis. These recently recognized cellular communicative roles of exosomes in health and disease provide a compelling rationale for the hypothesis that systemic diseases shed exosomes containing disease-specific biomarkers that function as signaling nano-vesicles targeting different body sites, including salivary glands and altered salivary biomarker profiles.
On the translational and clinical front, we have recently obtained proof-of-concept data that salivary biomarkers can detect pancreatic cancer (Zhang et al., 2010). A prospective sample collection and retrospective blinded validation (PRoBE-designed) study design was used to evaluate the performance and translational utilities of salivary transcriptomic for the non-invasive detection of pancreatic cancer. The Affymetrix HG U133 Plus 2.0 Array was used to profile transcriptomes and discover altered gene expression in salivary supernatants. Biomarkers discovered from the microarray study were subjected to clinical validation in an independent sample set of 30 individuals with pancreatic cancer, 30 with chronic pancreatitis, and 30 healthy control individuals. Twelve mRNA biomarkers were discovered and validated. The logistic regression model with the combination of 4 mRNA biomarkers (KRAS, MBD3L2, ACRV1, and DPM1) could differentiate pancreatic cancer patients from individuals without cancer (chronic pancreatitis and healthy control individuals), yielding a ROC-plot AUC value of 0.971 with 90.0% sensitivity and 95.0% specificity (Fig. 4). The salivary biomarkers possess discriminatory power for the detection of pancreatic cancer, with high specificity and sensitivity. This report provides the proof of concept of salivary biomarkers for the non-invasive detection of a systemic cancer and paves the way for prediction model validation studies followed by pivotal clinical validation. These early studies allow us to propose a plan to develop and definitively validate salivary biomarkers, engaging the PRoBE-design principles (prospective-specimen-collection and retrospective-blinded-evaluation) in preparation for FDA-level clinical validation studies (Pepe et al., 2001, 2008).
Figure 4.
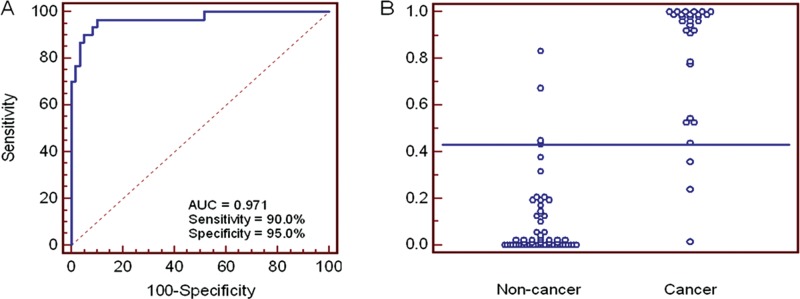
ROC curve and interactive dot diagram for the logistic regression model. (A) The logistic regression model using 4 biomarkers (KRAS, MBD3L2, ACRV1, and DPM1) yielded an AUC value of 0.971 (cut-off, 0.433). (B) Interactive dot diagram was based on the qPCR data of the non-cancer group (n = 60) and cancer group (n = 30).
Acknowledgments
The research of BJB and JEM is supported by the NIDCR Division of Intramural Research. The research of SS is supported by the NCI Division of Intramural Research. The research of DTWW is supported by NIDCR and NCI, grants RO1 DE15970 and R21 CA0126733. The research of JRY is supported by NIH, grant UO1 DE016267. The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article. The authors thank the American Association for Dental Research for hosting and sponsoring the Fall Focused Symposium.
Footnotes
Presented at the 3rd Fall Focused Symposium, November 12-13, 2010, Waterview Conference Center, Arlington, VA, sponsored by the American Association for Dental Research
References
- Abiko Y, Nishimura M, Kaku T. (2003). Defensins in saliva and the salivary glands. Med Electron Microsc 36:247-252 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambatipudi KS, Lu B, Hagen FK, Melvin JE, Yates JR. (2009). Quantitative analysis of age specific variation in the abundance of human female parotid salivary proteins. J Proteome Res 8:5093-5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerongen AV, Veerman EC. (2002). Saliva – the defender of the oral cavity. Oral Dis 8:12-22 [DOI] [PubMed] [Google Scholar]
- Baum BJ. (1993). Principles of saliva secretion. Ann NY Acad Sci 694:17-23 [DOI] [PubMed] [Google Scholar]
- Baum BJ, Costa PT, Jr, Izutsu KT. (1984). Sodium handling by aging human parotid glands is inconsistent with a two-stage secretion model. Am J Physiol 246(1 Pt 2):R35-R39 [DOI] [PubMed] [Google Scholar]
- Belbin TJ, Singh B, Smith RV, Socci ND, Wreesmann VB, Sanchez-Carbayo M, et al. (2005). Molecular profiling of tumor progression in head and neck cancer. Arch Otolaryngol Head Neck Surg 131:10-18 [DOI] [PubMed] [Google Scholar]
- Brogden KA, Heidari M, Sacco RE, Palmquist D, Guthmiller JM, Johnson GK, et al. (2003). Defensin-induced adaptive immunity in mice and its potential in preventing periodontal disease. Oral Microbiol Immunol 18:95-99 [DOI] [PubMed] [Google Scholar]
- Catalán MA, Nakamoto T, Melvin JE. (2009). The salivary gland fluid secretion mechanism. J Med Invest 56 (Suppl):192-196 [DOI] [PubMed] [Google Scholar]
- Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. (2010). Exosome release of beta-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol 190:1079-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C. (1966). The composition of saliva secreted in response to a gustatory stimulus and to pilocarpine. J Physiol 183:360-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C. (1972). Circadian rhythms in human salivary flow rate and composition. J Physiol 220:529-545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C. (1993). Considerations in the development of diagnostic tests on saliva. Ann NY Acad Sci 694:265-269 [DOI] [PubMed] [Google Scholar]
- Dawes C, Jenkins GN. (1964). The effects of different stimuli on the composition of saliva in man. J Physiol 170:86-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, et al. (2008). The proteomes of human parotid and submandibular/sublingual salivas collected as ductal secretions. J Proteome Res 7:1994-2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre HJ, Smith RA, Mettlin CJ. (2002). In Cancer Medicine e.5. Fifth ed. Hamilton, Ontario: Decker Inc. [Google Scholar]
- Feng Z, Prentice R, Srivastava S. (2004). Research issues and strategies for genomic and proteomic biomarker discovery and validation: a statistic perspective. Pharmacogenomics 5:709-719 [DOI] [PubMed] [Google Scholar]
- Fox PC, Heft MW, Herrera M, Bowers MR, Mandel ID, Baum BJ. (1987). Secretion of antimicrobial proteins from parotid glands of different aged healthy persons. J Gerontol 42:466-469 [DOI] [PubMed] [Google Scholar]
- Gao K, Zhou H, Zhang L, Lee JW, Zhou Q, Hu S, et al. (2009). Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS ONE 4:e5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez FE, Villegas J, Bourges H. (1991). An enzyme-linked immunosorbent assay for human secretory immunoglobulin A in parotid saliva. Rev Invest Clin 43:351-358 [PubMed] [Google Scholar]
- Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, et al. (2009). Proteomics analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J Proteome Res 8:1304-1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresz V, Kwon TH, Hurley PT, Varga G, Zelles T, Nielsen S, et al. (2001). Identification and localization of aquaporin water channels in human salivary glands. Am J Physiol Gastrointest Liver Physiol 281:G247-G254 [DOI] [PubMed] [Google Scholar]
- Guo T, Rudnick PA, Wang W, Lee CS, Devoe DL, Balgley BM. (2006). Characterization of the human salivary proteome by capillary isoelectric focusing/nanoreversed-phase liquid chromatography coupled with ESI-tandem MS. J Proteome Res 5:1469-1478 [DOI] [PubMed] [Google Scholar]
- Hardt M, Thomas LR, Dixon SE, Newport G, Agabian N, Prakobphol A, et al. (2005). Toward defining the human parotid gland salivary proteome and peptidome: identification and characterization using 2D SDS-PAGE, ultrafiltration, HPLC, and mass spectrometry. Biochemistry 8:2885-2899 [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, Oppenheim FG. (2007). Saliva: a dynamic proteome. J Dent Res 86:680-693 [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Kunkel LM. (1989). Dystrophin abnormalities in Duchenne/Becker muscular dystrophy. Neuron 2:1019-1029 [DOI] [PubMed] [Google Scholar]
- Hu S, Denny P, Denny P, Xie Y, Loo JA, Wolinsky LE, et al. (2004). Differentially expressed protein markers in human submandibular and sublingual secretions. Int J Oncol 5:1423-1430 [PubMed] [Google Scholar]
- Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, et al. (2005). Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics 5:1714-1728 [DOI] [PubMed] [Google Scholar]
- Huang CM. (2004). Comparative proteomic analysis of human whole saliva. Arch Oral Biol 49:951-962 [DOI] [PubMed] [Google Scholar]
- Jespersgaard C, Hajishengallis G, Russell MW, Michalek SM. (2002). Identification and characterization of a nonimmunoglobulin factor in human saliva that inhibits Streptococcus mutans glucosyltransferase. Infect Immun 70:1136-1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sagara J, Kurita H, Morifuji M, Ohishi M, Kurashina K, et al. (2004). Clinical significance of cellular distribution of moesin in patients with oral squamous cell carcinoma. Clin Cancer Res 10:572-580 [DOI] [PubMed] [Google Scholar]
- Ludwig JA, Weinstein J. (2005). Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer 5:845-885 [DOI] [PubMed] [Google Scholar]
- Mandel ID. (1989). The role of saliva in maintaining oral homeostasis. J Am Dent Assoc 119:298-304 [DOI] [PubMed] [Google Scholar]
- Mlecnik B, Scheideler M, Hackl H, Hartler J, Sanchez-Cabo F, Trajanoski Z. (2005). PathwayExplorer: Web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res 33(Web Server issue):W633-W637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Fujibuchi W, Kanehisa M. (1998). Computation with the KEGG pathway database. Biosystems 47:119-128 [DOI] [PubMed] [Google Scholar]
- Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ. (2007). Salivary proteome and its genetic polymorphisms. Ann NY Acad Sci 1098:22-50 [DOI] [PubMed] [Google Scholar]
- Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. (2010). Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE 5(1):e8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, et al. (2000). Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res 60:5954-5958 [PubMed] [Google Scholar]
- Pepe MS, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, et al. (2001). Phases of biomarker development for early detection of cancer. J Natl Cancer Inst 93:1054-1061 [DOI] [PubMed] [Google Scholar]
- Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. (2008). Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst 100:1432-1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff D. (2004). Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer 4:309-314 [DOI] [PubMed] [Google Scholar]
- Ransohoff D. (2005). Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer 5:142-149 [DOI] [PubMed] [Google Scholar]
- Ruzindana-Umunyana A, Weber JM. (2001). Interactions of human lacrimal and salivary cystatins with adenovirus endopeptidase. Antiviral Res 51:203-214 [DOI] [PubMed] [Google Scholar]
- Siqueira WL, Salih E, Wan DL, Helmerhorst EJ, Oppenheim FG. (2008). Proteome of human minor salivary gland secretion. J Dent Res 87:445-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Rasool HI, Palanisamy V, Mathisen C, Schmidt M, Wong DT, Gimzewski JK. (2010). Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 4:1921-1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ship JA, Fox PC, Baum BJ. (1991). How much saliva is enough? J Am Dent Assoc 122:63-69 [DOI] [PubMed] [Google Scholar]
- Sreebny LM, Vissink A. (2010). Sialometry: the measure of things with ease and reliability. In: Dry mouth, the malevolent symptom: a clinical guide. Sreebny LM, Vissink A, editors. Ames, IA: Wiley-Blackwell,pp. 64-76 [Google Scholar]
- Swensen SJ, Jett JR, Sloan JA, Midthun DE, Hartman TE, Sykes AM, et al. (2002). Screening for lung cancer with low-dose spiral computed tomography. Am J Respir Crit Care Med 165:508-513 [DOI] [PubMed] [Google Scholar]
- Takeshita H, Sato M, Shiwaku HO, Semba S, Sakurada A, Hoshi M, et al. (1999). Expression of the DMBT1 gene is frequently suppressed in human lung cancer. Jpn J Cancer Res 90:903-908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaysen JH, Thorn NA, Schwartz IL. (1954). Excretion of sodium, potassium, chloride and carbon dioxide in human parotid saliva. Am J Physiol 178:155-159 [DOI] [PubMed] [Google Scholar]
- Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, Tomer KB, Domingues PM, et al. (2004). Identification of human whole saliva protein components using proteomics. Proteomics 4:1109-1115 [DOI] [PubMed] [Google Scholar]
- Vitorino R, de Morais Guedes S, Ferreira R, Lobo MJ, Duarte J, Ferrer-Correia AJ, et al. (2006). Two-dimensional electrophoresis study of in vitro pellicle formation and dental caries susceptibility. Eur J Oral Sci 114:147-153 [DOI] [PubMed] [Google Scholar]
- Wagner P, Verma M, Srivastava S. (2004). Challenges for biomarkers in cancer detection. Ann NY Acad Sci 1022:9-16 [DOI] [PubMed] [Google Scholar]
- Walz A, Stuhler K, Wattenberg A, Hawranke E, Meyer HE, Schmalz G, et al. (2006). Proteome analysis of glandular parotid and submandibular-sublingual saliva in comparison to whole human saliva by two-dimensional gel electrophoresis. Proteomics 6:1631-1639 [DOI] [PubMed] [Google Scholar]
- Weir H, Thun M, Hankey B, Ries L, Howe HL, Wingo PA, et al. (2003). Annual report to the nation on the status of cancer, 1975-2000. J Natl Cancer Inst 95:1276-1299 [DOI] [PubMed] [Google Scholar]
- Wilmarth PA, Riviere MA, Rustvold DL, Lauten JD, Madden TE, David LL. (2004). Two-dimensional liquid chromatography study of the human whole saliva proteome. J Proteome Res 3:1017-1023 [DOI] [PubMed] [Google Scholar]
- Wu AJ, Atkinson JC, Fox PC, Baum BJ, Ship JA. (1993). Cross-sectional and longitudinal analyses of stimulated parotid salivary constituents in healthy, different-aged subjects. J Gerontol 48:M219-M224 [DOI] [PubMed] [Google Scholar]
- Wu W, Kemp BL, Proctor ML, Gazdar AF, Minna JD, Hong WK, et al. (1999). Expression of DMBT1, a candidate tumor suppressor gene, is frequently lost in lung cancer. Cancer Res 59:1846-1851 [PubMed] [Google Scholar]
- Yan W, Apweiler R, Balgley BM, Boontheung P, Bundy JL, Cargile BJ, et al. (2009). Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin Appl 3:116-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Berg EA, Costello CE, Troxler RF, Oppenheim FG. (2003). Identification of protein components in human acquired enamel pellicle and whole saliva using novel proteomics approaches. J Biol Chem 287:5300-5308 [DOI] [PubMed] [Google Scholar]
- Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH, et al. (2010). Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 138:949-957e1-e7 [DOI] [PMC free article] [PubMed] [Google Scholar]